Abstract
The study aimed to develop and validate a sepsis prediction model using structured electronic medical records (sEMR) and machine learning (ML) methods in emergency triage. The goal was to enhance early sepsis screening by integrating comprehensive triage information beyond vital signs. This retrospective cohort study utilized data from the MIMIC-IV database. Two models were developed: Model 1 based on vital signs alone, and Model 2 incorporating vital signs, demographic characteristics, medical history, and chief complaints. Eight ML algorithms were employed, and model performance was evaluated using metrics such as AUC, F1 Score, and calibration curves. SHapley Additive exPlanations (SHAP) and Local Interpretable Model-agnostic Explanations (LIME) methods were used to enhance model interpretability. The study included 189,617 patients, with 5.95% diagnosed with sepsis. Model 2 consistently outperformed Model 1 across most algorithms. In Model 2, Gradient Boosting achieved the highest AUC of 0.83, followed by Extra Tree, Random Forest, and Support Vector Machine (all 0.82). The SHAP method provided more comprehensible explanations for the Gradient Boosting algorithm. Modeling with comprehensive triage information using sEMR and ML methods was more effective in predicting sepsis at triage compared to using vital signs alone. Interpretable ML enhanced model transparency and provided sepsis prediction probabilities, offering a feasible approach for early sepsis screening and aiding healthcare professionals in making informed decisions during the triage process.
Keywords: Sepsis, Triage, Emergency, Interpretable machine learning, Warning mode
Subject terms: Fever, Bacterial infection, Experimental models of disease
Introduction
Despite global advances in sepsis management, early identification and treatment of sepsis remain critical challenges in emergency medicine, with persistently high morbidity and mortality rates. The 2021 American College of Emergency Physicians consensus statement emphasizes the importance of early recognition and intervention in suspected sepsis cases1. Emergency departments present a unique opportunity for early detection and intervention, as they are often the first point of contact for patients with severe infections. Timely implementation of goal-directed therapy in this setting can significantly improve patient outcomes. However, current approaches face significant limitations in reliably achieving early identification.
The urgency of early sepsis detection is underscored by a 7.6% decrease in survival for each hour of delayed treatment2,3. The 2018 Surviving Sepsis Campaign Bundle mandates critical interventions, including antibiotic administration and blood culture collection, within one hour of presentation4. However, achieving this target remains challenging due to difficulties in early identification. Epidemiological studies reveal that 86% of sepsis diagnoses occur at hospital admission5,6, with 75–80% of treatments initiated in emergency departments7,8. These findings highlight the critical role of triage in sepsis identification and the pressing need for reliable early warning tools that can be implemented at the initial point of patient contact.
Traditional scoring systems such as NEWS, MEWS, and qSOFA have played valuable roles in clinical decision-making, particularly in mitigating cognitive biases as demonstrated in recent studies9. However, these systems have shown limited effectiveness in sepsis prediction10–15, with qSOFA demonstrating particular weakness compared to NEWS and MEWS for early detection16–19. The Third International Consensus Definitions for Sepsis and the 2021 Sepsis Guidelines acknowledge these limitations20,21, recommending more comprehensive screening approaches for high-risk patients. While these scoring systems provide structured evaluation frameworks and reduce clinical judgment biases, their reliance on vital signs alone limits their utility as standalone tools for sepsis screening. Recent advances in healthcare technology, particularly in machine learning, offer the potential to retain the benefits of standardized assessment while significantly improving predictive accuracy.
Modern healthcare infrastructure, particularly structured electronic medical records (sEMR), enables the integration of comprehensive patient data—including vital signs, demographics, medical history, and chief complaints—into advanced analytical systems22,23. Machine learning (ML) models leveraging this rich data have demonstrated superior performance in critical illness prediction compared to traditional approaches22,24,25. Although the application of machine learning in sepsis prediction is an active area of research, most existing studies focus on inpatient or ICU patients rather than triage-stage data. Additionally, its “black box” nature and lack of interpretability remain significant barriers to clinical adoption26. Addressing these challenges is essential to ensuring that ML models provide actionable insights and are trusted by healthcare professionals.
This study aims to develop an interpretable ML framework that leverages comprehensive triage data to enhance sepsis prediction while ensuring clinical transparency. By incorporating SHapley Additive exPlanations (SHAP) and Local Interpretable Model-agnostic Explanations (LIME)27, the model not only improves predictive accuracy but also provides actionable insights into sepsis risk factors, empowering clinicians to make informed and timely decisions during critical triage processes.
Methods
Study design and data source
This was a retrospective cohort study designed according to the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis statements28. All data were obtained from the Medical Information Mart for Intensive Care IV (MIMIC-IV version 1.4) database, which contains the clinical information of patients in the emergency department, inpatient department, and intensive care unit of the Beth Israel Deaconess Medical Center, USA from 2008 to 2019. The database was developed by Massachusetts Institute of Technology’s Computational Physiology Laboratory and was approved by the Institutional Review Committees of Massachusetts Institute of Technology and Beth Israel Deaconess Medical Center. As the original data were anonymized and desensitized, the study was approved by the Hospital Ethics Committee. The data for this study were extracted by author ZL, who completed the examination of the training plan of the cooperative organization (Record ID 45797033).
Study population
Information on adult patients (age ≥ 18 years) in the emergency department was extracted from the MIMIC-IV-ED database. To ensure the accuracy and completeness of the patient information, we excluded patients who had no hospitalization information.
Outcomes
The outcome of the prediction model in this study was sepsis, and the diagnosis of sepsis was based on the 2016 international guidelines sepsis 3.0 criteria2. As the patients’ visit time in the MIMIC-IV database was from 2008 to 2019, the data extraction of sepsis in this study was not based on the International Classification of Diseases (ICD) code, but the diagnostic criteria of sepsis 3.0. The MIMIC-IV codebase provides the extracted code of sepsis 3.0, which can be obtained directly.
Predictor variables
The predictors collected in Supplementary Materials Box1 were based on all possible outcomes-related indicators of a patient at the time of emergency triage, including vital signs, demographic characteristics, medical history, and chief complaints. For vital signs, we extracted information on the body temperature, heart rate, respiratory rate, systolic blood pressure (SBP), diastolic blood pressure (DBP), and pain index. Regarding demographic characteristics, we extracted information on sex and age, and age was calculated by subtracting the date of birth from the date of hospitalization. The body mass index was not calculated because nearly 1/2 of the patients in the MIMIC database had missing height data, and in reality, most emergency departments could not obtain body mass index data during triage. For chronic diseases, we screened for common chronic diseases probably related to sepsis or having a poor prognosis according to clinical experience and guidelines29,30. For free-text chief complaints, we employed natural language processing techniques to address issues of spelling errors and language inconsistencies. This involved using spell-checking and text normalization techniques. For complex or ambiguous texts that could not be automatically corrected, we conducted manual reviews and made appropriate adjustments based on the specific context. The suspected infection was defined as the presence of fever, cough, diarrhea, sore throat, chills, cellulitis, and an external abscess visible to the naked eye.
Missing and extreme values
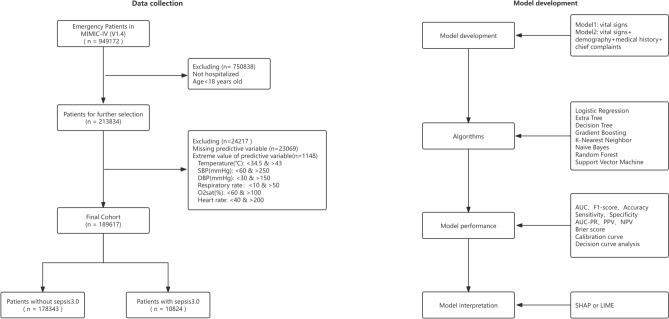
Only complete data were analyzed in this study. Because of the relatively small number of missing values and large sample size in this study, we removed all missing values. Similarly, we removed the extreme values of the numerical variables based on the clinical situation, as shown in Fig. 1. To improve the accuracy of the model, we used a normalization method to scale all the variables and map the data to the [0,1] interval.
Fig. 1.
Flow Chart. MIMIC Medical Information Mart for Intensive Care, SBP systolic blood pressure, DBP diastolic blood pressure, o2sat oxygen saturation, AUC area under the receiver operating characteristic curve, AUC-PR area under the precision-recall curve, PPV positive predictive value, NPV negative predictive value, SHAP SHapley Additive exPlanations, LIME Local Interpretable Model-agnostic Explanations.
Statistical analysis
In this study, continuous variables were described using the median and interquartile range, and the Mann–Whitney U test was used to determine differences between groups. Categorical variables were described using frequencies and percentages, and group comparisons were made using the chi-square test or Fisher’s exact test.
The data were randomly divided into a training set (80%) and test set (20%). We compared two models in our study: Model 1, which was based on vital signs alone, and Model 2, which included vital signs, demographic characteristics, medical history, and chief complaints. In our research, we selected eight widely-used and diverse machine learning algorithms based on their differing assumptions, learning mechanisms, and applicability to clinical prediction tasks:
Logistic Regression (LR): Chosen as the baseline linear model due to its simplicity, interpretability, and frequent use in medical research.
Decision Tree and Extra Tree: Tree-based models were included for their interpretability, ability to handle non-linear relationships, and complementary characteristics, with Extra Tree leveraging random splits for additional variability.
Gradient Boosting and Random Forest: Modern ensemble learning methods known for their robustness, strong predictive performance, and effectiveness in handling large datasets with complex patterns.
k-Nearest Neighbor (KNN) and Support Vector Machine (SVM): Non-linear algorithms were selected to evaluate their ability to capture local and non-linear relationships in the data.
Naive Bayes: Included for its probabilistic framework and computational efficiency, providing a contrasting approach to tree-based and ensemble models.
Class imbalance can lead to overfitting and suboptimal performance in machine learning models. In this dataset, the majority class (non-septic patients) accounts for 94.05%, whereas the minority class (septic patients) represents only 5.95%. To investigate the impact of class imbalance handling on model performance, we used the LR algorithm as an example and compared the performance of three resampling methods (SMOTE, SMOTE Tomek, and RandomUnderSampler) with the original, non-resampled data in Models 1 and 231. The model performance was evaluated using metrics including Precision, Sensitivity, and F1-Score, with a primary focus on improving the prediction ability for the minority class (septic patients). By adjusting the regularization parameters to control model complexity, we aimed to improve the model’s generalization ability and performance. The performance of different algorithms on the test set was evaluated using several metrics: F1 Score, Accuracy, Sensitivity, Specificity, Area Under the Precision-Recall Curve (AUC-PR), Positive Predictive Value (PPV), Negative Predictive Value (NPV), Area Under the ROC Curve (AUC), Brier Score, calibration curves, and Decision Curve Analysis (DCA) curves.
Based on the characteristics of the algorithms, either SHAP or LIME methods were used for interpretation. In our study, SHAP was applied to the Gradient Boosting and Random Forest models due to its ability to provide comprehensive feature importance insights for tree-based models. LIME enables rapid interpretation of any model, providing quick insights into model predictions. However, it falls short in effectively capturing the overall model performance and the complex interactions between features. SHAP’s slower computation speed in the context of Extra Tree and SVM limits its practical applicability in clinical settings.
Data extraction was conducted using Navicat Premium software (version 15.0), while statistical analysis and ML were carried out with R (version 4.1.3, The R Foundation for Statistical Computing, Vienna, Austria) and Python (version 3.8.5, Python Software Foundation). The main code and libraries used in the ML section of this study were detailed in the Supplementary Materials.
Results
Study population
This study included 189,617 patients: 178,343 (94.05%) non-septic patients and 10,824 (5.95%) septic patients (Table 1). Of the 171,584 patients without symptoms of infection, 9,095 (5.3%) were septic patients and of the 17,313 patients with symptoms of infection, 1,729 (10.0%) were septic patients (Supplementary Materials Table S1). In this study, most symptoms had a different proportion of septic patients (Supplementary Materials Table S1).
Table 1.
The relationship between sepsis as an outcome and each variable.
| Variable | Non-Sepsis (n = 178,343) | Sepsis (n = 10,824) | P-Value |
|---|---|---|---|
| Vital signs in triage | |||
| Oxygen saturation (%) | 98 (97, 100) | 96 (95, 99.) | < 0.001 |
| Heart rate (beats/min) | 84 (73, 97) | 93 (78, 110) | < 0.001 |
| Respiratory rate (times/min) | 17 (16, 18) | 19 (16, 20) | < 0.001 |
| Temperature range (℃) | 36.7 (36.4, 37.0) | 37.1 (36.5, 37.3) | < 0.001 |
| Systolic blood pressure (mmHg) | 133 (119, 150) | 122 (105, 141) | < 0.001 |
| Diastolic blood pressure (mmHg) | 76 (66, 86) | 69 (58, 80) | < 0.001 |
| Pain index | 3 (0, 7) | 0 (0, 7) | < 0.001 |
| Demographic characteristics | |||
| Age (year) | 58 (43, 71) | 66 (54, 77) | < 0.001 |
| Male [n (%)] | 86,666 (48.6%) | 5954 (55.0%) | < 0.001 |
| Past history [n (%)] | |||
| Smoke | 34,322 (19.2%) | 3158 (29.2%) | < 0.001 |
| Chronic obstructive pulmonary disease | 973 (0.5%) | 176 (1.6%) | < 0.001 |
| Chronic kidney diseases | 24,897 (14.0%) | 2808 (25.9%) | < 0.001 |
| Chronic heart failure | 5325 (3.0%) | 824 (7.6%) | < 0.001 |
| Coronary heart disease | 30,818 (17.3%) | 2950 (27.3%) | < 0.001 |
| Tumor | 5166 (2.9%) | 462 (4.3%) | < 0.001 |
| Hypertension | 31,405 (17.6%) | 1931 (17.8%) | 0.541 |
| Diabetes | 45,305 (25.4%) | 3939 (36.4%) | < 0.001 |
| Cirrhosis | 5701 (3.2%) | 1056 (9.8%) | < 0.001 |
| Chief complaints [n (%)] | |||
| Fever | 8597 (4.8%) | 1302 (12.0%) | < 0.001 |
| Chills | 360 (0.2%) | 42 (0.4%) | < 0.001 |
| Tachycardia | 1042 (0.6%) | 175 (1.6%) | < 0.001 |
| Weakness | 8043 (4.5%) | 704 (6.5%) | < 0.001 |
| Diarrhea | 2038 (1.1%) | 116 (1.1%) | 0.499 |
| Dizziness | 3900 (2.2%) | 118 (1.1%) | < 0.001 |
| Dyspepsia | 12,855 (7.2%) | 1372 (12.7%) | < 0.001 |
| Headache | 3476 (1.9%) | 134 (1.2%) | < 0.001 |
| Fatigue | 763 (0.4%) | 34 (0.3%) | 0.076 |
| Blood pressure low | 1090 (0.6%) | 485 (4.5%) | < 0.001 |
| Blood pressure high | 785 (0.4%) | 18 (0.2%) | < 0.001 |
| Lethargy | 941 (0.5%) | 229 (2.1%) | < 0.001 |
| Jaundice | 782 (0.4%) | 126 (1.2%) | < 0.001 |
| Syncope | 3374 (1.9%) | 112 (1.0%) | < 0.001 |
| Abscess | 756 (0.4%) | 48 (0.4%) | 0.761 |
| Mantal altered | 3665 (2.1%) | 729 (6.7%) | < 0.001 |
| Swelling | 4346 (2.4%) | 181 (1.7%) | < 0.001 |
| Anxiety | 1100 (0.6%) | 6 (0.1%) | < 0.001 |
| Palpitations | 1503 (0.8%) | 25 (0.2%) | < 0.001 |
| Hemoptysis | 429 (0.2%) | 79 (0.7%) | < 0.001 |
| Hematuria | 1016 (0.6%) | 62 (0.6%) | 0.967 |
| Nausea or vomiting | 9767 (5.5%) | 531 (4.9%) | 0.011 |
| Bradycardia | 342 (0.2%) | 22 (0.2%) | 0.791 |
| Cough | 3524 (2.0%) | 291 (2.7%) | < 0.001 |
| Sore throat | 761 (0.4%) | 35 (0.3%) | 0.107 |
| Dysuria | 806 (0.5%) | 34 (0.3%) | 0.036 |
| Cellulitis | 794 (0.4%) | 27 (0.2%) | 0.003 |
| Hyperglycemia | 1550 (0.9%) | 134 (1.2%) | < 0.001 |
| Hypoglycemia | 503 (0.3%) | 37 (0.3%) | 0.258 |
| Seizure | 1841 (1.0%) | 118 (1.1%) | 0.564 |
| Rash | 715 (0.4%) | 27 (0.2%) | 0.014 |
| Gastrointestinal bleeding | 1261 (0.7%) | 171 (1.6%) | < 0.001 |
| Slurred speech | 637 (0.4%) | 35 (0.3%) | 0.566 |
| Gait unsteady | 643 (0.4%) | 23 (0.2%) | 0.012 |
| Wound | 3375 (1.9%) | 175 (1.6%) | 0.040 |
| Lightheaded | 776 (0.4%) | 29 (0.3%) | 0.009 |
| Fall | 8521 (4.8%) | 382 (3.5%) | < 0.001 |
| Chest pain | 15,331 (8.6%) | 444 (4.1%) | < 0.001 |
| Abdominal pain | 24,282 (13.6%) | 1224 (11.3%) | < 0.001 |
| Back pain | 4832 (2.7%) | 180 (1.7%) | < 0.001 |
| Flank pain | 1880 (1.1%) | 76 (0.7%) | < 0.001 |
| Arm pain | 962 (0.5%) | 26 (0.2%) | < 0.001 |
| Arthralgia pain | 4505 (2.5%) | 101 (0.9%) | < 0.001 |
| Limb pain | 4876 (2.7%) | 150 (1.4%) | < 0.001 |
Impact of resampling on the data
The results showed that SMOTE, SMOTE Tomek, and RandomUnderSampler all significantly improved the Precision, Sensitivity, and F1-Score of the minority class compared to the original non-resampled data (Supplementary Materials Table S2), with their performances being relatively similar. Considering that the RandomUnderSampler relies solely on real data, it avoids the risk of overfitting associated with synthetic sample generation and requires fewer computational resources. Given the large-scale dataset and the need for multi-model analysis in this study, random undersampling was selected as the primary method to address class imbalance, achieving a balance between performance and computational efficiency.
Validation results of free-text preprocessing
To evaluate the performance of text preprocessing, we conducted validation on key steps of the workflow. For spell-checking, 500 randomly selected entries were manually reviewed, achieving a correction accuracy of 94.2% and a false positive rate of 4.5%, with most errors occurring in medical terms. For text normalization, a random sample of 300 entries showed a consistency improvement rate of 87.6%, with abbreviations such as “SOB” successfully standardized to "shortness of breath."
Performance of the models
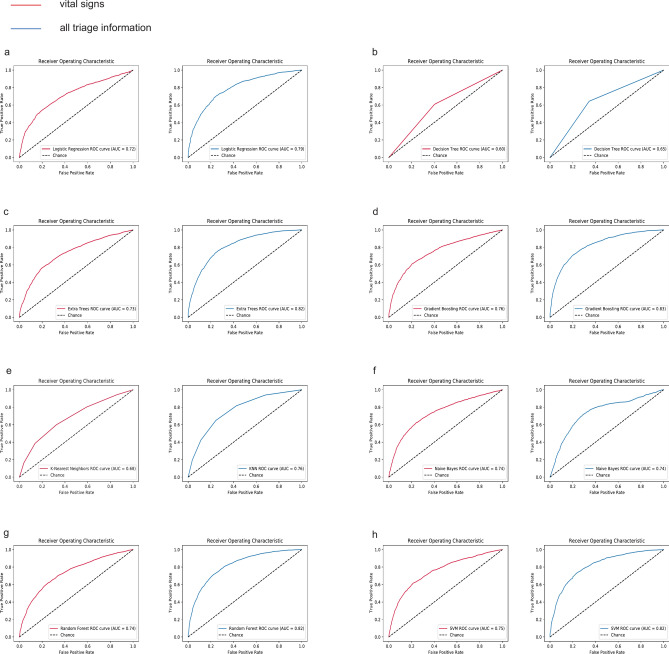
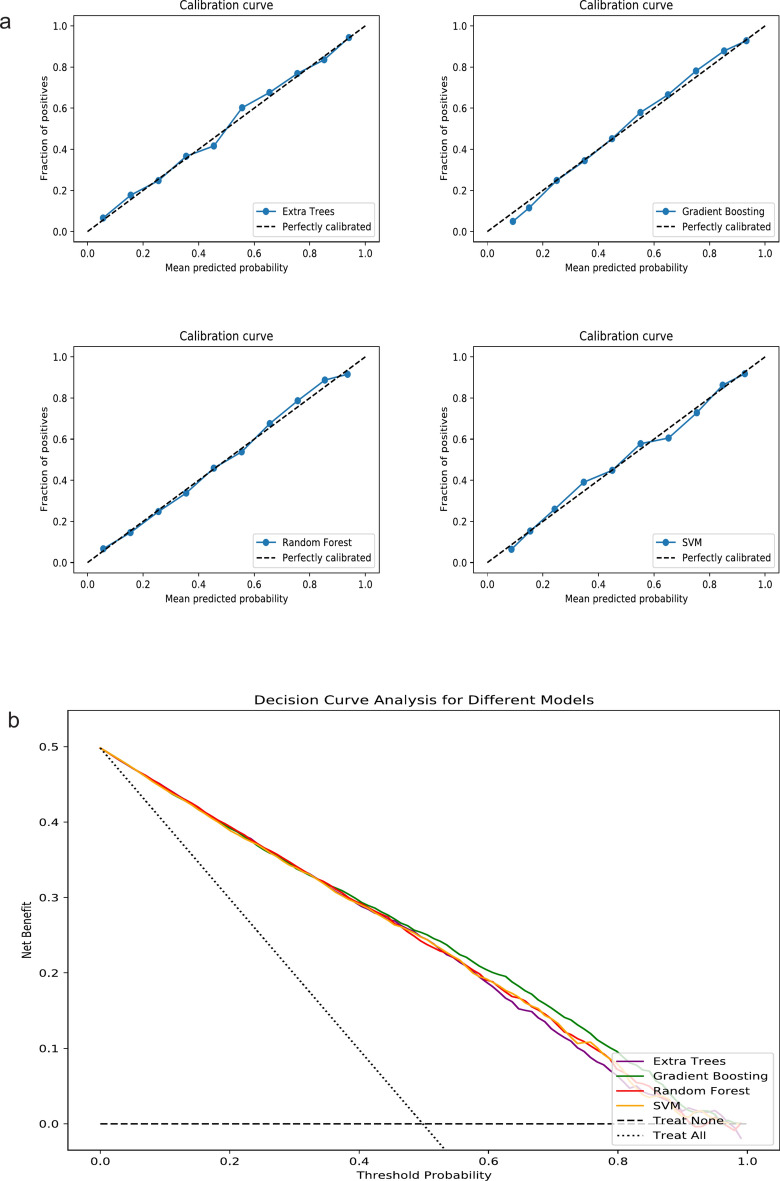
Figure 2 showed a comparison of AUC values for sepsis prediction using two models across eight algorithms. Except for Naive Bayes, Model 2 showed a significant improvement over Model 1 in all cases (Table 2). In Model 2, the AUC values of the eight algorithms, listed from highest to lowest, were as follows: Gradient Boosting (0.83), Extra Tree (0.82), Random Forest (0.82), SVM (0.82), LR (0.79), KNN (0.76), Naive Bayes (0.74), and Decision Tree (0.65). The training set performance metrics of Model 2 were presented in the Supplementary Materials (Table S3). By comparing the performance metrics between the training and testing sets, we found that most algorithms demonstrated consistent performance without significant overfitting. Table 3 presents the overall performance, discrimination, and calibration metrics of different algorithms across the two models. We selected the four ML algorithms with the best AUC performance in Model 2—Gradient Boosting, Extra Tree, Random Forest, and SVM—for further comparison. The calibration curves showed good performance for all four algorithms (Fig. 3a), and the DCA curves demonstrated that Gradient Boosting slightly outperformed the other three algorithms (Fig. 3b). Table 4 showed that Gradient Boosting achieved the highest net benefit (0.47) at the 5% threshold, outperforming Extra Tree (0.41), Random Forest (0.44), and SVM (0.44), with a consistent advantage observed across clinically relevant thresholds.
Fig. 2.
Comparison of ROC Curves of Different Algorithms on Two Models. (a) Logistic Regression; (b) Decision Tree; (c) Extra Tree; (d) Gradient Boosting; (e) k-Nearest Neighbor: (f) Naive Bayes; (g) Random Forest; (h) Support Vector Machine. ROC receiver operating characteristic curve, AUC area under the receiver operating characteristic curve.
Table 2.
DeLong test-based comparison of AUC between model 1 and model 2 across different algorithms.
| Algorithm | Model 1 (AUC) | Model 2 (AUC) | DeLong Test (z) | p-value |
|---|---|---|---|---|
| LR | 0.72 | 0.79 | − 4.3391 | < 0.001 |
| Decision Tree | 0.60 | 0.65 | − 4.2912 | < 0.001 |
| Extra Trees | 0.73 | 0.82 | − 5.8787 | < 0.001 |
| Gradient Boosting | 0.76 | 0.83 | − 4.6764 | < 0.001 |
| KNN | 0.68 | 0.76 | − 4.9441 | < 0.001 |
| Naive Bayes | 0.74 | 0.74 | − 0.3877 | 0.698 |
| Random Forest | 0.74 | 0.82 | − 5.9244 | < 0.001 |
| SVM | 0.75 | 0.82 | − 4.3505 | < 0.001 |
LR Logistic Regression, KNN K-Nearest Neighbors, SVM Support Vector Machine, AUC area under the receiver operating characteristic curve.
Table 3.
Performance of different algorithms in models 1 and 2.
| F1 Score | Accuracy | Sensitivity (Recall) | Specificity | AUC-PR | Precision (PPV) | NPV | Brier score | |
|---|---|---|---|---|---|---|---|---|
| Model 1 | ||||||||
| LR | 0.66 | 0.67 | 0.63 | 0.71 | 0.73 | 0.68 | 0.66 | 0.21 |
| Decision Tree | 0.60 | 0.60 | 0.60 | 0.59 | 0.70 | 0.60 | 0.60 | 0.39 |
| Extra Trees | 0.66 | 0.68 | 0.65 | 0.70 | 0.73 | 0.68 | 0.67 | 0.20 |
| Gradient Boosting | 0.68 | 0.70 | 0.65 | 0.75 | 0.76 | 0.72 | 0.68 | 0.19 |
| KNN | 0.62 | 0.64 | 0.59 | 0.67 | 0.69 | 0.64 | 0.62 | 0.24 |
| Naive Bayes | 0.60 | 0.67 | 0.49 | 0.85 | 0.73 | 0.76 | 0.62 | 0.22 |
| Random Forest | 0.67 | 0.68 | 0.65 | 0.70 | 0.73 | 0.68 | 0.67 | 0.20 |
| SVM | 0.67 | 0.70 | 0.62 | 0.77 | 0.75 | 0.73 | 0.67 | 0.20 |
| Model 2 | ||||||||
| LR | 0.72 | 0.73 | 0.71 | 0.73 | 0.79 | 0.72 | 0.72 | 0.18 |
| Decision Tree | 0.64 | 0.65 | 0.65 | 0.66 | 0.73 | 0.64 | 0.64 | 0.35 |
| Extra Trees | 0.74 | 0.75 | 0.74 | 0.76 | 0.80 | 0.75 | 0.74 | 0.17 |
| Gradient Boosting | 0.75 | 0.75 | 0.74 | 0.78 | 0.83 | 0.77 | 0.74 | 0.16 |
| KNN | 0.68 | 0.70 | 0.65 | 0.76 | 0.76 | 0.72 | 0.68 | 0.20 |
| Naive Bayes | 0.71 | 0.71 | 0.72 | 0.70 | 0.74 | 0.70 | 0.71 | 0.25 |
| Random Forest | 0.74 | 0.74 | 0.75 | 0.74 | 0.81 | 0.73 | 0.74 | 0.17 |
| SVM | 0.74 | 0.75 | 0.73 | 0.76 | 0.81 | 0.75 | 0.74 | 0.17 |
LR Logistic Regression, KNN K-Nearest Neighbors, AUC area under the receiver operating characteristic curve, AUC-PR area under the precision-recall curve, PPV positive predictive value, NPV negative predictive value, SVM Support Vector Machine.
Model 1 utilizes vital signs for modeling, Model 2 incorporates vital signs, demographics, medical history, and chief complaints. Note: Sensitivity and Recall refer to the same metric and are used interchangeably in the table. Similarly, Precision is equivalent to PPV.
Fig. 3.
Calibration Curves and Decision Curve Analysis Curves for the Four Best-Performing Algorithms in Model 2. (a) Calibration Curves; (b) Decision Curve Analysis Curves; SVM Support Vector Machine.
Table 4.
Comparison of net benefits of different algorithms in DCA at various threshold probabilities in model 2.
| Threshold (%) | Gradient boosting | Extra tree | Random forest | SVM |
|---|---|---|---|---|
| 5 | 0.47 | 0.41 | 0.44 | 0.44 |
| 10 | 0.35 | 0.32 | 0.33 | 0.35 |
| 20 | 0.20 | 0.18 | 0.19 | 0.19 |
SVM Support Vector Machine, DCA Decision Curve Analysis.
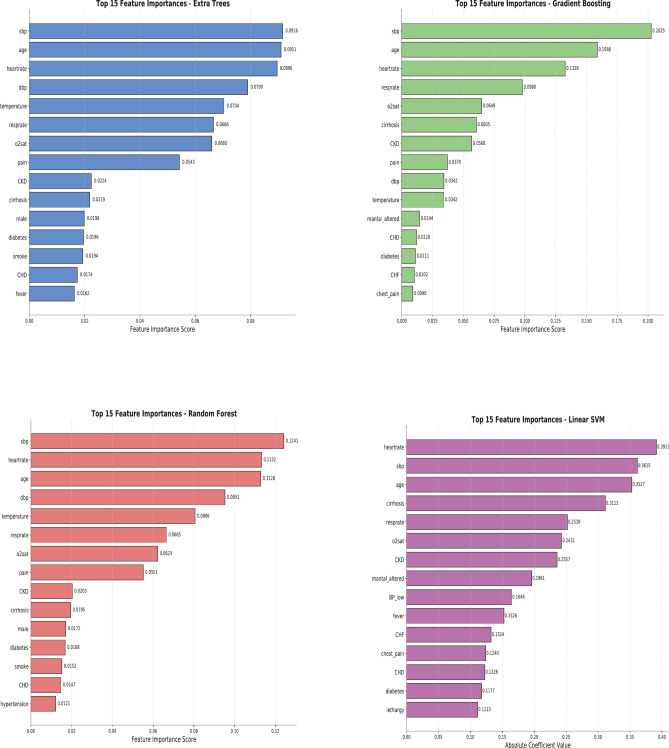
Feature importance and interpretation of the models
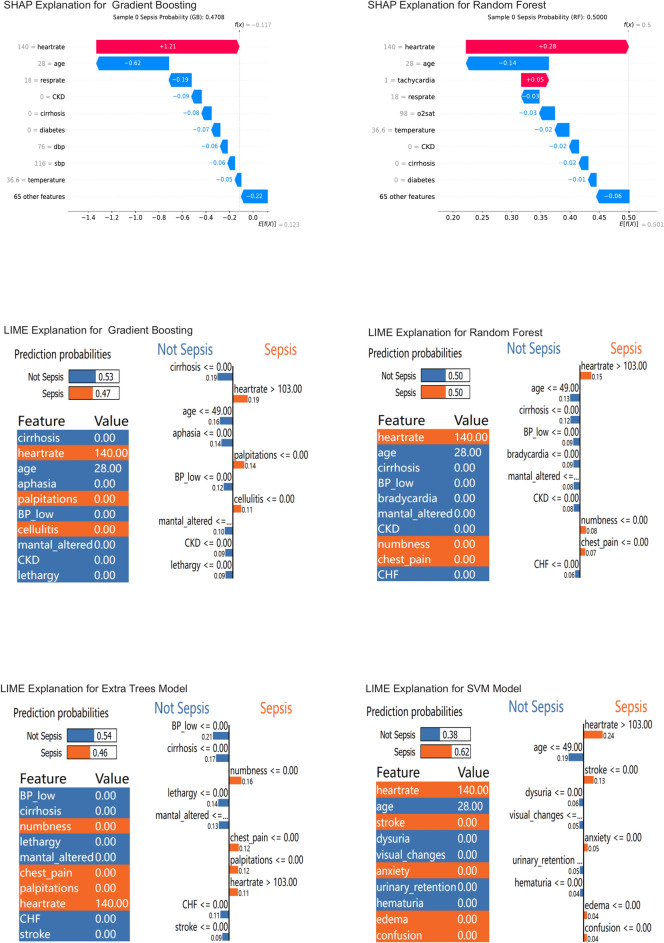
Figure 4 shows the feature importance for each of the top-performing four ML algorithms in Model 2. Using Case 1 as an example, we interpreted the predictions of the four algorithms and provided the probability of sepsis occurrence (Fig. 5). Generally, a probability of less than 20% indicates a low risk; 20%-50% suggests that observation might be appropriate; above 50% requires attention; and over 80% is highly suggestive of sepsis. The probabilities of sepsis predicted by the four algorithms were: Gradient Boosting (47%), Random Forest (50%), Extra Tree (46%), and SVM (64%). The explanations provided by the SHAP method were more comprehensible (Fig. 5).
Fig. 4.
Feature Importance of Four Algorithms in Model 2.
Fig. 5.
Interpretation of Four Algorithms in Model 2. SHAP SHapley Additive exPlanations, LIME Local Interpretable Model-agnostic Explanations. In the SHAP method, f (X) represented the final prediction result, which equaled the baseline value E [f (X)] plus the sum of all variable SHAP values. The SHAP values quantified the quantity and direction of each variable’s influence on predicting the outcome. Blue and red respectively represented decreases or increases in risk, with longer arrows indicating greater effects. The baseline value E [f (X)] was equivalent to the average risk in the dataset. The LIME method provided the overall prediction probability of the model and the prediction weight for each variable. Orange indicated an increase in risk, while blue indicated a decrease in risk.
Discussion
Our study found that modeling with more comprehensive triage information, rather than relying solely on vital signs, can more effectively predict sepsis at triage. The best-performing machine learning algorithm was Gradient Boosting, achieving an AUC of 0.83. The SHAP method enhanced the model’s transparency through improved interpretability.
The 2016 sepsis guidelines recommend screening for infections or suspected infections2. However, defining these terms is challenging, as early sepsis symptoms may not align with infection indicators. Our study found no international consensus, with definitions often based on physician experience. We identified suspected infections by symptoms like fever, cough, or visible abscesses. As shown in Table S1, 10% of patients with suspected symptoms and 5.3% without were septic. This suggests that screening based solely on suspected infections may miss cases. Early sepsis signs are often non-specific1,6–8,30, with many cases lacking fever, especially in older or immunocompromised individuals. Approximately one-third of sepsis cases lack fever, presenting instead with symptoms like hypothermia or altered mental status32, and about 20% of septic shock patients show no early infection signs30. Additionally, 20%-40% of suspected infections are non-infectious33,34. Therefore, sepsis screening should encompass all patients, not just those with suspected infections.
Sepsis is highly heterogeneous, making early prediction, particularly during triage, quite challenging. Furthermore, traditional warning models are designed to predict critical illness rather than sepsis, highlighting the need for remodeling. Additionally, these models convert vital signs into categorical variables for ease of application, which can somewhat diminish predictive efficiency. We initially explored the maximum efficacy of predicting sepsis based on triage vital signs using the AUC value. The best-performing algorithm was Gradient Boosting, with an AUC of 0.76, compared to the traditional LR algorithm, which had an AUC of 0.72 (Fig. 2a,d). Previous studies have demonstrated that certain demographic characteristics and medical histories are risk factors for sepsis, such as age ≥ 65 years, diabetes, chronic kidney disease, cirrhosis, and cancer30,35–37. Additionally, some symptoms have been shown to correlate with the occurrence of sepsis29,32. For instance, psychiatric symptoms are positively correlated37,38, while abdominal pain and chest pain are negatively correlated39. Demographic information, medical history, and chief complaints are structured data that can be obtained through sEMR during triage and analyzed using machine learning algorithms. In our Model 2, the best AUC value was 0.83 for Gradient Boosting, demonstrating a significant improvement over traditional models. While the improvement in AUC from 0.72 to 0.83 may appear modest, this enhancement represents a clinically meaningful advancement in sepsis prediction. Given that each hour of delayed treatment results in a 7.6% decrease in survival rate, even incremental improvements in early detection accuracy can translate to significant clinical benefits. Our model leverages existing electronic medical record infrastructure and readily available triage data, making implementation both feasible and cost-effective. Although traditional scoring systems (NEWS, MEWS, qSOFA) require minimal resources, their limited effectiveness in early sepsis detection may result in higher downstream costs due to delayed interventions. Furthermore, our model’s interpretability features provide clear, actionable insights that support clinical decision-making, potentially improving workflow efficiency in emergency settings. These advantages justify the implementation of our improved model, as the potential benefits in patient outcomes outweigh the modest resource requirements.
The differences in predicted sepsis probabilities among the algorithms (e.g., Gradient Boosting at 47% vs. SVM at 64%) can be attributed to the fundamental differences in their learning mechanisms and probability calibration. Tree-based models, such as Gradient Boosting and Random Forest, tend to provide more conservative and better-calibrated probability estimates due to ensemble smoothing, while SVM is more sensitive to features near decision boundaries, which can lead to higher or more variable probabilities. These discrepancies highlight the need for caution when interpreting probabilities, particularly in clinical settings. We compared eight common ML algorithms, and Gradient Boosting consistently performed the best across all metrics, including AUC and other model evaluation criteria. Gradient Boosting excels at capturing complex nonlinear interactions among diverse clinical features while reducing overfitting. Previous studies have consistently demonstrated that Gradient Boosting is among the best-performing algorithms for predicting critical illness and hospitalization rates across various clinical datasets and methodologies. For example, in a study predicting hospital mortality in ICU patients, Gradient Boosting exhibited superior performance compared to traditional scoring systems such as APACHE II, achieving an accuracy of 0.86 and an area under the ROC curve (AUC) of 0.8140. Similarly, Gradient Boosting Decision Trees were successfully employed in a population-based study to predict unplanned hospitalizations, achieving promising AUC values ranging from 0.789 to 0.80241. In the context of emergency department triage, a Gradient Boosting model stood out by predicting early mortality with an AUC of 0.962, highlighting its effectiveness in identifying high-risk patients42. These findings collectively underscore the robustness of Gradient Boosting algorithms in healthcare predictive analytics, particularly in critical care settings. In our study, the preference for Gradient Boosting aligns with its well-documented strengths in handling complex, non-linear relationships and datasets with missing or imbalanced variables, both of which are common challenges in sepsis prediction. Compared to alternative algorithms, Gradient Boosting also provided better-calibrated probabilities and feature importance metrics (as analyzed using SHAP values), thereby enhancing interpretability and actionable insights for clinical settings. These findings collectively underscore the robustness and adaptability of Gradient Boosting in healthcare predictive analytics, particularly in critical care and emergency contexts where timely and accurate predictions are crucial. The DCA demonstrated that Gradient Boosting achieved the highest net benefit across clinically relevant thresholds, particularly at the 5% threshold where early sepsis detection is critical. Its higher net benefit at lower thresholds reflects an optimal balance between sensitivity and specificity, effectively capturing more true positives while minimizing false positives. This is especially important for early intervention, which can significantly improve patient outcomes. Although net benefit decreased as thresholds increased, Gradient Boosting consistently outperformed other models, highlighting its robustness and potential to enhance clinical decision-making in sepsis risk prediction.
The purpose of interpretability in ML is to enhance model transparency, thereby effectively assisting healthcare professionals in decision-making. SHAP and LIME both have their pros and cons in explaining machine learning models. SHAP is theoretically robust and fairly allocates contribution values to each feature, explaining the difference between an individual sample’s predicted value and the model’s average. However, it can be computationally intensive. LIME, while lacking a strong theoretical foundation and not guaranteeing fair attribution of predicted values to features, is versatile and applicable to most models without requiring specific types43. In our study, the SHAP method provided explanations that were easier to understand and was highly compatible with the Gradient Boosting algorithm, eliminating concerns about computational speed. In scenarios where triage resources are limited, the high heterogeneity and atypical presentation of sepsis make early screening challenging yet highly valuable. We are the first to use interpretable ML to explore sepsis prediction based on more comprehensive triage information. By integrating sEMR with machine learning, we can quickly output sepsis prediction probabilities and explanations during triage, rather than just simple prediction outcomes. This approach offers feasibility for early sepsis screening and intervention in busy and resource-limited emergency settings. However, while our ML model demonstrates promising performance in sepsis prediction, its successful implementation in clinical practice still faces several challenges. Specifically, the integration of ML models into existing electronic medical record systems requires user-friendly interfaces to ensure predictions are presented in an intuitive and actionable format. Moreover, clinician education programs are essential to help healthcare professionals understand the model’s capabilities and properly interpret its outputs. Thus, future work should prioritize developing interfaces that seamlessly integrate with existing workflows and establishing training protocols to support effective model deployment in emergency department settings.
This study has several limitations that merit discussion. Firstly, the chief complaint content is unstructured data, and even with the use of natural language processing techniques, inevitable errors and inconsistencies may arise, potentially limiting the model’s accuracy and generalizability. Secondly, while the removal of missing and extreme values was implemented to improve data quality, this approach might have introduced bias or inadvertently excluded clinically significant outliers. Therefore, the application of advanced imputation techniques and sensitivity analyses in future studies could better evaluate the impact of these handling methods on model performance44. Additionally, although eight widely-used machine learning algorithms were employed, the selection process in this study was not as systematic as it could have been. Hence, future research could adopt a more structured approach to algorithm selection, including the exploration of newer methods and conducting thorough preliminary assessments to identify the most appropriate algorithms for specific clinical prediction tasks. Furthermore, to address the variability in predicted probabilities among different algorithms, combining predictions from multiple models (e.g., ensemble averaging) or applying advanced probability calibration techniques could improve the consistency and reliability of the outputs. However, this study does not explore these strategies in detail, and future studies should focus on incorporating and validating such approaches to enhance the interpretability and usability of predictive models in real-world clinical applications. Lastly, while the study demonstrated promising results, further validation is essential in real-world clinical settings using prospective data and diverse patient cohorts. Moreover, practical implementation of the model may also be influenced by factors such as existing workflows, resource availability, and other contextual considerations, which future studies should address to enhance the model’s applicability and reliability.
Conclusion
This study provided a feasible approach for early sepsis screening at triage. Our findings indicated that modeling with more comprehensive triage information using sEMR and ML methods was more effective in predicting sepsis at triage compared to relying solely on vital signs. Interpretable ML enhanced transparency and provided sepsis prediction probabilities, aiding healthcare professionals in making informed medical decisions during the triage process.
Supplementary Information
Acknowledgements
The authors would like to express their sincere gratitude to the MIMIC database for providing publicly available data, which has been instrumental in facilitating this research.
Author contributions
W.C. and Z.L. conceptualized the study. Z.L. and W.S. extracted and analyzed the data. Z.L. wrote the manuscript. T.L. and X.Z. verified the results. W.C. revised the manuscript. All the authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Education Department of Liaoning Province, China [Grant No. LJ232410159024].
Data availability
Publicly available datasets were analyzed in this study. These data can be found at https://mimic.mit.edu/. The datasets generated during and/or analysed during the current study are available in the figshare repository, “10.6084/m9.figshare.26098048.v1”.
Declarations
Competing interests
The authors declare no competing interests.
Ethics declarations
The data for this study came from a public database. The study design was approved by the appropriate ethics review board. Informed consent was not necessary because the database used was anonymized.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-85121-z.
References
- 1.Yealy, D. M. et al. Early care of adults with suspected sepsis in the emergency department and out-of-hospital environment: A consensus-based task force report. Ann. Emerg. Med.78, 1–19. 10.1016/j.annemergmed.2021.02.006 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Rhodes, A. et al. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock: 2016. Intensive Care Med.43, 304–377. 10.1007/s00134-017-4683-6 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Kalich, B. A. et al. Impact of an antibiotic-specific sepsis bundle on appropriate and timely antibiotic administration for severe sepsis in the emergency department. J. Emerg. Med.50, 79-88.e71. 10.1016/j.jemermed.2015.09.007 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Levy, M. M., Evans, L. E. & Rhodes, A. The Surviving Sepsis Campaign Bundle: 2018 update. Intensive Care Med.44, 925–928. 10.1007/s00134-018-5085-0 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Wang, H. E., Jones, A. R. & Donnelly, J. P. Revised national estimates of emergency department visits for sepsis in the United States. Crit. Care Med.45, 1443–1449. 10.1097/ccm.0000000000002538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhee, C. et al. Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009–2014. JAMA318, 1241–1249. 10.1001/jama.2017.13836 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang, H. E., Weaver, M. D., Shapiro, N. I. & Yealy, D. M. Opportunities for Emergency Medical Services care of sepsis. Resuscitation81, 193–197. 10.1016/j.resuscitation.2009.11.008 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Femling, J., Weiss, S., Hauswald, E. & Tarby, D. EMS patients and walk-in patients presenting with severe sepsis: Differences in management and outcome. South Med. J.107, 751–756. 10.14423/smj.0000000000000206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahmatinejad, Z. et al. Comparing in-hospital mortality prediction by senior emergency resident’s judgment and prognostic models in the emergency department. Biomed. Res. Int.2023, 6042762. 10.1155/2023/6042762 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wattanasit, P. & Khwannimit, B. Comparison the accuracy of early warning scores with qSOFA and SIRS for predicting sepsis in the emergency department. Am. J. Emerg. Med.46, 284–288. 10.1016/j.ajem.2020.07.077 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Oduncu, A. F., Kıyan, G. S. & Yalçınlı, S. Comparison of qSOFA, SIRS, and NEWS scoring systems for diagnosis, mortality, and morbidity of sepsis in emergency department. Am. J. Emerg. Med.48, 54–59. 10.1016/j.ajem.2021.04.006 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Sabir, L., Ramlakhan, S. & Goodacre, S. Comparison of qSOFA and Hospital Early Warning Scores for prognosis in suspected sepsis in emergency department patients: A systematic review. Emerg. Med. J. EMJ39, 284–294. 10.1136/emermed-2020-210416 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Usman, O. A., Usman, A. A. & Ward, M. A. Comparison of SIRS, qSOFA, and NEWS for the early identification of sepsis in the Emergency Department. Am. J. Emerg. Med.37, 1490–1497. 10.1016/j.ajem.2018.10.058 (2019). [DOI] [PubMed] [Google Scholar]
- 14.van der Woude, S. W., van Doormaal, F. F., Hutten, B. A., Nellen, F. J. & Holleman, F. Classifying sepsis patients in the emergency department using SIRS, qSOFA or MEWS. Netherlands J. Med.76, 158–166 (2018). [PubMed] [Google Scholar]
- 15.Brink, A. et al. Predicting mortality in patients with suspected sepsis at the Emergency Department; A retrospective cohort study comparing qSOFA, SIRS and National Early Warning Score. PLoS One14, e0211133. 10.1371/journal.pone.0211133 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loritz, M., Busch, H. J., Helbing, T. & Fink, K. Prospective evaluation of the quickSOFA score as a screening for sepsis in the emergency department. Intern. Emerg. Med.15, 685–693. 10.1007/s11739-019-02258-2 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez, R. M. et al. Comparison of qSOFA with current emergency department tools for screening of patients with sepsis for critical illness. Emerg. Med. J. EMJ35, 350–356. 10.1136/emermed-2017-207383 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Monclús Cols, E. et al. Comparison of the Quick Sepsis-related Organ Dysfunction score and severity levels assigned with the Andorran Triage Model in an urban tertiary care hospital emergency department. Emergencias revista de la Sociedad Espanola de Medicina de Emergencias30, 400–404 (2018). [PubMed] [Google Scholar]
- 19.Baig, M. A. et al. Comparison of qSOFA and SOFA score for predicting mortality in severe sepsis and septic shock patients in the emergency department of a low middle income country. Turk. J. Emerg. Med.18, 148–151. 10.1016/j.tjem.2018.08.002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans, L. et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Intens. Care Med.47, 1181–1247. 10.1007/s00134-021-06506-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans, L. et al. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock 2021. Crit. Care Med.49, e1063–e1143. 10.1097/ccm.0000000000005337 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Fernandes, M. et al. Clinical decision support systems for triage in the emergency department using intelligent systems: A review. Artif. Intell. Med.102, 101762. 10.1016/j.artmed.2019.101762 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Mueller, B., Kinoshita, T., Peebles, A., Graber, M. A. & Lee, S. Artificial intelligence and machine learning in emergency medicine: A narrative review. Acute Med. Surg.9, e740. 10.1002/ams2.740 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yun, H., Choi, J. & Park, J. H. Prediction of critical care outcome for adult patients presenting to emergency department using initial triage information: An XGBoost algorithm analysis. JMIR Med. Inform.9, e30770. 10.2196/30770 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Hond, A. et al. Machine learning for developing a prediction model of hospital admission of emergency department patients: Hype or hope?. Int. J. Med. Inform.152, 104496. 10.1016/j.ijmedinf.2021.104496 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Christodoulou, E. et al. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J. Clin. Epidemiol.110, 12–22. 10.1016/j.jclinepi.2019.02.004 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Jin, Y. & Kattan, M. W. Methodologic issues specific to prediction model development and evaluation. Chest164, 1281–1289. 10.1016/j.chest.2023.06.038 (2023). [DOI] [PubMed] [Google Scholar]
- 28.Collins, G. S., Reitsma, J. B., Altman, D. G. & Moons, K. G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. BMJ (Clin. Res. Ed.)350, g7594. 10.1136/bmj.g7594 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Freitag, A., Constanti, M., O’Flynn, N. & Faust, S. N. Suspected sepsis: Summary of NICE guidance. BMJ (Clin. Res. Ed.)354, i4030. 10.1136/bmj.i4030 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Filbin, M. R. et al. Challenges and opportunities for emergency department sepsis screening at triage. Sci. Rep.8, 11059. 10.1038/s41598-018-29427-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahmatinejad, Z. et al. A comparative study of explainable ensemble learning and logistic regression for predicting in-hospital mortality in the emergency department. Sci. Rep.14, 3406. 10.1038/s41598-024-54038-4 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaboli, A. et al. Triage of patients with fever: The Manchester triage system’s predictive validity for sepsis or septic shock and seven-day mortality. J. Crit. Care59, 63–69. 10.1016/j.jcrc.2020.05.019 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Heffner, A. C., Horton, J. M., Marchick, M. R. & Jones, A. E. Etiology of illness in patients with severe sepsis admitted to the hospital from the emergency department. Clin. Infect. Dis.50, 814–820. 10.1086/650580 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein Klouwenberg, P. M. et al. Likelihood of infection in patients with presumed sepsis at the time of intensive care unit admission: A cohort study. Crit. Care19, 319. 10.1186/s13054-015-1035-1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molnár, G. et al. Differentiating sepsis from similar groups of symptoms at triage level in emergency care. Physiol. Int.10.1556/2060.2021.00005 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Smyth, M. A. et al. Derivation and internal validation of the screening to enhance prehospital identification of sepsis (SEPSIS) score in adults on arrival at the emergency department. Scand. J. Trauma Resuscitat. Emerg. Med.27, 67. 10.1186/s13049-019-0642-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, B. et al. Development and internal validation of a simple prognostic score for early sepsis risk stratification in the emergency department. BMJ Open11, e046009. 10.1136/bmjopen-2020-046009 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petruniak, L., El-Masri, M. & Fox-Wasylyshyn, S. Exploring the Predictors of Emergency Department Triage Acuity Assignment in Patients With Sepsis. Can. J. Nurs. Res. Revue canadienne de recherche en sciences infirmieres50, 81–88. 10.1177/0844562118766178 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Shibata, J. et al. Risk factors of sepsis among patients with qSOFA<2 in the emergency department. Am. J. Emerg. Med.50, 699–706. 10.1016/j.ajem.2021.09.035 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Luo, Y., Wang, Z. & Wang, C. Improvement of APACHE II score system for disease severity based on XGBoost algorithm. BMC Med. Inform. Decis. Mak.21, 237. 10.1186/s12911-021-01591-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olza, A., Millán, E. & Rodríguez-Álvarez, M. X. Development and validation of predictive models for unplanned hospitalization in the Basque Country: Analyzing the variability of non-deterministic algorithms. BMC Med. Inform. Decis. Mak.23, 152. 10.1186/s12911-023-02226-z (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klug, M. et al. A gradient boosting machine learning model for predicting early mortality in the emergency department triage: Devising a nine-point triage score. J. General Intern. Med.35, 220–227. 10.1007/s11606-019-05512-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ali, S. et al. The enlightening role of explainable artificial intelligence in medical & healthcare domains: A systematic literature review. Comput. Biol. Med.166, 107555. 10.1016/j.compbiomed.2023.107555 (2023). [DOI] [PubMed] [Google Scholar]
- 44.Rahmatinejad, Z. et al. Comparison of six scoring systems for predicting in-hospital mortality among patients with SARS-COV2 presenting to the emergency department. Indian J. Crit. Care Med.27, 416–425. 10.5005/jp-journals-10071-24463 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found at https://mimic.mit.edu/. The datasets generated during and/or analysed during the current study are available in the figshare repository, “10.6084/m9.figshare.26098048.v1”.