Abstract
This study utilizes the Breast Ultrasound Image (BUSI) dataset to present a deep learning technique for breast tumor segmentation based on a modified UNet architecture. To improve segmentation accuracy, the model integrates attention mechanisms, such as the Convolutional Block Attention Module (CBAM) and Non-Local Attention, with advanced encoder architectures, including ResNet, DenseNet, and EfficientNet. These attention mechanisms enable the model to focus more effectively on relevant tumor areas, resulting in significant performance improvements. Models incorporating attention mechanisms outperformed those without, as reflected in superior evaluation metrics. The effects of Dice Loss and Binary Cross-Entropy (BCE) Loss on the model’s performance were also analyzed. Dice Loss maximized the overlap between predicted and actual segmentation masks, leading to more precise boundary delineation, while BCE Loss achieved higher recall, improving the detection of tumor areas. Grad-CAM visualizations further demonstrated that attention-based models enhanced interpretability by accurately highlighting tumor areas. The findings denote that combining advanced encoder architectures, attention mechanisms, and the UNet framework can yield more reliable and accurate breast tumor segmentation. Future research will explore the use of multi-modal imaging, real-time deployment for clinical applications, and more advanced attention mechanisms to further improve segmentation performance.
Keywords: Breast tumor segmentation, UNet, Grad-CAM, Non-local attention, Attention mechanisms
Introduction
As one of the main causes of cancer-related mortality among women, breast cancer remains a serious worldwide health concern1–4. Increasing the rate of survival and improving treatment results are dependent on early and precise diagnosis of breast cancers. Because imaging techniques like ultrasonography are non-invasive, reasonably priced, and capable of differentiating between solid and cystic tumors, they are frequently utilized in breast cancer screening programs. However, because of the low contrast, noise, and irregular forms of the tumors, one of the main issues in ultrasound imaging is the difficulty of effectively segmenting breast cancers. Radiologists’ manual segmentation is labor-intensive, time-consuming, and sensitive to subjectivity, notwithstanding its effectiveness5–8.
Intending to produce dependable, consistent, and effective segmentation findings, automated breast tumor segmentation algorithms have become an essential tool to overcome these constraints. Deep learning-based methods, in particular Convolutional Neural Networks (CNNs), have shown great potential among the various techniques investigated for this goal9–11. CNNs have proven to perform better in tasks related to image categorization, detection, and segmentation12–14. The CNN-based UNet design, which is renowned for its symmetric encoder-decoder architecture, is a frequently utilized structure in medical image segmentation. The decoder reconstructs the segmentation mask, which maps the tumor regions in the input image, while the encoder is made to capture high-level information. Despite its effectiveness, accurately capturing intricate tumor features in breast ultrasound images remains challenging due to the significant variability in tumor size, shape, and texture. These complexities highlight the need for enhanced architectural modifications and additional mechanisms to address these limitations effectively15–19.
A two-step deep learning framework was suggested by Li et al.20 to segment breast tumors in breast ultrasound (BUS) images with a minimal amount of manual labeling. Breast anatomy decomposition is accomplished in the first step using a semi-supervised semantic segmentation method. The input BUS image is divided into four anatomical features in this step: layers of fat, muscle, mammary gland, and thorax. The layers of fat and mammary glands are employed as a restricted zone to reduce the search space for tumor segmentation. In a weakly supervised learning scenario, when only image-level labels are provided, the second phase addresses breast tumor segmentation. A classification network first detects tumors, and then the suggested class activation mapping and deep level set (CAM-DLS) technique is used to segment the tumors. This novel method efficiently increases segmentation precision while reducing the need for laborious manual labeling.
In order to create new images from the input mammogram, Ranjbarzadeh et al.21 suggested a breast cancer recognition system that uses multiple encoding techniques. Each encoded image emphasizes distinct features essential for accurate texture detection, enabling the system to capture a broader range of relevant information for precise analysis. The pectoral muscle is then removed using these encoded images, which solves a common problem in mammography analysis. For pixel-wise classification, the technique applies 11 different encoded images to a shallow cascade CNN. For classification, this CNN concatenates the extracted features into a vertical vector by processing local patches from the encoded images. The vertical vector is then fed into fully connected layers. The suggested architecture improves the system’s capability to evaluate textures effectively by utilizing several mammography representations without depending on a deep CNN model. Comprehensive tests conducted on two publicly available datasets show that this strategy delivers competitive performance in comparison to many baseline approaches.
An enhanced selective kernel convolution method for breast tumor segmentation was presented by Chen et al.22. This technique combines many feature map region representations and dynamically modifies their weights in both the channel and spatial dimensions. The model may now minimize the impact of less significant areas while emphasizing high-value regions owing to this recalibration. With the integration of the improved selective kernel convolution into a deep supervision U-Net architecture, tumor feature representation becomes adaptive. Experiments on three publicly available datasets of breast ultrasounds showed that the method works well, outperforming a number of the most advanced segmentation methods.
Wang et al.23 introduced a Progressive Dual Priori Network (PDPNet) for the purpose of segmenting breast lesions from dynamic contrast-enhanced magnetic resonance images (DCE-MRI) acquired from multiple centers. Using a coarse-segmentation module, PDPNet first locates tumor locations. It then gradually improves the tumor mask by adding weak semantic priors and cross-scale correlation knowledge. This multi-stage method increases shape recognition for irregular tumors and sharpens the model’s attention on tumors, particularly small or low-contrast ones. Experimental comparisons against state-of-the-art techniques revealed notable gains: across multi-center datasets, PDPNet achieved at least 5.13% and 7.58% better DSC and HD95, respectively. Further ablation investigations shown that the dual priors optimize segmentation accuracy by fine-tuning tumor focus and shape awareness, while the localization module efficiently eliminates interference from normal tissues, improving model generalization.
Advanced approaches, including attention mechanisms, have been incorporated into the UNet architecture to significantly enhance tumor segmentation performance. By strengthening the network’s capacity to concentrate on the most pertinent regions of the image, attention modules are intended to increase the precision of feature extraction and segmentation24–26. This study investigates the integration of two types of attention mechanisms within the UNet framework: the Convolutional Block Attention Module (CBAM) and Non-Local Attention, to meet the issues of variable tumor sizes and fuzzy borders. CBAM employs both spatial and channel-wise attention to refine feature maps, enabling the model to focus on significant characteristics in both dimensions effectively. In contrast, Non-Local Attention captures long-range dependencies and contextual relationships within the image, allowing the model to consider global information. This capability is predominantly essential for accurately segmenting complex and irregular tumor boundaries, where local features alone may not provide sufficient context.
When training segmentation models, choosing an applicable loss function is as critical as incorporating attention mechanisms, as different loss functions emphasize distinct aspects of segmentation tasks. In this study, we compared the performance of models trained with two widely utilized loss functions in medical image segmentation: Binary Cross-Entropy (BCE) Loss and Dice Loss. BCE Loss, traditionally employed for binary classification tasks, effectively guides the network in distinguishing between tumor and non-tumor regions. However, medical segmentation tasks often face a significant class imbalance, as tumor areas typically occupy only a small fraction of the image. Dice Loss addresses this issue by directly optimizing the overlap between ground-truth masks and predicted segmentation masks, making it particularly suitable for improving segmentation accuracy in imbalanced datasets.
In the medical field, the interpretability of models is becoming more and more important, going beyond conventional performance indicators like Precision, Recall, F1 Score, Intersection over Union (IoU), and Dice Coefficient1,9,27,28. To ensure clinical applicability and win over the medical community, it is imperative to comprehend how models make judgments. To provide more insight into the behavior of the models, this work uses two interpretability techniques: Gradient-weighted Class Activation Mapping (Grad-CAM) and Receiver Operating Characteristic (ROC) analysis. A more sophisticated understanding of model performance is provided by ROC analysis, which assesses the trade-offs between true positive and false positive rates across various categorization levels. In contrast, Grad-CAM highlights the sections of the input image that most influenced the model’s conclusion, providing visual explanations and assisting in determining whether the model is concentrating on clinically significant regions of the image.
This work advances the field of breast tumor segmentation by exploring the integration of state-of-the-art CNN encoders—ResNet-18, DenseNet-121, and EfficientNet-B0—with attention mechanisms and different loss functions to enhance segmentation performance. A comprehensive comparison of various model designs is presented, with an emphasis on both qualitative and quantitative outcomes to ensure interpretability. Through an in-depth analysis of segmentation performance and model interpretability, this study provides valuable insights into the application of deep learning (DL) methods for ultrasound image-based breast tumor segmentation, highlighting their potential for clinical relevance and enhanced diagnostic accuracy.
The rest of this essay is organized as follows: The dataset, the UNet-based model architecture, the use of Depthwise Separable Convolutions, and the incorporation of attention mechanisms, such as Non-Local Attention and the CBAM Module, are all covered in Section “Methodology” (Methodology). Additionally, it describes the loss functions used to maximize model performance. The training process, assessment measures, and in-depth analyses utilizing ROC curves and Grad-CAM for interpretability are covered in Section “Experiments setup”. The experiment results are presented and the performance of the different models is analyzed in Section “Results and discussion”. Lastly, a summary of the results and recommendations for further research are provided in Section “Conclusion and future works”.
Methodology
Dataset
The Breast Ultrasound Image (BUSI) dataset29, a commonly used publically available dataset for breast tumor segmentation tasks, was employed in this study. This dataset includes a diverse collection of breast ultrasound scans, accompanied by ground truth masks that precisely define tumor borders within the images. Additionally, each image is labeled as either benign or malignant, making the dataset appropriate for both segmentation and classification tasks. This dual-purpose labeling provides a comprehensive resource for developing and evaluating models aimed at tumor detection and characterization. Because of the inherent diversity in ultrasound imaging, which includes differences in tumor size, form, texture, and intensity in addition to noise, the BUSI dataset presents unique challenges.
The dataset was split into three sections for the sake of this study: a test set, a validation set, and a training set29:
Training samples: The training set is made up of 624 resized images (224 × 224 pixels) and the segmentation masks that go with them. The DL model is trained on this set, which enables the algorithm to pick up on the intricate patterns and characteristics connected to breast tumor segmentation.
Validations samples: The validation set consists of 78 scaled images (224 × 224 pixels) together with the masks that go with them. This collection allows hyperparameter adjustment and model selection by tracking the model’s performance throughout training. When making changes to the model without exposing it to test data, the validation set acts as a checkpoint.
Test samples: The test set consists of 78 scaled images (224 × 224 pixels) together with the masks that go with them. This set is only used to assess how well the final model performs following training, giving an accurate indication of how well the model generalizes to fresh, untested data.
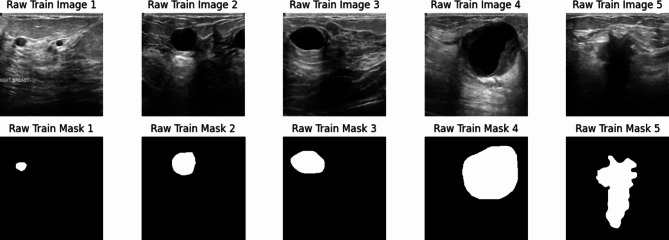
By dividing the dataset into three distinct components—training, validation, and test sets—this study minimizes the risk of overfitting and ensures a robust assessment of the model. This methodology enhances confidence in the model’s ability to generalize effectively to real-world ultrasound images, providing a reliable evaluation of its performance. In breast tumor segmentation tasks, well-balanced data distribution across training, validation, and testing sets contributes to robust and dependable outcomes. Some samples of the BUSI dataset are displayed in the Fig. 1.
Fig. 1.
Some samples from the BUSI dataset.
Model architecture
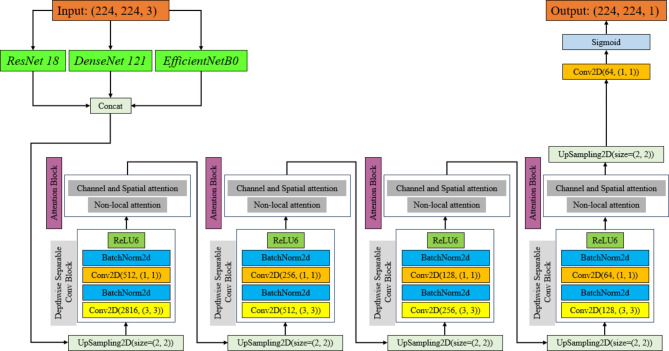
The UNet framework serves as the foundation for the model architecture used in this work, which is improved using several encoder networks and sophisticated attention mechanisms to better breast tumor segmentation30–32. The encoder section of the network makes use of the well-known rich, multi-scale feature extraction algorithms ResNet-1833,34, DenseNet-12135–37, and EfficientNet-B038,39. The CBAM and Non-Local Attention modules are included in the decoder to capture long-range dependencies and concentrate on pertinent spatial and channel-wise information to refine the feature maps40–46. To lower the computational complexity of the model without sacrificing performance, Depthwise Separable Convolutions (DSC) are employed47–49. Both BCE and Dice Losses are used in the model’s training to account for class imbalance and guarantee precise segmentation50–53. High-quality segmentation results are ensured via skip connections between the encoder and decoder, which assist preserve significant spatial features. Figure 2 exhibit the block diagram of the proposed model with different configurations.
Fig. 2.
Block diagram of the suggested model with different configurations. The ReLU layer is replaced with ReLU6 layer to improve the performance of the model.
Depthwise separable convolutions
In this study, DSC are essential for keeping the model’s robust feature extraction capabilities while lowering its computing cost. With this kind of convolution, the usual convolution operation is divided into two easier steps: pointwise convolution, which combines the output from the Depthwise step, and the Depthwise convolution, which applies a single filter to each input channel47–49.
Depthwise Convolution: Compared to typical convolutions, which apply a convolutional filter to every input channel, this step applies a convolutional filter to each input channel independently, greatly lowering the number of computations.
Pointwise Convolution: A 1 × 1 convolution (pointwise convolution) combines the feature maps from each of the input channels after Depthwise convolution, resulting in a final feature map that requires less computing power to rebuild.
Specifically, this architecture makes use of the ReLU6 activation function to increase the model’s efficiency and stability. ReLU6 is an activation function variation of the conventional ReLU that prevents possible problems with extremely large activations by capping its output at 6. This is especially helpful in situations when numerical stability is crucial, such as medical image segmentation, or when training deep models with high learning rates54–56. ReLU6 is used in conjunction with DSC to optimize the model’s efficiency and performance. This combination is especially useful for addressing the complicated nature of breast ultrasound images for tumor segmentation because it not only minimizes the number of parameters but also guarantees numerical stability during training.
Attention mechanisms
Attention mechanisms have become an essential component of DL models, specifically for complex image segmentation tasks. In medical image segmentation, these mechanisms allow models to filter out noisy or irrelevant information and focus on the most critical areas of the image, such as tumor boundaries. This capability significantly improves the precision and reliability of segmentation outcomes, especially in challenging scenarios like breast ultrasound imaging. Attention modules concentrate on crucial areas and improve segmentation accuracy by dynamically balancing important spatial and channel-wise characteristics57–59. This is particularly helpful for handling difficult medical images, such as ultrasound imaging of the breast, where cancers can differ greatly in size, shape, and intensity.
Non-local attention
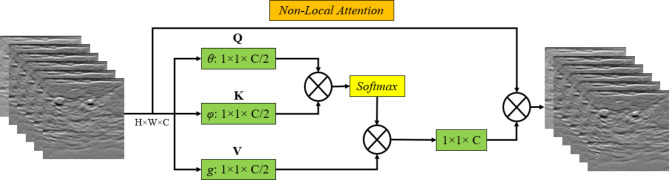
Using relationships between far-off pixels, Non-Local Attention is a strong mechanism that catches long-range dependencies in image data. Non-local attention enables the model to globally assess and link features throughout the entire image, in contrast to standard convolutional procedures, which are restricted to local receptive fields40–42. This is especially crucial for medical image segmentation, like that of breast tumors, where the tumor boundaries might be erratic and delicate, necessitating the inclusion of information from distant portions of the image for the model to produce reliable predictions.
Non-Local Attention allows the model to better understand the entire structure and context of the tumor by computing interactions between all pixel pairs. This can result in segmentation results that are more accurate and consistent60–62. By concentrating on wider spatial linkages in addition to local aspects, which may be essential for identifying complicated tumor forms, this mechanism enhances other attention modules such as CBAM. The structure of employed Non-Local Attention is demonstrated in Fig. 3.
Fig. 3.
The structure of employed Non-Local Attention block.
Convolutional block attention module
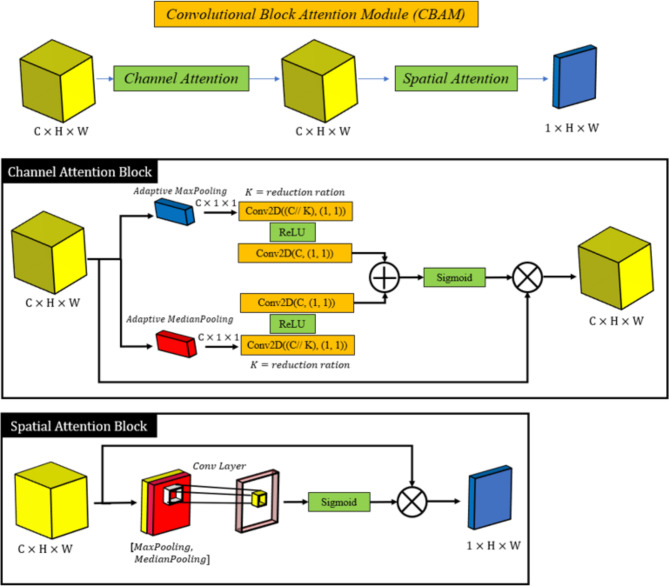
Convolutional Block Attention Module (CBAM) is an attention mechanism that applies attention at both spatial and channel levels to improve convolutional neural networks’ feature extraction performance. By concentrating on the most crucial data in these dimensions, CBAM refines feature maps in two successive stages43–46,63: Channel Attention and Spatial Attention.
Channel Attention: This section emphasizes the significance of various feature channels. In order to help the model prioritize the most relevant features while suppressing less important ones, it gives higher weights to channels that contain more critical information for the task.
Spatial Attention: Spatial attention is used to highlight the most significant areas of the feature maps after channel-wise features have been improved. The model can concentrate on important parts of the image, like tumor regions, and ignore unimportant background information by weighting spatial positions.
The CBAM module offers a notable performance boost with little computational overhead because it is lightweight and simple to incorporate into current CNN architectures. By focusing on the most important features and regions in the ultrasound images, CBAM helps our model to capture finer details and increase segmentation accuracy. The structure of employed CBAM module is demonstrated in Fig. 4.
Fig. 4.
The structure of employed Convolutional Block Attention module.
Loss functions
When training DL models, loss functions are essential, particularly when the task at hand is medical image segmentation, where the objective is precise pixel-level label prediction. Two main loss functions—BCE and Dice Losses—are used in this study. Each loss function has a distinct effect on how the model learns.
BCE Loss: BCE Loss is frequently applied at the pixel level in segmentation tasks and is utilized for binary classification issues. It computes the discrepancy between the ground truth label and the estimated likelihood that a pixel is a part of the tumor. Because BCE handles each pixel separately, it works well in segmentation situations where the classification of each pixel is important. This loss function effectively guides the model in distinguishing between non-tumor and tumor regions, making it a fundamental choice for binary segmentation tasks. Class imbalance, a common challenge in medical image segmentation, arises when tumor areas are meaningfully smaller than the background. This imbalance can affect the performance of BCE Loss, as it may prioritize the majority background class, leading to under-segmentation of the tumor areas. Addressing this imbalance often requires additional strategies, such as complementary loss terms or weighted loss functions, to ensure that the model accurately focuses on the minority tumor areas50,51.
Dice Loss: Dice Loss is especially helpful in managing class imbalance and is specifically made for image segmentation. Optimizing the Dice coefficient—a measure of how similar two sets are—it directly maximizes the overlap between the predicted segmentation mask and the ground truth. In situations where the tumor occupies a smaller portion of the image, dice loss helps to improve the overlap between the predicted tumor regions and the actual tumor, resulting in more accurate segmentation52,53,64.
Experiments setup
Training procedure
Configuring the model, establishing the training loop, and implementing a systematic training loop with early stopping to prevent overfitting are all part of the training process for the breast tumor segmentation model. The main steps in the training process are outlined below:
Device Setup: The implementation of our model was carried out employing Python with Keras and TensorFlow libraries. To guarantee effective training and assessment, an NVIDIA A100 GPU provided by Google Colab was used. The A100 GPU, with its large memory and high computational capacity, allowed us to train the model effectively on the BUSI dataset, handling attention mechanisms and the complex architecture without bottlenecks.
Optimizer Setup: During training, the Adam optimizer was used to minimize the loss function. Adam was chosen because of its capacity to adjust learning rates and deliver strong practice results with little tweaking. To enable stable convergence, the learning rate is set to 10 e-4.
Training Configuration: The model was trained for a maximum of 50 epochs. If, after a predetermined number of epochs (patiently 10, for example), validation performance did not improve, training was stopped early. By stopping the model early, the best-performing model based on the validation loss was preserved and the model was kept from overfitting to the training set.
Training Loop: The model was trained using a standard training loop, in which the training data was fed into the model at each epoch, and backpropagation was used to calculate and minimize the loss. After each epoch, the validation loss was tracked to evaluate the performance of the model. The model was saved if the validation loss gets better; if it didn’t get better after ten consecutive epochs, training was terminated early.
Hyperparameter tuning is essential for enhancing the performance of DL models. This work employed a grid search methodology to determine the best hyperparameters for training. The learning rate was evaluated across several values, specifically 10 e-5, 10 e-4, and 10 e-3, with 10 e-4 demonstrating optimal convergence and stability. The batch sizes of 8, 16, and 32 were assessed, with a batch size of 16 achieving the optimal equilibrium between computing efficiency and model performance. Dropout rates ranged from 0.2 to 0.5 to mitigate overfitting, with 0.3 yielding the most consistent outcomes. The Adam optimizer was used for its variable learning rate features, and early stopping with a patience of 10 epochs was utilized to mitigate overfitting. The ultimate hyperparameters were selected based on validation set performance, guaranteeing that the model attained strong generalization without undue computational burden. These details are essential for the reproducibility of the study and for directing future research in analogous segmentation tasks. This process guarantees stable optimization, effective hardware utilization, and avoidance of overfitting, culminating in a well-trained model for breast tumor segmentation.
Assessment metrics
A set of reliable metrics is needed to assess a breast tumor segmentation model’s performance in terms of how well it predicts the tumor regions in the ultrasound images. The segmentation performance is evaluated quantitatively in this study using the following assessment metrics3,5,65–69:
 |
1 |
 |
2 |
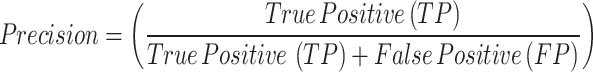 |
3 |
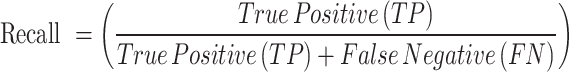 |
4 |
 |
5 |
Out of all the pixels predicted as tumors, precision quantifies the percentage of correctly predicted positive pixels (tumor regions). It illustrates how well the model can evade false positives. Out of all the actual positive pixels, recall quantifies the percentage of correctly predicted positive pixels (tumor regions). It illustrates how well the model can identify every tumor pixel. The F1 Score is a single score that provides a balance between both metrics, derived from the harmonic mean of Precision and Recall. When the dataset is unbalanced, as in medical segmentation, where the tumor region is frequently smaller than the background, it is especially helpful.
The overlap between the ground truth mask and the predicted segmentation mask is measured by IoU. It is the proportion of the union area—the entire area covered by both masks—to the intersection area, which is made up of shared pixels between the true and predicted masks. Greater accuracy in segmentation is indicated by a higher IoU. Another overlap metric that is frequently used in medical image segmentation and is closely related to IoU is the Dice Coefficient. The similarity between the ground truth mask and the predicted mask is measured. It has a range of 0 to 1, similar to IoU, where 1 denotes perfect overlap.
ROC curve analysis
The Receiver Operating Characteristic (ROC) curve is a visual aid that shows how diagnostic a classification model can be at different threshold values. The ROC curve plots the True Positive Rate (TPR), or Recall, against the False Positive Rate (FPR) for various thresholds in the context of breast tumor segmentation. By altering the classification threshold, it sheds light on the model’s capacity to discriminate between tumor and non-tumor regions70,71.
Sensitivity (Recall) or True Positive Rate (TPR): Indicates the percentage of real tumor pixels that the model correctly identified.
False Positive Rate (FPR): Indicates the percentage of pixels that are not tumors but are mistakenly identified as such.
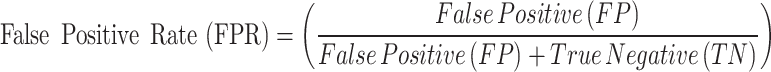 |
6 |
The ROC curve is produced by assessing the model’s predictions across a range of threshold values. The trade-off between the True and False Positive rates is shown as the threshold varies. A model should ideally have a low FPR combined with a high TPR, which would produce a curve that slopes toward the upper-left corner of the plot. A scalar value representing the model’s overall performance across all potential thresholds is called the Area Under the ROC Curve (AUC). The range of an AUC score is 0 to 1:
AUC = 1: Demonstrates flawless model performance, meaning that the model can accurately distinguish between areas with and without tumors.
AUC = 0.5: This shows that the model has no discriminatory power; it is equivalent to random guessing.
AUC > 0.7: Higher values indicate better performance, and this range is generally regarded as acceptable.
The ROC curve aids in the analysis of the trade-off between sensitivity (accurately identifying tumors) and false positive rate (misclassifying normal tissue as tumor). This trade-off is relevant to breast tumor segmentation. The model’s capacity to distinguish between tumor and non-tumor regions across a range of thresholds can be measured by looking at the AUC. In medical applications, where a low false positive rate (to prevent needless interventions) and high sensitivity (to detect as many tumors as possible) are essential, this analysis is especially significant.
Grad-CAM analysis
Highly effective for visualizing and analyzing DL models, Grad-CAM is especially useful for image-based tasks such as breast tumor segmentation. Grad-CAM creates a heatmap that shows which areas are most important to the model’s decision by highlighting the areas of an image that the model uses to make predictions. This serves as a visual confirmation that the segmentation corresponds with the ground truth in the context of breast tumor segmentation, helping to guarantee that the model is focusing on the appropriate tumor regions. This is critical for medical applications where clinical use depends on prediction accuracy and relevance72–74.
Additionally, error analysis and model refinement are aided by Grad-CAM. Grad-CAM heatmaps can indicate whether the model concentrated on noisy or irrelevant regions of the image when it makes inaccurate predictions, offering insights into possible model flaws. Through comprehension of the model’s “looking” during segmentation, researchers can modify the architecture or training procedure to enhance performance. Building confidence in the model’s judgments requires this degree of interpretability, particularly in crucial applications like tumor detection and medical imaging segmentation.
Results and discussion
The evaluation results of the breast tumor segmentation models are shown in this section, with an emphasis on the effects of various encoder architectures, attention mechanisms, and loss functions. Several critical metrics, including Precision, Recall, F1 Score, IoU, and Dice Coefficient, were employed to evaluate the segmentation’s accuracy and robustness. Furthermore, ROC curve analysis and Grad-CAM visualizations were used to offer a more comprehensive comprehension of the discriminatory power and interpretability of the models.
Comparison with different backbones and loss functions
The results demonstrate how well attention mechanisms and well-chosen loss functions can be integrated to improve breast tumor segmentation performance on ultrasound images by comparing different configurations. Tables 1, 2, 3, and 4 denote the acquired values of the models’ performance during the different configurations.
Table 1.
Performance of different segmentation models for breast tumors using different combinations of encoder architectures, loss functions, and attention mechanisms.
| Model | Precision | Recall | F1 score | IoU | Dice |
|---|---|---|---|---|---|
|
CBAM Attention + DSC + ResNet + DenseNet + EfficientNet + BCE Loss (Model 1) |
0.6194 | 0.6378 | 0.6140 | 0.5305 | 0.6140 |
|
Total params: 23,881,989 Trainable params: 23,881,989 Non-trainable params: 0 | |||||
|
CBAM Attention + DSC + ResNet + DenseNet + EfficientNet + Dice Loss (Model 2) |
0.6349 | 0.5946 | 0.5942 | 0.5083 | 0.5942 |
|
Total params: 23,881,989 Trainable params: 23,881,989 Non-trainable params: 0 | |||||
|
Non-Local Attention + DSC + ResNet + DenseNet + EfficientNet + BCE Loss (Model 3) |
0.6303 | 0.5980 | 0.5943 | 0.5094 | 0.5943 |
|
Total params: 24,493,277 Trainable params: 24,493,277 Non-trainable params: 0 | |||||
|
Non-Local Attention + DSC + ResNet + DenseNet + EfficientNet + Dice Loss (Model 4) |
0.6284 | 0.5045 | 0.5358 | 0.4534 | 0.5358 |
|
Total params: 24,493,277 Trainable params: 24,493,277 Non-trainable params: 0 | |||||
|
Frozen Layers + DSC + ResNet + DenseNet + EfficientNet + Dice Loss (Model 5) |
0.5608 | 0.4206 | 0.4623 | 0.3713 | 0.4623 |
|
Total params: 23,794,557 Trainable params: 1,656,641 Non-trainable params: 22,137,916 | |||||
|
Frozen Layers + DSC + ResNet + DenseNet + EfficientNet + BCE Loss (Model 6) |
0.5296 | 0.4414 | 0.4559 | 0.3708 | 0.4559 |
|
Total params: 23,794,557 Trainable params: 1,656,641 Non-trainable params: 22,137,916 | |||||
|
DSC + ResNet + DenseNet + EfficientNet + BCE Loss (Model 7) |
0.6336 | 0.5744 | 0.5856 | 0.5076 | 0.5856 |
|
Total params: 23,794,557 Trainable params: 23,794,557 Non-trainable params: 0 | |||||
|
DSC + ResNet + DenseNet + EfficientNet + Dice Loss (Model 8) |
0.6653 | 0.5234 | 0.5710 | 0.4885 | 0.5710 |
|
Total params: 23,794,557 Trainable params: 23,794,557 Non-trainable params: 0 | |||||
|
Non-Local Attention + Frozen Layers + DSC + ResNet + DenseNet + EfficientNet + Dice Loss (Model 9) |
0.5072 | 0.4222 | 0.4183 | 0.3269 | 0.4183 |
|
Total params: 24,493,277 Trainable params: 2,355,361 Non-trainable params: 22,137,916 | |||||
|
CBAM Attention + Frozen Layers + DSC + ResNet + DenseNet + EfficientNet + Dice Loss (Model 10) |
0.5764 | 0.4284 | 0.459 | 0.3660 | 0.4593 |
|
Total params: 23,881,989 Trainable params: 1,744,073 Non-trainable params: 22,137,916 | |||||
|
Non-Local Attention + Frozen Layers + DSC + ResNet + DenseNet + EfficientNet + BCE Loss (Model 11) |
0.5216 | 0.4051 | 0.4256 | 0.3425 | 0.4256 |
|
Total params: 24,493,277 Trainable params: 2,355,361 Non-trainable params: 22,137,916 | |||||
|
CBAM Attention + Frozen Layers + DSC + ResNet + DenseNet + EfficientNet + BCE Loss (Model 12) |
0.5109 | 0.4887 | 0.4660 | 0.3685 | 0.4660 |
|
Total params: 23,881,989 Trainable params: 1,744,073 Non-trainable params: 22,137,916 | |||||
|
Frozen Layers + DSC + DenseNet + EfficientNet + Dice Loss (Model 13) |
0.5572 | 0.4391 | 0.4544 | 0.3620 | 0.4544 |
|
Total params: 12,350,269 Trainable params: 1,388,865 Non-trainable params: 10,961,404 | |||||
|
Frozen Layers + DSC + DenseNet + EfficientNet + BCE Loss (Model 14) |
0.5489 | 0.4058 | 0.4377 | 0.3487 | 0.4377 |
|
Total params: 12,350,269 Trainable params: 1,388,865 Non-trainable params: 10,961,404 | |||||
|
Frozen Layers + DSC + ResNet + EfficientNet + BCE Loss (Model 15) |
0.4548 | 0.2851 | 0.3181 | 0.2466 | 0.3181 |
|
Total params: 16,305,149 Trainable params: 1,121,089 Non-trainable params: 15,184,060 | |||||
|
Frozen Layers + DSC + ResNet + EfficientNet + Dice Loss (Model 16) |
0.4553 | 0.3899 | 0.3829 | 0.3007 | 0.3829 |
|
Total params: 16,305,149 Trainable params: 1,121,089 Non-trainable params: 15,184,060 | |||||
|
Frozen Layers + DSC + ResNet + DenseNet + BCE Loss (Model 17) |
0.0757 | 0.7948 | 0.1261 | 0.0757 | 0.1261 |
|
Total params: 19,117,569 Trainable params: 987,201 Non-trainable params: 18,130,368 | |||||
|
Frozen Layers + DSC + ResNet + DenseNet + Dice Loss (Model 18) |
0.0757 | 0.7948 | 0.1261 | 0.0757 | 0.1261 |
|
Total params: 19,117,569 Trainable params: 987,201 Non-trainable params: 18,130,368 | |||||
|
DSC + DenseNet + EfficientNet + BCE Loss (Model 19) |
0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
|
Total params: 12,350,296 Trainable params: 12,350,296 Non-trainable params: 0 | |||||
|
DSC + DenseNet + EfficientNet + Dice Loss (Model 20) |
0.0757 | 0.7948 | 0.1261 | 0.0757 | 0.1261 |
|
Total params: 12,350,296 Trainable params: 12,350,296 Non-trainable params: 0 | |||||
Precision, recall, F1 Score, IoU, and dice coefficient are among the metrics.
Table 2.
Area Under the Curve (AUC) for different models across training, validation, and test datasets is displayed through ROC curve analysis. While frozen-layer models performed poorly and produced classification results that were almost random, attention-mechanism models (such as CBAM and Non-Local Attention) had the highest AUC scores.
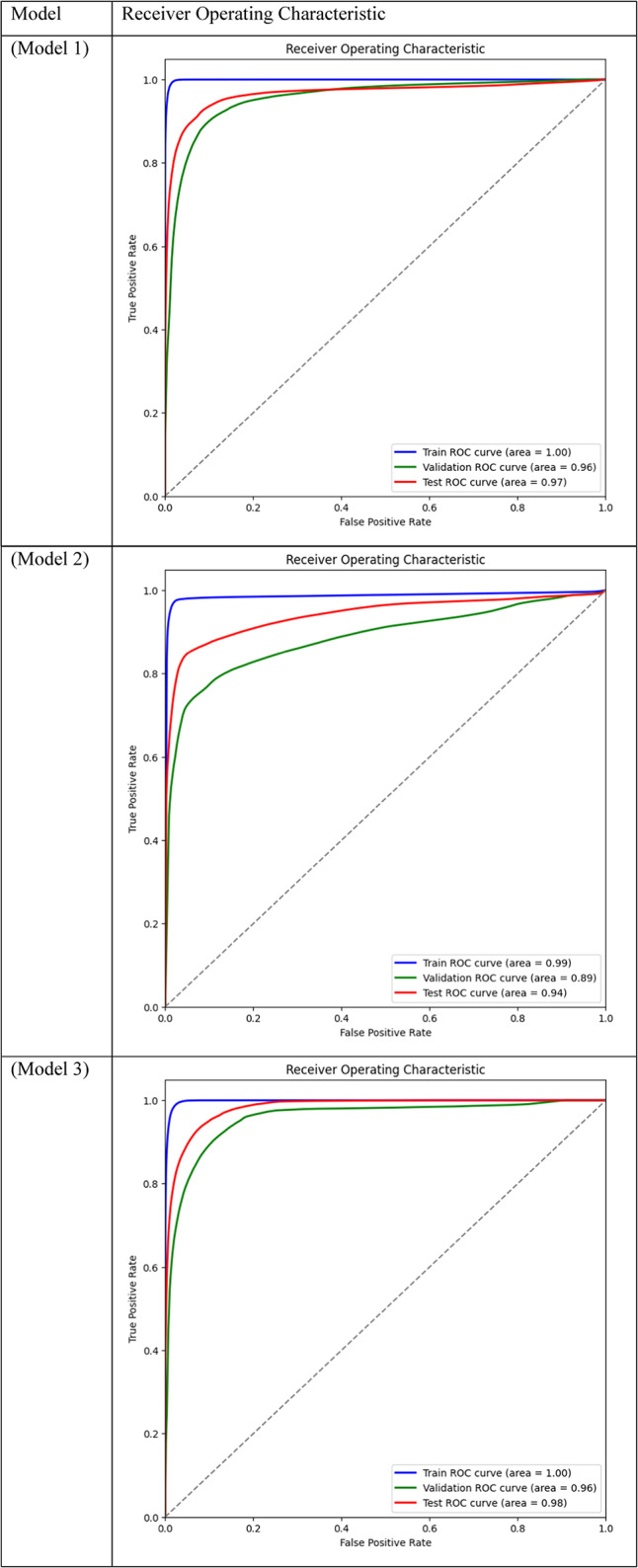
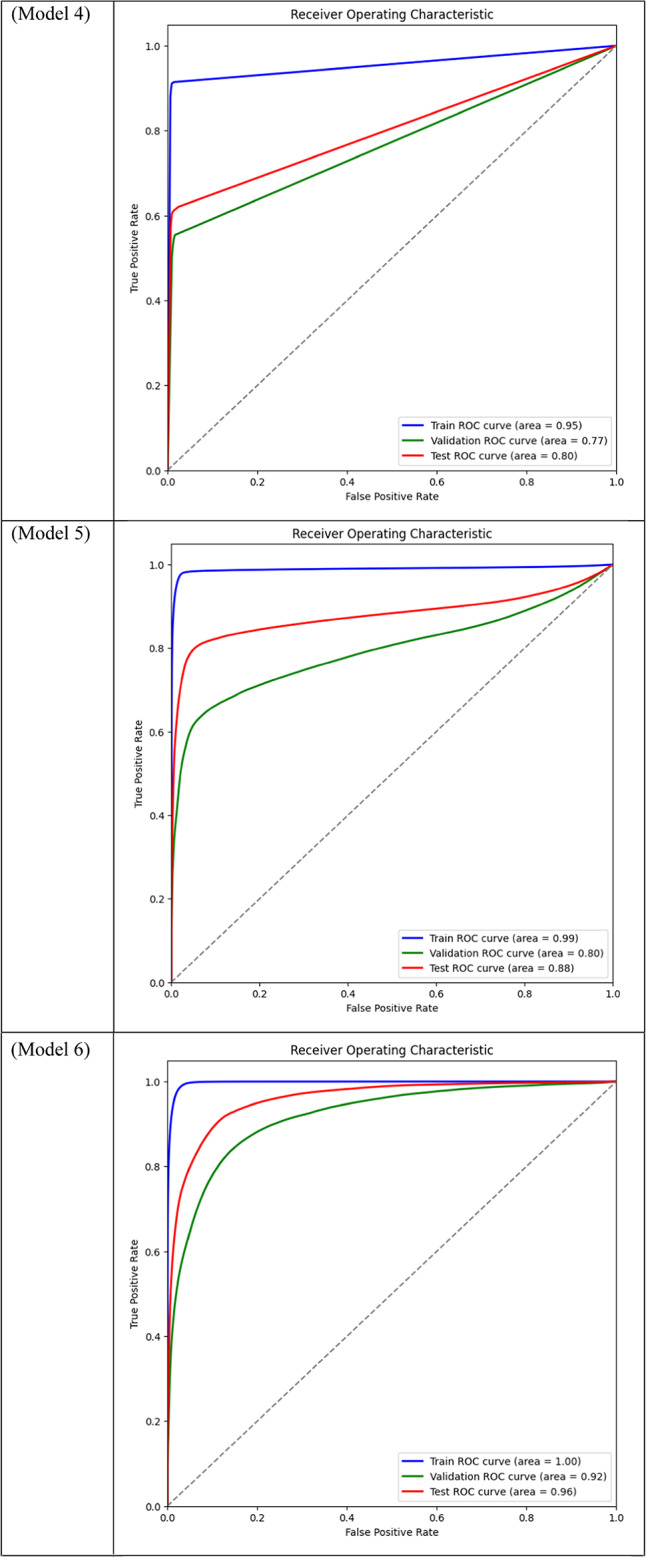
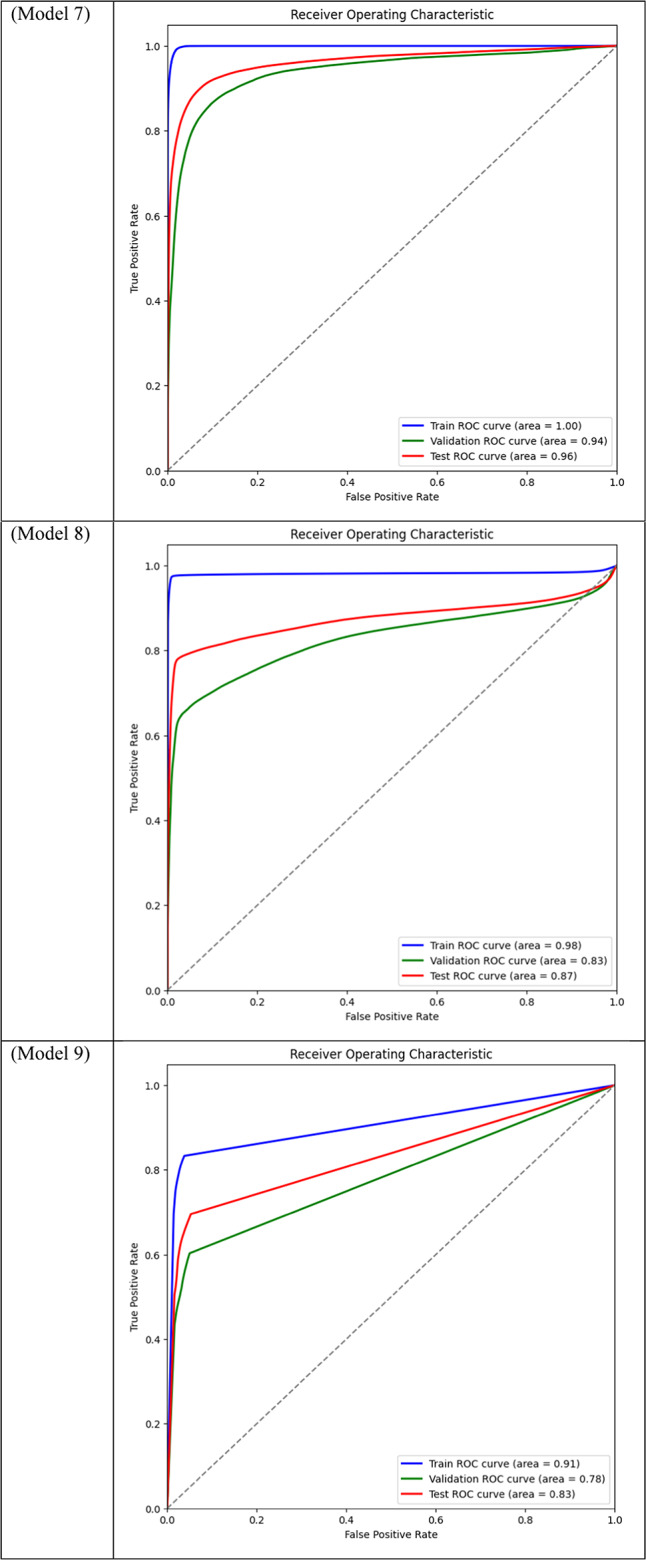
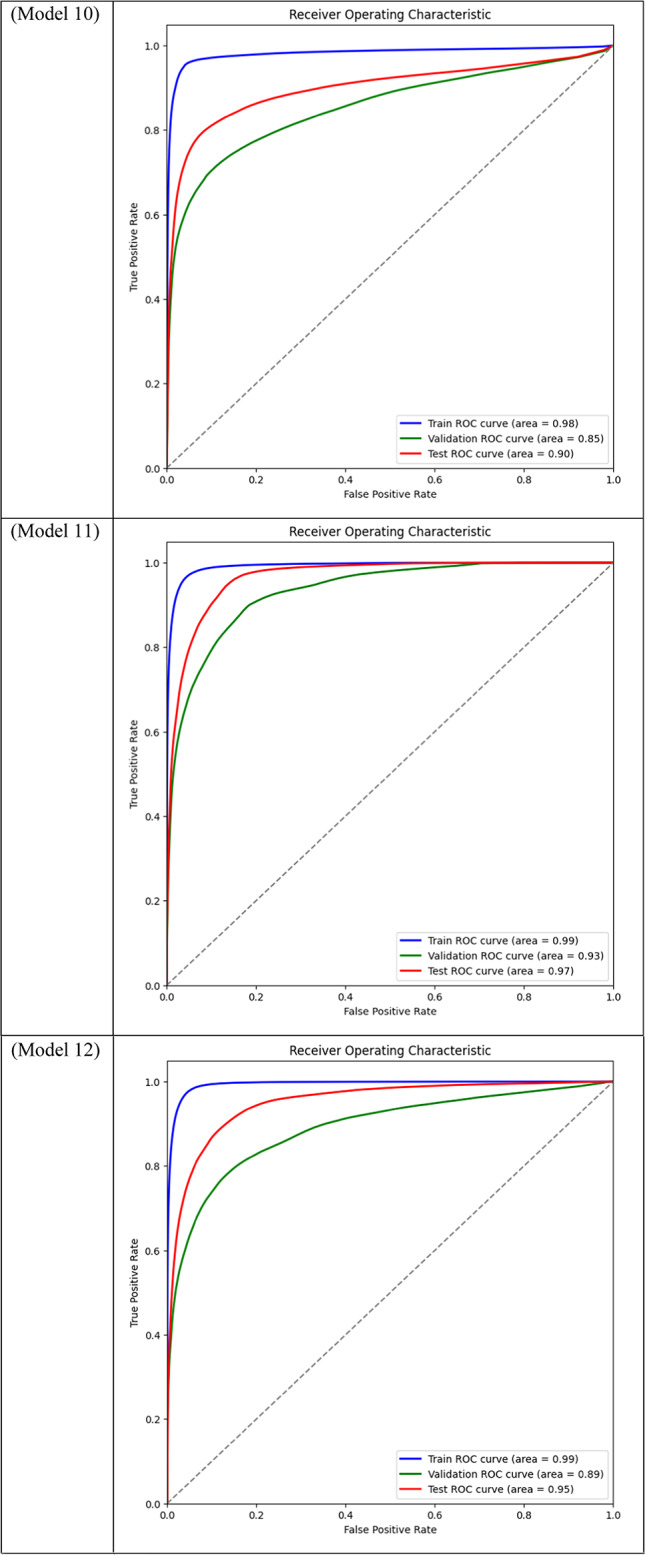
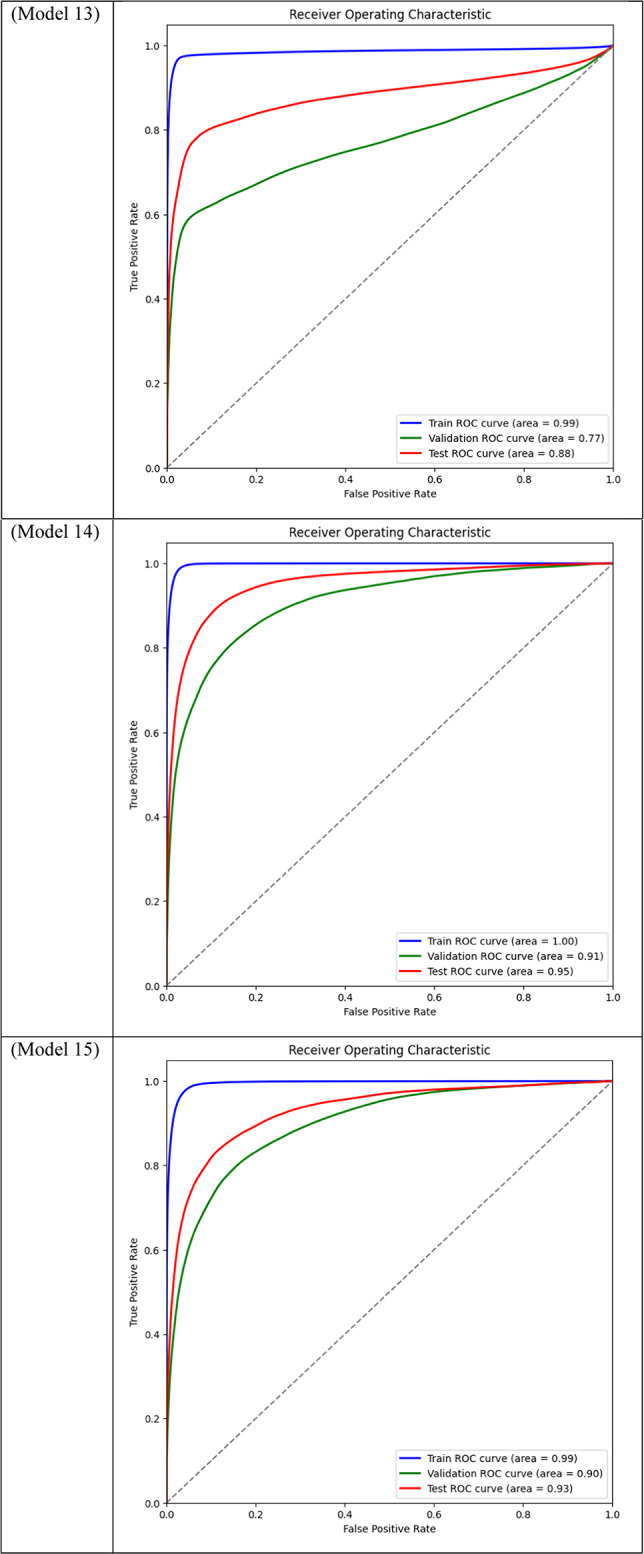
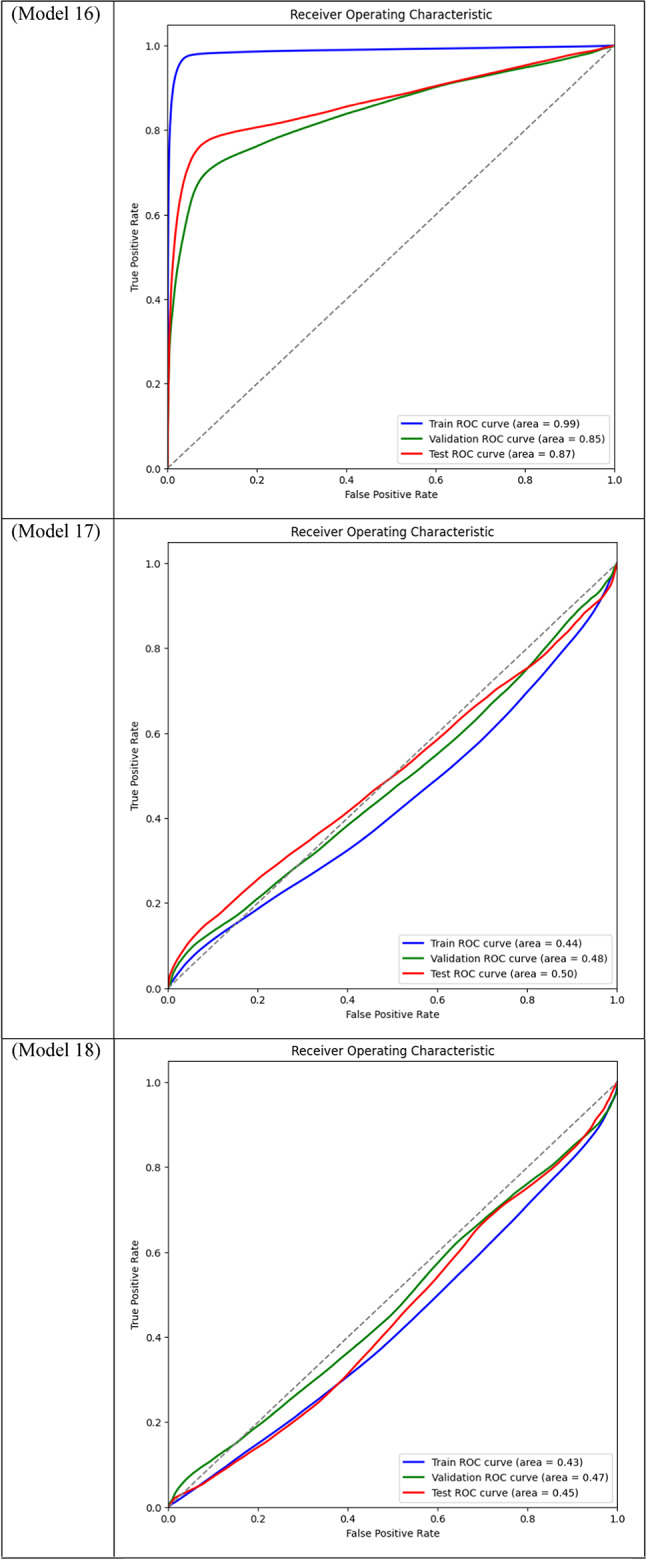
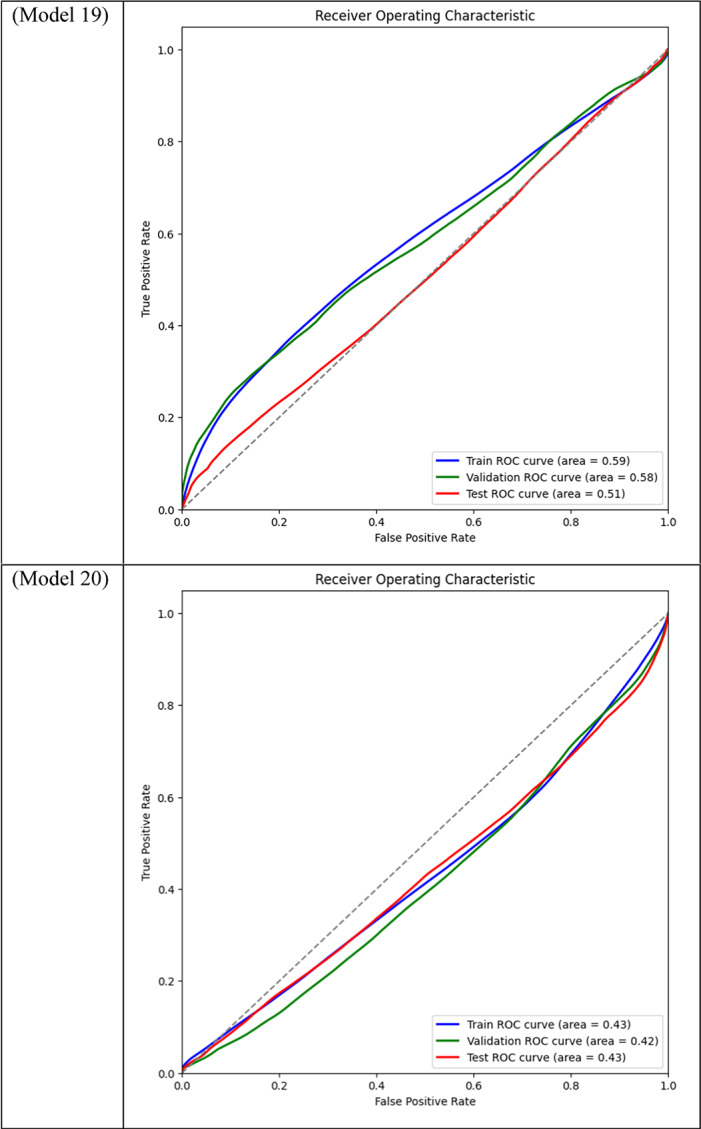
Table 3.
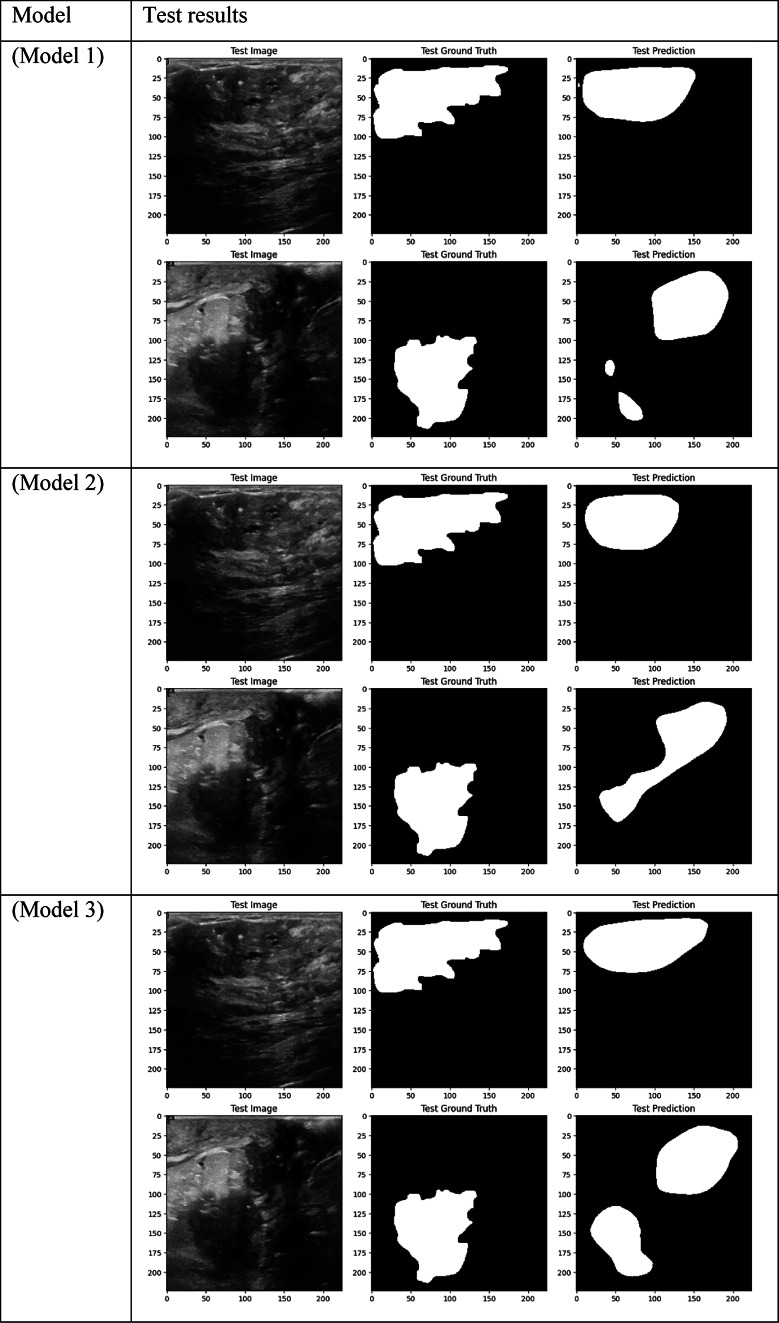
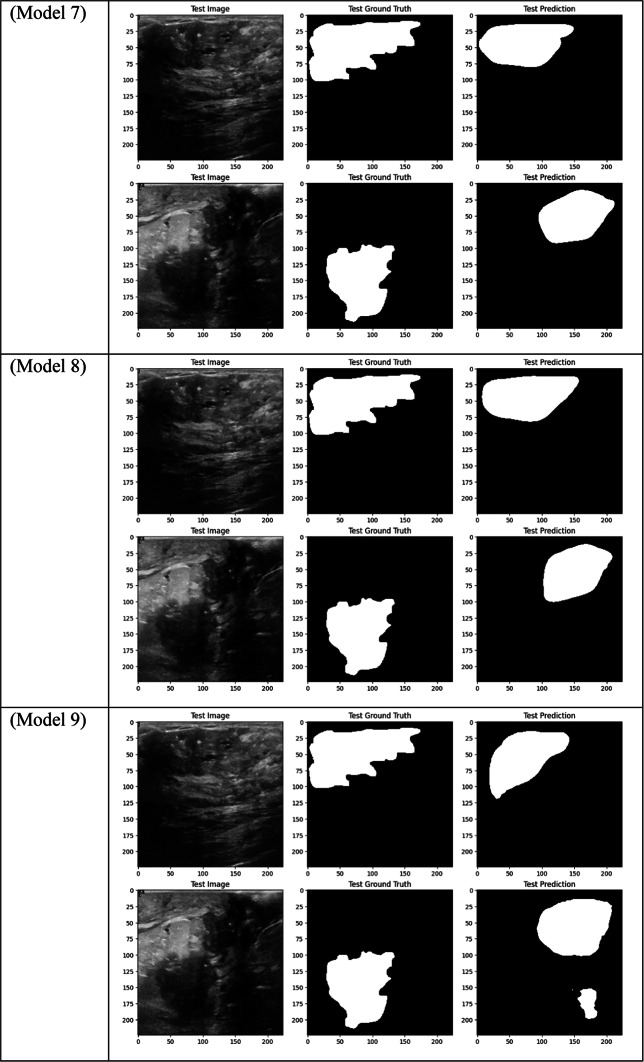
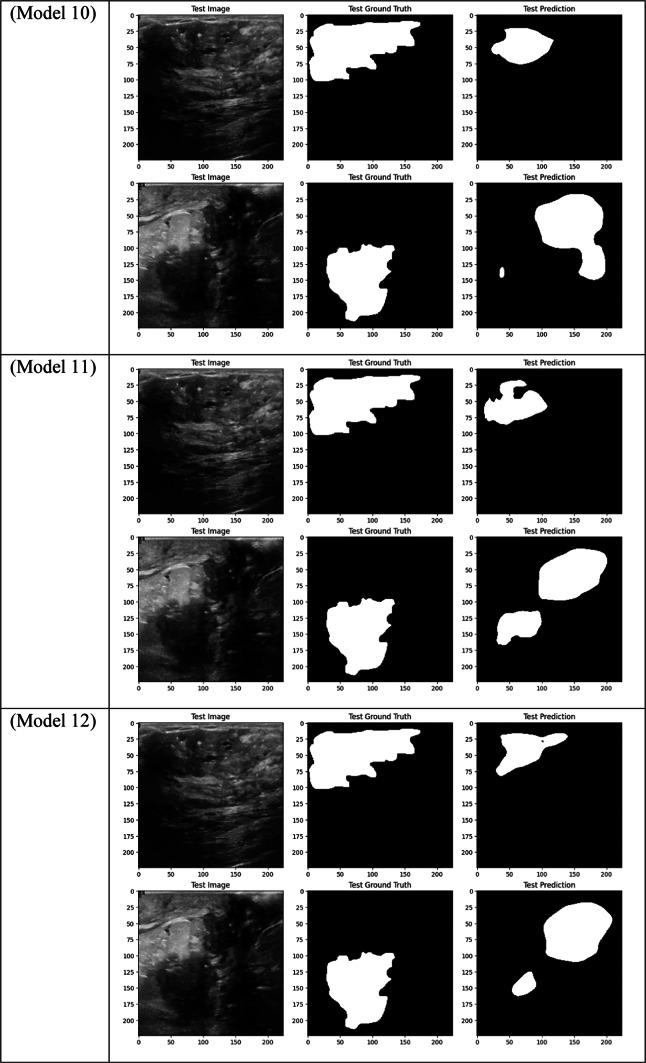
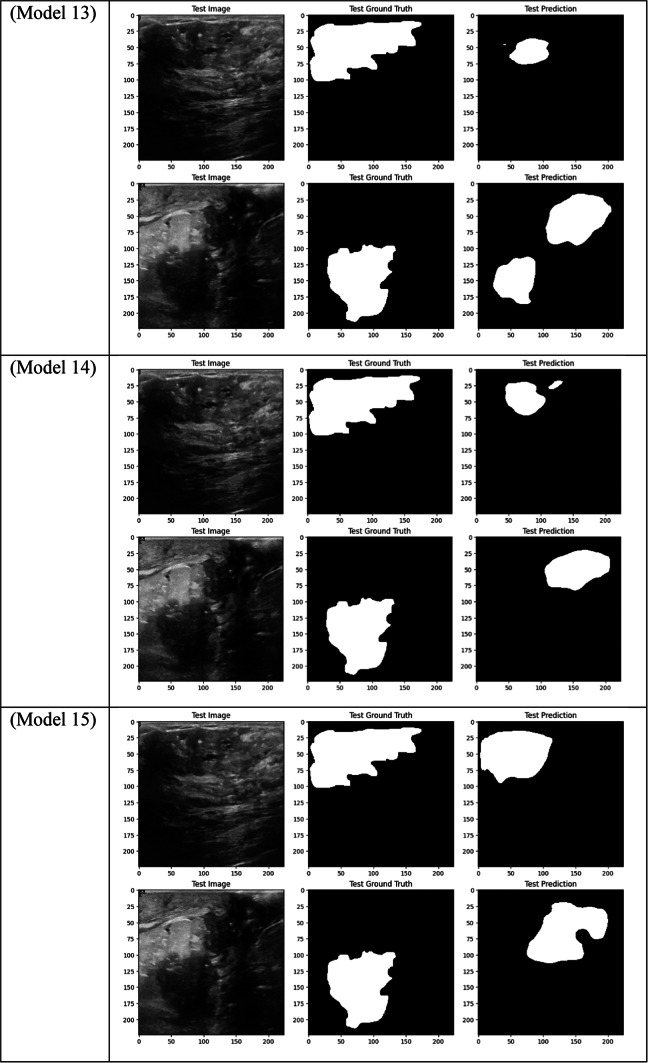
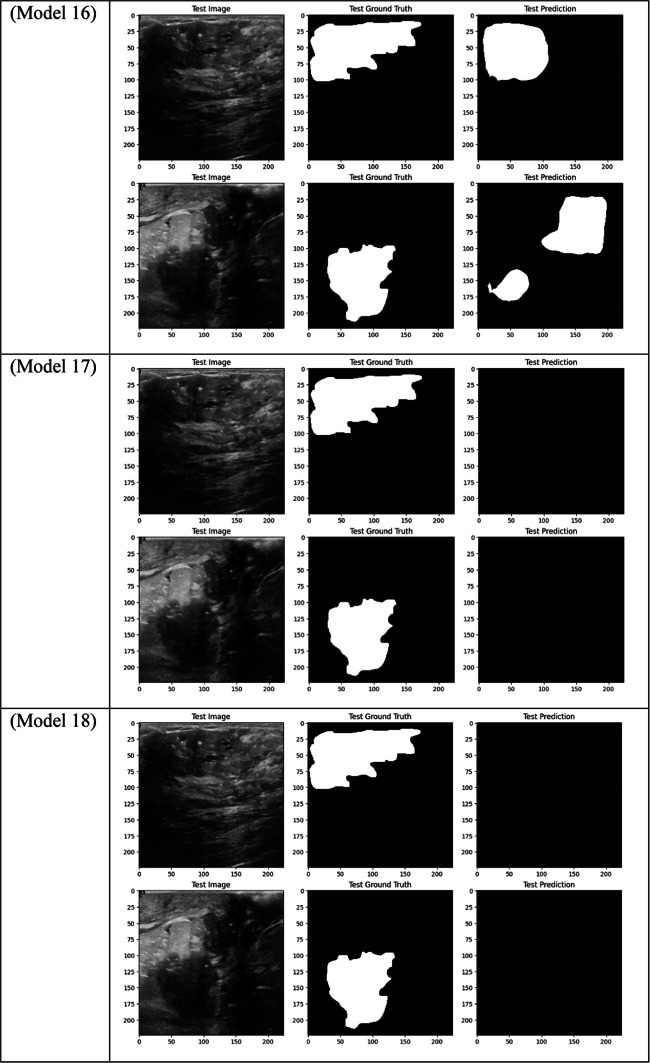
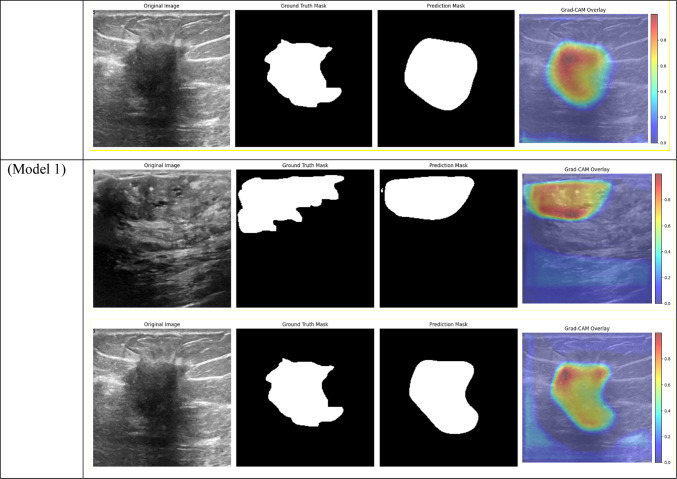
Visual comparison of segmentation outcomes produced by diverse models on the test dataset.
Table 4.
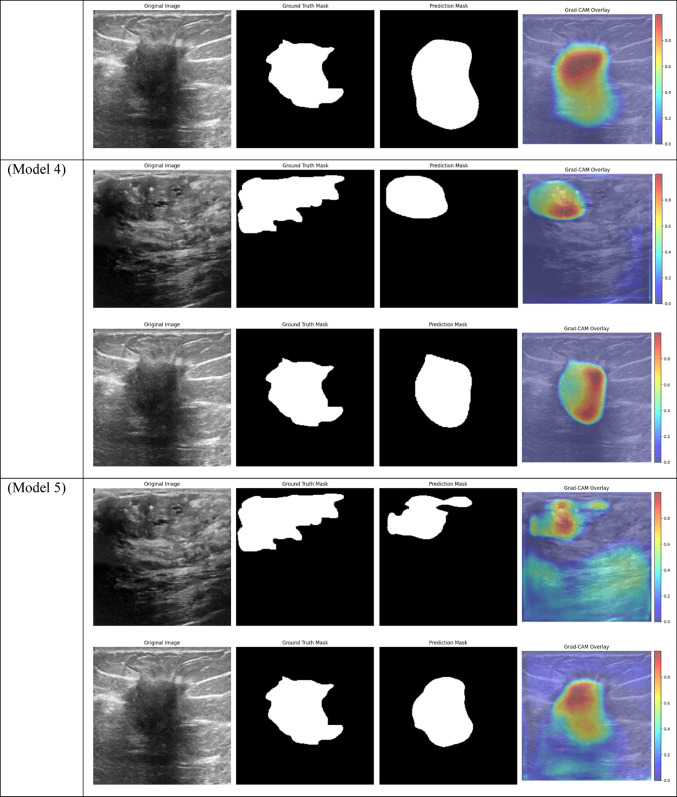
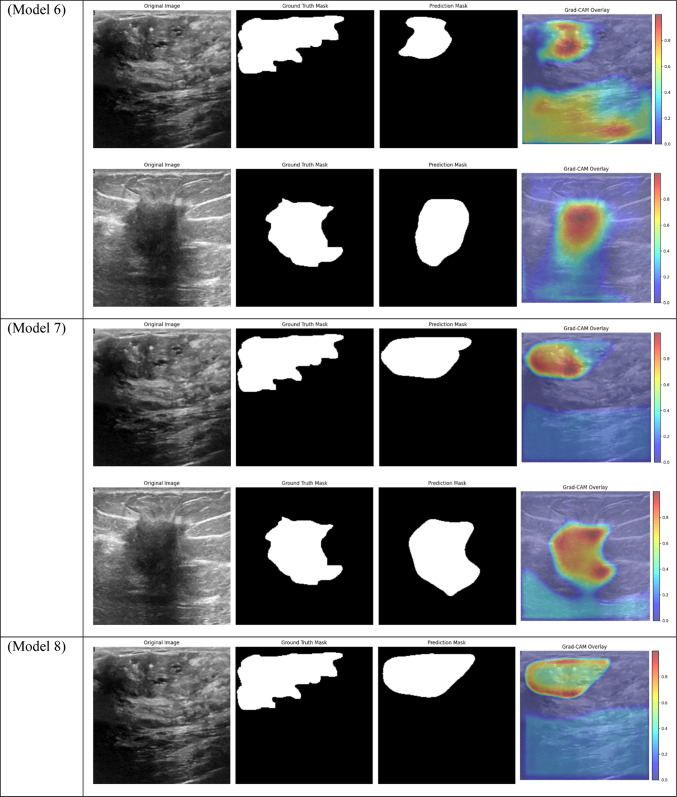
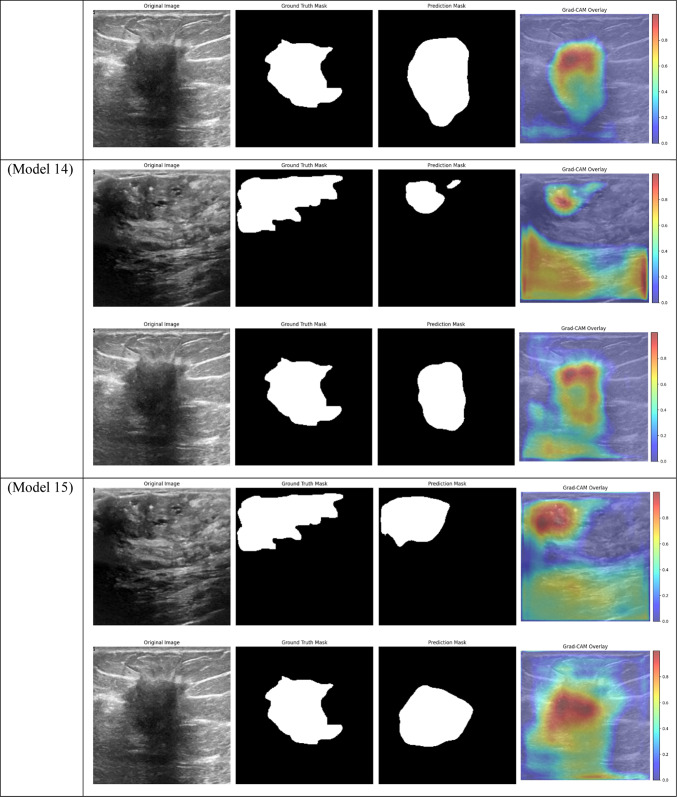
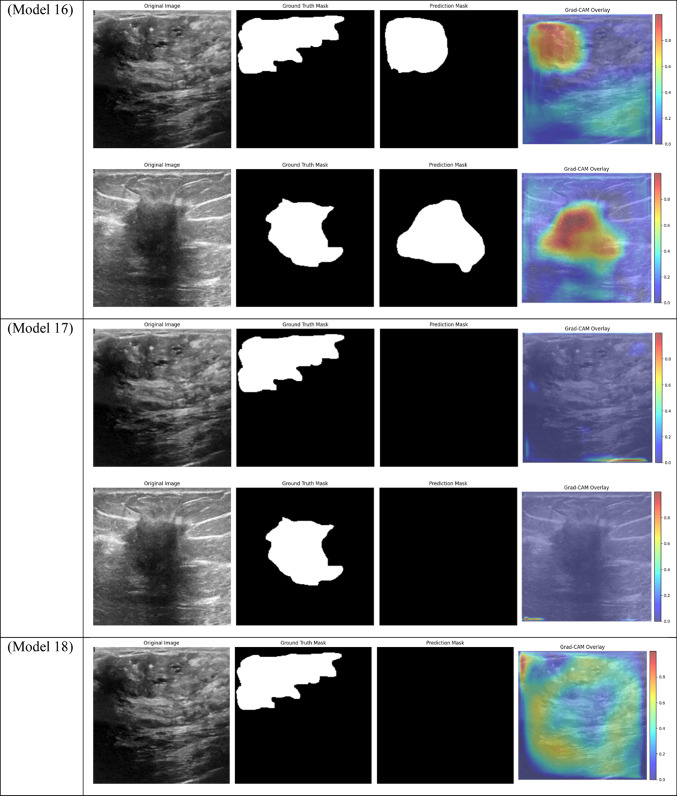
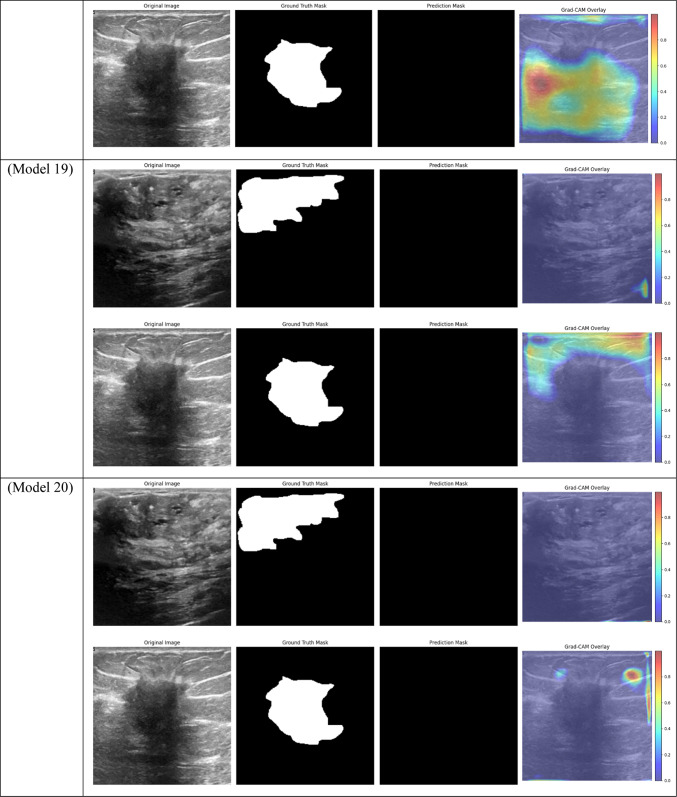
Grad-CAM visualizations of segmentation outcomes from diverse models on the test dataset.
The models that performed best in the breast tumor segmentation task are clearly shown by the performance results, especially when considering the Dice score—a crucial metric for assessing segmentation overlap. With an overall Dice score of 0.6140, Model 1, which combines BCE Loss with CBAM Attention, DSC, ResNet, DenseNet, and EfficientNet encoders, achieved one of the best results. This outcome demonstrates how well CBAM can refine spatial and channel-wise features, enabling the model to concentrate on the most pertinent tumor regions. Model 1 improved the overlap with the ground truth by capturing fine-grained details in the segmentation masks by utilizing CBAM’s dual attention mechanisms.
Model 2, which employs Dice Loss rather than BCE, likewise performed admirably, obtaining a Dice score of 0.5942. Although Dice Loss maximizes segmentation overlap directly, its Recall (0.5946) was marginally lower than that of Model 1, indicating that although the model was successful in optimizing overlap, it might have overlooked certain tumor regions, resulting in a decrease in overall sensitivity. Model 3, which combines BCE Loss and Non-Local Attention, produced similar results, scoring 0.5943 on the Dice scale. In more complex cases where local features alone might not be sufficient, Non-Local Attention helped improve segmentation accuracy by capturing long-range dependencies. It did not, however, outperform models based on CBAM.
However, contrary to what was first believed, Model 8, which uses DSC and a combination of ResNet, DenseNet, and EfficientNet encoders with Dice Loss, did not produce the best Dice results. Despite this, it had good Precision (0.6653) performance, which makes it a good option for tasks where reducing false positives is a top priority. Though it performed well in accurately classifying positive tumor regions, Models 1, 2, and 3’s overall overlap between predicted and true masks was stronger due to their attention mechanisms, as evidenced by their higher Dice score of 0.5710.
With a Dice score of 0.1261, Models 17, 18, and 20 generated identical results, indicating their poorest performance. Also, Model 19 obtains zero values in all metrics. Models 17, and 18, which combined different combinations of ResNet and DenseNet encoders with frozen layers and DSC, were unable to segment the tumor regions efficiently. Their low Dice scores show how poorly the predicted segmentation masks and the ground truth overlap, underscoring the significant limitation of freezing encoder layers on the model’s capacity to learn and adjust to the subtleties of the dataset. This demonstrates how crucial it is to allow trainable encoder layers, especially for challenging tasks like medical image segmentation.
All CBAM Attention metrics, including Dice, showed consistently strong performance from the models, with Model 1 leading the pack. Model 1’s combination of CBAM and BCE Loss produced one of the highest Dice scores, demonstrating how attention mechanisms help the model focus on important features while also enhancing the overlap between the true and predicted masks. Non-local attention models, like Model 3, on the other hand, performed similarly but fell short of CBAM-based models in terms of segmentation accuracy, especially when it came to the Dice score. This suggests that CBAM is more effective at refining spatial and channel-wise features in ultrasound images, while Non-Local Attention aids in capturing broader contextual information.
Models 5 and 6, which are frozen-layer models, also did poorly on the Dice score, with respective scores of 0.4559 and 0.4623. The models’ capacity to adjust to the particular features of the dataset was hampered by freezing the encoder layers, which led to worse segmentation quality. The findings support the requirement for fully trainable networks in medical image segmentation tasks, where precise segmentation depends on optimizing the feature extraction procedure.
As fewer parameters are updated during the backpropagation process, using frozen layers may result in a reduction in training time, which is one of its main benefits. Keeping the general-purpose knowledge from a large pre-trained dataset can also help prevent overfitting in scenarios where the available dataset is small. Nevertheless, the incapacity to modify these pre-trained features turns into a drawback in breast tumor segmentation tasks, where the tumor regions frequently display distinct and intricate patterns. The inability to fine-tune the encoder’s features resulted in incredibly poor segmentation results, with the models failing to capture the pertinent features of the tumor regions, as demonstrated in Models 17, 18, and 20, all of which had frozen layers and received the lowest Dice score of 0.1261.
Furthermore, in specific medical tasks, the inadequate performance of frozen-layer models emphasizes the necessity for fully trainable networks. For the purpose of breast tumor segmentation, the model must concentrate on particular regions of the ultrasound images that might not match the generic features discovered by pre-trained encoders on sizable natural image datasets (like ImageNet). Layers that are frozen prevent the model from adjusting to the unique anatomical features, texture, and contrast of tumors in medical images, which results in less-than-ideal performance. Model 9 provides further evidence that even sophisticated attention mechanisms are unable to make up for the lack of adaptability in the frozen encoder layers. Non-local attention combined with frozen layers in this model results in a Dice score of just 0.4183.
However, fully trainable models fared much better, with a Dice score of 0.6140 attained by Model 1 (CBAM Attention with BCE Loss). Higher segmentation accuracy was achieved by this model’s ability to modify each of its layers, including the encoder, which improved its ability to extract pertinent information from the breast ultrasonography images. Similarly, with a Dice score of 0.5943, Model 3 (Non-Local Attention with BCE Loss) proved the usefulness of trainable encoders. These findings highlight how crucial it is to fine-tune the entire model, particularly for tasks requiring accurate segmentation of intricate anatomical structures.
With a train AUC of 1.00, validation AUC of 0.96, and test AUC of 0.97, Model 1, which combines CBAM Attention with DSC and BCE Loss, performed quite well across all datasets, as shown in Table 2. The small difference between training and test set performance suggests that the model is highly capable of differentiating between tumor and non-tumor regions, with low overfitting. Strong cross-dataset generalization is ensured by the model’s ability to concentrate on the most important features owing to the addition of CBAM.
Model 3, which uses Non-Local Attention, was the best-performing model in terms of test set AUC, likewise performing well with a Train AUC of 1.00, Validation AUC of 0.96, and Test AUC of 0.98. This shows that Non-Local Attention is especially good at identifying long-range dependencies in the images, which improves the model’s capacity to effectively generalize to new data. This high test AUC demonstrates even more how Non-Local Attention enhances the model’s performance on challenging segmentation tasks. In contrast, Model 2, which employs Dice Loss rather than BCE, performed marginally worse on the validation set, as evidenced by its Validation AUC of 0.89 and Test AUC of 0.94. Even though the model’s performance was still good, its AUC was lower than that of Models 1 and 3, indicating that while Dice Loss was a good option for maximizing overlap in the segmentation task, it may not have been as effective at generalizing as BCE Loss. However, the test set performance is relatively strong, suggesting that Model 2 is still a competitive option.
Models 17, 18, 19, and 20, on the other hand, performed the worst across all datasets, with AUC values close to or below 0.50, which suggests almost random performance. Model 17’s Train AUC of 0.44, Validation AUC of 0.48, and Test AUC of 0.50, for example, demonstrate the model’s inability to distinguish between tumor and non-tumor regions. These models made use of DSC and frozen layers; the incredibly low results indicate that freezing layers in these situations severely restricted the model’s capacity to learn from the data, resulting in subpar segmentation performance. Model 4, with a Validation AUC of 0.77 and a Test AUC of 0.80, performed worse than the other models as well. The comparatively wide discrepancy between the test results and the Train AUC (0.95) indicates that this model had trouble generalizing, maybe as a result of overfitting. This problem could be the result of the model being trained with an excessive amount of focus on particular image regions due to the use of Dice Loss in combination with Non-Local Attention.
While they performed fairly well, the mid-performing models—Models 5 and 8 in particular—showed less generalization to the test set. For instance, Model 8’s Train AUC of 0.98, Validation AUC of 0.83, and Test AUC of 0.87 show that, despite learning well from the training set, it performed poorly on the test set that was not shown to it. This decline is probably the result of the absence of attention mechanisms, which made it harder for the model to concentrate on important areas of the pictures and produced less reliable generalizations.
Comparison with the state-of-the-art approaches
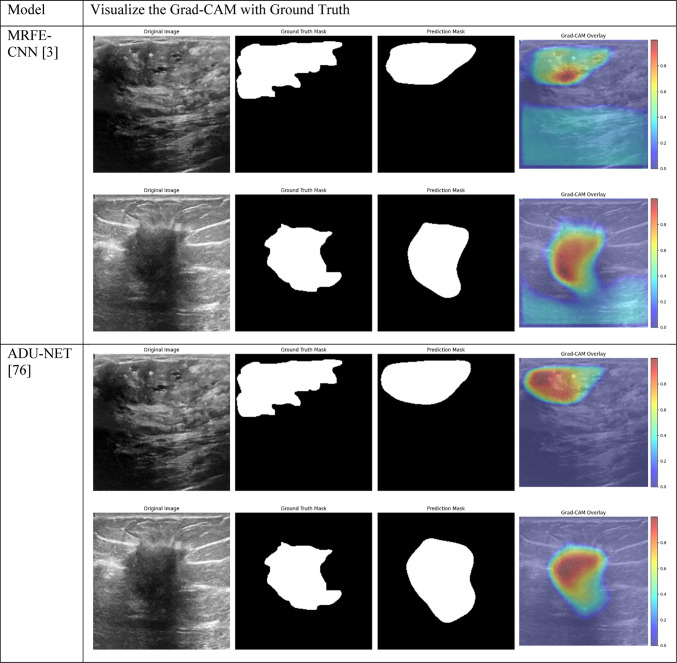
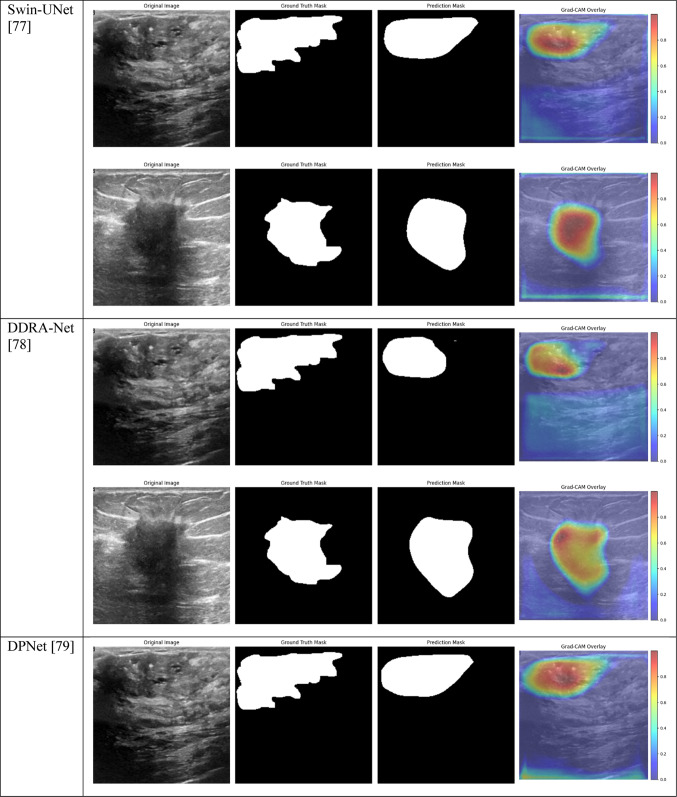
To evaluate the performance of the proposed model, we compared it against five state-of-the-art segmentation models: MRFE-CNN3, ADU-NET75, Swin-UNet76, DDRA-Net77, and DPNet78. Each baseline model represents a different method to medical image segmentation, with unique architectural components, attention mechanisms, or feature extraction approaches. These models were implemented and evaluated on the same BUSI dataset to ensure a fair comparison.
The outcomes in the Tables 5 and 6 highlight the superior performance of the suggested model (Model 1) compared to five state-of-the-art models for breast tumor segmentation. Among all metrics, Model 1 achieved the highest IoU of 0.5305 and Dice score of 0.6140, indicating its ability to effectively capture and segment tumor areas. While ADU-NET and DDRA-Net also performed well, achieving Dice scores of 0.5856 and 0.5872, respectively, they fell short in IoU, implying relatively less overlap between predicted and ground-truth segmentation masks. Furthermore, Swin-UNet and MRFE-CNN demonstrated competitive precision but lower recall, which suggests these models struggled with accurately identifying all tumor regions.
Table 5.
Performance comparison of the suggested model with state-of-the-art techniques in breast tumor segmentation on the BUSI dataset.
| Model | Precision | Recall | F1 score | IoU | Dice |
|---|---|---|---|---|---|
| MRFE-CNN3 | 0.6624 | 0.5318 | 0.5690 | 0.4783 | 0.5690 |
| ADU-NET75 | 0.6081 | 0.5964 | 0.5856 | 0.5019 | 0.5856 |
| Swin-UNet76 | 0.6165 | 0.5639 | 0.5693 | 0.4825 | 0.5693 |
| DDRA-Net77 | 0.6664 | 0.5529 | 0.5872 | 0.4996 | 0.5872 |
| DPNet78 | 0.5421 | 0.5611 | 0.5257 | 0.4433 | 0.5257 |
|
CBAM Attention + DSC + ResNet + DenseNet + EfficientNet + BCE Loss (Model 1) |
0.6194 | 0.6378 | 0.6140 | 0.5305 | 0.6140 |
The maximum values for each metric are highlighted in bold for clarity.
Table 6.
Grad-CAM visualizations of segmentation outcomes of the suggested model with state-of-the-art techniques in breast tumor segmentation on the BUSI dataset.
From a recall and precision perspective, the suggested model demonstrates an optimal balance. It achieved a recall of 0.6378, outperforming all other models, which reveals its ability to identify more tumor areas correctly. Although DDRA-Net had a slightly higher precision at 0.6664, its recall was significantly lower at 0.5529, indicating a trade-off between identifying tumors and avoiding false positives. On the other hand, models such as DPNet, which attained the lowest IoU and Dice scores, struggled to segment tumor areas accurately, likely due to limitations in feature extraction and attention mechanisms.
Overall, the results highlight the advantages of integrating Non-Local Attention, CBAM, and multiple encoder designs in the proposed model. Compared to transformer-based Swin-UNet, which demonstrated strong performance in some metrics, the suggested model provided a more balanced and computationally efficient solution, better suited for clinical deployment. These findings underscore the robustness and effectiveness of our architecture for breast tumor segmentation tasks, providing significant improvements over existing techniques.
Conclusion and future works
Using the BUSI dataset, this study investigated the use of DL techniques for breast tumor segmentation, with an emphasis on the integration of attention mechanisms, various encoder architectures, and loss functions. Higher Dice scores, IoU, and AUC values consistently indicated superior performance in tumor region segmentation for models that included attention mechanisms like CBAM and Non-Local Attention. More precise segmentation was achieved by these attention-based models because they were better able to concentrate on important elements present in the ultrasound images. On the other hand, models lacking attention mechanisms or with frozen layers typically performed worse, underscoring the significance of fully trainable networks and sophisticated attention mechanisms in medical image segmentation.
Although both Dice Loss and BCE Loss produced good model performance, the choice of loss function affected how well recall and precision were balanced. While Dice Loss maximized the overlap between the predicted and ground truth segmentation masks, BCE Loss typically produced higher recall. By demonstrating that attention-based models consistently focused on the appropriate tumor regions while non-attention models tended to focus on irrelevant areas, the Grad-CAM visualizations further supported the significance of attention mechanisms. Overall, the study showed that the segmentation performance of breast tumors can be greatly enhanced by combining deep feature extraction via sophisticated encoders, attention mechanisms, and suitable loss functions.
Utilizing sophisticated encoder architectures, attention mechanisms, and modified loss functions, the model can automate the segmentation procedure with high accuracy, diminishing the dependence on laborious manual delineation by radiologists. This automation can reduce subjectivity in tumor border delineation and yield consistent outcomes across diverse imaging settings. Additionally, the interpretability of the model is improved by Grad-CAM visualizations, which allows clinicians to trust the segmentation results by comprehending the image regions that the model prioritizes. The computational demands of the suggested method may present difficulties in resource-limited settings; however, forthcoming refinements, including model compression and streamlined inference pipelines, could render real-time deployment achievable. Integrating this framework into clinical workflows could provide radiologists with a second opinion, improve diagnostic accuracy, and ultimately improve patient outcomes by enabling the earlier and more reliable detection of breast malignancies.
Future research could go in several directions to improve breast ultrasound image segmentation performance even more. Investigating more sophisticated attention mechanisms, such as transformer-based attention or self-attention modules, is one possible avenue to pursue to enhance the model’s capacity to comprehend the global context in intricate medical images. Furthermore, experimenting with hybrid loss functions—which combine the advantages of Dice Loss and BCE—might produce results that are more evenly distributed in terms of segmentation overlap, recall, and precision.
To increase the segmentation model’s adaptability, additional research is being done on the integration of multi-modal imaging data, which combines ultrasound images with those from MRIs and mammograms. To further generalize the model and lower the likelihood of overfitting, the dataset should be expanded to include a wider variety of breast ultrasonography images. Ultimately, the integration of DL-based segmentation systems into medical practice may be facilitated by the real-time application of these models for clinical use, together with interpretability tools like Grad-CAM to foster confidence among clinicians.
Author contributions
Shokofeh Anari: Conceptualization, Investigation, Methodology, Software. Soroush Sadeghi: Formal analysis, Software, Validation. Ghazaal Sheikhi : Methodology, Investigation. Ramin Ranjbarzadeh: Supervisor, Writing- Reviewing and Editing, Validation. Malika Bendechache: Supervisor, Writing- Reviewing and Editing, Validation.
Data availability
The dataset is available at: https://www.kaggle.com/datasets/sabahesaraki/breast-ultrasound-images-dataset/data
Declarations
Competing interests
The authors declare that there are no conflicting or financial interests.
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhao, X. et al. BreastDM: A DCE-MRI dataset for breast tumor image segmentation and classification. Comput. Biol. Med.164, 107255. 10.1016/J.COMPBIOMED.2023.107255 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Ranjbarzadeh, R. et al. Breast tumor localization and segmentation using machine learning techniques: Overview of datasets, findings, and methods. Comput. Biol. Med.152, 106443. 10.1016/J.COMPBIOMED.2022.106443 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Ranjbarzadeh, R. et al. MRFE-CNN: Multi-route feature extraction model for breast tumor segmentation in Mammograms using a convolutional neural network. Ann. Oper. Res. 1–22. 10.1007/S10479-022-04755-8 (2022).
- 4.Srivastava, U. P., Vaidehi, V., Koirala, T. K. & Ghosal, P. Performance analysis of an ANN-based model for breast cancer classification using wisconsin dataset. In Proceedings of 2023 International Conference on Intelligent Systems, Advanced Computing and Communication, ISACC 2023 (2023). 10.1109/ISACC56298.2023.10083642.
- 5.Anari, S., de Oliveira, G. G., Ranjbarzadeh, R., Alves, A. M., Vaz, G. C. & Bendechache, M. EfficientUNetViT: Efficient breast tumor segmentation utilizing U-Net architecture and pretrained vision transformer (2024). 10.20944/PREPRINTS202408.1015.V1. [DOI] [PMC free article] [PubMed]
- 6.Mahmood, T. et al. A brief survey on breast cancer diagnostic with deep learning schemes using multi-image modalities. IEEE Access8, 165779–165809. 10.1109/access.2020.3021343 (2020). [Google Scholar]
- 7.Benhammou, Y., Achchab, B., Herrera, F. & Tabik, S. BreakHis based breast cancer automatic diagnosis using deep learning: Taxonomy, survey and insights. Neurocomputing375, 9–24. 10.1016/j.neucom.2019.09.044 (2020). [Google Scholar]
- 8.Borah, N., Varma, P. S. P., Datta, A., Kumar, A., Baruah, U. & Ghosal, P. Performance analysis of breast cancer classification from mammogram images using vision transformer. In 2022 IEEE Calcutta Conference, CALCON 2022 - Proceedings, 238–243 (2022). 10.1109/CALCON56258.2022.10060315.
- 9.Peng, C. et al. LMA-Net: A lesion morphology aware network for medical image segmentation towards breast tumors. Comput. Biol. Med.147, 105685. 10.1016/J.COMPBIOMED.2022.105685 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Yan, Y. et al. Accurate segmentation of breast tumors using AE U-net with HDC model in ultrasound images. Biomed. Signal Process. Control72, 103299. 10.1016/J.BSPC.2021.103299 (2022). [Google Scholar]
- 11.Tagnamas, J., Ramadan, H., Yahyaouy, A. & Tairi, H. Multi-task approach based on combined CNN-transformer for efficient segmentation and classification of breast tumors in ultrasound images. Vis. Comput. Ind. Biomed. Art7(1), 1–15. 10.1186/S42492-024-00155-W/TABLES/8 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarbakhsh, P. Spatial attention mechanism and cascade feature extraction in a U-Net model for enhancing breast tumor segmentation. Appl. Sci.13(15), 8758. 10.3390/APP13158758 (2023). [Google Scholar]
- 13.Ranjbarzadeh, R., Zarbakhsh, P., Caputo, A., Tirkolaee, E. B. & Bendechache, M. Brain tumor segmentation based on an optimized convolutional neural network and an improved chimp optimization algorithm (2022). 10.21203/RS.3.RS-2203596/V1. [DOI] [PubMed]
- 14.Bagherian Kasgari, A., Ranjbarzadeh, R., Caputo, A., Baseri Saadi, S. & Bendechache, M. Brain tumor segmentation based on zernike moments, enhanced ant lion optimization, and convolutional neural network in MRI images 345–366 (2023). 10.1007/978-3-031-42685-8_10.
- 15.Iqbal, A. & Sharif, M. PDF-UNet: A semi-supervised method for segmentation of breast tumor images using a U-shaped pyramid-dilated network. Expert Syst. Appl.221, 119718. 10.1016/J.ESWA.2023.119718 (2023). [Google Scholar]
- 16.Lachinov, D., Vasiliev, E. & Turlapov, V. Glioma segmentation with cascaded Unet. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Vol. 11384 LNCS, 189–198 (2019). 10.1007/978-3-030-11726-9_17/COVER/.
- 17.Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N. & Liang, J. UNet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging39(6), 1856–1867. 10.1109/TMI.2019.2959609 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, W., Liu, B., Peng, S., Sun, J. & Qiao, X. S3D-UNET: Separable 3D U-Net for brain tumor segmentation. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Vol. 11384 LNCS 358–368 (2019). 10.1007/978-3-030-11726-9_32/COVER/.
- 19.Agarwal, R., Ghosal, P., Sadhu, A. K., Murmu, N. & Nandi, D. Multi-scale dual-channel feature embedding decoder for biomedical image segmentation. Comput. Methods Programs Biomed.257, 108464. 10.1016/J.CMPB.2024.108464 (2024). [DOI] [PubMed] [Google Scholar]
- 20.Li, Y., Liu, Y., Huang, L., Wang, Z. & Luo, J. Deep weakly-supervised breast tumor segmentation in ultrasound images with explicit anatomical constraints. Med. Image Anal.76, 102315. 10.1016/J.MEDIA.2021.102315 (2022). [DOI] [PubMed] [Google Scholar]
- 21.Ranjbarzadeh, R. et al. ME-CCNN: Multi-encoded images and a cascade convolutional neural network for breast tumor segmentation and recognition. Artif. Intell. Rev.2023, 1–38. 10.1007/S10462-023-10426-2 (2023). [Google Scholar]
- 22.Chen, G. et al. ESKNet: An enhanced adaptive selection kernel convolution for ultrasound breast tumors segmentation. Expert Syst. Appl.246, 123265. 10.1016/J.ESWA.2024.123265 (2024). [Google Scholar]
- 23.Wang, L. et al. Progressive dual priori network for generalized breast tumor segmentation. IEEE J. Biomed. Health Inform.10.1109/JBHI.2024.3410274 (2024). [DOI] [PubMed] [Google Scholar]
- 24.Ru, J. et al. Attention guided neural ODE network for breast tumor segmentation in medical images. Comput. Biol. Med.159, 106884. 10.1016/J.COMPBIOMED.2023.106884 (2023). [DOI] [PubMed] [Google Scholar]
- 25.Luo, Y., Huang, Q. & Li, X. Segmentation information with attention integration for classification of breast tumor in ultrasound image. Pattern Recognit.124, 108427. 10.1016/J.PATCOG.2021.108427 (2022). [Google Scholar]
- 26.Li, S., Dong, M., Du, G. & Mu, X. Attention dense-U-Net for automatic breast mass segmentation in digital mammogram. IEEE Access7, 59037–59047. 10.1109/ACCESS.2019.2914873 (2019). [Google Scholar]
- 27.Zhang, J., Saha, A., Zhu, Z. & Mazurowski, M. A. Hierarchical convolutional neural networks for segmentation of breast tumors in MRI With application to radiogenomics. IEEE Trans. Med. Imaging38(2), 435–447. 10.1109/TMI.2018.2865671 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Zhang, S. et al. Fully automatic tumor segmentation of breast ultrasound images with deep learning. J. Appl. Clin. Med. Phys.24(1), e13863. 10.1002/ACM2.13863 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Dhabyani, W., Gomaa, M., Khaled, H. & Fahmy, A. Dataset of breast ultrasound images. Data Brief.28, 104863. 10.1016/J.DIB.2019.104863 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar, A., Ghosal, P., Kundu, S. S., Mukherjee, A. & Nandi, D. A lightweight asymmetric U-Net framework for acute ischemic stroke lesion segmentation in CT and CTP images. Comput. Methods Programs Biomed.226, 107157. 10.1016/J.CMPB.2022.107157 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Lew, C. O. et al. A publicly available deep learning model and dataset for segmentation of breast, fibroglandular tissue, and vessels in breast MRI. Sci. Rep.14(1), 1–10. 10.1038/s41598-024-54048-2 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bin Yang, K., Lee, J. & Yang, J. Multi-class semantic segmentation of breast tissues from MRI images using U-Net based on Haar wavelet pooling. Sci. Rep.13(1), 1–12. 10.1038/s41598-023-38557-0 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sunnetci, K. M., Kaba, E., Beyazal Çeliker, F. & Alkan, A. Comparative parotid gland segmentation by using ResNet-18 and MobileNetV2 based DeepLab v3+ architectures from magnetic resonance images. Concurr. Comput.35(1), e7405. 10.1002/CPE.7405 (2023). [Google Scholar]
- 34.Kang, Z., Xiao, E., Li, Z. & Wang, L. Deep learning based on ResNet-18 for classification of prostate imaging-reporting and data system category 3 lesions. Acad. Radiol.31(6), 2412–2423. 10.1016/J.ACRA.2023.12.042 (2024). [DOI] [PubMed] [Google Scholar]
- 35.Kausalya, K., Chandra, B., Kumar, S. B. & Riyaz, A. M. Enhancing chronic kidney disease diagnosis through Densenet-121 approach. In Proceedings—2024 4th International Conference on Pervasive Computing and Social Networking, ICPCSN 2024, 55–62 (2024). 10.1109/ICPCSN62568.2024.00017.
- 36.Kaushik, P., Rathore, R., Kumar, A., Goshi, G. & Sharma, P. Identifying melanoma skin disease using convolutional neural network DenseNet-121. In 2024 IEEE International Conference on Interdisciplinary Approaches in Technology and Management for Social Innovation, IATMSI 2024 (2024). 10.1109/IATMSI60426.2024.10502880.
- 37.Bello, A., Ng, S.-C. & Leung, M.-F. Skin cancer classification using fine-tuned transfer learning of DENSENET-121. Appl. Sci.14(17), 7707. 10.3390/APP14177707 (2024). [Google Scholar]
- 38.Kansal, K., Chandra, T. B. & Singh, A. ResNet-50 vs. EfficientNet-B0: Multi-centric classification of various lung abnormalities using deep learning. Procedia Comput. Sci.235, 70–80. 10.1016/J.PROCS.2024.04.007 (2024). [Google Scholar]
- 39.S. A. A, L. S. G, M. S. M, & J. R. I. A deep learning model for diagnosing diabetic retinopathy using EfficientNet-B0. In 2024 IEEE Students Conference on Engineering and Systems (SCES), 1–5 (2024). 10.1109/SCES61914.2024.10652422.
- 40.Li, S., Tian, Y., Wang, C., Wu, H. & Zheng, S. Cross spectral and spatial scale non-local attention-based unsupervised pansharpening network. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens.16, 4858–4870. 10.1109/JSTARS.2023.3278296 (2023). [Google Scholar]
- 41.Wang, H., Zhai, D., Liu, X., Jiang, J. & Gao, W. Unsupervised deep exemplar colorization via pyramid dual non-local attention. IEEE Trans. Image Process.32, 4114–4127. 10.1109/TIP.2023.3293777 (2023). [DOI] [PubMed] [Google Scholar]
- 42.Wu, X., Zhang, K., Hu, Y., He, X. & Gao, X. Multi-scale non-local attention network for image super-resolution. Signal Process.218, 109362. 10.1016/J.SIGPRO.2023.109362 (2024). [Google Scholar]
- 43.Woo, S., Park, J., Lee, J.-Y. & Kweon, I. S. CBAM: Convolutional block attention module (2018).
- 44.Wang, L. et al. Deep learning driven real time topology optimization based on improved convolutional block attention (Cba-U-Net) model. Eng. Anal. Bound. Elem.147, 112–124. 10.1016/J.ENGANABOUND.2022.11.034 (2023). [Google Scholar]
- 45.Ricciardi, C. et al. On the use of a convolutional block attention module in deep learning-based human activity recognition with motion sensors. Diagnostics13(11), 1861. 10.3390/DIAGNOSTICS13111861 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng, X., Ma, Y., Zhang, X. & Zhu, H. Convolutional block attention module-based neural network for enhanced IQ imbalance estimation in low signal-to-noise ratio environments. IEEE International Conference on Communications, 4066–4071 (2024). 10.1109/ICC51166.2024.10622606.
- 47.Dong, H. et al. A malicious code detection method based on stacked depthwise separable convolutions and attention mechanism. Sensors23(16), 7084. 10.3390/S23167084 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batool, A. & Byun, Y. C. Lightweight EfficientNetB3 model based on depthwise separable convolutions for enhancing classification of leukemia white blood cell images. IEEE Access11, 37203–37215. 10.1109/ACCESS.2023.3266511 (2023). [Google Scholar]
- 49.Ouzar, Y., Djeldjli, D., Bousefsaf, F. & Maaoui, C. X-iPPGNet: A novel one stage deep learning architecture based on depthwise separable convolutions for video-based pulse rate estimation. Comput. Biol. Med.154, 106592. 10.1016/J.COMPBIOMED.2023.106592 (2023). [DOI] [PubMed] [Google Scholar]
- 50.Guo, Q., Wang, C., Xiao, D. & Huang, Q. A novel multi-label pest image classifier using the modified Swin Transformer and soft binary cross entropy loss. Eng. Appl. Artif. Intell.126, 107060. 10.1016/J.ENGAPPAI.2023.107060 (2023). [Google Scholar]
- 51.Bruch, S., Wang, X., Bendersky, M. & Najork, M.An analysis of the softmax cross entropy loss for learning-to-rank with binary relevance. In ICTIR 2019—Proceedings of the 2019 ACM SIGIR International Conference on Theory of Information Retrieval, New York, NY, USA: Association for Computing Machinery, Inc, Sep. 75–78 (2019). 10.1145/3341981.3344221.
- 52.Ming, Q. & Xiao, X. Towards accurate medical image segmentation with gradient-optimized dice loss. IEEE Signal Process. Lett.31, 191–195. 10.1109/LSP.2023.3329437 (2024). [Google Scholar]
- 53.Yeung, M. et al. Calibrating the dice loss to handle neural network overconfidence for biomedical image segmentation. J. Digit Imaging36(2), 739–752. 10.1007/S10278-022-00735-3/FIGURES/5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan, J. et al. EFFC-Net: Lightweight fully convolutional neural networks in remote sensing disaster images. Geo-spatial Inf. Sci.10.1080/10095020.2023.2183145 (2023). [Google Scholar]
- 55.Shanmuga, S. M. & Bhambri, P. Bone Marrow cancer detection from leukocytes using neural networks. Computational Intelligence and Blockchain in Biomedical and Health Informatics, 307–319 (2024). 10.1201/9781003459347-21/BONE-MARROW-CANCER-DETECTION-LEUKOCYTES-USING-NEURAL-NETWORKS-SUNDARI-SHANMUGA-PANKAJ-BHAMBRI.
- 56.Nguyen, D. L., Vo, X. T., Priadana, A. & Jo, K. H. Vehicle detector based on improved YOLOv5 architecture for traffic management and control systems. IECON Proc. (Industrial Electronics Conference)10.1109/IECON51785.2023.10311764 (2023). [Google Scholar]
- 57.Guo, M. H. et al. Attention mechanisms in computer vision: A survey. Comput. Vis. Media (Beijing)8(3), 331–368. 10.1007/S41095-022-0271-Y/METRICS (2022). [Google Scholar]
- 58.Hsu, C. Y., Hu, R., Xiang, Y., Long, X. & Li, Z. Improving the Deeplabv3+ model with attention mechanisms applied to eye detection and segmentation. Mathematics10(15), 2597. 10.3390/MATH10152597 (2022). [Google Scholar]
- 59.Pacal, I., Celik, O., Bayram, B. & Cunha, A. Enhancing EfficientNetv2 with global and efficient channel attention mechanisms for accurate MRI-based brain tumor classification. Cluster Comput, 1–26. 10.1007/S10586-024-04532-1/TABLES/4 (2024).
- 60.Xiang, S. & Liang, Q. Remote sensing image compression with long-range convolution and improved non-local attention model. Signal Process.209, 109005. 10.1016/J.SIGPRO.2023.109005 (2023). [Google Scholar]
- 61.Fan, Q., Zou, E. B., Tai, Y., Lai, R. & He, Y. Z. A cross-fusion of non-local attention network for infrared small target tracking. Infrared Phys. Technol.141, 105453. 10.1016/J.INFRARED.2024.105453 (2024). [Google Scholar]
- 62.Chen, D. Q. Down-scale simplified non-local attention networks with application to image denoising. Signal Image Video Process18(1), 47–54. 10.1007/S11760-023-02708-7/TABLES/5 (2024). [Google Scholar]
- 63.Ghosal, P. et al. Compound attention embedded dual channel encoder-decoder for ms lesion segmentation from brain MRI. Multimed. Tools Appl.2024, 1–33. 10.1007/S11042-024-20416-3 (2024). [Google Scholar]
- 64.Mei, C. et al. Semi-supervised image segmentation using a residual-driven mean teacher and an exponential Dice loss. Artif. Intell. Med.148, 102757. 10.1016/J.ARTMED.2023.102757 (2024). [DOI] [PubMed] [Google Scholar]
- 65.Pezeshki, H. Breast tumor segmentation in digital mammograms using spiculated regions. Biomed. Signal Process. Control76, 103652. 10.1016/J.BSPC.2022.103652 (2022). [Google Scholar]
- 66.Balaji, K. Image augmentation based on variational autoencoder for breast tumor segmentation. Acad. Radiol.30, S172–S183. 10.1016/J.ACRA.2022.12.035 (2023). [DOI] [PubMed] [Google Scholar]
- 67.Moghbel, M., Ooi, C. Y., Ismail, N., Hau, Y. W. & Memari, N. A review of breast boundary and pectoral muscle segmentation methods in computer-aided detection/diagnosis of breast mammography. Artif. Intell. Rev.53(3), 1873–1918. 10.1007/s10462-019-09721-8 (2020). [Google Scholar]
- 68.Wang, S. et al. Breast tumor segmentation in DCE-MRI with tumor sensitive synthesis. IEEE Trans. Neural Netw. Learn. Syst.10.1109/TNNLS.2021.3129781 (2021). [DOI] [PubMed] [Google Scholar]
- 69.Anari, S., Tataei Sarshar, N., Mahjoori, N., Dorosti, S. & Rezaie, A. Review of deep learning approaches for thyroid cancer diagnosis. Math. Probl. Eng.2022, 1–8. 10.1155/2022/5052435 (2022). [Google Scholar]
- 70.Zha, H. et al. Preoperative ultrasound-based radiomics score can improve the accuracy of the Memorial Sloan Kettering Cancer Center nomogram for predicting sentinel lymph node metastasis in breast cancer. Eur. J. Radiol.135, 109512. 10.1016/J.EJRAD.2020.109512 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Movahedi, F., Padman, R. & Antaki, J. F. Limitations of receiver operating characteristic curve on imbalanced data: Assist device mortality risk scores. J. Thorac. Cardiovasc. Surg.165(4), 1433-1442.e2. 10.1016/J.JTCVS.2021.07.041 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muhammad, D. & Bendechache, M. Unveiling the black box: A systematic review of explainable artificial intelligence in medical image analysis. Comput. Struct. Biotechnol. J.24, 542–560. 10.1016/J.CSBJ.2024.08.005 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R., Parikh, D. & Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization (2017). Accessed: Oct. 22, 2021. [Online]. Available: http://gradcam.cloudcv.org
- 74.Zhang, Y. et al. Grad-CAM helps interpret the deep learning models trained to classify multiple sclerosis types using clinical brain magnetic resonance imaging. J. Neurosci. Methods353, 109098. 10.1016/J.JNEUMETH.2021.109098 (2021). [DOI] [PubMed] [Google Scholar]
- 75.Ejiyi, C. J. et al. Attention-enriched deeper UNet (ADU-NET) for disease diagnosis in breast ultrasound and retina fundus images. Prog. Artif. Intell.13(4), 351–366. 10.1007/S13748-024-00340-1/TABLES/6 (2024). [Google Scholar]
- 76.Cao, H. et al. Swin-Unet: Unet-like pure transformer for medical image segmentation. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 13803 LNCS, 205–218 (2023). 10.1007/978-3-031-25066-8_9/TABLES/6.
- 77.Sun, J. et al. DDRA-Net: Dual-channel deep residual attention UPerNet for breast lesions segmentation in ultrasound images. IEEE Access12, 43691–43703. 10.1109/ACCESS.2024.3373551 (2024). [Google Scholar]
- 78.Liu, S., Liu, D. & Lin, Y. DPNet: A dual-attention patching network for breast tumor segmentation in an ultrasound image. Multimed. Syst.30(6), 1–12. 10.1007/S00530-024-01562-Y/TABLES/6 (2024). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset is available at: https://www.kaggle.com/datasets/sabahesaraki/breast-ultrasound-images-dataset/data