Abstract
γ-Radiation resistance is a major obstacle to the success of radiotherapy in colorectal cancer. Antioxidant-related factors contribute to resistance to radiation therapy and, therefore, are targets for improving the therapeutic response. In this study, we evaluated the molecular mechanisms underlying γ-radiation resistance using the colorectal cancer cell line SNUC5 and γ-radiation-resistant variant SNUC5/RR, including analyses of the role of nuclear factor erythroid 2-related factor 2 (NRF2), a transcription factor that regulates antioxidant enzymes, and related epigenetic regulators. Reactive oxygen species (ROS) levels, antioxidant enzyme expression, NRF2 expression, and nuclear translocation were higher in SNUC5/RR cells irradiated with or without 8 Gy than in SNUC5 cells. The DNA demethylase ten-eleven translocation 1 (TET1) expression and TET1 binding to the NRF2 promoter in SNUC5/RR cells were stronger than those in SNUC5 cells, indicating lower methylation of CpG islands in the NRF2 promoter. TET1 knockdown in SNUC5/RR cells suppressed NRF2 expression significantly. Additionally, histone mixed-lineage leukemia (MLL), a histone methyltransferase, was upregulated, leading to increased trimethylation of histone H3 lysine 4, whereas enhancer of zeste homolog 2 (EZH2), a histone methyltransferase, was downregulated, leading to decreased trimethylation of histone H3 lysine 27. Histone deacetylase (HDAC) and histone acetyltransferase (HAT) levels were lower and higher in SNUC5/RR cells than in SNUC5 cells, respectively. MLL and HAT knockdown in SNUC5/RR cells irradiated with or without 8 Gy decreased levels of NRF2 and heme-oxygenase 1, resulting in enhanced γ-radiation sensitivity. These findings support NRF2 as a target for improving the response to radiotherapy in patients with colorectal cancer.
Keywords: γ-Radiation resistance, Colorectal cancer, Epigenetic alteration
INTRODUCTION
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide, accounting for approximately 11% of all cancer-related deaths, and ranks as the third leading cause of cancer mortality (Ryu et al., 2024). Despite advancements in diagnosis and treatment, including surgery, chemotherapy, radiotherapy, and the emerging use of immunotherapy, CRC treatment outcomes remain suboptimal in some patients (Kim et al., 2021b). However, the therapeutic efficacy of immunotherapy for CRC has been inconsistent, warranting further investigation.
Radiotherapy, the cornerstone of CRC treatment, employs high-energy ionizing radiation (IR) to damage malignant cells, primarily through the radiolysis of water molecules. This process generates ROS, such as superoxide anions and hydroxyl radicals, which induce DNA damage and trigger apoptosis (Lal and Gupta, 2016). However, CRC cells often exhibit resistance to IR, which significantly compromises the effectiveness of radiotherapy (Häfner and Debus, 2016; Jin et al., 2016). This phenomenon, termed radioresistance, arises through the interplay between multiple genes, signaling pathways, and adaptive cellular mechanisms (Geng and Wang, 2017; Tang et al., 2018). Radioresistance is one of the main obstacles to successful treatment of CRC, often resulting in poor prognosis and reduced survival rates (Kim et al., 2015a). Consequently, radiosensitizers have received considerable attention for mitigating this issue and enhancing the therapeutic efficacy of radiotherapy.
ROS homeostasis, regulated by the redox system, has been implicated in the development of radioresistance in various malignant cancers (Kim et al., 2015b; Chaiswing et al., 2018). Cancer cells resist IR-induced damage by either reducing ROS production or activating antioxidant defense systems to scavenge ROS. Moreover, the exposure of colon cancer cells to high-dose radiation (10 Gy) upregulates antioxidant enzyme expression and promotes radioresistance (McCann et al., 2021).
Nuclear factor erythroid 2-related factor 2 (NRF2) is a key transcription factor that regulates the expression of antioxidant enzymes and promotes oxidative stress resistance (He et al., 2020). In CRC, NRF2 overexpression has been directly linked to increased resistance to radiotherapy. Therefore, understanding the molecular mechanisms by which NRF2 modulates radioresistance may provide new opportunities to improve therapeutic response to radiotherapy.
Epigenetic modifications are emerging as critical contributors to radioresistance in CRC and other cancers (Kim et al., 2013; Wang et al., 2022). For instance, overexpression of the long noncoding RNA LINCE00630 has been shown to mediate CRC radioresistance through epigenetic suppression of the tumor suppressor gene brain-expressed X-linked gene 1 (Liu et al., 2020). These findings suggest the potential utility of epigenetic agents in overcoming radioresistance and improving clinical outcomes in patients with CRC undergoing radiotherapy. Cancer cells that adapt to oxidative stress via the Keap1/NRF2 pathway or other antioxidative mechanisms, such as serum- and glucocorticoid-inducible kinase 1, often exhibit enhanced radioresistance (Talarico et al., 2016; Bonura et al., 2022). Additionally, ROS induces epigenetic alterations, including DNA methylation and histone modification, which play pivotal roles in carcinogenesis (Kietzmann et al., 2017; García-Guede et al., 2020; Jing et al., 2022; Liu et al., 2023). For example, in nasopharyngeal carcinoma, DNMT3B-mediated methylation of p53 and p21 promotes radioresistance (Wu et al., 2020), whereas H3K27Me3 contributes to radioresistance by inhibiting 2’-5’-oligoadenylate synthetase 1 (Zhou et al., 2024).
NRF2 overexpression is associated with the progression and therapeutic resistance of various cancers (Lee et al., 2020). A meta-analysis revealed that high NRF2 expression correlates with increased aggressiveness in non-small cell lung cancer (Wang et al., 2020). Furthermore, NRF2 activation suppresses estrogen-related receptor α in breast cancer cells by inducing the expression of key proteins, such as Rho, focal adhesion kinase 1, and Rho-associated coil-containing protein kinase 1, which contribute to the proliferation and metastasis of cancer cells (Zimta et al., 2019).
Despite these insights, the specific molecular mechanisms through which NRF2 modulates redox homeostasis and contributes to radioresistance in CRC remain unclear. In this study, we aimed to elucidate the role of NRF2 in γ-radiation resistance in CRC to identify potential strategies for improving the efficacy of radiotherapy.
MATERIALS AND METHODS
Cell culture
To establish IR-resistant SNUC5 colon cancer cells (Korean Cell Line Bank, Seoul, Korea), cells were exposed to 30 cycles at 2 Gy. The total dose was 60 Gy administered over approximately 6-8 months.
Cell viability assay
The cells were seeded in a 96-well plate at a density of 2×103 cells/well and treated with 8 Gy. After culturing for 72 h, 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT; Sigma-Aldrich Co., Ltd., St. Louis, MO, USA), and trypan blue assays (Sigma-Aldrich Co., Ltd.) were performed according to previously reported methods (Bumah et al., 2021).
Colony formation assay
Five hundred cells were seeded and cultured for 10 days. Colonies with more than 50 cells were counted under a microscope using a Diff-Quick staining kit (Sysmex, Kobe, Japan), following the procedure described by Ke et al. (2023).
Detection of apoptosis
To detect apoptotic bodies, the cells were observed under a fluorescence microscope (Cool SNAP-Pro color digital camera; Media Cybernetics, Silver Spring, MD, USA) after Hoechst 33342 staining (Sigma-Aldrich Co., Ltd.) at 37°C for 10 min. To detect the sub-G1 cell population, the cells were fixed in 70% ethanol, stained with a solution containing 100 μg/mL propidium iodide (PI), 100 μg/mL ribonuclease A, and 2 mM EDTA in PBS, and incubated for 1 h at 37°C. The sub-G1 analysis was performed using a flow cytometer and FACSDiva™ 6.0 software (FACS Canto II; Becton Dickinson, Mountain View, CA, USA), following the protocols described in a previous study (McClelland et al., 2021).
Western blot analysis
Cell lysates were electrophoresed, transferred onto a membrane, and incubated with primary antibodies (Table 1), followed by incubation with secondary immunoglobulin G-horseradish peroxidase conjugates (Pierce, Rockford, IL, USA). Protein bands were visualized using a luminescent image analyzer (Bio-Rad Laboratories, Hercules, CA, USA), following previously reported methods (Sormunen et al., 2023).
Table 1.
Lists of primary antibodies
| Antibodies | Company/City/State/Country |
|---|---|
| Actin | Santa Cruz Biotechnology (Santa Cruz, CA, USA) |
| Bcl-2 associated X protein (Bax) B-cell lymphoma protein (Bcl-2) |
|
| Catalase (CAT) | |
| Glutathione synthetase (GSS) | |
| Ten-eleven translocation (TET) 2 | |
| TET3 | |
| Caspase-3 Caspase-9 Enhancer of zeste homolog 2 (EZH2) Heme oxygenase (HO-1) Histone acetyltransferase 1 (HAT1) Histone deacetylase 1 (HDAC1) H3K9Ac H3K4Me3 H3K27Me3 |
Cell Signaling Technology (Beverly, MA, USA) |
| DNA methyltransferase 1 (DNMT1) DNMT3A DNMT3B Glutamate-cysteine ligase catalytic subunit (GCLG) Mixed-lineage leukemia 1 (MLL1) Nuclear factor erythroid 2-related factor 2 (NRF2) TATA-box binding protein (TBP) TET1 |
Abcam (Cambridge, MA, USA) |
Assessment of reactive oxygen species (ROS)
Approximately 3×105 cells/well were stained using 25 µM 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA; Molecular Probes, Eugene, OR, USA), and the DCF intensity was detected using a flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) or confocal laser scanning microscopy (LSM 5 PASCAL version 3.5; Carl Zeiss, Jena, Germany) following the protocols described in a previous study (Kim and Xue, 2020).
Next-generation sequencing (NGS) assay
RNA sequencing was performed by Macrogen Inc. (Seoul, Korea). Briefly, 1 μg of total RNA from each cell sample (n=2) was used to synthesize cDNA using the SMARTer ultra-low RNA kit for sequencing (Clontech Laboratories, Mountain View, CA, USA), according to the manufacturer’s instructions. RNA integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). NGS was performed using an Illumina HiSeq-2000 RNA-seq platform (50 cycles, single-end). Sequences were assessed for quality and aligned against the human genome using the TopHat aligner (http://tophat.cbcb.umd.edu/). Differential gene expression levels between SNUC5 and SNUC5/RR cells were compared using Cuffdiff (http://cufflinks.cbcb.umd.edu/), following the protocol described by Kim et al. (2021a). The p-values for differential expression were adjusted for multiple testing using the false discovery rate method to ensure the robustness of significance determination.
Immunocytochemistry
Fixed cells were incubated with the primary antibody, followed by incubation with a FITC-conjugated secondary antibody (1:500; Santa Cruz Biotechnology). The cells were observed under a confocal microscope using LSM 510 (Carl Zeiss) with a mounting medium containing DAPI (Vector Laboratories, Burlingame, CA, USA). The assay was performed according to the procedure described by Castillo et al. (2019).
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
qRT-PCR was performed using the Bio-Rad iQ5 real-time PCR detection system (Bio-Rad Laboratories) under the following conditions: pre-denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, and 60°C for 1 min. The primers used are listed in Table 2.
Table 2.
NRF2 and actin primer sequences
| Gene | Primer | Sequence (5′–3′) |
|---|---|---|
| NRF2 | Forward | GAGAGCCCAGTCTTCATTGC |
| Reverse | TTGGCTTCTGGACTTGGAAC | |
| Actin | Forward | CACCTTCTACAATGAGCTGCGTGT |
| Reverse | CACAGCCTGGATAGCAACGTACA |
Methylation-specific PCR (MSP) and quantitative MSP (qMSP)
The NRF2 promoter region was examined for potential CpG islands 1176 bp upstream of the translation start site using the NCBI database. Two CpG-rich islands were identified within the NRF2 promoter region: −505 to −254 and −252 to +65. PCR primers covering 11 CpG sites in the promoter region from −479 to −342 bp were designed using MetPrimer. Bisulfate modification of DNA was performed using a Methylamp™ DNA modification kit (Epigentek, Pittsburgh, PA, USA) according to the manufacturer’s instructions. To analyze the methylation of NRF2 DNA, MSP and qMSP were performed using the Epitect MSP kit (Qiagen, Hilden, Germany) and EpiScope® MSP kit (TaKaRa Bio, Shiga, Japan), respectively. MSP products on the gel were visualized under ultraviolet light after staining with ethidium bromide. qMSP was performed using a CFX96TM real-time system (Bio-Rad) as described previously (Ohtsubo et al., 2022). The MSP and qMSP primers are listed in Table 3.
Table 3.
MSP and qMSP primer sequences
| NRF2 | Primer | Sequence (5′–3′) |
|---|---|---|
| Unmethylated | Forward | GGAGGTGTAGTTTTTATATTAATGT |
| Reverse | ACCAACTAAAATCCCAACAAACA | |
| Methylated | Forward | AGGGAGGCGTAGTTTTTATATTAAC |
| Reverse | AACTAAAATCCCAACAAACGAA |
Measurement of 5-methylcytosine (5-mC) and 5-hydroxymethylcytosine (5-hmC)
Fixed cells were incubated with anti-5-mC and anti-5-hmC antibodies, followed by incubation with a FITC-conjugated secondary antibody (1:500; Santa Cruz Biotechnology). Fluorescence intensity was detected using a confocal microscope or flow cytometer, as described by Rocha et al. (2019).
Chromatin immunoprecipitation (ChIP) and quantitative PCR
ChIP assay was performed using a simple ChIPTM kit (Cell Signaling Technology) according to the manufacturer’s instructions. The DNA recovered from the immunoprecipitated complexes was subjected to qPCR. Primers for the NRF2 region with TET1-, MLL1-, and HAT1-binding sites were as follows: forward primer 5′-TGAGATATTTTGCACATCCGATA-3′ and reverse primer 5′-ACTCTCAGGGTTCCTTTACACG-3′ (Kang et al., 2019).
Transfection of siRNA
Cells were transfected with 10-20 nM control siRNA and siRNAs against TET1, MLL1, and HAT1 (Santa Cruz Biotechnology) using Lipofectamine RNAiMax (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions (Berardo et al., 2019).
Statistical analysis
All results are expressed as the mean ± standard error. Data were analyzed using analysis of variance and Tukey’s test, and statistical significance was set at p<0.05.
RESULTS
Sensitivity of SNUC5 and SNUC5/RR cells to γ-radiation
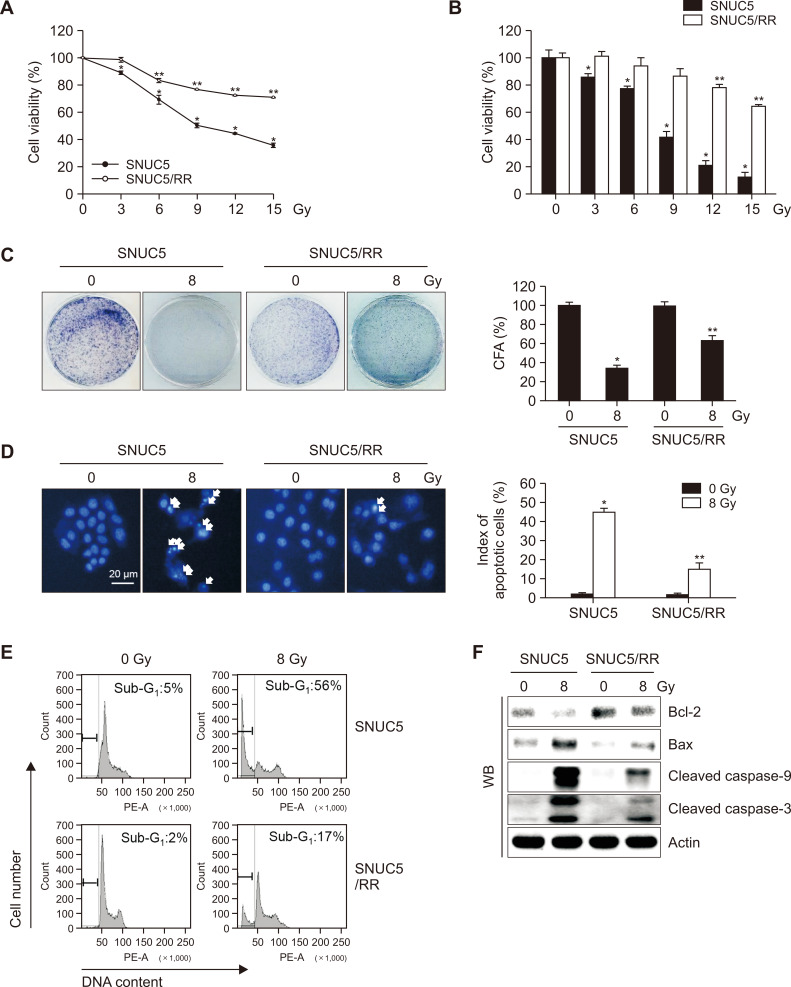
The cytotoxic effects of γ-radiation on SNUC5 and its γ-radiation-resistant variant SNUC5/RR were compared by examining cell viability, colony formation, and apoptosis rates. The concentrations of γ-radiation that yielded 50% growth inhibition (IC50) were 8 Gy in SNUC5 cells and >15 Gy in SNUC5/RR cells (Fig. 1A), as confirmed using a trypan blue assay (Fig. 1B). Sensitivity to 8 Gy radiation was higher in SNUC5 cells than that in SNUC5/RR cells, as evidenced by colony formation assays, Hoechst 33342 staining, and analyses of the sub-G1 population (Fig. 1C-1E). In addition, at 8 Gy, the expression levels of pro-apoptotic proteins Bax, active caspase-9, and active caspase-3 were higher in SNUC5 cells than those in SNUC5/RR cells. In contrast, the expression of the anti-apoptotic protein, Bcl-2, was lower in SNUC5 cells than that in SNUC5/RR cells (Fig. 1F).
Fig. 1.
Sensitivity of SNUC5 and SNUC5/RR cells to γ-radiation. Cells were exposed to a dose of 8 Gy. (A, B) Cell viability assessed after three days of culture using (A) MTT and (B) trypan blue assays. (C) Colony-forming ability evaluated after 10 days of culture using a colony staining kit. (D) Apoptosis detected using Hoechst 33342 staining. (E) Apoptotic sub-G1 cells identified using flow cytometry. (F) Expression of apoptosis-related proteins assessed using western blotting. (A-D) *p<0.05 vs. untreated SNUC5 cells; **p<0.05 vs. untreated SNUC5/RR cells (n=3).
Intracellular ROS levels in SNUC5 and SNUC5/RR cells
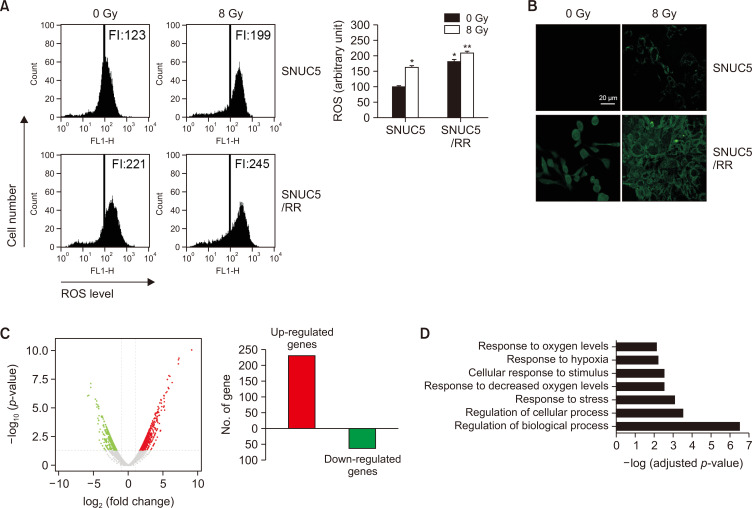
ROS levels in SNUC5 and SNUC5/RR cells in response to γ-radiation were measured using H2DCFDA staining. Flow cytometry revealed higher levels of ROS in SNUC5/RR cells than those in SNUC5 cells, with or without exposure to 8 Gy (Fig. 2A). These findings were confirmed by confocal microscopy, which showed that the intensity of green fluorescence generated by ROS was higher in SNUC5/RR cells than in SNUC5 cells (Fig. 2B).
Fig. 2.
Intracellular ROS levels in SNUC5 and SNUC5/RR cells. (A, B) ROS levels detected using (A) flow cytometry and (B) confocal imaging after staining with H2DCFDA. FI indicates the fluorescence intensity of DCF. *p<0.05 vs. untreated SNUC5 cells; **p<0.05 vs. untreated SNUC5/RR cells (n=3). (C) Differentially expressed genes between SNUC5 and SNUC5/RR cells identified using RNA-Seq. Volcano plot presents differentially expressed genes between SNUC5 and SNUC5/RR cells. Red dots represent upregulated genes, and green dots represent downregulated genes. (D) Gene ontology enrichment analysis of the upregulated genes in SNUC5/RR and SNUC5 cells. The lengths of the bars indicate -log (adjusted p-value).
We further screened for global mRNA changes following irradiation in SNUC5 and SNUC5/RR cells using RNA sequencing (Fig. 2C). The volcano plot revealed significantly differentially expressed genes in SNUC5 and SNUC5/RR cells (Fig. 2C, left). Based on a two-fold expression change, 297 radiation-responsive genes differentially expressed between SNUC5/RR and SNUC5 cells were identified, of which 231 were upregulated and 66 were downregulated (Fig. 2C, right). The upregulated genes were associated with the regulation of biological and cellular processes, stress responses, decreased oxygen levels, and stimulus responses (Fig. 2D).
Expression of NRF2 in SNUC5 and SNUC5/RR cells
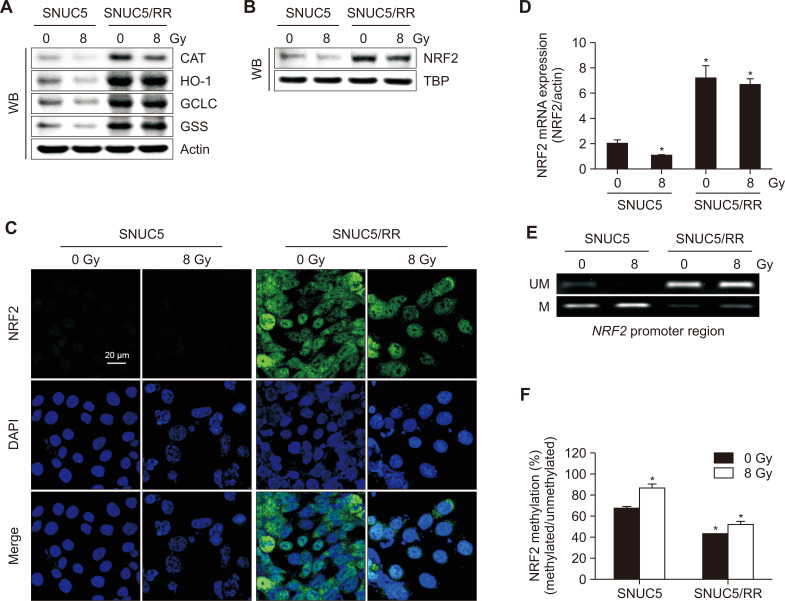
To investigate the role of NRF2 in SNUC5 and SNUC5/RR cells, we analyzed the expression of NRF2 and related antioxidant enzymes. The expression levels of CAT, HO-1, GCLC, and GSS were higher in SNUC5/RR cells than those in SNUC5 cells with or without radiation at 8 Gy (Fig. 3A). Nuclear NRF2 was expressed at higher levels in SNUC5/RR cells than in SNUC5 cells, treated with or without 8 Gy radiation, at both the mRNA and protein levels (Fig. 3B-3D). MSP and qMSP were performed to assess the methylation status of the NRF2 promoter. Specific CpG sites in the promoter region of NRF2 were hypermethylated in SNUC5 cells compared to those in SNUC5/RR cells with or without 8 Gy exposure (Fig. 3E, 3F).
Fig. 3.
NRF2 expression in SNUC5 and SNUC5/RR cells. (A) Antioxidant-related proteins detected using western blotting. (B, C) Nuclear NRF2 protein expression assessed using (B) western blotting and (C) confocal microscopy. The TBP antibody was used as a loading control for nuclear proteins. Green fluorescence indicates NRF2 expression and blue fluorescence indicates nuclei. The merged image indicates the nuclear localization of NRF2. (D) NRF2 mRNA expression assessed using qRT-PCR. *p<0.05 vs. untreated SNUC5 cells (n=3). (E) Genomic DNA isolated from SNUC5 and SNUC5/RR cells was subjected to an MSP analysis of the NRF2 promoter. Methylation was detected using methylation-specific primers. Unmethylated detection was detected by amplifying the product using unmethylation-specific primers. UM: unmethylated DNA; M: methylated DNA. (F) Real-time quantitative MSP analysis of NRF2 in SNUC5 and SNUC5/RR cells. Bars represent the methylation percentages of the CpG islands. *p<0.05 vs. untreated SNUC5 cells (n=3).
DNA demethylation in SNUC5 and SNUC5/RR cells
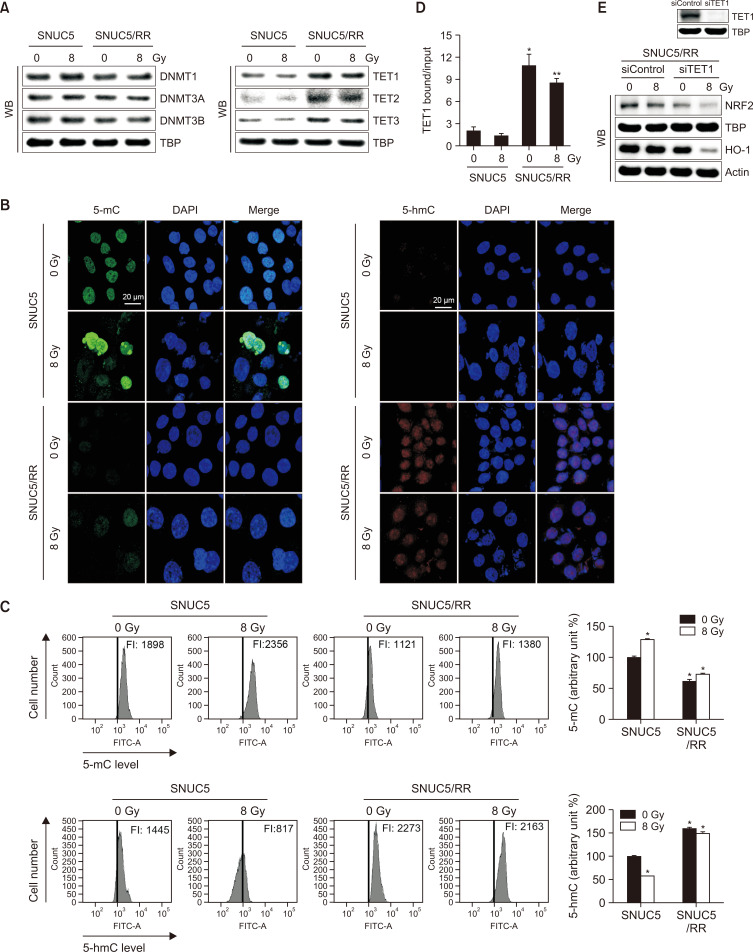
The expression levels of proteins involved in epigenetic modifications, specifically DNA methyltransferases (DNMTs) and demethylases (TETs), were evaluated. DNMT1, DNMT3A, and DNMT3B expression levels did not differ significantly between SNUC5 and SNUC5/RR cells, whereas TET1, TET2, and TET3 expression levels were higher in SNUC5/RR cells than those in SNUC5 cells irrespective of radiation (8 Gy) exposure (Fig. 4A). TET activity was assessed by measuring 5-mC and 5-hmC levels. The levels of 5-mC were lower in SNUC5/RR cells than in SNUC5 cells with or without 8 Gy exposure, whereas 5-hmC levels were higher in SNUC5/RR cells than in SNUC5 cells, as shown by confocal imaging and flow cytometry (Fig. 4B, 4C). TET1 binding to the NRF2 promoter, as assessed by the ChIP assay, was significantly increased in SNUC5/RR cells compared to that in SNUC5 cells, with or without 8 Gy exposure (Fig. 4D). Furthermore, TET1 knockdown in SNUC5/RR cells decreased the expression of NRF2 and its target protein, HO-1 with or without 8 Gy exposure (Fig. 4E). These results suggest that TET1 positively regulates NRF2 activation in SNUC5/RR cells.
Fig. 4.
DNA methylation-related proteins in SNUC5 and SNUC5/RR cells. (A) Expression of DNA methylases DNMT1, 3A, and 3B and DNA demethylases TET1, 2, and 3 determined using western blotting. (B, C) TET-catalyzed conversion of 5-mC to 5-hmC assessed using (B) confocal imaging and (C) flow cytometry. *p<0.05 vs. untreated SNUC5 cells (n=3). (D) Anti-TET1 antibodies and primers used to amplify the NRF2 promoter region used for a ChIP analysis. *p<0.05 vs. untreated SNUC5 cells; **p<0.05 vs. untreated SNUC5/RR cells (n=3). (E) Expression levels of NRF2 and HO-1 in SNUC5/RR cells transfected with siControl or siTET1 for 24 h determined using western blotting.
Expression of histone modification-related proteins in SNUC5 and SNUC5/RR cells
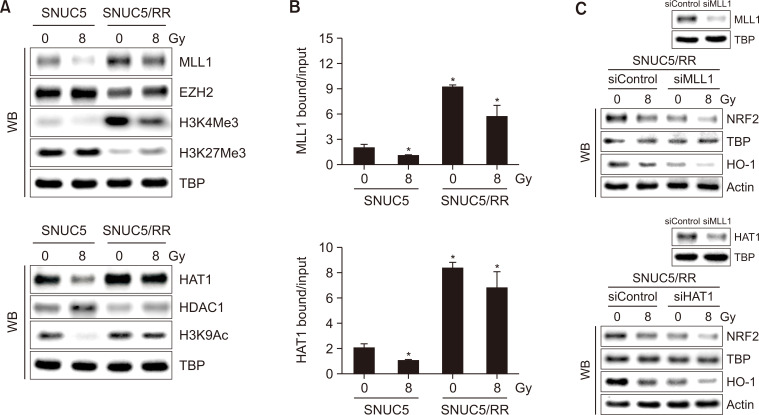
TET-dependent DNA demethylation upregulated NRF2 expression in SNUC5/RR cells. We further investigated the expression of histone methylation- and acetylation-related proteins in SNUC5 and SNUC5/RR cells. Levels of histone methyltransferase MLL1 and trimethylation of its target protein H3K4 (H3K4Me3) were higher in SNUC5/RR cells than those in SNUC5 cells with or without 8 Gy exposure, whereas levels of EZH2 and trimethylation of its target protein H3K27(H3K27Me3) were lower in SNUC5/RR cells (Fig. 5A). In addition to histone methylation, HAT1 expression was higher and HDAC1 expression was lower in SNUC5/RR cells than that in SNUC5 cells, with or without 8 Gy exposure, resulting in increased H3K9 acetylation (H3K9Ac) (Fig. 5A). The binding of MLL1 or HAT1 to the NRF2 promoter was significantly higher in SNUC5/RR cells with or without 8 Gy exposure (Fig. 5B). Furthermore, knockdown of MLL1 or HAT1 in SNUC5/RR cells decreased the expression levels of NRF2 and HO-1 (Fig. 5C). These results suggest that MLL1 and HAT1 positively regulate NRF2 activation in SNUC5/RR cells.
Fig. 5.
Histone modification-related protein expression in SNUC5 and SNUC5/RR cells. (A) Expression of histone methylation-related proteins, MLL1, EZH2, H3K4Me3, and H3K27Me3, and histone acetylation-related proteins, HAT1, HDAC1, and H3K9Ac, determined using western blotting. (B) Anti-MLL1 or -HAT1 antibodies and primers targeting the NRF2 promoter region used for the ChIP analysis. *p<0.05 vs. untreated SNUC5 cells (n=3). (C) NRF2 and HO-1 protein expression levels in SNUC5/RR cells transfected with siMLL1 or siHAT1 RNA for 24 h determined using western blotting.
Sensitivity to γ-radiation in SNUC5/RR cells with TET1, MLL1, and HAT1 knockdown
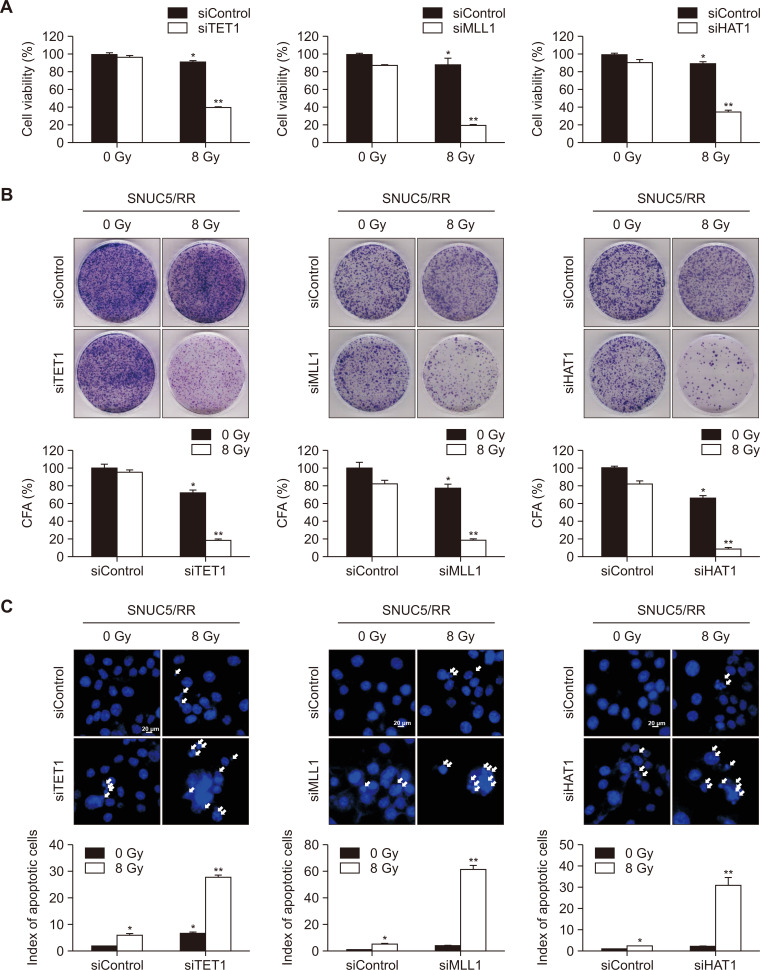
The putative roles of TET1, MLL1, and HAT1 in the sensitivity of SNUC5/RR cells to γ-radiation were investigated using siRNA-mediated silencing in vitro. siTET1-, siMLL1-, or siHAT1-transfected SNUC5/RR cells showed significantly lower viability than siControl-transfected SNUC5/RR cells exposed to 8 Gy (Fig. 6A). In addition, the ability of the cells to form colonies was decreased in siTET1-, siMLL1-, or siHAT1-transfected SNUC5/RR cells with 8 Gy exposure compared to siControl-transfected cells, indicating a reduced proliferative capacity (Fig. 6B). Moreover, apoptotic rates were higher in siTET1-, siMLL1-, or siHAT1-transfected SNUC5/RR cells exposed to 8 Gy than in siControl-transfected cells exposed to 8 Gy (Fig. 6C).
Fig. 6.
Sensitivity of SNUC5/RR cells with TET1, MLL1, and HAT1 knockdown to γ-radiation. (A) Cell viability determined using the MTT assay. (B) For the colony formation assay, cells were cultured for 10 days. Colonies containing more than 50 cells were counted. (C) Apoptotic cells determined using the Hoechst 33342 assay. (A-C) *p<0.05 vs. untreated SNUC5 cells; **p<0.05 vs. untreated SNUC5/RR cells (n=3).
DISCUSSION
Radiotherapy is a widely used treatment for cancer as it induces DNA damage and promotes cell death in malignant cells. However, many colon cancer cells develop resistance mechanisms that reduce the effectiveness of treatment (Geng and Wang, 2017; Tang et al., 2018). In particular, key genes involved in apoptosis and DNA damage repair are often silenced by epigenetic alterations, such as DNA hypermethylation and histone modifications, which contribute to IR resistance (Wang et al., 2022; Laanen et al., 2023).
In the present study, we observed that SNUC5/RR cells exhibited a higher IC50 value than the parental SNUC5 cells, indicating greater resistance to radiation. This increased resistance was associated with alterations in the expression of apoptosis markers, with reduced levels of pro-apoptotic proteins (Bax, active caspase-9, and active caspase-3) and increased levels of the anti-apoptotic protein (Bcl-2) in SNUC5/RR cells. These findings suggest that dysregulation of apoptotic pathways is a key feature of radioresistance in CRC. Furthermore, SNUC5/RR cells showed elevated levels of ROS and enhanced expression of antioxidant enzymes, which were mediated by increased NRF2 activity. High ROS levels are likely to trigger adaptive antioxidant mechanisms, enabling cells to resist IR-induced damage. These findings indicate that NRF2 is a protective factor in IR-resistant CRC cells, likely by facilitating cellular defense mechanisms against oxidative stress.
The present study showed that NRF2 expression and activation were modulated by DNA demethylation and histone modifications. Furthermore, the demethylase TET1 was upregulated in SNUC5/RR cells, leading to hypomethylation of the NRF2 promoter and enhanced NRF2 expression. Additionally, histone modifiers, such as MLL1 and HAT1, boost NRF2 expression by modifying histones H3K4 and H3K9. These results highlight the critical role of epigenetic changes in radioresistance in CRC.
We also demonstrated that knockdown of TET1, MLL1, or HAT1 in SNUC5/RR cells significantly reduced NRF2 expression and increased IR sensitivity by reducing NRF2 levels. This suggests that targeting these epigenetic regulators may be a promising strategy for overcoming radioresistance in CRC. The results of our study highlight the potential of targeting epigenetic modifications as a therapeutic approach to enhance radiotherapy efficacy.
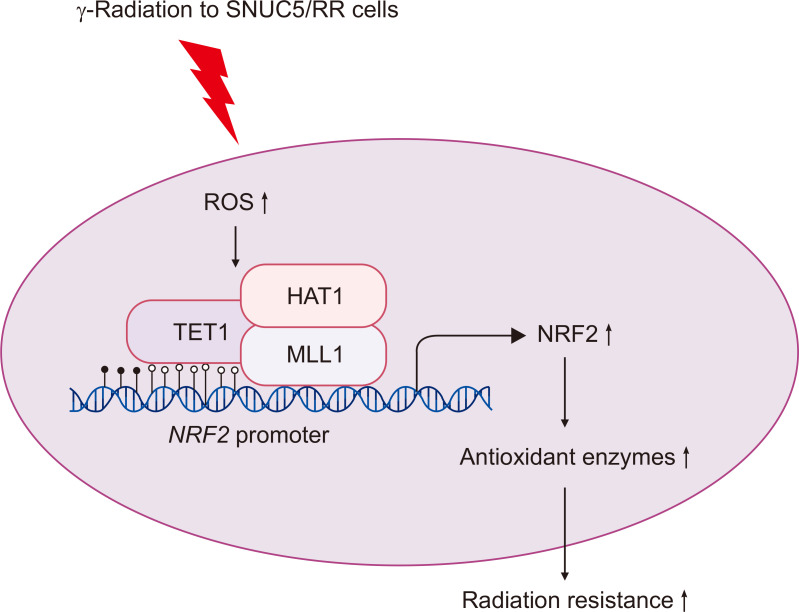
In conclusion, our findings suggest that resistance to γ-radiation in CRC is driven by alterations in NRF2 expression, which is mediated by both DNA demethylation and histone modifications (Fig. 7). Based on these findings, we propose that combining radiotherapy with epigenetic modulators could serve as an effective strategy to overcome IR resistance and improve the therapeutic outcomes in patients with CRC. This approach could lead to more personalized treatment strategies based on the specific epigenetic landscape of individual cancers.
Fig. 7.
Proposed model of regulation of NRF2 in γ-radiation resistance through epigenetic mechanisms. γ-Radiation increases intracellular ROS levels and the expression of DNA demethylase TET1, as well as histone modifying factors MLL1 and HAT1 in SNUC5/RR cells. TET1, MLL1, and HAT1 subsequently bind to the NRF2 promoter, enhancing NRF2 expression, which consequently induces the expression of antioxidant enzymes targeted by NRF2, leading to increased resistance to γ-radiation. Nuclear factor erythroid 2-related factor 2 (NRF2), Reactive oxygen species (ROS), ten-eleven translocation 1 (TET1), mixed-lineage leukemia (MLL), histone acetyltransferase (HAT).
ACKNOWLEDGMENTS
This work was supported by a research grant from the Jeju National University Hospital in 2023.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- Berardo C., Siciliano V., Di Pasqua L. G., Richelmi P., Vairetti M., Ferrigno A. Comparison between lipofectamine RNAiMAX and GenMute transfection agents in two cellular models of human hepatoma. Eur. J. Histochem. 2019;63:3048. doi: 10.4081/ejh.2019.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonura A., Giacomarra M., Montana G. The Keap1 signaling in the regulation of HSP90 pathway. Cell Stress Chaperones. 2022;27:197–204. doi: 10.1007/s12192-022-01253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumah V. V., Masson-Meyers D. S., Awosika O., Zacharias S., Enwemeka C. S. The viability of human cells irradiated with 470 nm light at various radiant energies in vitro. Lasers Med. Sci. 2021;36:1661–1670. doi: 10.1007/s10103-021-03250-z. [DOI] [PubMed] [Google Scholar]
- Castillo F., Mackenzie T. A., Cautain B. Immunofluorescence analysis by confocal microscopy for detecting endogenous FOXO. Methods Mol. Biol. 2019;1890:143–149. doi: 10.1007/978-1-4939-8900-3_12. [DOI] [PubMed] [Google Scholar]
- Chaiswing L., St Clair W. H., St Clair D. K. Redox paradox: A novel approach to therapeutics-resistant cancer. Antioxid. Redox Signal. 2018;29:1237–1272. doi: 10.1089/ars.2017.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Guede Á., Vera O., Ibáñez-de-Caceres I. When oxidative stress meets epigenetics: implications in cancer development. Antioxidants. 2020;9:468. doi: 10.3390/antiox9060468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L., Wang J. Molecular effectors of radiation resistance in colorectal cancer. Precis. Radiat. Oncol. 2017;1:27–33. doi: 10.1002/pro6.5. [DOI] [Google Scholar]
- Häfner M. F., Debus J. Radiotherapy for colorectal cancer: current standards and future perspectives. Visc. Med. 2016;32:172–177. doi: 10.1159/000446486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Ru X., Wen T. NRF2, a transcription factor for stress response and beyond. Int. J. Mol. Sci. 2020;21:4777. doi: 10.3390/ijms21134777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Gao S., Guo H., Ren S., Ji F., Liu Z., Chen X. Re-sensitization of radiation resistant colorectal cancer cells to radiation through inhibition of AMPK pathway. Oncol. Lett. 2016;11:3197–3201. doi: 10.3892/ol.2016.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing M., Zhang H., Wei M., Tang Y., Xia Y., Chen Y., Shen Z., Chen C. Reactive oxygen species partly mediate DNA methylation in responses to different heavy metals in pokeweed. Front. Plant Sci. 2022;13:845108. doi: 10.3389/fpls.2022.845108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K. A., Piao M. J., Hyun Y. J., Zhen A. X., Cho S. J., Ahn M. J., Yi, Hyun J. W. Luteolin promotes apoptotic cell death via upregulation of Nrf2 expression by DNA demethylase and the interaction of Nrf2 with p53 in human colon cancer cells. Exp. Mol. Med. 2019;51:1–14. doi: 10.1038/s12276-019-0238-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X., Chen Z., Wang X., Kang H., Hong S. Quercetin improves the imbalance of Th1/Th2 cells and Treg/Th17 cells to attenuate allergic rhinitis. Autoimmunity. 2023;56:2189133. doi: 10.1080/08916934.2023.2189133. [DOI] [PubMed] [Google Scholar]
- Kietzmann T., Petry A., Shvetsova A., Gerhold J. M., Görlach A. The epigenetic landscape related to reactive oxygen species formation in the cardiovascular system. Br. J. Pharmacol. 2017;174:1533–1554. doi: 10.1111/bph.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. M., Hong Y., Lee S., Liu P., Lim J. H., Lee Y. H., Lee T. H., Chang K. T., Hong Y. Therapeutic implications for overcoming radiation resistance in cancer therapy. Int. J. Mol. Sci. 2015a;16:26880–26913. doi: 10.3390/ijms161125991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Xue X. Detection of total reactive oxygen species in adherent cells by 2',7'-dichlorodihydrofluorescein diacetate staining. J. Vis. Exp. 2020;160:10. doi: 10.3791/60682-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I. A., Hur J. Y., Kim H. J., Park J. H., Hwang J. J., Lee S. A., Lee S. E., Kim W. S., Lee K. Y. Targeted next-generation sequencing analysis for recurrence in early-stage lung adenocarcinoma. Ann. Surg. Oncol. 2021a;28:3983–3993. doi: 10.1245/s10434-020-09276-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Kim C. W., Shin A., Kang H., Jung S. J. Effect of chemotherapy and radiotherapy on cognitive impairment in colorectal cancer: evidence from Korean National Health Insurance database Cohort. Epidemiol. Health. 2021b;43:e2021093. doi: 10.4178/epih.e2021093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Youn H., Kang C., Youn B. Inflammation-induced radioresistance is mediated by ROS-dependent inactivation of protein phosphatase 1 in non-small cell lung cancer cells. Apoptosis. 2015b;20:1242–1252. doi: 10.1007/s10495-015-1141-1. [DOI] [PubMed] [Google Scholar]
- Laanen P., Cuypers A., Saenen E., Horemans N. Flowering under enhanced ionising radiation conditions and its regulation through epigenetic mechanisms. Plant Physiol. Biochem. 2023;196:246–259. doi: 10.1016/j.plaphy.2023.01.049. [DOI] [PubMed] [Google Scholar]
- Lal M., Gupta D. Studies on radiation sensitization efficacy by silymarin in colon carcinoma cells. Discoveries. 2016;4:e56. doi: 10.15190/d.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Kim S., Lee Y., Lee H., Lee Y., Park H., Nahm J.H., Ahn S., Yu S.J., Lee K., Kim H. The clinicopathological and prognostic significance of Nrf2 and Keap1 expression in hepatocellular carcinoma. Cancers. 2020;12:2128. doi: 10.3390/cancers12082128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Huang W., Hong J., Cai C., Zhang W., Zhang J., Kang Z. Long noncoding RNA LINC00630 promotes radioresistance by regulating BEX1 gene methylation in colorectal cancer cells. IUBMB Life. 2020;72:1404–1414. doi: 10.1002/iub.2263. [DOI] [PubMed] [Google Scholar]
- Liu Z. Y., Song K., Tu B., Lin L. C., Sun H., Zhou Y., Li R., Shi Y., Yang J. J., Zhang Y., Zhao J. Y., Tao H. Crosstalk between oxidative stress and epigenetic marks: new roles and therapeutic implications in cardiac fibrosis. Redox Biol. 2023;65:102820. doi: 10.1016/j.redox.2023.102820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann E., O'Sullivan J., Marcone S. Targeting cancer-cell mitochondria and metabolism to improve radiotherapy response. Transl Oncol. 2021;14:100905. doi: 10.1016/j.tranon.2020.100905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland R. D., Culp T. N., Marchant D. J. Imaging flow cytometry and confocal immunofluorescence microscopy of virus-host cell interactions. Front. Cell. Infect. Microbiol. 2021;11:749039. doi: 10.3389/fcimb.2021.749039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K., Miyake K., Arai S., Fukuda K., Suzuki C., Kotani H., Tanimoto A., Nishiyama A., Nanjo S., Yamashita K., Takeuchi S., Yano S. Methylation of tumor suppressive miRNAs in plasma from patients with pancreaticobiliary diseases. Cancer Diagn. Progn. 2022;2:378–383. doi: 10.21873/cdp.10120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha M. A., Veronezi G. M. B., Felisbino M. B., Gatti M. S. V., Tamashiro W. M. S. C., Mello M. L. S. Sodium valproate and 5-aza-2'-deoxycytidine differentially modulate DNA demethylation in G1 phase-arrested and proliferative HeLa cells. Sci. Rep. 2019;9:18236. doi: 10.1038/s41598-019-54848-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H. S., Kim H. J., Ji W. B., Kim B. C., Kim J. H., Moon S. K., Kang S. I., Kwak H. D., Kim E. S., Kim C. H., Kim T. H., Noh G. T., Park B. S., Park H. M., Bae J. M., Bae J. H., Seo N. E., Song C. H., Ahn M. S., Eo J. S., Yoon Y. C., Yoon J. K., Lee K. H., Lee K. H., Lee K. Y., Lee M. S., Lee S. H., Lee J. M., Lee J. E., Lee H. H., Ihn M. H., Jang J. H., Jeon S. K., Chae K. J., Choi J. H., Pyo D. H., Ha G. W., Han K. S., Hong Y. K., Hong C. W., Kwak J. M. Colon cancer: the 2023 Korean clinical practice guidelines for diagnosis and treatment. Ann. Coloproctol. 2024;40:89–113. doi: 10.3393/ac.2024.00059.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormunen A., Koivulehto E., Alitalo K., Saksela K., Laham-Karam N., Ylä-Herttuala S. Comparison of automated and traditional western blotting methods. Methods Protoc. 2023;6:43. doi: 10.3390/mps6020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talarico C., Dattilo V., D'Antona L., Menniti M., Bianco C., Ortuso F., Alcaro S., Schenone S., Perrotti N., Amato R. SGK1, the new player in the game of resistance: chemo-radio molecular target and strategy for inhibition. Cell Physiol. Biochem. 2016;39:1863–1876. doi: 10.1159/000447885. [DOI] [PubMed] [Google Scholar]
- Tang L., Wei F., Wu Y., He Y., Shi L., Xiong F., Gong Z., Guo C., Li X., Deng H., Cao K., Zhou M., Xiang B., Li X., Li Y., Li G., Xiong W., Zeng Z. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J. Exp. Clin. Cancer Res. 2018;37:87. doi: 10.1186/s13046-018-0758-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Xu L., Wang G., Chen L., Li C., Jiang X., Gao H., Yang B., Tian W. Prognostic and clinicopathological significance of NRF2 expression in non-small cell lung cancer: a meta-analysis. PLoS One. 2020;15:e0241241. doi: 10.1371/journal.pone.0241241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Han Y., Jin Y., He Q., Wang Z. The advances in epigenetics for cancer radiotherapy. Int. J. Mol. Sci. 2022;23:5654. doi: 10.3390/ijms23105654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Guo E., Ming J., Sun W., Nie X., Sun L., Peng S., Luo M., Liu D., Zhang L., Mei Q., Long G., Hu G., Hu G. Radiation-induced DNMT3B promotes radioresistance in nasopharyngeal carcinoma through methylation of p53 and p21. Mol. Ther. Oncolytics. 2020;17:306–319. doi: 10.1016/j.omto.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Xiao Y., Liu H., Chen Q., Zhu L., Zeng L., Liu X., Pan Y., Zhang J., Fu J., Shao C. Elevation of H3K27me3 level contributes to the radioresistance of nasopharyngeal carcinoma by inhibiting OAS1 expression. Am. J. Physiol. Cell Physiol. 2024;326:C60–C73. doi: 10.1152/ajpcell.00358.2023. [DOI] [PubMed] [Google Scholar]
- Zimta A. A., Cenariu D., Irimie A., Magdo L., Nabavi S. M., Atanasov A. G., Berindan-Neagoe I. The role of Nrf2 activity in cancer development and progression. Cancers. 2019;11:1755. doi: 10.3390/cancers11111755. [DOI] [PMC free article] [PubMed] [Google Scholar]