Abstract
The ETS transcription factor family is characterized by a conserved ETS DNA-binding domain and its members have been implicated in a plethora of biological processes, including development, cell transformation and metastasis. ER71 is a testis-specific ETS protein that is not homologous to any other protein outside its ETS domain, suggesting that it fulfills a unique physiological role. Here, we report that ER71 is a constitutively nuclear protein whose intracellular localization is dependent on a portion of the ETS domain, namely ER71 amino acids 276–315. Furthermore, the DNA binding activity is intramolecularly regulated, as the N-terminus of ER71 has a negative effect on DNA binding while the C-terminus dramatically enhances this activity. We also demonstrate that ER71 possesses an extremely potent N-terminal transactivation domain comprised of amino acids 1–157. Finally, we show that ER71 is capable of directly activating both an E74 site-driven and the matrix metalloproteinase-1 promoter. Altogether, these data represent the first functional characterization of ER71, which may perform important functions in the developing and adult testis as well as in testicular germ cell tumorigenesis.
INTRODUCTION
Proteins that belong to the ETS family of transcription factors share a conserved DNA-binding region of ∼85 amino acids which binds to target sequences encompassing the purine-rich motif 5′-GGA(A/T)-3′. ETS factors have been implicated in a wide array of biological processes, including skeletal development, neural synapse formation, hematopoiesis, immunomodulation, tumorigenesis and metastasis (1–3).
One of the most prevalent questions about ETS transcription factors is that of functional specificity: how do ETS proteins manage to achieve functional specificity if their DNA-binding domains and, as a consequence, the sequences that they target are highly conserved? One answer is that gene regulation by ETS proteins is highly dependent on protein–protein interactions with co-activators and other transcription factors (1,2,4). An example is provided by the interaction of the ETS factor GABP-α with its partner GABP-β, which results in the formation of a tetramer that is capable of recognizing DNA target sequences with high affinity (5,6). Similarly, the ETS proteins Sap-1 and Elk-1 must physically interact with the serum response factor in order to bind to and activate the c-fos protooncogene (7,8). In a comparable fashion, PU.1 collaborates with Pip/IRF-4 to stimulate the immunoglobulin light chain λ enhancer and up-regulates transcription of the CD20 gene (9–11). Perhaps the best example of how protein partnerships alter the specificity of ETS factors is the cooperative DNA binding of Ets-1 with Pax-5: upon forming a ternary complex with DNA and Pax-5, Ets-1 undergoes a conformational change that enables it to interact with a non-consensus DNA- binding site (12,13).
Furthermore, the tissue expression pattern of a given ETS factor largely determines its biological role. While some ETS proteins, such as Ets-1, are ubiquitously expressed, others are only found in a limited number of tissues. For instance, PU.1 expression is restricted to hematopoietic cell lineages, where PU.1 is essential for differentiation of monocytes and B cells (14,15). Moreover, connections between sensory and motor neurons that control a common muscle are defined by the differential expression of two related ETS proteins, PEA3 and ER81, during neuronal development (16).
Finally, the selectivity of an ETS factor for a specific promoter also depends on the context in which an ETS-binding site is found, as different ETS proteins have a different affinity for an ETS-binding site depending on the sequences that flank the 5′-GGA(A/T)-3′ core motif. For instance, whereas Ets-1 prefers to bind to 5′-ACCGGA(A/T)G(T/C)-3′ consensus sequences, the ETS protein PU.1 binds preferentially to 5′-A(G/C)(A/C/G)GGAA(G/C)T-3′ (17,18).
Targeted gene disruption and overexpression experiments in mice have expanded our understanding of the biological roles played by specific ETS proteins, demonstrating that different ETS factors perform distinct biological functions. For example, lack of PU.1 activity leads to severe impairment of lymphocyte development (14,15). Similarly, mice without the ETS protein Spi-B have hampered B cell responses (19). PEA3 null male mice exhibit sexual dysfunction, thought to reflect an underlying neurological defect (20), and functional connections between group Ia sensory afferent and motor neurons fail to develop in mice lacking the closely related ETS protein ER81 (21). On the other hand, overexpression of certain ETS proteins has been associated with various diseases. For instance, transgenic mouse studies have shown that Ets-2 overexpression leads to a phenotype that is reminiscent of the one observed in Down’s syndrome individuals, consistent with the location of the human Ets-2 gene on chromosome 21 (22). Furthermore, mice that overexpress the ETS factor Fli-1 are affected by a systemic lupus erythematosus-like disorder (23). Despite these studies, the precise role for most ETS factors remains to be elucidated.
ER71 is a member of the ETS factor family that, until now, has not been studied in detail. It is a 336 amino acid protein with a calculated molecular weight of 37.2 kDa whose expression appears to be testis-specific (6). ER71 is a remarkably unique protein; the degree of identity between its ETS domain and those of other factors is at most 67%, while the amino acid sequences that lie outside its ETS domain do not share homology with any other protein in current databases. As a first step in understanding the role of ER71 and its mechanisms of action, we functionally dissected this ETS transcription factor in this study.
MATERIALS AND METHODS
Plasmids
Tagged versions of truncations of murine ER71 and the full-length protein were obtained by cloning into the eukaryotic expression vectors pEVRF0-HA, pEVRF1-HA or pEVRF2-HA and pCS3+-6 Myc, which resulted in the N-terminal fusion of either an HA (hemagglutinin) or a hexa-Myc epitope sequence. Untagged constructs were obtained by cloning ER71 into pEV3S (24). For in vitro transcription and translation purposes, we generated a KS+ vector (Stratagene) with a modified multiple cloning site providing a consensus Kozak sequence (KS+K), into which different ER71 clones were inserted. We used the pAB-Gal4-linker plasmid (25) in order to generate chimeric proteins consisting of the DNA-binding domain of the yeast GAL4 protein and C-terminally fused ER71 amino acids. As reporter constructs, we utilized GAL42–tk80–Luc, E743–tk80–Luc and pGL2B-MMP-1 (–525/+15) plasmids, all of which have been described before (26,27). The pEGFP-C1 plasmid (Clontech) was utilized to generate the green fluorescent protein (GFP) fusions, GFP–ER71276–283 and GFP–ER71276–315.
Transfections
293T (human embryonic kidney fibroblasts), MLTC-1 (mouse Leydig tumor cells), Mv1Lu (mink lung) or RK13 (rabbit kidney) cells were transiently transfected by the calcium phosphate co-precipitation method. Briefly, the desired amount of the appropriate ER71 plasmid was mixed with 1 µg (or 2.5 µg in the case of MTLC-1 cells) of a reporter construct and 0.2 µg β-galactosidase expression plasmid pEQ176, together with sufficient KS+ to bring the total amount of DNA to 9 µg. Subsequently, 200 µl of 250 mM CaCl2 and 200 µl of 275 mM NaCl, 42 mM HEPES, 9.6 mM KCl, 1.5 mM Na2HPO4, pH 7.14, were added and the resulting mix distributed onto cells. Transfection was allowed to occur for 8–12 h at 37°C in 3% CO2. The cells were then washed twice with phosphate-buffered saline (PBS) and incubated in fresh medium for another 24–36 h at 37°C in 10% CO2.
Reporter gene assays
Thirty-six hours after transfection, cells were washed once with PBS and lysed in 25 mM Tris–HCl pH 7.8, 2 mM EDTA, 10% glycerol, 1% Triton X-100, 2 mM DTT for 5 min. Plates were then scraped and the extract was collected and centrifuged at 20 800 g for 1 min. The pellet was discarded and the cleared supernatant was assayed for luciferase activity. β-Galactosidase activity was used to monitor transfection efficiency (28).
Immunostaining
Cells were seeded onto coverslips and transiently transfected as described above with 4.5 µg of the indicated ER71 construct. Twenty-four hours after transfection the cells were washed with PBS, incubated with 1% sucrose, 3.7% formaldehyde in PBS for 10 min, washed once with 0.1 M glycine in PBS, permeabilized with PBS containing 2% normal donkey serum, 0.4% Triton X-100 for 20 min, washed three times in washing buffer (0.2% bovine serum albumin, 0.1% Triton X-100, PBS) and incubated for 1 h with anti-HA (12CA5) or anti-Myc (9E10) murine monoclonal antibodies at room temperature. After incubation, cells were washed four times with washing buffer and incubated with fluorescein isothiocyanate-coupled donkey anti-mouse antibodies in the dark. The coverslips were then washed twice in washing buffer, stained with 4′,6-diamidino-2-phenylindole (0.5 µg/ml) for 5 min, washed twice in PBS, mounted onto slides and visualized under an inverted fluorescence microscope. To determine the percentage of nuclear localization, at least 100 transfected cells were counted. We classified the staining of each cell as mostly nuclear (>70% of nuclear staining), mostly cytoplasmic (>70% cytoplasmic staining) or evenly distributed throughout the cell.
In vitro transcription and translation
Transcription and translation reactions were carried out using a Promega T3 coupled TNT kit resulting in synthesized proteins labeled with [35S]methionine. Quantitation of radioactive proteins on SDS–PAGE gels was done using a phosphorimager. Normalization of protein levels was done by taking into account the methionine content of each protein.
Electrophoretic mobility shift assays
Equimolar amounts of in vitro transcribed and translated ER71 constructs were incubated with 32P-labeled probe for 1 h on ice in 10% glycerol, 15 mM Tris–HCl pH 8, 0.5 mM MgCl2, 1 mM EDTA, 25 mM NaCl, 0.1 µg/µl bovine serum albumin, 0.1 µg/µl poly(dI–dC)·poly(dI–dC). After incubation, samples were loaded onto a 5% polyacrylamide gel and protein–DNA complexes resolved by electrophoresis and visualized by autoradiography. Quantitation of radioactivity present in each DNA–protein complex was done with the help of a phosphorimager system. The 35S signal was filtered out by inserting a transparent film between the dried gel and the phosphorimager screen. We then expressed these results as relative DNA binding activity in comparison with wild-type ER71. All results represent mean values derived from at least three independent experiments.
32P-labeled probes were generated by annealing complementary oligonucleotide pairs and subsequently radiolabeled by filling in the resulting overhangs by incubation with Klenow enzyme and [α-32P]dATP for 30 min at room temperature. Following incubation, the reaction was stopped with EDTA and the probes were purified on NucTrap columns (Stratagene) according to the manufacturer’s instructions. Synthetic oligonucleotides derived from the E74-binding site contained wild-type (sense, 5′-AGCTTCTCTAGCTGAATAACCGGAAGTAACTCATCG-3′; antisense, 5′-TCGACGATGAGTTACTTCCGGTTATTCAGCTAGAGA-3′) or a mutant ETS-binding site (sense, 5′-AGCTTCTCTAGCTGAATAACCCCAAGTAACTCATCG-3′; antisense, 5′-TCGACGATGAGTTACTTGGGGTTATTCAGCTAGAGA-3′). In addition, a probe was derived from the MMP-1 promoter (sense, 5′-TAATCAAGAGGATGTTATAAAGCATGAGTCAGAC-3′; antisense, 5′-CTGTCTGACTCATGCTTTATAACATCCTCTTG-3′). Consensus ETS-binding sites are shown in bold, with mutated residues underlined.
Site-directed mutagenesis
ER71 mutants were generated by site-directed mutagenesis using Stratagene’s QuikChange kit. We confirmed mutations by sequencing the entire coding region. After confirmation, we subcloned each mutant into the appropriate vector in order to ensure that no mistakes were inadvertently generated in the vector during the mutagenesis reaction.
RESULTS
ER71 is a constitutively nuclear protein
A prerequisite of transcription factors is that they are present in the nucleus in order to exert their function. To investigate the intracellular localization of ER71, Mv1Lu cells were transiently transfected with expression vectors of Myc-tagged, full-length and truncated ER71, and their localization within the cell assessed by indirect immunofluorescence microscopy. Full-length ER71 (amino acids 1–336) was exclusively nuclear in ∼94% of the transfected cells (Fig. 1, top left), with <6% of the transfected cells displaying variable degrees of cytoplasmic staining in addition to nuclear staining. Nuclear localization of the C-terminal truncation ER711–206 (Fig. 1, top right) was significantly disrupted, with at least 60% of the cells having equal nuclear and cytoplasmic staining. The staining pattern of two other truncations, ER71208–336 and ER711–315, was identical to wild-type ER71 (Fig. 1, bottom panels). These results indicate that the residues responsible for nuclear localization of ER71 are present between amino acids 208 and 315, which encompass the ETS domain. Similar results were obtained with another cell line, RK13, and with HA-tagged ER71 (data not shown).
Figure 1.
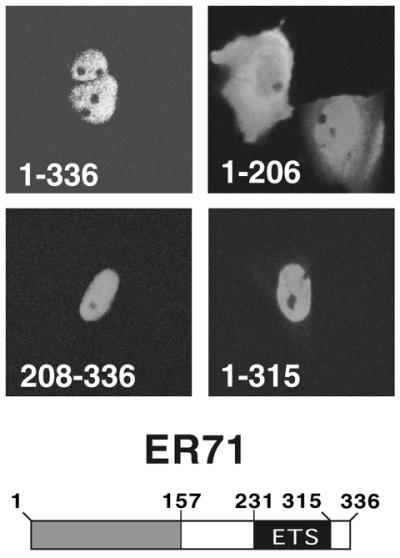
ER71 is a nuclear protein. Mv1Lu cells were transfected with the indicated Myc-tagged ER71 constructs and stained with anti-Myc antibodies. Please note that upon resequencing of the murine ER71 cDNA, an additional codon, which is missing in the originally published sequence, was noticed at position 24, resulting in an extra glutamic acid residue. Thus, the total number of amino acids in ER71 is 336, and not 335.
Amino acids 276–315 are important for ER71 nuclear localization
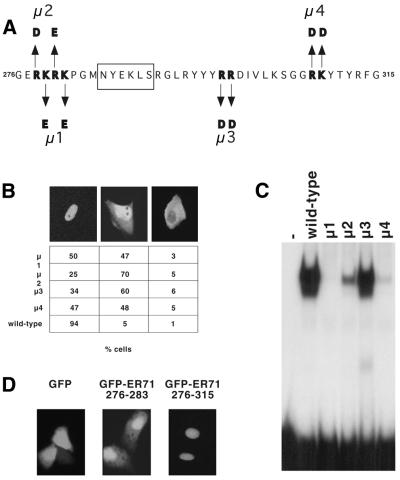
Nuclear localization signals often consist of a cluster of basic amino acids flanked by non-polar residues (29). Upon closer inspection of the ETS domain of ER71, we found that amino acids 276–283 may fit this standard. Additional clustered basic amino acids are between positions 284 and 315 and may be part of a bipartite nuclear localization signal (29). Thus, several mutants (µ1–µ4, Fig. 2A) were generated in order to determine whether these basic residues are required for nuclear localization of full-length ER71. Nuclear localization of all these ER71 mutants was significantly disrupted (Fig. 2B). Mutants µ2 and µ3 exhibited the strongest disruption in nuclear localization, with ≥60% of the cells displaying comparable nuclear and cytoplasmic staining. Similarly, the staining of about half of all cells transfected with either the µ1 or µ4 mutant was evenly distributed between the nucleus and the cytoplasm. This is clearly distinct from wild-type ER71, in which ∼94% of cells displayed exclusive nuclear staining.
Figure 2.
Amino acids 276–315 target ER71 to the nucleus. (A) Amino acid changes in the respective µ1–µ4 mutants of ER71. Residues crucial for DNA binding are boxed. (B) The different ER711–336 mutants were transfected into Mv1Lu cells and immunostained. At least 100 transfected cells were counted and their staining categorized as either predominantly nuclear, evenly distributed or predominantly cytoplasmic. Shown are representative photographs of each category (left, middle and right, respectively). The percentage of cells with respective staining is given below each photograph. (C) Wild-type or the indicated ER71 mutants were incubated with radiolabeled E74 oligonucleotide and the resulting DNA–protein complexes resolved in a native gel. (D) RK13 cells were transfected with 4.5 µg GFP, GFP–ER71276–283 or GFP–ER71276–315. Green fluorescence was observed upon excitation with 488 nm light with a fluorescence microscope.
The amino acid changes introduced into mutants µ1–µ4 are within the ETS DNA-binding domain. Conceivably, disruption of nuclear localization observed with µ1–µ4 may be due to loss of DNA binding activity, which may normally sequester ER71 to the chromosomes. In order to exclude this possibility, we tested the DNA binding properties of the ER71 mutants in electrophoretic mobility shift assays with a radioactively labeled E74 probe (Fig. 2C), which is specifically bound by ER71, as shown in Figure 3B. Mutant µ1 lost its ability to bind DNA. This finding was not unexpected, since both of the mutated residues in µ1 (K279 and K281) are highly conserved in ETS proteins and known to contact the phosphate backbone of DNA (2). Furthermore, the µ2 and µ4 mutants displayed severely impaired DNA binding (Fig. 2C). However, the µ3 mutant of ER71 was capable of binding DNA relatively well (∼50% of wild-type DNA binding activity), implying that nuclear localization of ER71 is not due to sequestration by chromosomal ETS-binding sites.
Figure 3.
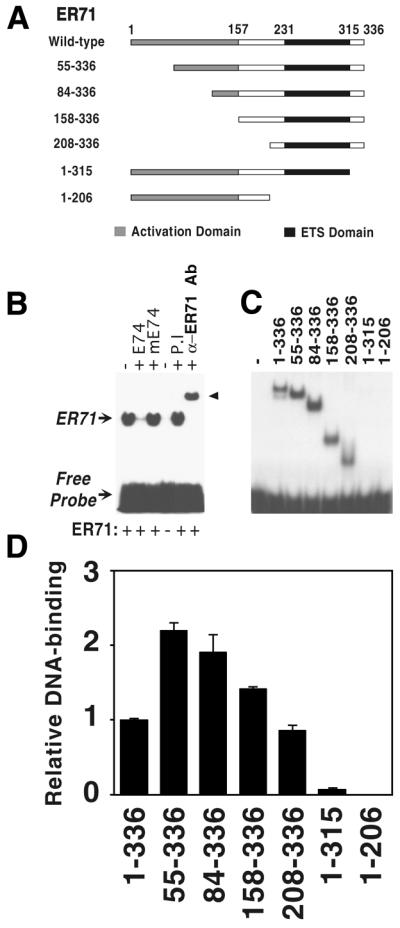
DNA binding properties of ER71. (A) Schematic representation of the ER71 truncations used. (B) The ability of full-length ER71 to retard the migration of 32P-labeled E74 probe in a non-denaturing acrylamide gel was assessed. Where indicated, an excess of non-labeled E74 or mutated E74 oligonucleotide, preimmune (P.I.) rabbit serum or a specific anti-ER71 antibody (α-ER71 Ab) was included in the DNA binding reaction mixture. (C) Equal protein amounts of the different ER71 constructs were incubated as in (B) and protein–DNA interactions visualized by autoradiography. In the original autoradiogram, a faint band can be seen in the lane corresponding to ER711–315. (D) Quantitation of relative DNA binding activity of the ER71 truncations.
To further confirm that residues found within positions 276–315 of ER71 are important in nuclear targeting, we fused ER71 amino acids 276–283 and 276–315 to the C-terminus of GFP. Due to poor expression of GFP–ER71 constructs in Mv1Lu cells, these were transiently transfected into RK13 cells. GFP alone was evenly distributed throughout the cell, while both GFP–ER71276–283 and GFP–ER71276–315 displayed a primarily nuclear staining (Fig. 2D). Although ER71 amino acids 276–283 are sufficient for some nuclear targeting, a more pronounced, nearly exclusive nuclear localization was observed with GFP–ER71276–315, suggesting that amino acids within positions 284–315 may enhance the strength of the nuclear localization signal of the 276–283 region. Altogether, these experiments demonstrate that ER71 is efficiently targeted to the nucleus by amino acids 276–315.
DNA binding properties of ER71
ETS transcription factors bind to specific sequences within promoter elements that contain the core motif 5′-GGA(A/T)-3′ (2). Consistently, ER71 has been shown to be capable of binding oligonucleotides containing this motif (6). Here, we established that ER71 can also bind to the E74 site, which has been documented to be bound by several ETS factors with very high affinity (28,30,31). As shown in Figure 3B, in vitro transcribed and translated full-length ER71 was able to bind to the radioactively labeled E74 oligonucleotide and retard its migration in a native gel. This interaction was abolished by addition of an excess of unlabeled E74 competitor, while a similar probe with a mutated ETS-binding site (mE74) was unable to disrupt this interaction. Incubation with a specific anti-ER71 antibody (raised against amino acids 161–191), but not with corresponding preimmune serum, resulted in a supershift, further corroborating that ER71 was indeed responsible for retarding migration of the E74 probe (Fig. 3B). In conclusion, ER71 specifically binds to the E74 site.
Because intramolecular modulation of DNA binding has been demonstrated to be an important regulatory mechanism of transcription factors, including several ETS proteins (31–34), we decided to investigate whether ER71 possesses domains that modulate DNA binding. To this end, we in vitro transcribed and translated several ER71 truncations (see Fig. 3A) and measured their ability to bind the E74 site in electrophoretic mobility shift assays. Truncation of 54 or 83 amino acids from the N-terminus of ER71 enhanced DNA binding ∼2-fold over full-length ER71 (Fig. 3C and D). Further deletion of N-terminal amino acids abrogated this effect, with ER71158–336 and ER71208–336 binding DNA comparably with the wild-type. Surprisingly, the DNA binding ability of ER711–315, which lacks the last 21 amino acids but retains an intact ETS domain, was almost completely abolished. ER711–206, which lacks the ETS domain, was included as a negative control and was not able to bind DNA. Altogether, these data show that the C-terminus of ER71 (amino acids 316–336) is required for efficient DNA binding by the ETS domain, while amino acids 1–54 have a small negative impact on the DNA binding function of the ETS domain.
The ER71 transactivation domain
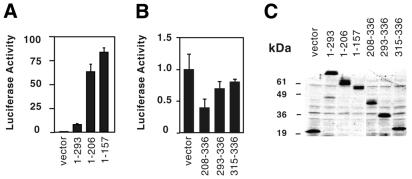
We next set out to elucidate whether ER71 contains regions with transactivation potential. To this end, we fused the DNA-binding domain of the yeast GAL4 protein to several regions of ER71. Plasmids encoding these GAL4–ER71 chimeras were transfected into 293T cells in order to assess their ability to activate a GAL4-binding site-driven luciferase reporter plasmid. Amino acids 1–293 fused to GAL4 activated transcription 8.5-fold more than GAL4 alone (Fig. 4A). Deletion of further C-terminal amino acids in GAL4–ER711–206 and GAL4–ER711–157 led to much stronger levels of activation (up to 84-fold over GAL4), however, amino acids 158–206 appear not to play a significant role, since the levels of activation mediated by GAL4–ER711–206 and GAL4–ER711–157 were quite comparable. Control western blots indicate that these effects were not due to different levels of protein expression (Fig. 4C). Thus, our results indicate the presence of an extremely potent N-terminal transactivation domain in ER71 spanning amino acids 1–157, which appears to be negatively regulated by amino acids C-terminal of position 206.
Figure 4.
Identification of a transactivation domain in ER71. 293T cells were transiently co-transfected with 1 µg GAL42–tk80–Luc reporter and the indicated expression vector for either progressive C-terminal (A) or N-terminal truncations (B) of ER71 fused to the GAL4 DNA-binding domain. The following amounts of expression vectors were employed: 1 ng GAL4, 50 ng GAL4–ER711–293, 50 ng GAL4–ER711–206, 80 ng GAL4–ER711–157, 1 ng GAL4–ER71208–336, 4 ng GAL4–ER71293–336 and 2.2 ng GAL4–ER71315–336. (C) Immunoblot with antibodies directed against the GAL4 DNA-binding domain demonstrating comparable levels of expression of the different GAL4–ER71 fusions. In order to be above the detection limit of our western blotting procedure, the amounts of expression vector were all raised 100-fold over those employed in (A) and (B).
We then tested the impact of these C-terminal amino acids of ER71 on gene transcription. Indeed, GAL4–ER71208–336 was unable to activate transcription, rather repressing it by more than half (Fig. 4B). Deletion of a great part or the whole of the ETS domain in GAL4–ER71293–336 or GAL4–ER71315–336 alleviated this repression. Again, these effects appear not to be due to different levels of protein expression (Fig. 4C). We conclude that amino acids 208–293 are capable of repressing transcription.
ER71 activates transcription from the E74 site
We next analyzed the impact of ER71 expression on an artificial promoter consisting of three copies of the E74 site fused to a basal promoter and the luciferase gene (E743–tk80–Luc reporter). Full-length ER71 was able to activate transcription ∼3-fold (Fig. 5A), whereas it failed to significantly activate the tk80–Luc reporter containing no E74 site (Fig. 5C). Consistent with the data obtained with the GAL4–ER71 chimeras, progressive truncation of amino acids from the N-terminus (84–336, 158–336 and 208–336) resulted in a concomitant loss of transactivation potential (Fig. 5A). ER711–206, which lacks a DNA-binding domain, was unable to activate transcription. Finally, the ability of ER711–315 to activate transcription was significantly impaired when compared with the wild-type, probably as a result of its decreased DNA binding ability. Since comparable levels of wild-type and truncated ER71 were expressed (Fig. 5B), the differences in transactivation potential are not due to different protein levels.
Figure 5.
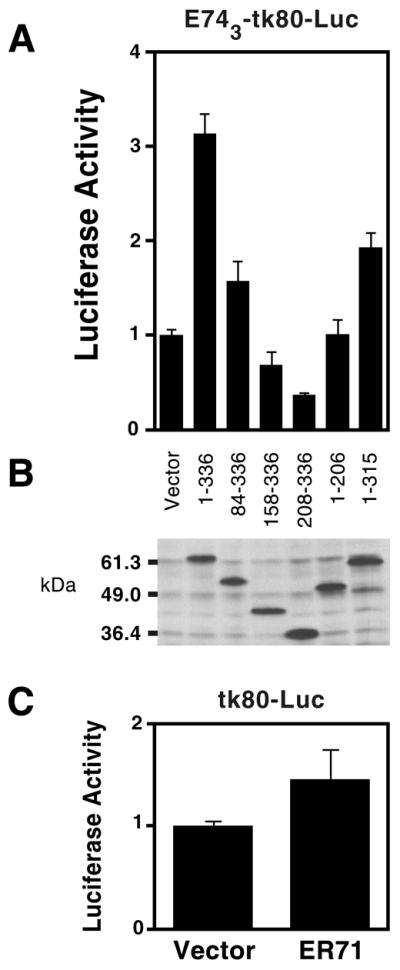
ER71 activates the E743–tk80–Luc reporter construct. (A) Mv1Lu cells were transfected with 150 ng Myc-tagged ER71 constructs (2 µg in the case of ER711–315) and total amount of DNA was adjusted to 2 µg with empty vector. Activation of the E74 site-driven reporter is depicted. (B) Corresponding anti-Myc western blot with cell extracts derived from the transfected Mv1Lu cells. (C) Regulation of the tk80–Luc reporter that is devoid of E74 sites by ER71.
Activation of the matrix metalloproteinase-1 (MMP-1) promoter by ER71
Next, we wanted to assess the ability of ER71 to activate transcription in a more natural context. To this end, we utilized the promoter of the MMP-1 gene, whose regulation by ETS factors has been previously shown to occur primarily through an ETS site centered around positions –88 to –85 (35,36). The respective sequence is 5′-AGAGGATGTT-3′ and matches relatively well the optimal DNA-binding site for ER71, 5′-(G/C)(G/C)(C/A)GGA(A/T)(G/A)(T/C)C-3′ (6).
Indeed, ER71 was able to bind a radioactively labeled oligonucleotide containing the –88 to –85 ETS core site of the MMP-1 promoter (Fig. 6). This interaction of ER71 with the wild-type MMP-1 oligonucleotide could be abolished by incubation with unlabeled E74 competitor, but not by the mutated E74 oligonucleotide, and addition of an anti-ER71 antibody supershifted the ER71–DNA complex. As expected, ER711–206, which lacks an ETS domain, failed to interact with the MMP-1 promoter. Two non-specific bands were also observed (Fig. 6, asterisks), due to complexes formed between the MMP-1 oligonucleotide and proteins found in the reticulocyte lysate used to produce ER71 in vitro. These results show that ER71 specifically binds to the –88 to –85 ETS site in the MMP-1 promoter.
Figure 6.
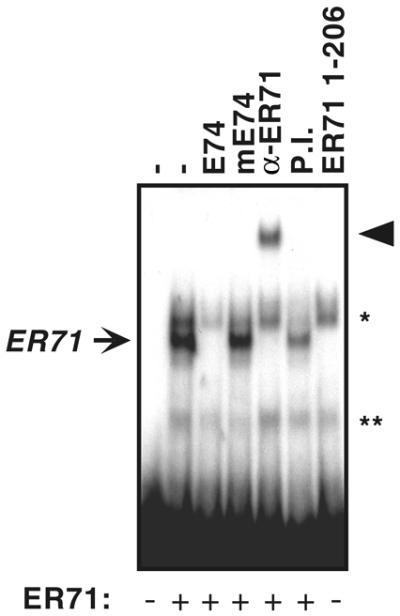
ER71 binds to the MMP-1 promoter. In vitro transcribed and translated ER71 was incubated with a 32P-labeled probe derived from the MMP-1 promoter. DNA–protein complexes were resolved by electrophoretic mobility shift assays. Where indicated, excess unlabeled E74, mutated E74, anti-ER71 antibodies or preimmune (P.I.) serum was included during the DNA binding reaction. Non-specific bands are marked by asterisks.
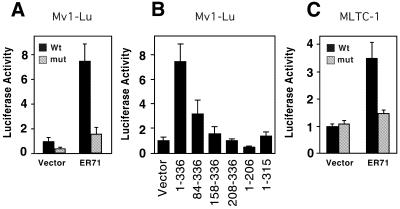
In order to investigate whether ER71 could activate the MMP-1 promoter, we co-transfected Mv1Lu cells with an ER71 expression vector and either a wild-type MMP-1 luciferase reporter construct or one containing a mutated ETS-binding site. ER71 was able to activate the wild-type MMP-1 promoter ∼7-fold (Fig. 7A, black bars). Mutation of the –88 to –85 ETS core in the MMP-1 promoter severely reduced this activation (Fig. 7A, grey bars), yet did not abolish it. This residual transcriptional activation mediated by ER71 is most likely due to additional sequences in the MMP-1 promoter that can act as secondary ETS-binding sites and accommodate (low affinity) ER71 binding (35). Altogether, these observations clearly indicate that ER71 activates the MMP-1 promoter predominantly through the –88 to –85 ETS-binding site.
Figure 7.
ER71 strongly activates the MMP-1 promoter. (A) Mv1Lu cells were co-transfected with a MMP-1 promoter-driven luciferase reporter construct and 1 µg ER71 expression vector or empty vector. Comparison of activation of wild-type MMP-1 promoter (black bars) or one whose ETS-binding site at positions –88 to –85 was mutated from GGAT to AAAT (grey bars). (B) Similarly, regulation of the wild-type MMP-1 promoter by the indicated ER71 truncations in Mv1Lu cells. Amounts of expression vectors used were identical to those in Figure 5A. (C) Activation of the MMP-1 promoter (2.5 µg) by ER71 in MLTC-1 cells. 1.5 µg expression vector for ER71 or empty vector were employed.
We next wished to correlate the ability of ER71 to stimulate the MMP-1 promoter with the presence of the N-terminal activation domain. Indeed, partial or complete deletion of the N-terminal activation domain reduced the ability of ER71 to activate transcription: the activity of ER7184–336 was less than half of full-length ER71, and ER71158–336 and ER71208–336 transcription levels were further lowered to those obtained with vector alone (Fig. 7B). Similarly, ER711–315 was unable to raise transcription levels, probably due to its inability to efficiently bind DNA. Furthermore, the ER711–206 protein slightly repressed transcription, which may be due to the sequestration of limiting basal transcription factors by the strong N-terminal activation domain of ER71.
Finally, we assessed the ability of ER71, which is testis specifically expressed (6), to activate the MMP-1 promoter in a testis cell line, MLTC-1, which is derived from murine Leydig cells. Similarly as for Mv1Lu cells, ER71 was able to activate the MMP-1 luciferase reporter in MLTC-1 cells, and mutation of the –88 to –85 ETS core site abrogated this effect (Fig. 7C). Altogether, our results demonstrate that ER71 is capable of binding to and activating the MMP-1 promoter and that this activation is dependent on the presence of its strong N-terminal activation domain.
DISCUSSION
In this report, we have functionally dissected the ETS protein ER71 and demonstrated that it is a constitutively nuclear protein, possesses a transactivation domain and is capable of binding to and activating transcription from both the E74 site and the MMP-1 promoter.
Nuclear targeting of ER71 is mediated by amino acids 276–315, which are part of the ETS domain. In particular, mutation of four consecutive basic amino acids in the sequence 276GERKRKPG283, which is similar to the paradigmatic SV40 large T antigen nuclear localization signal, PKKRKV (29,37,38), disrupts nuclear localization, suggesting that these eight amino acids constitute a nuclear localization signal. Consistently, fusing these amino acids to GFP results in nuclear accumulation of GFP. Interestingly, these eight amino acids are also homologous to motifs that have been implicated in nuclear targeting of Ets-1 (GKRKNKPK) and Elk-1 (GLRKNKTN) (30,39), suggesting that they may be a conserved feature of ETS proteins. However, nuclear localization is not only determined by amino acids 276–283, since mutation of basic residues within positions 284–315 also disrupts nuclear localization. Consistently, whereas GFP–ER71276–283 is not exclusively nuclear, GFP–ER71276–315 is completely localized to the nucleus, indicating that regions 276–283 and 284–315 collaborate in nuclear localization of ER71.
It is possible that amino acids 276–283 and 284–315 form a bipartite nuclear localization signal, which has been observed in nucleoplasmin, DNA helicase Q1, HIV-1 integrase and pRB (29,40). The existence of a bipartite nuclear localization signal would be supported by our analysis of the published crystal structure of Ets-1 with the aid of RasMol software, revealing that basic residues homologous to those found within ER71 amino acids 276–315 are roughly arranged into two clusters separated by the DNA-binding motif (not shown). Recently, it has been suggested that some nuclear localization signals may be tripartite in nature (41) or may be rather large, as is the case of the BIB domain responsible for nuclear import of ribosomal proteins (42). Whichever is the case for ER71, the relevant sequence and structural requirements for nuclear localization of ER71 are contained within amino acids 276–315.
Some ETS factors can autoregulate binding of their ETS domain to DNA (31,34). In line with this, truncation of ER71 amino acids 1–54 resulted in an ∼2-fold enhancement of DNA binding. More drastically, deleting the C-terminal amino acids 316–336 almost abolished the DNA binding activity of ER71, in stark contrast to observations made with the ETS factors Ets-1 and PEA3, where either truncation of amino acids C-terminal of the ETS domain or incubation with an antibody directed towards this region enhanced DNA binding (31,34,43,44).
Analysis of GAL4–ER71 fusions revealed the presence of a highly potent N-terminal activation domain spanning amino acids 1–157 capable of activating transcription 84-fold. The transactivation potency of this domain is comparable with that of GAL4–Herpes virus protein 16, the paradigm of potent transcriptional activators (45,46). The high effectiveness of the N-terminal ER71 activation domain is unusual, since ETS factors in general possess weak activation domains, although some are considerably activated by mitogenic or stress signals (1–3). Previous reports show that the DNA-binding domain of several ETS proteins (28,47–50) or regions outside their ETS domain (44,50,51) negatively impact on transactivation. Similarly, GAL4–ER711–293 is ∼10-times less active than GAL4–ER711–157. Consistently, our results indicate that amino acids 208–293, which encompass the majority of the ETS domain of ER71, constitute a repression domain. It is possible to envisage a situation where binding of ER71 to a target sequence as well as to interacting proteins derepresses the transactivating region, as has been suggested for the ETS proteins Elk-1, Sap-1a and Ets-1 (13,52).
ER71 was able to strongly activate the MMP-1 promoter as well as an artificial reporter driven by three E74 sites. Indeed, ER71 is capable of binding specifically to the E74 site as well as to an ETS core site at position –88 to –85 of the MMP-1 promoter, which has been previously shown to be crucial for MMP-1 activation by the ETS proteins Erg, Ets-1, Fli-1 and ER81 (35,36,53). The ability of ER71 to activate the MMP-1 promoter was dependent on its N-terminal activation domain. On the other hand, the MMP-1 promoter was slightly repressed by the ER711–206 truncation, which encompasses the strong N-terminal activation domain but lacks the ETS DNA-binding domain. It is highly possible that this repressive effect is due to the sequestration of limiting co-factors by the N-terminal activation domain, thus suppressing basal MMP-1 transcription. This mechanism of squelching, which was initially identified with the GAL4 protein (54), has also been observed with other transcription factors, including nuclear hormone receptors, p53, E2F and Ets-2 (55–58).
ER71 is a remarkably unique ETS factor; searches of current databases show that, aside from its ETS domain, ER71 has no homology to any other known protein. This observation, in conjunction with the fact that expression of ER71 appears to be restricted to testes in the adult mouse (6), raises the exciting possibility of an important, unique function for ER71. Indeed, our results showing the ability of ER71 to activate the MMP-1 promoter points to a possible role of ER71 during spermatogenesis, when extensive tissue remodeling, which is dependent on matrix metalloproteinase activity, occurs (59,60). In addition, ER71-mediated up-regulation of the MMP-1 gene may play a role in seminoma cells, where enhanced MMP-1 expression is correlated with their metastatic potential (61).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Thomas Brown for providing the ER71 cDNA. This work was supported by the Mayo Foundation, by a scholarship (to L.D.H.) from the Mayo Graduate School and by a scholarship (to R.J.) from the Sidney Kimmel Foundation for Cancer Research.
REFERENCES
- 1.Sharrocks A.D., Brown,A.L., Ling,Y. and Yates,P.R. (1997) The ETS-domain transcription factor family. Int. J. Biochem. Cell Biol., 29, 1371–1387. [DOI] [PubMed] [Google Scholar]
- 2.Graves B.J. and Petersen,J.M. (1998) Specificity within the ets family of transcription factors. Adv. Cancer Res., 75, 1–55. [DOI] [PubMed] [Google Scholar]
- 3.Sementchenko V.I. and Watson,D.K. (2000) Ets target genes: past, present and future. Oncogene, 19, 6533–6548. [DOI] [PubMed] [Google Scholar]
- 4.Crepieux P., Coll,J. and Stehelin,D. (1994) The Ets family of proteins: weak modulators of gene expression in quest for transcriptional partners. Crit. Rev. Oncogen., 5, 615–638. [PubMed]
- 5.Thompson C.C., Brown,T.A. and McKnight,S.L. (1991) Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science, 253, 762–768. [DOI] [PubMed] [Google Scholar]
- 6.Brown T.A. and McKnight,S.L. (1992) Specificities of protein-protein and protein-DNA interaction of GABP alpha and two newly defined ets-related proteins. Genes Dev., 6, 2502–2512. [DOI] [PubMed] [Google Scholar]
- 7.Dalton S. and Treisman,R. (1992) Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell, 68, 597–612. [DOI] [PubMed] [Google Scholar]
- 8.Hipskind R.A., Rao,V.N., Mueller,C.G., Reddy,E.S. and Nordheim,A. (1991) Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature, 354, 531–534. [DOI] [PubMed] [Google Scholar]
- 9.Brass A.L., Zhu,A.Q. and Singh,H. (1999) Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J., 18, 977–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Himmelmann A., Riva,A., Wilson,G.L., Lucas,B.P., Thevenin,C. and Kehrl,J.H. (1997) PU.1/Pip and basic helix loop helix zipper transcription factors interact with binding sites in the CD20 promoter to help confer lineage- and stage-specific expression of CD20 in B lymphocytes. Blood, 90, 3984–3995. [PubMed] [Google Scholar]
- 11.Eisenbeis C.F., Singh,H. and Storb,U. (1995) Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev., 9, 1377–1387. [DOI] [PubMed] [Google Scholar]
- 12.Garvie C.W., Hagman,J. and Wolberger,C. (2001) Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell, 8, 1267–1276. [DOI] [PubMed] [Google Scholar]
- 13.Pufall M.A. and Graves,B.J. (2002) Ets-1 flips for new partner pax-5. Structure, 10, 11–14. [DOI] [PubMed] [Google Scholar]
- 14.McKercher S.R., Torbett,B.E., Anderson,K.L., Henkel,G.W., Vestal,D.J., Baribault,H., Klemsz,M., Feeney,A.J., Wu,G.E., Paige,C.J. and Maki,R.A. (1996) Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J., 15, 5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 15.Scott E.W., Simon,M.C., Anastasi,J. and Singh,H. (1994) Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science, 265, 1573–1577. [DOI] [PubMed] [Google Scholar]
- 16.Lin J.H., Saito,T., Anderson,D.J., Lance-Jones,C., Jessell,T.M. and Arber,S. (1998) Functionally related motor neuron pool and muscle sensory afferent subtypes defined by coordinate ETS gene expression. Cell, 95, 393–407. [DOI] [PubMed] [Google Scholar]
- 17.Nye J.A., Petersen,J.M., Gunther,C.V., Jonsen,M.D. and Graves,B.J. (1992) Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev., 6, 975–990. [DOI] [PubMed] [Google Scholar]
- 18.Ray-Gallet D., Mao,C., Tavitian,A. and Moreau-Gachelin,F. (1995) DNA binding specificities of Spi-1/PU.1 and Spi-B transcription factors and identification of a Spi-1/Spi-B binding site in the c-fes/c-fps promoter. Oncogene, 11, 303–313. [PubMed] [Google Scholar]
- 19.Su G.H., Chen,H.M., Muthusamy,N., Garrett-Sinha,L.A., Baunoch,D., Tenen,D.G. and Simon,M.C. (1997) Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J., 16, 7118–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laing M.A., Coonrod,S., Hinton,B.T., Downie,J.W., Tozer,R., Rudnicki,M.A. and Hassell,J.A. (2000) Male sexual dysfunction in mice bearing targeted mutant alleles of the PEA3 ets gene. Mol. Cell. Biol., 20, 9337–9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arber S., Ladle,D.R., Lin,J.H., Frank,E. and Jessell,T.M. (2000) ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell, 101, 485–498. [DOI] [PubMed] [Google Scholar]
- 22.Sumarsono S.H., Wilson,T.J., Tymms,M.J., Venter,D.J., Corrick,C.M., Kola,R., Lahoud,M.H., Papas,T.S., Seth,A. and Kola,I. (1996) Down’s syndrome-like skeletal abnormalities in Ets2 transgenic mice. Nature, 379, 534–537. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L., Eddy,A., Teng,Y.T., Fritzler,M., Kluppel,M., Melet,F. and Bernstein,A. (1995) An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Mol. Cell. Biol., 15, 6961–6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthias P., Muller,M.M., Schreiber,E., Rusconi,S. and Schaffner,W. (1989) Eukaryotic expression vectors for the analysis of mutant proteins. Nucleic Acids Res., 17, 6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baniahmad A., Kohne,A.C. and Renkawitz,R. (1992) A transferable silencing domain is present in the thyroid hormone receptor, in the v-erbA oncogene product and in the retinoic acid receptor. EMBO J., 11, 1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janknecht R., Ernst,W.H., Pingoud,V. and Nordheim,A. (1993) Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J., 12, 5097–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korzus E., Nagase,H., Rydell,R. and Travis,J. (1997) The mitogen-activated protein kinase and JAK-STAT signaling pathways are required for an oncostatin M-responsive element-mediated activation of matrix metalloproteinase 1 gene expression. J. Biol. Chem., 272, 1188–1196. [DOI] [PubMed] [Google Scholar]
- 28.Bredemeier-Ernst I., Nordheim,A. and Janknecht,R. (1997) Transcriptional activity and constitutive nuclear localization of the ETS protein Elf-1. FEBS Lett., 408, 47–51. [DOI] [PubMed] [Google Scholar]
- 29.Jans D.A., Xiao,C.Y. and Lam,M.H. (2000) Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays, 22, 532–544. [DOI] [PubMed] [Google Scholar]
- 30.Janknecht R., Zinck,R., Ernst,W.H. and Nordheim,A. (1994) Functional dissection of the transcription factor Elk-1. Oncogene, 9, 1273–1278. [PubMed] [Google Scholar]
- 31.Greenall A., Willingham,N., Cheung,E., Boam,D.S. and Sharrocks,A.D. (2001) DNA binding by the ETS-domain transcription factor PEA3 is regulated by intramolecular and intermolecular protein.protein interactions. J. Biol. Chem., 276, 16207–16215. [DOI] [PubMed] [Google Scholar]
- 32.Hupp T.R., Meek,D.W., Midgley,C.A. and Lane,D.P. (1992) Regulation of the specific DNA binding function of p53. Cell, 71, 875–886. [DOI] [PubMed] [Google Scholar]
- 33.Grimm S. and Baeuerle,P.A. (1993) The inducible transcription factor NF-kappa B: structure-function relationship of its protein subunits. Biochem. J., 290, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jonsen M.D., Petersen,J.M., Xu,Q.P. and Graves,B.J. (1996) Characterization of the cooperative function of inhibitory sequences in Ets-1. Mol. Cell. Biol., 16, 2065–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buttice G., Duterque-Coquillaud,M., Basuyaux,J.P., Carrere,S., Kurkinen,M. and Stehelin,D. (1996) Erg, an Ets-family member, differentially regulates human collagenase1 (MMP1) and stromelysin1 (MMP3) gene expression by physically interacting with the Fos/Jun complex. Oncogene, 13, 2297–2306. [PubMed] [Google Scholar]
- 36.Westermarck J., Seth,A. and Kahari,V.M. (1997) Differential regulation of interstitial collagenase (MMP-1) gene expression by ETS transcription factors. Oncogene, 14, 2651–2660. [DOI] [PubMed] [Google Scholar]
- 37.Kalderon D., Roberts,B.L., Richardson,W.D. and Smith,A.E. (1984) A short amino acid sequence able to specify nuclear location. Cell, 39, 499–509. [DOI] [PubMed] [Google Scholar]
- 38.Kalderon D., Richardson,W.D., Markham,A.F. and Smith,A.E. (1984) Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature, 311, 33–38. [DOI] [PubMed] [Google Scholar]
- 39.Boulukos K.E., Pognonec,P., Rabault,B., Begue,A. and Ghysdael,J. (1989) Definition of an Ets1 protein domain required for nuclear localization in cells and DNA-binding activity in vitro. Mol. Cell. Biol., 9, 5718–5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robbins J., Dilworth,S.M., Laskey,R.A. and Dingwall,C. (1991) Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell, 64, 615–623. [DOI] [PubMed] [Google Scholar]
- 41.Pokorska A., Drevet,C. and Scazzocchio,C. (2000) The analysis of the transcriptional activator PrnA reveals a tripartite nuclear localisation sequence. J. Mol. Biol., 298, 585–596. [DOI] [PubMed] [Google Scholar]
- 42.Jakel S. and Gorlich,D. (1998) Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J., 17, 4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown L.A., Amores,A., Schilling,T.F., Jowett,T., Baert,J.L., de Launoit,Y. and Sharrocks,A.D. (1998) Molecular characterization of the zebrafish PEA3 ETS-domain transcription factor. Oncogene, 17, 93–104. [DOI] [PubMed] [Google Scholar]
- 44.Bojovic B.B. and Hassell,J.A. (2001) The PEA3 Ets transcription factor comprises multiple domains that regulate transactivation and DNA binding. J. Biol. Chem., 276, 4509–4521. [DOI] [PubMed] [Google Scholar]
- 45.Sadowski I., Ma,J., Triezenberg,S. and Ptashne,M. (1988) GAL4-VP16 is an unusually potent transcriptional activator. Nature, 335, 563–564. [DOI] [PubMed] [Google Scholar]
- 46.Rossow K.L. and Janknecht,R. (2001) The Ewing’s sarcoma gene product functions as a transcriptional activator. Cancer Res., 61, 2690–2695. [PubMed] [Google Scholar]
- 47.Schneikert J., Lutz,Y. and Wasylyk,B. (1992) Two independent activation domains in c-Ets-1 and c-Ets-2 located in non-conserved sequences of the ets gene family. Oncogene, 7, 249–256. [PubMed] [Google Scholar]
- 48.Rao V.N., Ohno,T., Prasad,D.D., Bhattacharya,G. and Reddy,E.S. (1993) Analysis of the DNA-binding and transcriptional activation functions of human Fli-1 protein. Oncogene, 8, 2167–2173. [PubMed] [Google Scholar]
- 49.Siddique H.R., Rao,V.N., Lee,L. and Reddy,E.S. (1993) Characterization of the DNA binding and transcriptional activation domains of the erg protein. Oncogene, 8, 1751–1755. [PubMed] [Google Scholar]
- 50.Janknecht R., Ernst,W.H. and Nordheim,A. (1995) SAP1a is a nuclear target of signaling cascades involving ERKs. Oncogene, 10, 1209–1216. [PubMed] [Google Scholar]
- 51.Janknecht R. (1996) Analysis of the ERK-stimulated ETS transcription factor ER81. Mol. Cell. Biol., 16, 1550–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Treisman R., Marais,R. and Wynne,J. (1992) Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J., 11, 4631–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bosc D.G., Goueli,B.S. and Janknecht,R. (2001) HER2/Neu-mediated activation of the ETS transcription factor ER81 and its target gene MMP-1. Oncogene, 20, 6215–6224. [DOI] [PubMed] [Google Scholar]
- 54.Gill G. and Ptashne,M. (1988) Negative effect of the transcriptional activator GAL4. Nature, 334, 721–724. [DOI] [PubMed] [Google Scholar]
- 55.Kamei Y., Xu,L., Heinzel,T., Torchia,J., Kurokawa,R., Gloss,B., Lin,S.C., Heyman,R.A., Rose,D.W., Glass,C.K. and Rosenfeld,M.G. (1996) A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell, 85, 403–414. [DOI] [PubMed] [Google Scholar]
- 56.Lopez G.N., Webb,P., Shinsako,J.H., Baxter,J.D., Greene,G.L. and Kushner,P.J. (1999) Titration by estrogen receptor activation function-2 of targets that are downstream from coactivators. Mol. Endocrinol., 13, 897–909. [DOI] [PubMed] [Google Scholar]
- 57.Magae J., Illenye,S., Tejima,T., Chang,Y.C., Mitsui,Y., Tanaka,K., Omura,S. and Heintz,N.H. (1997) Transcriptional squelching by ectopic expression of E2F-1 and p53 is alleviated by proteasome inhibitors MG-132 and lactacystin. Oncogene, 15, 759–769. [DOI] [PubMed] [Google Scholar]
- 58.Foos G. and Hauser,C.A. (2000) Altered Ets transcription factor activity in prostate tumor cells inhibits anchorage-independent growth, survival and invasiveness. Oncogene, 19, 5507–5516. [DOI] [PubMed] [Google Scholar]
- 59.Mruk D., Zhu,L.J., Silvestrini,B., Lee,W.M. and Cheng,C.Y. (1997) Interactions of proteases and protease inhibitors in Sertoli-germ cell cocultures preceding the formation of specialized Sertoli-germ cell junctions in vitro. J. Androl., 18, 612–622. [PubMed] [Google Scholar]
- 60.Gronning L.M., Wang,J.E., Ree,A.H., Haugen,T.B., Tasken,K. and Tasken,K.A. (2000) Regulation of tissue inhibitor of metalloproteinases-1 in rat Sertoli cells: induction by germ cell residual bodies, interleukin-1alpha and second messengers. Biol. Reprod., 62, 1040–1046. [DOI] [PubMed] [Google Scholar]
- 61.Koshida K., Konaka,H., Kato,H., Miyagi,T., Egawa,M., Uchibayashi,T. and Namiki,M. (2000) Correlation between expression of metastasis-related genes and lymph node metastasis in testicular cancer [in Japanese]. Hinyokika Kiyo, 46, 775–781. [PubMed] [Google Scholar]