Abstract
DNA nanostructures are ordered oligonucleotide arrangements that have applications for DNA computers, crystallography, diagnostics and material sciences. Peptide nucleic acid (PNA) is a DNA/RNA mimic that offers many advantages for hybridization, but its potential for application in the field of DNA nanotechnology has yet to be thoroughly examined. We report the synthesis and characterization of tethered PNA molecules (bisPNAs) designed to assemble two individual DNA molecules through Watson–Crick base pairing. The spacer regions linking the PNAs were varied in length and contained amino acids with different electrostatic properties. We observed that bisPNAs effectively assembled oligonucleotides that were either the exact length of the PNA or that contained overhanging regions that projected outwards. In contrast, DNA assembly was much less efficient if the oligonucleotides contained overhanging regions that projected inwards. Surprisingly, the length of the spacer region between the PNA sequences did not greatly affect the efficiency of DNA assembly. Reasons for inefficient assembly of inward projecting DNA oligonucleotides include non-sequence-specific intramolecular interactions between the overhanging region of the bisPNA and steric conflicts that complicate simultaneous binding of two inward projecting strands. These results suggest that bisPNA molecules can be used for self-assembling DNA nanostructures provided that the arrangement of the hybridizing DNA oligonucleotides does not interfere with simultaneous hybridization to the bisPNA molecule.
INTRODUCTION
Nanotechnology involves assembly of small molecules into complex architectures for higher function (1). The canonical Watson–Crick base pairing of adenine to thymine and guanine to cytosine is ideal for organizing biomolecules in a highly predictable fashion. Numerous reports on using oligonucleotides to build higher order structures include DNA matrices based on subunits of fixed Holliday junctions (2), streptavidin–DNA fragment nanoparticle networks (3) and DNA dendrimer formations for drug delivery (4; for reviews see 5–8).
Peptide nucleic acids (PNAs) (Fig. 1A) are a promising connector for the assembly of DNA-based nanostructures. PNAs are synthetic DNA analogs containing a neutral 2-aminoethylglycine backbone (9) and hybridize sequence specifically to complementary DNA and RNA oligonucleotides (10). Binding occurs with high affinity (10,11), high sensitivity to mismatch discrimination (12) and is unaffected by the ionic strength (10). Because PNAs possess a neutral amide backbone they bind less to proteins than do oligomers with negatively charged linkages (13,14) and are nuclease and protease resistant (15). PNAs have an exceptional ability to hybridize to sequences within duplex DNA by strand invasion, suggesting that PNA molecules should also be superior agents for binding to single-stranded DNA at regions that contain intramolecular structure (16,17). An important practical advantage is that methods for PNA synthesis are compatible with peptide synthesis, allowing PNAs to be easily modified with amino acids and other moieties (18,19). High affinity binding by PNAs has already been used for nanostructure assembly, with applications for labeling of DNA (20,21) and strand invasion into DNA hairpins and tetraloop motifs (22; for reviews see 23,24).
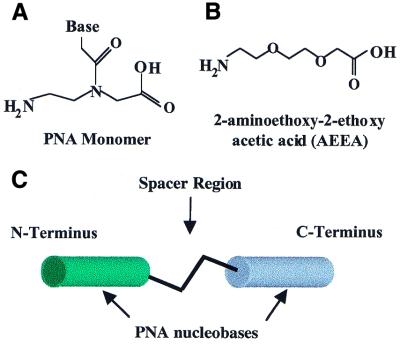
Figure 1.
(A) Structure of a PNA monomer. (B) Structure of AEEA, the linker molecule used to join PNA strands. (C) Model of a bisPNA showing the N- and C-terminal PNAs connected by a spacer region that may include amino acids or one or more AEEA molecules.
Here we describe the synthesis of bisPNAs that contain spacer regions that differ in length and amino acid substitution. We characterize the ability of bisPNAs to assemble DNA and observe that the capacity of a bisPNA molecule to hybridize to two oligonucleotides is primarily dependent on the arrangement of the DNA oligonucleotides being assembled. These results suggest that bisPNA molecules with simple chemical modifications can be used in generating DNA:bisPNA:DNA ‘units’ for nanotechnology and DNA nanostructure assembly, but that proper orientation of the assembled DNA oligonucleotides is essential.
MATERIALS AND METHODS
PNA synthesis
PNA monomers (Fig. 1), 2-aminoethoxy-2-ethoxy acetic acid (AEEA) and other reagents for PNA synthesis were obtained from PE Biosystems (Foster City, CA). Fmoc-amino acids were obtained from Advanced Chemtech (Louisville, KY) or Calbiochem-Novabiochem Corp. (La Jolla, CA). PNAs were prepared by automated synthesis using an Expedite 8909 synthesizer (PE Biosystems) as previously described (25).
PNAs were analyzed and purified by reverse phase high performance liquid chromatography (RP-HPLC) and mass spectral analysis by matrix-assisted laser desorption ionization time of flight (MALDI-TOF) as previously described (25). Briefly, RP-HPLC analysis and purification of bisPNAs involved a running buffer of dH2O/0.1% trifluoroacetic acid and an elution buffer of acetonitirile/0.1% trifluoroacetic acid from a C18 column inside a water jacket maintained at 55°C. Elution of PNA was monitored at 260 nm on a Dynamax UV-1 absorbance detector (Varian, Walnut Creek, CA). After purification, PNAs were lyophilized and resuspended in dH2O. Mass spectrometry was performed by MALDI-TOF on a Voyager-DE Workstation (Applied Biosystems, Foster City, CA) using either α-cyano-4-hydroxycinnamic acid or sinapinic acid solution as a matrix (Sigma-Aldrich, St Louis, MO). The concentration of PNA solutions was measured using a Cary 100Bio UV-Visible spectrophotometer (Varian) at 260 nm and room temperature, using an extinction coefficient of 241.4 ml µmol–1 cm–1 (all of the bisPNA sequences are the same). The extinction coefficients of PNA 24 and PNA 26 are 127.8 ml µmol–1 cm–1 and 113.6 ml µmol–1 cm–1, respectively.
Preparation of DNA oligonucleotides
DNA oligonucleotides (Invitrogen-Life Technologies, Carlsbad, CA) were radiolabeled using [γ-32P]ATP (Amersham Pharmacia Biotech, Piscataway, NJ) using T4 polynucleotide kinase (Sigma-Aldrich). Unincorporated [γ-32P]ATP was removed by passing the oligonucleotides through a Bio-Spin 6 column (Bio-Rad Laboratories, Hercules, CA) that had been pre-equilibrated with distilled water. Equal amounts of unlabeled oligonucleotides were treated similarly and purified by Bio-Spin 6 column in parallel. These unlabeled oligonucleotides were used to estimate the concentration of the radiolabeled oligomers that had been treated similarly. Unlabeled oligonucleotides were also used for gel shift experiments. The concentration of each unlabeled DNA oligonucleotide was calculated on a Cary 100Bio UV-Visible spectrophotometer (Varian) at 260 nm using the extinction coefficient given by the manufacturer. For each 40mer and 50mer oligonucleotide the nucleobases were randomized, except for the complementary 12mer target sequences, to minimize the potential for formation of specific secondary structures.
Melting temperature (Tm) analysis
Melting temperature (Tm) experiments were performed on a Cary 100Bio UV-Visible spectrophotometer (Varian) at 260 nm. BisPNA and DNA oligonucleotides were suspended in Na2HPO4 buffer (100 µM, pH 7.5) at 2 µM each. The temperature was ramped from 95 to 15°C and back up to 95°C at a rate of 5°C/min with a 12 s hold at each reading. Cary WinUV software was used to determine the Tm for each combination.
Polyacrylamide gel analysis of bisPNA:DNA hybridizations
PNAs tend to aggregate upon storage, so each working solution of bisPNA was heated to 80°C for 5 min and then cooled to room temperature prior to use. bisPNA (150 nM final concentration) was mixed with 32P-radiolabeled DNA oligonucleotides (150 nM final concentration) and unlabeled DNA oligonucleotide (0–25 µM) in a 50 µM Tris–HCl buffer (pH 8.0) in a 20 µl reaction. Oligonucleotide strands were annealed using a PE 9600 Thermocycler (Perkin Elmer, Norwalk, CT) (95°C for 5 min, cooled to 4°C over a period of 60 min). The samples were then flash frozen in an ethanol and dry ice bath and stored at –20°C until use. On ice, 10 µl of a 30% glycerol tracking dye (0.05% bromophenol blue, 0.05% xylene cyanol and 0.05% orange G) was added, followed by a quick spin, before loading into a non-denaturing 10% (19:1) polyacrylamide gel (Bio-Rad). The gel was run at 4°C and 250 V for 3.5 h. The gel was analyzed by autoradiography using a Molecular Dynamics model 425F phosphorimager (Sunnyvale, CA).
RESULTS AND DISCUSSION
Design of DNA oligonucleotides and bisPNAs
We designed bisPNA molecules containing two PNA strands linked by spacer regions of varied lengths (Fig. 1A–C). Previous reports have demonstrated that polypyrimidine bisPNAs can form four-stranded complexes capable of invading duplex DNA (26). The PNA sequences used in our studies, however, contained mixed purine and pyrimidine sequences because they were designed to bind and assemble two different single-stranded DNA oligonucleotides.
To test the effects of linker length and amino acid substitution within the spacer region between the two PNA strands we varied the number of AEEA linker molecules (Fig. 1B and Table 1) and number and identity of amino acids. Each AEEA molecule is ∼11 Å in length, water soluble and highly flexible. Amino acids were included in some of the spacer regions to increase the distance between the PNA sequences and to test the effect of amino acid charge and steric bulk on the ability of the bisPNA to assembly both DNA oligonucleotides.
Table 1. bisPNA sequences, expected and observed masses, and melting temperature values (Tm) for hybridization to oligonucleotide complements.
| bisPNA | Sequence (N→C) | Mass (Da) (found/calculated) | Tma (˚C) |
|---|---|---|---|
| PNA 1 | tcttcacctaga-lys | 3332.70/3332.60 | 50.4 |
| PNA 2 | gatacatatttg-lys | 3412.19/3411.62 | 47.8 |
| AEEA1 | tcttcacctaga-(aeea)1-gatacatatttg-lys | 6738.01/6744.42 | 52.7/48.3 |
| AEEA3 | tcttcacctaga-(aeea)3-gatacatatttg-lys | 7032.80/7034.82 | 49.5/45.6 |
| AEEA6 | tcttcacctaga-(aeea)6-gatacatatttg-lys | 7476.20/7470.42 | 49.1/46.3 |
| AEEA6asp5 | tcttcacctaga-(aeea-asp)5(aeea)-gatacatatttg-lys | 8023.45/8028.92 | 48.4/46.7 |
| AEEA6lys5 | tcttcacctaga-(aeea-lys)5(aeea)-gatacatatttg-lys | 8092.93/8093.42 | 49.1/47.8 |
| AEEA6phe5 | tcttcacctaga-(aeea-phe)5(aeea)-gatacatatttg-lys | 8495.29/8496.35 | 50.3/50.5 |
aFor bisPNAs, the temperature on the right is the Tm of the N-terminal half of the bisPNA (analogous to PNA 1), while the temperature on the left is the Tm of the C-terminal half of the bisPNA (analogous to PNA 2).
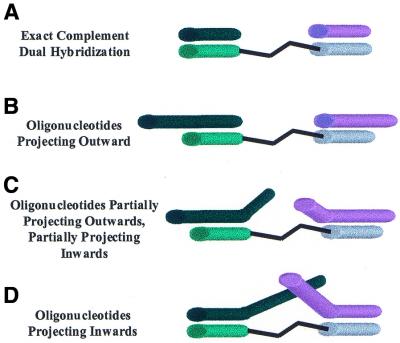
We designed DNA oligonucleotides to hybridize to bisPNAs in four different arrangements (Fig. 2A–D). The oligonucleotides were either exactly complementary to the PNA strands (Fig. 2A) or longer, so that overhanging single-stranded regions were created (Fig. 2B–D). The longer oligonucleotides could project outwards (DNA-1outward and DNA-2outward), project partially outwards and partially inwards (DNA-1mid and DNA-2mid) or project inwards (DNA-1inward and DNA-2inward) relative to the bisPNA (Fig. 2B–D and Table 1). Except for the target sequence, each base of the DNA oligonucleotide was randomized to minimize secondary structure and DNA:DNA interactions. We studied the ability of PNAs to assemble DNA oligomers with overhanging bases because such overhangs can be used to bind additional nucleic acids and permit complex structures to be built up.
Figure 2.
Models of bisPNA hybridized to DNA oligonucleotides. (A) DNA oligonucleotides that are exactly complementary to the PNA and do not have overhanging regions. (B) DNA oligonucleotides that have overhanging regions projecting outwards. (C) DNA oligonucleotides that have overhanging regions partially projecting inwards and partially projecting outwards. (D) DNA oligonucleotides that have overhanging regions projecting inwards.
Synthesis and characterization of PNA and bisPNA molecules
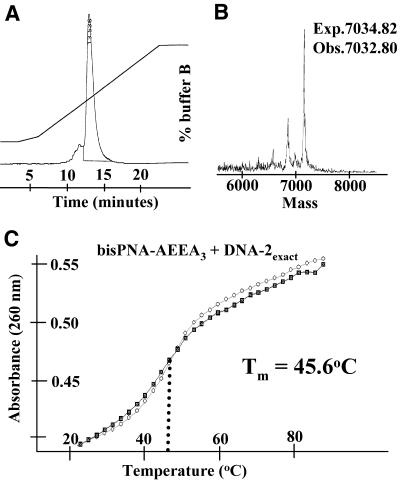
The automated synthesis of bisPNA molecules involving at least 25 couplings required additional precautions to ensure complete synthesis of the molecule. Typically, when a PNA sequence contains three or more consecutive purines or pyrimidines synthesis efficiency decreases. To minimize this problem we repeated coupling steps for addition of the third consecutive base and any that followed. We found that the spacer molecule AEEA was difficult to couple efficiently, so each of these couplings was repeated prior to addition of the next molecule. Repeated coupling was also done for each amino acid. With these precautions, automated synthesis of bisPNAs with extensive spacer regions was routine. We analyzed and purified bisPNAs by C18 RP-HPLC (Fig. 3A). Full-length PNAs were retained on the C18 column longer than truncated products from failed syntheses. Correct synthesis was confirmed by mass spectrometry (Fig. 3B). Both HPLC and mass spectral analysis routinely indicated that the main product was the desired one.
Figure 3.
(A) RP-HPLC purification of bisPNA-AEEA3. Purification conditions are described in Materials and Methods. (B) MALDI-TOF analysis of bisPNA-AEEA3 with expected and observed molecular weights noted. (C) Tm analysis of bisPNA-AEEA3 reversibly hybridizing to complementary DNA-2exact. Closed squares represent association during decreasing temperatures. Open circles representdissociation during increasing temperatures.
Melting temperature (Tm) analysis of exact complement DNA oligonucleotides to bisPNAs
We determined the melting temperature (Tm) values of the bisPNAs and complementary DNA oligonucleotides to characterize their potential for stable and selective hybridization (Fig. 3C). The Tm values for hybridization of bisPNAs with exactly complementary oligonucleotides were nearly the same as that of the individual 12 base PNAs corresponding to each half of the bisPNA (Table 1). This similarity in Tm values demonstrates that neither the molecules that make up the spacer region nor the unbound half of the bisPNA molecule significantly affects the temperature dependence of hybridization of the PNA and DNA oligonucleotides that lack an overhanging region. As we describe below, the interactions of bisPNAs with DNA oligonucleotides that do contain overhanging regions increase the Tm values in some cases (Table 2).
Table 2. Sequences of exact complement and overhanging oligonucleotides, melting temperatures (Tm) with bisPNA-AEEA3 and differences in Tm between exact complements and overhanging oligonucleotides.
| Name | Sequence (5′→3′) | Tm (°C) | ΔTm (°C) | |
|---|---|---|---|---|
| Exact complements | DNA-1exact | tctaggtgaaga | 49.5 | - |
| DNA-2exact | caaatatgtatc | 45.6 | - | |
| Projecting outward | DNA-1outward | tctaggtgaaga(n)38 | 49.3 | –0.2 |
| DNA-2outward | (n)28caaatatgtatc | 48.1 | 2.5 | |
| Partial inward/partial outward | DNA-1mid DNA-2mid | (n)19tctaggtgaaga(n)19 (n)14caaatatgtatc(n)14 | 54 52.8 | 4.5 6.2 |
| Projecting inward | DNA-1inward | (n)38tctaggtgaaga | 54.4 | 4.9 |
| DNA-2inward | caaatatgtatc(n)28 | 51.9 | 5.4 |
n, a randomized nucleotide.
Assembly of short, complementary oligonucleotides by bisPNA
We analyzed the hybridization of bisPNAs to DNA oligonucleotides by monitoring the ability of the PNAs to shift the mobility of 32P-radiolabeled DNA upon non-denaturing polyacrylamide gel electrophoresis. In all of the experiments described below, we established the mobility of labeled DNA alone, the mobility of labeled DNA bound to one PNA strand and the mobility of a mixture of two labeled DNA oligonucleotides directed to different PNA strands. 32P-labeled and unlabeled oligonucleotides were added to bisPNAs at the same time.
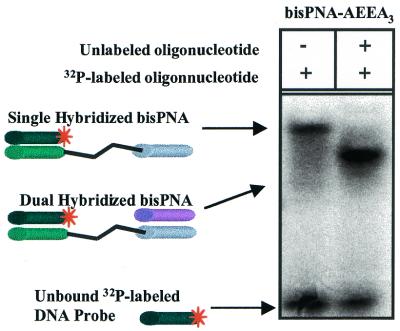
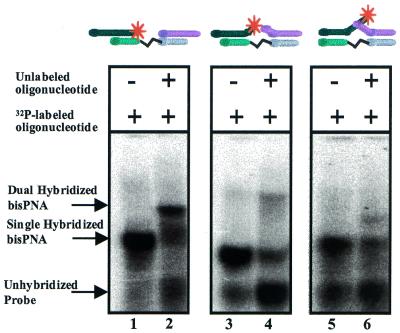
A bisPNA containing three AEEA linkers (PNA-AEEA3, Table 1) readily assembled short complementary DNA oligonucleotides, DNA-1exact and DNA-2exact (Table 2). Surprisingly, when bisPNA-AEEA3 hybridized to both 12mer DNA oligonucleotides migration was faster than a single hybridized bisPNA-AEEA3 (Fig. 4). The faster mobility of the DNA:bisPNA:DNA was not observed with the longer DNA sequences used in these studies. The increased mobility of the larger complex may be due to the dual hybridized bisPNA-AEEA3 having twice the negative charge (from the second oligonucleotide phosphate backbone) and to formation of a more compact structure than the single hybridized bisPNA.
Figure 4.
Non-denaturing polyacrylamide gel electrophoresis of bisPNA-AEEA3 with 32P-labeled DNA-1exact and unlabeled DNA-2exact. Left lane, bisPNA-AEEA3 and 32P-labeled DNA-1exact are present in a 1:1 ratio (150:150 nM). Right lane, bisPNA-AEEA3 and 32P-labeled DNA-1exact and unlabeled DNA-2exact are present in a 1:1:3.3 ratio (150:150:500 nM).
Assembly of oligonucleotides that project beyond the bisPNA
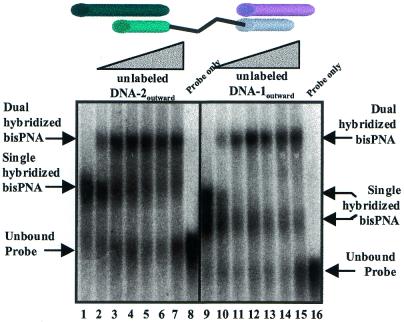
We next tested the ability of bisPNA-AEEA3 (Table 1) to hybridize longer DNA oligonucleotides that extended past one or both PNA termini. Characterizing assembly of these longer oligonucleotides is important because the extended DNA sequences provide the potential for additional base pairing necessary for formation of higher order structures. When bisPNA was incubated with DNA oligonucleotides designed to project outwards we observed that the bisPNA readily assembled both DNA strands with similar results regardless of which DNA strand was labeled with 32P (Fig. 5). The efficiency of hybridization was not affected by increasing the concentration of the unlabeled strands. In contrast to the ability of bisPNAs to successfully assemble short DNA oligonucleotides or DNA oligonucleotides with overhangs that project outwards (Figs 5 and 6, lanes 2), assembly of DNA oligonucleotides that project inwards upon hybridization was less apparent (Fig. 6, lanes 4 and 6). To improve binding of inward facing oligomers, we varied annealing conditions, but our attempts to bind two DNA oligonucleotides with inward projecting overhanging sequences invariably yielded only a small fraction of bisPNA bound to both DNA strands.
Figure 5.
Comparison of each half of bisPNA-AEEA3 for hybridization to one or two outward projecting DNA oligonucleotides. Lanes 1–7 show hybridization of a 1:1 (150:150 nM) ratio of bisPNA-AEEA3 and 32P-labeled DNA-1outward. For lanes 2–7, unlabeled DNA-2outward was added in increasing concentrations (equivalents relative to bisPNA): 0.33, 1.66, 3.33, 16.6, 33.3 and 166 equiv. Lane 8 is 32P-labeled DNA-1outward only. Lanes 9–15 show hybridization of a 1:1 ratio (150:150 nM) of bisPNA-AEEA3 and 32P-labeled DNA-2outward. For lanes 10–15, unlabeled DNA-1outward was added in increasing concentrations (equivalents relative to bisPNA): 0.33, 1.66, 3.33, 16.6, 33.3 and 166 equiv. Lane 16 is 32P-labeled DNA-2outward only.
Figure 6.
Comparison of bisPNA-AEEA3 hybridization to DNA oligonucleotides that project outwards, partially project inwards and project inwards. Lane 1, bisPNA-AEEA3 and 32P-labeled DNA-1outward at 1:1 ratios (150:150 nM). Lane 2, bisPNA-AEEA3 and 32P-labeled DNA-1outward and unlabeled DNA-2outward at 1:1:33 ratios (150:150:5000 nM). Lane 3, bisPNA-AEEA3 and 32P-labeled DNA-1mid at 1:1 ratios (150:150 nM). Lane 4, bisPNA-AEEA3 and 32P-labeled DNA-1mid and unlabeled DNA-2mid at 1:1:33 ratios (150:150:5000 nM). Lane 5, bisPNA-AEEA3 and 32P-labeled DNA-1inward at 1:1 ratios (150:150 nM). Lane 6, bisPNA-AEEA3 and 32P-labeled DNA-1inward and unlabeled DNA-2inward at 1:1:33 ratios (150:150:5000 nM).
DNA assembly by bisPNAs connected by spacers of differing lengths
Inefficient assembly of the inward facing DNA oligonucleotides suggested that the first oligonucleotide bound to the bisPNA blocked binding of the second. One solution for overcoming this obstacle was to increase the length of the spacer region between the two PNA strands. In theory, this would position the PNA strands farther apart, making them less susceptible to intramolecular interactions that might block hybridization.
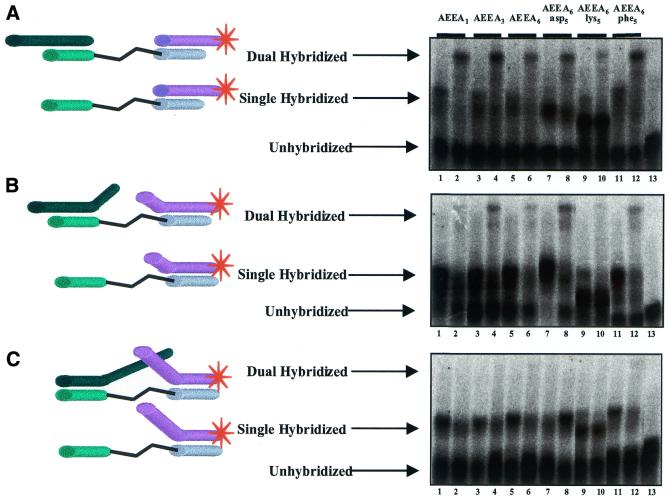
To investigate the effect of changing the spacer region on PNA assembly we compared bisPNAs with one, three or six AEEA linkers (bisPNAs AEEA1, AEEA3 and AEEA6, Table 1). Spacers also alternated five amino acids (aspartic acid, lysine or phenylalanine) within six AEEA molecules (AEEA6asp5, AEEA6lys5 or AEEA6phe5, Table 1). Aspartic acid, lysine and phenylalanine were selected based on their varied electrostatic and steric properties, allowing the effects of substantial chemical alteration to be investigated. In all cases we observed hybridization by DNA oligonucleotides that projected outwards (Fig. 7A). Hybridization by bisPNA-AEEA6lys5 was the least efficient, suggesting that electrostatic interactions between lysine and DNA may be blocking hybridization of the second DNA strand.
Figure 7.
Comparison of oligonucleotide assembly by bisPNAs that contain spacers with varying numbers of AEEA spacers and amino acids. The DNA oligonucleotides project outwards, partially inwards and inwards. (A) bisPNA hybridization to DNA oligonucleotides that project outwards. (B) bisPNA hybridization to DNA oligonucleotides that project partially inwards. (C) bisPNA hybridization to DNA oligonucleotides that project inward. Lanes 1 and 2 represent bisPNA-AEEA1, lane 3 and 4 represent bisPNA-AEEA3, lanes 5 and 6 represent bisPNA-AEEA6, lanes 7 and 8 represent bisPNA-AEEA6asp5, lanes 9 and 10 represent bisPNA-AEEA6lys5 and lanes 11 and 12 represent bisPNA-AEEA6phe5. For each hybridization reaction, bisPNA and 32P-labeled DNA oligonucleotide are at a 1:1 ratio (150:150 nM). Even numbered lanes include an additional 10 equiv. of unlabeled oligonucleotide (1500 nM). Lane 13 is 32P-labeled DNA oligonucleotide only.
BisPNA hybridization to partially inward projecting DNA oligonucleotides (Fig. 7B) did not occur as readily as to outward projecting DNA oligonucleotides. The DNA oligonucleotides containing a short region projecting inward were assembled by the two bisPNAs with the linker regions containing aspartic acid or phenylalanine (Fig. 7B, lanes 8 and 12) but not by bisPNA-AEEA6lys5 that contained lysine residues (Fig. 7B, lanes 2 and 10). In contrast, DNA oligonucleotides with long regions that projected inward exhibited virtually no assembly by bisPNAs regardless of the length or chemical properties of the linker (Fig. 7C). These same results were observed under a variety of annealing conditions.
In this study we have focused on making increasingly flexible linkers. It is possible that rigid linkers with defined geometries might be able to hold the two strands of the PNAs far apart and allow more efficient assembly of complex DNA oligonucleotides. Shi and Bergstrom have described the synthesis of such linkers and have used them to bridge two single-stranded DNA oligonucleotides for nanostructure assembly (27).
Reasons for inefficient assembly of oligonucleotides that project inward
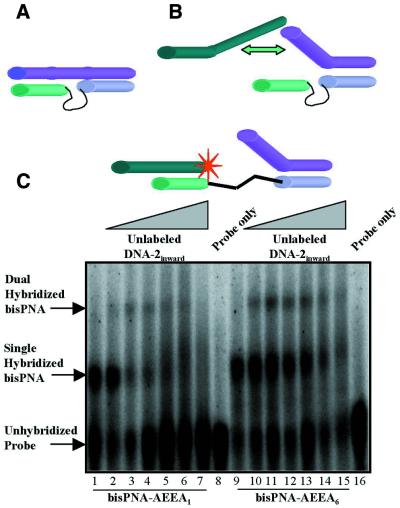
One reason for the inefficient assembly of DNA oligonucleotides containing inward overhanging regions is that once the DNA is bound to the complementary PNA strand, the inward facing overhanging region may associate with the second PNA strand through non-Watson–Crick interactions (Fig. 8A). Normally such interactions would be weak, but in this case they may be more important because they would be effectively intramolecular. If this type of non-specific association were to occur, it would be reflected in elevated Tm values for the inward facing DNA oligonucleotides relative to DNA oligonucleotides that are direct complements or that contain regions that are outward facing. Support for the belief that non-specific PNA–DNA contacts can be important is provided by Raney and co-workers, who noted that PNAs have a high propensity for interactions with single-stranded DNA that promote aggregation (28).
Figure 8.
(A) Interference with dual hybridization by an inward facing oligonucleotide that makes Watson–Crick base pairs with a complementary PNA strand, and forms non-Watson–Crick interactions with the non-complementary PNA strand. (B) Interfence with dual hybridization by steric conflicts between two inward facing oligomers. The explanations for the inefficient dual hybridization portrayed in (A) and (B) are not mutually exclusive. (C) Comparison of bisPNA-AEEA1 and bisPNA-AEEA6 for their ability to hybridize to both an inward projecting and an outward projecting DNA oligonucleotide. Lanes 1–7, bisPNA-AEEA1 and 32P-labeled DNA-1outward in 1:1 ratios (150:150 nM). For lanes 2–7, unlabeled DNA-2inward was added in increasing equivalents (relative to bisPNA-AEEA1): 0.33, 1.66, 3.33, 16.6, 33.3 and 166 equiv. Lane 8 is 32P-labeled DNA-1outward only. Lanes 9–15, bisPNA-AEEA6 and 32P-labeled DNA-1outward in 1:1 ratios (150:150 nM). For lanes 10–15, unlabeled DNA-2inward was added in increasing equivalents (relative to bisPNA-AEEA6: 0.33, 1.66, 3.33, 16.6, 33.3 and 166 equiv. Lane 16 is 32P-labeled DNA-1outward only.
DNA oligonucleotides with overhanging regions that projected outwards (DNA-1outward and DNA-2outward) had melting temperatures nearly equal to that of short 12mer DNA oligonucleotides (Table 2). However, the DNA oligonucleotides that projected inwards or partially inwards had significantly higher Tm values relative to the exact complement DNA oligonucleotides, increasing by 4.5–6.2°C. These data are consistent with the suggestion that non-Watson–Crick interactions between PNA and DNA oligonucleotides can form and may contribute to the inability of the second DNA oligonucleotide to efficiently bind PNA.
An explanation for inefficient assembly of DNA oligonucleotides that project inwards is that the two overhanging regions physically interfere with one another and block dual hybridization (Fig. 8B). If this steric conflict were preventing hybridization, it should be possible for PNAs to assemble a DNA oligonucleotide with an inward projecting region and a DNA strand that is exactly complementary or that contains an overhanging region that projects outward. To test this hypothesis we hybridized both inward projecting and outward projecting DNA oligonucleotides to PNA-AEEA1 or PNA-AEEA6.
In contrast to the inability of two inward projecting oligonucleotides to hybridize simultaneously (Fig. 7C), we observed that PNA-AEEA1 and PNA-AEEA6 were able to assembly an inward and an outward projecting oligomer (Fig. 8C). Assembly by PNA-AEEA1 (Fig. 8C, lanes 1–7) was not as efficient as was assembly by PNA-AEEA6 (Fig. 8C, lanes 9–15), suggesting that longer spacers can promote assembly. The finding that an inward and an outward facing DNA can be assembled supports the conclusion that conflicts between overhanging strands were one factor contributing to the lack of dual hybridization by two inward projecting DNA oligonucleotides observed in Figure 7C. It is likely that assembly is complicated by both steric conflicts between the overhanging regions and non-sequence-specific interactions with the second PNA strand.
Applications of PNAs for the assembly of DNA structures
PNAs may have many applications in the field of nanotechnology and one of the purposes of this study was to determine the rules guiding use of PNAs for the assembly of DNA structures. There have been recent reports of the use of DNA oligonucleotides to assemble nanostructures, most of which describe systems that could use PNAs as a facilitator for construction. Seeman and co-workers constructed 2- and 3-dimensional DNA arrays based on the DNA branched junction motif and ‘sticky ends’ to ligate individual ‘units’ together (29). bisPNAs could be used similarly, with the tether molecules forming the corners between the PNA:DNA duplexes on either side in a 2-dimensional arrangement. Furthermore, trisPNAs can be synthesized (C.J.Nulf, unpublished results), capable of hybridizing to three DNA oligonucleotides and forming the vertices of a 3-dimensional cube.
PNAs could also be used for joining the ends to generate circular oligonucleotides or concatamers. Circular DNA oligonucleotides have unique functions and interesting hybridization properties (30). They have been synthesized by ligating two ends of a linear oligonucleotide after bringing the two ends together with a connecting oligonucleotide (31). The bisPNAs used in our studies could be used to stably join DNA ends without the need for ligation and the resultant circular constructs could be used to build higher order structures.
A third possible application of PNAs in nanotechnology is the development of electrical-sensitive nanoprobes. Recently, Mirkin and co-workers have functionalized gold nanoparticles with DNA oligonucleotides that create conductivity changes associated with target–probe binding events (32). Similarly, Josephson and co-workers have determined by magnetic resonance imaging that monodispersed oligonucleotide-modified magnetic nanoparticles exhibit a decrease in relaxation time of adjacent water molecules when hybridized to a complementary oligonucleotide-modified magnetic nanoparticle (33). The superior hybridization properties of PNAs might make them ideal for these applications, and their neutrality might be useful for the probing and optimization of electrical sensors.
Importantly, all of the applications discussed above involve hybridization to the terminal ends of DNA oligonucleotides, an ability that our studies indicate is possessed by bisPNAs. Other applications, especially those that require assembly within living cells, may be more challenging. PNAs can efficiently invade duplex DNA (21–23), but the assembly of two duplex DNAs is likely to be complicated because of the problems that we have uncovered with joining inward overhanging sequences. We have attempted to use bisPNAs to join two DNA plasmids but have not been successful (C.J.Nulf, unpublished results). Another interesting biological application would be the use of PNAs to bridge RNA sequences to alter splicing or expression. This tactic would also require that bisPNAs doubly bind RNA overhanging sequences, but since this hybridization would be intramolecular (i.e. the PNA strands would bind to different sites on the same RNA), it might occur more readily than the intermolecular assembly attempted in our studies.
CONCLUSIONS
We show here that bisPNAs can be used to assemble DNA oligomers, but that this assembly depends on the relative geometries of the DNAs being assembled. Assembly occurs readily when the DNAs are exactly complementary to the PNA strands or contain overhanging regions that project outwards. Steric conflicts between the DNA oligonucleotides and non-Watson–Crick association between DNA and PNA complicate assembly of DNA oligonucleotides that contain regions that project inwards. Since most applications of DNA for nanostructure assembly use oligonucleotides that project outwards from a central connection, our results indicate that the bisPNAs can be used for nanotechnology applications and that their favorable characteristics may lead to improved assemblies.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to express our thanks to Ahmet Canizoglu for his important contributions during the early stages of this work and to Dr Dwaine Braasch for assistance with PNA synthesis. This work was support by grants from the National Institutes of Health (GM-60642) and the Robert A. Welch Foundation (I-1244).
REFERENCES
- 1.Feynman R.P. (1961) There is plenty of room at the bottom. In Gilbert,H.D. (ed.), Miniaturization. Reinhold, New York, NY, pp. 282–296.
- 2.Winfree E., Liu,F., Wenzler,L.A. and Seeman,N.C. (1998) Design and self-assembly of two-dimensional DNA crystals. Nature, 394, 539–544. [DOI] [PubMed] [Google Scholar]
- 3.Niemeyer C.M., Adler,M., Gao,S. and Chi,L.F. (2001) Nanostructured DNA-protein aggregates consisting of covalent oligonucleotide-streptavidin conjugates. Bioconjugate Chem., 12, 364–371. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Boros,P., Liu,J., Qin,L., Bai,Y., Bielinska,A.U., Kukowska-Latallo,J.F., Baker,J.R. and Bromberg,J.S. (2000) DNA/dendrimer complexes mediate gene transfer into murine cardiac transplants ex vivo. Mol. Ther., 2, 602–608. [DOI] [PubMed] [Google Scholar]
- 5.Seeman N.C. (1998) DNA nanotechnology: novel DNA constructions. Annu. Rev. Biophys. Biomol. Struct., 27, 225–248. [DOI] [PubMed] [Google Scholar]
- 6.Seeman N.C. (1999) DNA engineering and its application to nanotechnology. Trends Biotechnol., 17, 437–443. [DOI] [PubMed] [Google Scholar]
- 7.Seeman N.C. (1998) Nucleic acid nanostructures and topology. Angew. Chem. Int. Ed., 37, 3220–3238. [DOI] [PubMed] [Google Scholar]
- 8.Niemeyer C.M. (2001) Nanoparticles, proteins and nucleic acids: biotechnology meets materials science. Angew. Chem. Int. Ed., 40, 4128–4158. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen P.E., Egholm,M., Berg,R.H. and Buchardt,O. (1991) Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science, 254, 497–500. [DOI] [PubMed] [Google Scholar]
- 10.Egholm M., Buchardt,O., Christensen,L., Behrens,C., Freier,S.M., Driver,D.A., Berg,R.H., Kim,S.K., Norden,B. and Nielsen,P.E. (1993) PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature, 365, 566–568. [DOI] [PubMed] [Google Scholar]
- 11.Jensen K.K., Orum,H., Nielsen,P.E. and Norden,B. (1997) Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry, 36, 5072–5077. [DOI] [PubMed] [Google Scholar]
- 12.Orum H., Nielsen,P.E., Egholm,M., Berg,R.H., Buchardt,O. and Stanley,C. (1993) Single base pair mutation analysis by PNA directed PCR clamping. Nucleic Acids Res., 21, 5332–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mischiati C., Borgatti,M., Bianchi,N., Rutigliano,C., Tomassetti,M., Feriotto,G. and Gambari,R. (1999) Interaction of the human NF-kappaB p52 transcription factor with DNA-PNA hybrids mimicking the NF-kappaB binding sites of the human immunodeficiency virus type 1 promoter. J. Biol. Chem., 274, 33114–33122. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton S.E., Iyer.,M., Norton,J.C. and Corey,D.R. (1996) Specific and nonspecific inhibition of RNA synthesis by DNA, PNA and phosphorothioate promoter analog duplexes. Bioorg. Med. Chem. Lett., 6, 2897–2900. [Google Scholar]
- 15.Demidov V.V., Potaman,V.N., Frank-Kamenetskii,M.D., Egholm,M., Buchard,O., Sonnichsen,S.H.and Nielsen,P.E. (1994) Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol., 48, 1310–1313. [DOI] [PubMed] [Google Scholar]
- 16.Kushon S.A., Jordan,J.P., Seifert,J.L., Nielsen,P.E. and Armitage,B.A. (2001) Effect of secondary structure on the thermodynamics and kinetics of PNA hybridization to DNA hairpins. J. Am. Chem. Soc., 123, 10805–10813. [DOI] [PubMed] [Google Scholar]
- 17.Bentin T. and Nielsen,P.E. (1996) Enhanced peptide nucleic acid binding to supercoiled DNA: possible implications for DNA “breathing” dynamics. Biochemistry, 35, 8863–8869. [DOI] [PubMed] [Google Scholar]
- 18.Koch T., Naesby,M., Wittung,P., Jorgensen,M., Larsson,C., Buchardt,O., Stanley,C.J., Norden,B., Nielsen,P.E. and Orum,H. (1995) PNA-peptide chimerae. Tetrahedron Lett., 36, 6933–6936. [Google Scholar]
- 19.Braasch D.A. and Corey,D.R. (2001) Synthesis, analysis, purification and intracellular delivery of peptide nucleic acids. Methods, 23, 97–107. [DOI] [PubMed] [Google Scholar]
- 20.Zelphati O., Liang,X., Hobart,P. and Felgner,P.L. (1999) Gene chemistry: functionally and conformationally intact fluorescent plasmid DNA. Hum. Gene Ther., 10, 15–24. [DOI] [PubMed] [Google Scholar]
- 21.Demidov V.V., Kuhn,H., Lavrentieva-Smolina,I.V. and Frank-Kamenetskii,M.D. (2001) Peptide nucleic acid-assisted topological labeling of duplex DNA. Methods, 23, 123–131. [DOI] [PubMed] [Google Scholar]
- 22.Datta B. and Armitage,B.A. (2001) Hybridization of PNA to structured DNA targets: quaduplex invasion and the overhang effect. J. Am. Chem. Soc., 123, 9612–9619. [DOI] [PubMed] [Google Scholar]
- 23.Demidov V.V., Bukanov,N.O. and Frank-Kamenetskii,M.D. (2001) Duplex DNA capture. Curr. Issues Mol. Biol., 2, 31–35. [PubMed] [Google Scholar]
- 24.Nielsen P.E. (2001) Peptide nucleic acid: a versatile tool in genetic diagnostics and molecular biology. Curr. Opin. Biotechnol., 12, 16–20. [DOI] [PubMed] [Google Scholar]
- 25.Mayfield L.D. and Corey,D.R. (1999) Automated synthesis of peptide nucleic acids and peptide nucleic acid-peptide conjugates. Anal. Biochem., 268, 401–404. [DOI] [PubMed] [Google Scholar]
- 26.Hansen G.I., Bentin,T., Larsen,H.J. and Nielsen,P.E. (2001) Structural isomers of bis-PNA bound to a target in duplex DNA. J. Mol. Biol., 307, 67–74. [DOI] [PubMed] [Google Scholar]
- 27.Shi J.F. and Bergstrom,D.E. (1997) Assembly of novel DNA cycles with rigid tetrahedral linkers. Angew. Chem. Int. Ed., 36, 111–113. [Google Scholar]
- 28.Tackett A.J., Corey,D.R. and Raney,K.D. (2002) Non-Watson–Crick interactions between PNA and DNA inhibit the ATPase activity of bacteriophage T4Dda helicase. Nucleic Acids Res., 30, 950–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi J., Li,X.J., Yang,X.P. and Seeman,N.C. (1996) Ligation of triangles built from bulged 3-arm DNA branched junctions. J. Am. Chem. Soc., 118, 6121–6130. [Google Scholar]
- 30.Kool E.T. (1996) Circular oligonucleotides—new concepts in oligonucleotide design. Annu. Rev. Biophys. Biomol. Struct., 25, 1–28. [DOI] [PubMed] [Google Scholar]
- 31.Wang S. and Kool,E.T. (1994) Circular RNA oligonucleotides. Synthesis, nucleic acid binding properties and a comparison with circular DNAs. Nucleic Acids Res., 22, 2326–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park S.J., Taton,T.A. and Mirkin,C.A. (2002) Array-based electrical detection of DNA with nanoparticle probes. Science, 295, 1503–1506. [DOI] [PubMed] [Google Scholar]
- 33.Perez J.M., O’Loughin,T., Simeone,F.J., Weissleder,R. and Josephson,L. (2002) DNA-based magnetic nanoparticle assembly acts as a magnetic relaxation nanoswitch allowing screening of DNA-cleaving agents. J. Am. Chem. Soc., 124, 2856–2857. [DOI] [PubMed] [Google Scholar]