Abstract
The mitochondrial respiratory chain is a source of reactive oxygen species (ROS) that are responsible for oxidative modification of biomolecules, including proteins. Due to its association with mitochondrial DNA, DNA polymerase γ (pol γ) is in an environment to be oxidized by hydrogen peroxide and hydroxyl radicals that may be generated in the presence of iron ions associated with DNA. We tested whether human pol γ was a possible target of ROS with H2O2 and investigated the effect on the polymerase activities and DNA binding efficiency. A 1 h treatment with 250 µM H2O2 significantly inhibited DNA polymerase activity of the p140 subunit and lowered its DNA binding efficiency. Addition of p55 to the p140 catalytic subunit prior to H2O2 treatment offered protection from oxidative inactivation. Oxidatively modified amino acid residues in pol γ resulting from H2O2 treatment were observed in vitro as well as in vivo, in SV40-transfected human fibroblasts. Pol γ was detected as one of the major oxidized mitochondrial matrix proteins, with a detectable decline in polymerase activity. These results suggest pol γ as a target of oxidative damage, which may result in a reduction in mitochondrial DNA replication and repair capacities.
INTRODUCTION
Oxidative damage to mitochondrial DNA (mtDNA) has been correlated with the aging process and can produce mutations that lead to degenerative diseases (1,2). Reactive oxygen species (ROS) and other free radicals are generated in the mitochondrial membranes by electron transport during oxidative phosphorylation. As much as 1–5% of the oxygen consumed in the mitochondria is released as superoxide or hydrogen peroxide (3). Hydrogen peroxide is the most stable oxygen species but the toxic effects are partially a result of its ability to form damaging radicals. Hydrogen peroxide can react with transition state metals to form hydroxide ion and hydroxyl radicals via the Fenton reaction. mtDNA is located near the site of free radical generation in the mitochondrial inner membrane and suffers more oxidative damage than nuclear DNA (4–7). Early reports comparing nucleotide substitutions in mtDNA from somatic tissues of different primates revealed a 10-fold higher rate of evolution for mtDNA relative to the nuclear genome (8,9), implying a relatively high mutation rate for mtDNA. Oxidative damage of mtDNA is more extensive and persists when cells are treated with prolonged exposure to hydrogen peroxide as compared to nuclear DNA damage (10).
Besides DNA, hydroxyl radicals also attack lipids and proteins. Protein oxidation results mainly in introduction of carbonyl groups, which can be easily detected (11). Proteins bearing carbonyl groups are generally dysfunctional since these moieties may alter both structure and function of the protein. Hydrogen peroxide, apart from indirectly causing oxidation of proteins by being a source of highly reactive free radicals, can also directly modify several amino acids, such as cysteine, methionine, tryptophan and tyrosine (12–15), leading to significant loss of protein function (16–20).
The DNA backbone is associated with iron ions which are thought to account for oxidation of the DNA via Fenton chemistry which generates hydroxyl radicals (21). Additionally, DNA-binding proteins, such as replication proteins, are also in this environment for oxidation via DNA-associated iron molecules. DNA polymerase γ (pol γ) is the sole DNA polymerase in the mitochondria and is responsible for replication and repair of the mtDNA. Due to its association with mtDNA and the location within the mitochondria, pol γ is exposed to various ROS, including hydrogen peroxide. Oxidation of pol γ may be in part responsible for slower DNA replication or repair. Additionally, oxidative inactivation of pol γ may, at least in part, set forth a cascade of further oxidative stress due to the loss of mtDNA replication and subsequent energy decline.
We and others have cloned and overexpressed the human gene encoding the catalytic subunit of pol γ (22–24). The full-length human cDNA for the 55 kDa accessory subunit has been isolated and overexpressed (25–27) and the three-dimensional crystal structure of this protein has been determined (28). When associated with the catalytic subunit, the accessory subunit confers high processivity to pol γ through enhanced DNA binding (25,26). In the course of purification we have noted that pol γ is sensitive to inactivation in the absence of reducing agents. In this paper we examined the effect of hydrogen peroxide on pol γ: its activity and DNA binding efficiency as well as the possible impact on the enzyme activity in living cells.
MATERIALS AND METHODS
Reagents
Poly(rA)·oligo(dT)12–18, poly(dA)·oligo(dT)12–18 and unlabelled dTTP (10 mM stock) were purchased from Pharmacia. The radioisotopes [α-32P]dTTP and [γ-32P]ATP were from Amersham. AG 501-X8(D) resin, 20–50 mesh, was from Bio-Rad. Oligonucleotides were purchased from Oligo Etc. Hydrogen peroxide (30%) and the protease inhibitor cocktail were from Sigma. The OxyBlot protein oxidation detection kit was purchased from Intergen. SuperSignal West Pico chemiluminescent substrate, an enhanced chemiluminescent substrate for detection of horseradish peroxidase, was from Pierce. Autoradiogram films were from Kodak. Minimum essential medium (MEM) with and without l-glutamine, MEM non-essential and essential amino acids solutions, MEM vitamin solution, l-glutamine and trypsin–EDTA were from Life Technologies. Fetal bovine serum was from Gemini Bio-Products.
Cell culture
The simian virus 40 (SV40)-transformed human fibroblast cell line GM00637E (NIGMS Human Genetic Mutant Cell repository, Coriell Institute, Camden, NJ) was cultured in Earle’s modified Eagle’s medium supplemented with 15% fetal bovine serum, MEM essential and non-essential amino acids as well as 2.0 mM l-glutamine at 37°C, under 95% air:5% CO2 in a humidified atmosphere.
Enzymes
The recombinant wild-type histidine-tagged human pol γ and the exonuclease-deficient form (Exo–p140) were purified to homogeneity from baculovirus-infected insect cells as described (23). The accessory subunit (p55) was purified to homogeneity from Escherichia coli and the heterodimeric forms of the polymerase were reconstituted as described (25). Human DNA polymerase α (pol α) was purified from baculovirus-infected insect cells as described (29,30). Human DNA polymerase β (pol β) was purchased from Chimerx. Catalase was obtained from Pharmacia. The DPg polyclonal antisera against the catalytic subunit of human pol γ has been described (23).
In vitro protein oxidation
The pol γ subunits p140 and p55, as well as pol α and β, were treated with various concentrations of hydrogen peroxide (0.05, 0.1, 0.25, 0.5, 1, 1.5, 2 and 2.5 mM) for 60 min at 37°C, in buffer containing 50 mM Tris–HCl, pH 7.5, 10% glycerol, 1 mM EDTA and 0.01% NP-40. After the treatment the excess H2O2 was removed by 5 min incubation at room temperature with 500 U catalase (1 U catalase decomposes 1 µM H2O2/min at 25°C, pH 7). Oxidized enzymes were then used for activity determination.
Polymerase assays
Polymerase activity on poly(dA)·oligo(dT) was measured in 50 µl reaction mixtures containing 25 mM HEPES–KOH, pH 8.0, 5 µg acetylated BSA, 2.5 mM 2-mercaptoethanol, 2 mM MgCl2, 10 µM [α-32P]dTTP (1000–3000 c.p.m./pmol), 2.5 µg poly(dA)·oligo(dT)12–18 and 75 mM NaCl. Reverse transcriptase activity in mitochondrial extracts was determined in 50 µl reaction mixtures containing 25 mM HEPES–KOH, pH 8.0, 2.5 mM 2-mercaptoethanol, 10 µg acetylated BSA, 0.5 mM MnCl2, 10 µM [α-32P]dTTP (1000–3000 c.p.m./pmol), 2.5 µg poly(rA)·oligo(dT)12–18 and 75 mM NaCl as previously described (23). In both cases, after 15 min incubation at 37°C, TCA-insoluble radioactivity was determined by liquid scintillation counting. One unit is the amount of enzyme required to catalyze the incorporation of 1 pmol dTMP into TCA-precipitable DNA in 1 h at 37°C.
The processivity of pol γ after treatment with H2O2 was determined as previously described (25). Exonuclease activity of H2O2-treated exonuclease-proficient p140 alone, as well as in the presence of the p55 accessory subunit, was determined as previously described (23).
Gel mobility shift assay
A 38mer oligonucleotide primer (5′-TTA TCG CAC CTA CGT TCA ATA TTA CAG GCG AAC ATA CT-3′) was 5′-end-labeled and hybridized to a 1.3 molar excess of a complementary 34mer (5′-GTA TGT TCG CCT GTA ATA TTG AAC GTA GGT GCG A-3′) to generate a primer–template substrate with a 3′-base recessed 3′-end. Prior to DNA binding, Exo–p140, p55 and Exo–p140/p55 complex (1:1.1 molar ratio) were incubated for 30 min at 37°C with H2O2 (0.05, 0.1, 0.15, 0.2, 0.25, 0.5, 1.0, 1.5, 2 and 2.5 mM) in buffer containing 10 mM HEPES–KOH, pH 8.0, 1 mM EDTA and 10% glycerol in a final volume of 10 µl. To remove the remaining H2O2, 1 µl of catalase (25 ng, 20 U/ng) was added to each sample and incubated for 5 min at room temperature. Binding mixtures (20 µl) contained 10 mM HEPES–KOH, pH 8.0, 0.5 mM DTT, 4 µg acetylated BSA (200 µg/ml), 250 fmol end-labeled 38mer–34mer primer–template and 250 fmol either H2O2-treated Exo–p140, p55 or Exo–p140/p55 complex. After 5 min preincubation at room temperature the mixtures were resolved by electrophoresis on 6% polyacrylamide gels in 20 mM HEPES–KOH, pH 8.0, and 0.1 mM EDTA, pH 8.0. After 3 h at 100 V, 4°C, the gels were dried, exposed to a PhosphorImager screen and analyzed on a Storm 860 PhosphorImager (Molecular Dynamics).
Detection of protein oxidation
As a result of protein oxidation, carbonyl groups are introduced into protein side chains by a site-specific mechanism. We used an OxyBlot kit (Intergen) to immunodetect these carbonyl groups in oxidatively modified p140, p55, pol α and pol β as described by the manufacturer. Briefly, after incubation in the presence of 0.1, 0.5, 1 or 2.5 mM H2O2 the proteins (in equal amounts of 6 µg/sample) were treated with 2,4-dinitrophenylhydrazine (DNP-hydrazine), in order to derivatize the carbonyl groups to 2,4-dinitrophenylhydrazone (DNP-hydrazone). They were then separated on 4–20% polyacrylamide gels and transferred to Immobilon-P membranes (Millipore). The membranes were probed with first antibody, specific to the DNP moiety of the proteins. The next step was incubation with horseradish peroxidase–antibody conjugate directed against the primary antibody. Membranes were then treated with SuperSignal West Pico chemiluminescent substrate and exposed to autoradiography film. The method applied for detection of the oxidized catalytic subunit of pol γ in mitochondrial lysates from GM00637E cells treated with H2O2 was similar, however, with few modifications enabling immunoprecipitation (see below).
Hydrogen peroxide treatments
Hydrogen peroxide stock (30%) was diluted with phosphate-buffered saline (PBS) and the concentration determined by absorbance at 240 nm as described (31). Cells, ∼80% confluent, were washed twice with 1× PBS and exposed to 100, 250 or 400 µM H2O2 in serum-free MEM for 2 h. A control cell culture was mock-treated with serum-free medium alone. After treatment, the medium was discarded and cells were immediately harvested by brief treatment with trypsin–EDTA (0.25%) for mitochondria preparation.
Mitochondrial preparation
Harvested SV40-transformed human fibroblasts were washed twice with PBS, centrifuged (5000 r.p.m., 5 min) and supernatants were discarded. The pellets were weighed and gently resuspended in HDB buffer (5 mM KPO4, pH 7.5, 2 mM MgCl2, 1 mM 2-mercaptoethanol), at 9× volume of the wet weight of the cells. Protease inhibitor cocktail (1:100 v/v) was added and the cells were left on ice for 1 h. Once the cells were swollen (as judged with 5 µl on a slide with a 20/40 objective lens) they were broken by Dounce homogenization for 20–30 strokes until only 0–2 intact cells/microscope field remained. Mannitol–sucrose buffer (2.5×; 0.525 M mannitol, 0.175 M sucrose, 5 mM Tris–HCl, 5 mM EDTA, 5 mM MgCl2, pH 7.5) was added to a 1× concentration. To remove nuclei the cells were centrifuged for 5 min at 2000 r.p.m. and 4°C and the supernatants with remaining mitochondria were collected. They were then centrifuged for 30 min at 15 000 r.p.m. The pellets containing mitochondria were washed three times with 1× mannitol–sucrose buffer, in order to remove remaining nuclear DNA and proteins. Mitochondria pellets were resuspended in mitochondrial lysis buffer containing 1% NP-40, 0.3 M NaCl, 10% glycerol, 20 mM Tris–HCl, pH 8.0, 14 mM 2-mercaptoethanol and proteinase inhibitors and left on ice for 10 min. The lysates were then centrifuged for 2 h at 14 000 r.p.m. Supernatants containing pol γ were frozen in 25 µl aliquots in liquid nitrogen and stored at –80°C. These mitochondrial lysates were used for determination of either reverse transcriptase activity or level of pol γ catalytic subunit oxidation.
Immunoprecipitation of DNA polymerase γ from crude mitochondrial lysates
Two milliliters of DPg polyclonal antiserum (∼10 mg/ml) were mixed with 1 ml of protein A–Sepharose for 1 h in 100 mM Tris–HCl, pH 8.0, at 4°C. The Sepharose beads were then extensively washed with 100 mM Tris–HCl, pH 8.0, followed by 10 mM Tris–HCl, pH 8.0. To detect oxidatively modified proteins in mitochondrial lysates we used an OxyBlot kit (Intergen) as described above. Briefly, protein extracts (∼950 µg/sample) were treated with DNP-hydrazine, in order to derivatize the carbonyl groups to DNP-hydrazone. Then the buffer was exchanged for 10 mM Tris–HCl, pH 8.0, (40–50 min centrifugation, 4°C, Microcon 30) to enable immunoprecipitation; final sample volumes were 50 µl. Fifty microliters of antibody–protein A–Sepharose resin were mixed for 1 h at 4°C, end over end, with DNP-hydrazine-modified proteins in 10 mM Tris–HCl, pH 8.0, buffer. The Sepharose beads were then washed extensively with the same buffer and 25 µl samples were resolved on 4–20% polyacrylamide gels and proteins electrotransferred to an Immobilon-P membrane (Millipore). Anti-DNP antibodies from the OxyBlot kit were used to detect DNP moieties of the oxidatively modified pol γ catalytic subunit as described above.
Other methods
Deionized water used for all buffer preparations was purified on AG 501-X8(D) resin in order to remove any remaining metal ions. Protein concentration was determined according to Bradford (32), with BSA as the standard. For immunoblots, mitochondrial lysates were resolved by SDS–PAGE, electrotransferred to Immobilon-P membranes (Millipore) in 0.025 M Tris, 0.192 M glycine, probed with anti-DPg polyclonal antisera (23) and visualized with alkaline phosphatase-conjugated second antibody and Western Blue reagent (Promega).
RESULTS
In vitro oxidation of pol γ and effect on activities
To address whether pol γ can be modified by ROS and what effect oxidative modification has on the activity of the polymerase we measured the modification and inactivation of the polymerase by treatment with hydrogen peroxide. Reducing agents such as 2-mercaptoethanol or dithiothreitol and BSA were omitted from the H2O2 reaction, thereby enhancing the accessibility of enzyme treatment. After incubation with H2O2 the remaining hydrogen peroxide was removed from reactions by incubation with catalase for 5 min. Oxidatively modified enzymes were then characterized for activity and oxidation.
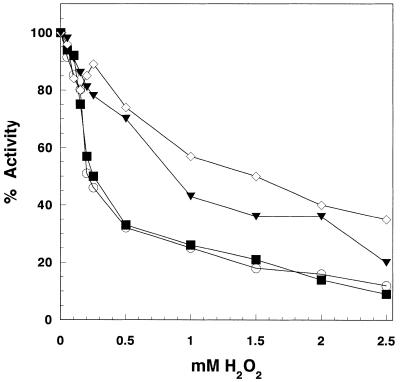
To examine in vitro hydrogen peroxide influence on human mtDNA pol γ activity we treated both the catalytic (p140) and accessory (p55) subunits separately, as well as in complex, with increasing H2O2 concentrations. Reduction in polymerase activity by this treatment was measured on poly(dA)·oligo(dT)12–18 as substrate. Polymerase activity of the pol γ p140 catalytic subunit (Exo–p140) proved to be moderately sensitive to hydrogen peroxide (Fig. 1). We observed 50% activity loss following its preincubation with 250 µM H2O2 for 60 min. When in complex with the p55 accessory subunit the polymerase activity was preserved and the complex remained 50% active even when the H2O2 concentration exceeded 1.5 mM. However, when p140 was oxidized prior to complex formation, p55 addition did not prevent significant activity loss. When p55 alone was subjected to hydrogen peroxide prior to association with p140, the polymerase activity in the complex was affected to a much lesser extent than treatment of p140 alone and this activity was only slightly lower than the p140/p55 complex subjected to oxidation after complex formation. This result indicates that the oxidized p55 did not significantly lower polymerase activity. These results also suggest that the p55 accessory subunit, when in complex with p140, may protect the catalytic subunit from oxidative modification by H2O2.
Figure 1.
Inhibition of pol γ DNA polymerase activity by hydrogen peroxide. Exo–p140 (0.2 pmol), either alone or with p55 (1:1 molar ratio), was preincubated for 60 min at 37°C with increasing concentrations of H2O2. Subsequently DNA polymerase activity on poly(dA)·oligo(dT) was measured as described in Materials and Methods. Open circles, Exo–p140; filled squares, Exo–p140 preincubated with H2O2 + p55; open diamonds, Exo–p140/p55 complex; filled triangles, Exo–p140 + p55 preincubated with H2O2 prior to adding to Exo–p140.
We also tested the effect of oxidizing the accessory subunit on the processivity of p140/p55 complex. Treatment of p55 with up to 1 mM H2O2 prior to association with p140 had no effect on the processivity of the complex when compared to the untreated p140/p55 complex and demonstrated DNA synthesis in excess of several thousand nucleotides in length (data not shown) and was similar to what was previously observed for the complex (25).
Hydrogen peroxide treatment reduced DNA binding activity in pol γ
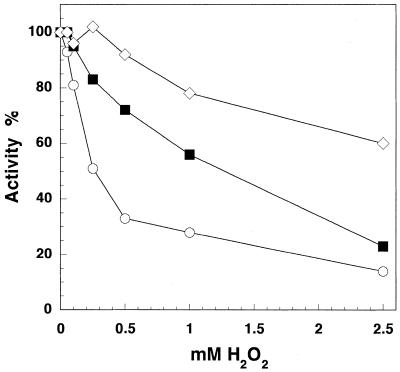
In order to compare the sensitivity to hydrogen peroxide among DNA polymerases from different polymerase classes, we treated two nuclear polymerases, human pol α (B class) and human pol β (X class), with H2O2, in exactly the same manner as we did with both subunits of pol γ, and compared their activities using the same substrate, poly(dA)·oligo(dT) (Fig. 2). The catalytic subunit of pol γ was more susceptible to hydrogen peroxide than the two nuclear DNA polymerases. Pol γ activity was reduced by 50% in 250 µM H2O2, whereas at the same H2O2 concentration pol α and pol β retained ∼80% and almost 100% of their polymerase activity, respectively. Pol α showed significant (>50%) activity loss at H2O2 concentrations >1 mM. To observe similar activity loss for pol β we had to preincubate the polymerase with 5 mM H2O2.
Figure 2.
Effect of hydrogen peroxide on DNA polymerase activities of Exo–p140, pol β and pol α. All three enzymes were treated with H2O2 for 60 min and their DNA polymerase activities on poly(dA)·oligo(dT) were measured as described in Materials and Methods. Reaction mixtures contained ∼48 ng (0.4 pmol) Exo–p140 (open circles), 17 ng (0.4 pmol) pol β (open diamonds), ∼66 ng (0.4 pmol) pol α (filled squares), respectively.
The effect of H2O2 on the intrinsic exonucleolytic activity of pol γ was also examined. Preincubation with hydrogen peroxide at concentrations up to 1 mM had little effect on exonuclease activity of the wild-type p140 and almost no effect on exonucleolytic activity of the wild-type p140/p55 complex (data not shown).
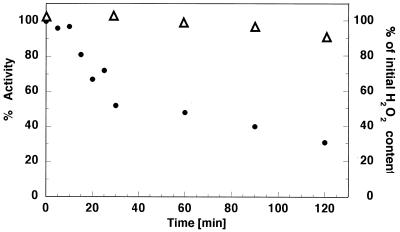
We also examined the influence of H2O2 on p140 activity over time (Fig. 3). For that purpose the enzyme was preincubated at 37°C in the presence of 250 µM H2O2. At selected time intervals (0–120 min) portions of p140 (H2O2-treated and an untreated control) were taken and incubated for 5 min with catalase to remove remaining H2O2. Polymerase activity in all collected samples was then determined. Samples of p140 that were not treated with H2O2 were used as relative controls for corresponding time point samples of H2O2-treated enzyme. When exposed to 250 µM H2O2 over time p140 lost 50% of its activity during the first 30–40 min; after that time activity loss was slower and 30% activity remained after 2 h of treatment. After the 2 h incubation the hydrogen peroxide concentration decreased by only 12% as compared to 0 min (Fig. 3).
Figure 3.
Time course of pol γ catalytic subunit polymerase activity inhibition by 200 µM hydrogen peroxide. Exo–p140 was incubated either alone or with 200 µM H2O2 at 37°C. At the time intervals indicated samples were taken (0.2 pmol) and DNA polymerase activity (closed circles, left axis) on poly(dA)·oligo(dT) was measured as described in Materials and Methods. Percent activity at each time point was calculated relative to the control Exo–p140 samples collected at corresponding time points and used as the 100% reference. The rRight axis and open triangles depict the amount of H2O2 remaining after the incubation time relative to the zero time point.
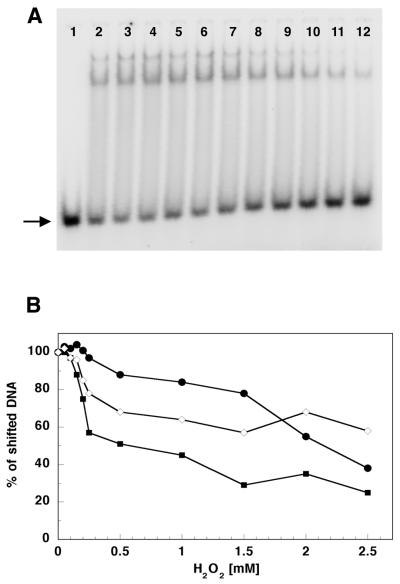
In order to examine H2O2 influence on p55, p140 and p140/p55 DNA binding efficiency we performed gel mobility shift assays with a single-stranded, 32P-labeled oligonucleotide annealed to its complementary sequence (Fig. 4A and B). Hydrogen peroxide affected the ability of p140, p55 and p140/p55 complex to bind to double-stranded (ds)DNA template, however to a lesser extent than it affected the activity of these proteins; again p140 proved to be most sensitive, losing ∼50% of its dsDNA ‘binding capacity’ when preincubated for 30 min with >0.5 mM H2O2. Also, the ability of p140/p55 complex to bind dsDNA was affected by preincubation with H2O2, but to a lesser extent than that of p140; even when preincubated with 2.5 mM H2O2 the complex retained ∼60% ‘binding capacity’ (Fig. 4B).
Figure 4.
Hydrogen peroxide affects the ability of p140, p55 and p140/p55 complex to bind to dsDNA. (A) An example of an autoradiogram of a gel mobility shift experiment showing p140 with no treatment (lane 2) or treatment with 0.05, 0.1, 0.15, 0.2, 0.25, 0.50, 1.0, 1.5, 2.0 and 2.5 mM H2O2 (lanes 3–12, respectively). Lane 1 contains the oligonucleotide substrate without addition of protein. Oligonucleotide substrate preparation and reaction conditions are described in Materials and Methods. Products were separated on a 15% polyacrylamide non-denaturating gel and quantitated on a Molecular Dynamics PhosphorImager as described in Materials and Methods. Two shifted products were observed corresponding to either one or two p140 molecules bound per oligonulceotide substrate. (B) DNA binding efficiency of p140 (filled squares), p55 (closed circles) and p140/p55 complex (open diamonds) after preincubation at increasing H2O2 concentrations. The arrow indicates the unshifted dsDNA substrate.
Detection of oxidized amino acids in pol γ
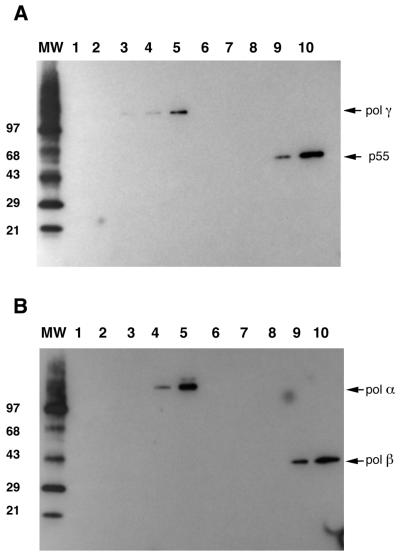
The next step was to examine whether the inhibitory effect of hydrogen peroxide on activity and DNA binding efficiency of the proteins analyzed is a result of oxidative modifications introduced into the protein molecule itself. We used an OxyBlot kit (Intergen), specially designed for the detection of oxidatively modified proteins. Protein oxidation mainly results in introduction of carbonyl groups into protein side chains by a site-specific mechanism. After treatment with DNP-hydrazine these carbonyl groups are derivatized to DNP-hydrazone, which can be detected with specific antibodies against the DNP moiety of the protein. We treated both the pol γ catalytic and accessory subunits as well as pol α and pol β with increasing concentrations of hydrogen peroxide (0.1, 0.5, 1 and 2.5 mM) and developed DNP-hydrazone groups as described in Materials and Methods. Oxidation of all four proteins was detected with the anti-DNP moiety antibodies (Fig. 5A and B), but only in samples preincubated with the higher concentrations of H2O2 (1 and 2.5 mM). These results demonstrate oxidative modification of the proteins by treatment with hydrogen peroxide.
Figure 5.
In vitro oxidation of Exo–p140, p55, pol β and pol α by hydrogen peroxide. Equal amounts of protein (6 µg) were incubated in the presence of increasing H2O2 concentrations as indicated. After 60 min incubation the excess H2O2 was removed by a 5 min incubation with catalase. Proteins were then separated by SDS–PAGE on 4–20% polyacrylamide gels, electrotransferred to an Imobilon P membrane (Millipore) and probed with anti-DNP antisera (Intergen) against derivatized carbonyl groups in oxidized proteins as described in Materials and Methods. (A) Oxidation of Exp–p140 and p55. Lanes 1–5 depict the oxidation of Exo–p140 with no treatment (lane 1) or treatment with 0.1, 0.5, 1 and 2.5 mM H2O2, respectively. Lanes 6–10 depict the oxidation of Hp55 with no treatment (lane 6) or treatment with 0.1, 0.5, 1 and 2.5 mM H2O2, respectively. (B) Oxidation of pol α and pol β. Lanes 1–5 depict the oxidation of pol α with no treatment (lane 1) or treatment with 0.1, 0.25, 0.5 and 1 mM H2O2, respectively. Lanes 6–10 depict the oxidation of pol β with no treatment (lane 6) or treatment with 0.1, 0.25, 0.5 and 1 mM H2O2, respectively. MW represents the oxidized molecular weight markers (Intergen) in kDa in both panels.
Pol γ is oxidized in vivo
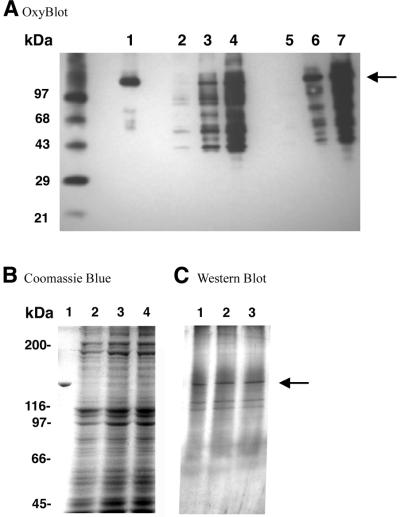
In order to examine the in vivo effects of H2O2 on pol γ we treated SV40-transformed human fibroblasts with hydrogen peroxide and assayed mitochondrial lysates for reverse transcriptase activity and protein oxidation assessment. The reverse transcriptase assay allowed us to distinguish between pol γ and other polymerase activities that may be contaminating the mitochondrial lysates. Mitochondria were isolated and mitochondrial lysates made after 2 h treatment of SV40-transformed human fibroblasts with 100 and 250 µM H2O2. Oxidized proteins in these lysates and in pol γ immunoprecipitates were detected by western blot using the anti-DNP antibodies (Fig. 6). We were able to detect oxidized proteins in the soluble mitochondrial lysates (Fig. 6, lanes 2–4) and in p140 immunoprecipitates from mitochondrial lysates (Fig. 6, lanes 5–7). One of the more prominent oxidized polypeptides observed in the mitochondrial lysate co-migrated with the p140 catalytic subunit of pol γ and represented ∼11% of all the oxidized polypeptides above 30 kDa (Fig. 6, lanes 2–4). This polypeptide was confirmed to be the pol γ catalytic subunit by enrichment of the DNP-positive polypeptide in the immunoprecipitate with polyclonal antibodies against the catalytic subunit (Fig. 6, lanes 5–7). This enriched 140 kDa polypeptide was also positive in a western blot using polyclonal antibodies against the catalytic subunit of human pol γ (data not shown). Lane 2 displays the oxidized proteins from untreated mitochondria and represents the endogenous level of oxidized mitochondrial proteins. The 140 kDa polypeptide is barely visible as part of the five DNP-positive polypeptides in the untreated control.
Figure 6.
In vivo oxidation of pol γ catalytic subunit. (A) Crude mitochondrial lysates from SVNF cells treated with H2O2 were preincubated with DNP-hydrazine to derivatize carbonyl residues occurring in proteins as a result of their oxidation. After derivatization, polyclonal DPg antisera against the catalytic subunit of pol γ were used to immunoprecipitate p140. Both crude mitochondrial lysates and immunoprecipitates were resolved on 4–20% polyacrylamide gels and electrotransferred to an Imobilon P membrane. Derivatized moieties were then recognized by anti-DNP antibodies as described in Materials and Methods. MW, oxidized molecular weight markers; lane 1, 0.5 µg oxidized Exo–p140 as control; lanes 2–4, 150 µg protein from crude mitochondrial lysate from non-treated cells (lane 2) and from cells treated with 100 (lane 3) and 250 µM (lane 4) H2O2; lanes 5–7, immunoprecipitate from mitochondrial lysates of non-treated cells (lane 5) and from cells treated with 100 (lane 6) and 250 µM (lane 7) H2O2. (B) Coomassie Blue staining of the crude mitochondrial lysates represented in lanes 2–4 of (A). Lane 1, 0.5 µg Exo–p140 control; lanes 2–4, crude mitochondrial lysates (∼150 µg) from non-treated cells (lane 2) and from cells incubated with 100 (lane 3) and 250 µM (lane 4) H2O2. Protein molecular weight markers are indicated on the left. (C) Immunoblot utilizing polyclonal DPg antisera against the catalytic subunit of pol γ. Lanes 1–3, immunoprecipitates from mitochondrial lysates from non-treated cells (lane 1) and from cells treated with 100 (lane 2) and 250 µM (lane 3) H2O2. The arrow indicates the position of the pol γ catalytic subunit.
Although we readily detected oxidation of the pol γ catalytic subunit in H2O2-treated cells, pol γ activity in these mitochondrial lysates was reduced by only 10 and 15% on treatment with 250 and 400 µM H2O2, respectively. This is most likely due to the fact that in mitochondria in vivo the majority of the pol γ catalytic subunit may be present in complex with the accessory subunit, and is at least partially protected from oxidative modification and activity loss. The protection by p55 probably only blocks the oxidation of a minority of amino acids, some of which are involved in polymerase function or substrate binding, while leaving the rest of the solvent-accessible amino acids in p140 available for oxidative modification. These latter amino acids, although readily oxidized, do not result in inactivation of polymerase activity. This would explain that while we readily detected p140 oxidation in vivo, most of the pol γ was still active. The observed slight activity loss may correspond to inactivation of this p140 fraction that remains unbound to p55.
DISCUSSION
Electron leakage from the electron transport chain during oxidative phosphorylation in the mitochondria is known to generate ROS and is believed to be the largest source of endogenous oxidative damage (3). Because of the proximity of the intra-mitochondrial matrix to the electron transport complexes, the possibility arises that pol γ is a target for oxidative modification. We tested this hypothesis by studying the ability of pol γ to be oxidized in vitro and assessed the effect on DNA polymerase activity and DNA binding. We also observed in mitochondrial lysates from cells treated with H2O2 that pol γ was one of the more abundant proteins to be oxidized.
In vitro treatment of the single catalytic subunit of pol γ readily inactivated polymerase activity without a significant effect on exonuclease activity. The same treatment of the nuclear polymerases, human pol α and β, had little effect on activity. Western blot analysis of these three polymerases with antibodies specific for DNP-hydrazine-derivatized carbonyl groups verified that all three polymerases were modified at similar levels. Thus, the sensitivity of the pol γ catalytic subunit to H2O2 relative to that of pol α and β indicates that a solvent-accessible amino acid(s) in pol γ becomes modified and that this amino acid plays a key role in the polymerase activity, either directly in catalysis or in binding of the substrates. Due to the strong conservation of amino acids in the active site between polymerase families A (pol γ) and B (pol α), direct oxidation of a catalytic residue seems less likely in pol γ. Thus, human pol γ may contain sensitive sites that participate in substrate binding directly or indirectly which when oxidized inhibit normal substrate binding. The decrease in DNA binding activity upon oxidation further suggests a role of these oxidized amino acids in substrate binding.
Interestingly, addition of p55 prior to H2O2 treatment protected the p140 catalytic subunit from this oxidation. Treatment of the accessory subunit alone had little effect on the polymerase activity or processivity of the complex, suggesting that the function of the accessory subunit is not compromised when oxidized. We have observed a similar scenario for N-ethylmaleimide (NEM) inhibition of pol γ where the Hp55 accessory subunit protected the catalytic subunit from inhibition (25). NEM is known to modify solvent-accessible sulfhydryl groups such as cysteine residues. Since no cysteine residue is known to participate in DNA polymerase catalysis among family A polymerases, the modification of a cysteine by NEM may reflect the function of this residue in substrate binding. It is interesting that a similar protection by Hp55 from H2O2 oxidation was observed in this study and raises the possibility that the same cysteine residue protected by Hp55 from NEM treatment is oxidized by H2O2 but also protected by Hp55. Further experiments need to be carried out to address this possibility.
The modification of pol γ by H2O2 was also observed in vivo in cells treated with H2O2. Oxidized protein residues became detectable at much lower hydrogen peroxide concentrations than were required to detect in vitro oxidation of purified proteins (see Fig. 5A and B). This can be explained by the fact that all possible factors (e.g. metal ions) that would participate in hydrogen peroxide conversion to other, potentially more reactive, ROS (OH·) were mostly eliminated in the in vitro assay. When in living cells, hydrogen peroxide interacts with cellular biomolecules contributing to an increase in the steady-state concentrations of various reactive species within the cell. The final observed oxidation effect is most probably due to those secondary events. Mitochondrial lysates showed several proteins modified by H2O2 treatment and specifically a polypeptide that migrates with the p140 catalytic subunit of pol γ. This polypeptide appeared to be one of the more abundant proteins modified. Immunoprecipitation with polyclonal antibodies against pol γ confirmed that this band was indeed the pol γ catalytic subunit. Thus, the pol γ catalytic subunit is oxidized in vivo and may represent one of the major targets in the mitochondrial matrix. This oxidation was accompanied by a small but significant decline in activity, suggesting that the accessory subunit was protecting the polymerase from most of the deleterious effects on DNA replication activity.
Although the p55 subunit offered protection from H2O2 during DNA synthesis and its function in DNA replication necessitates the high processivity that p55 offers, the role of p55 in DNA repair is unknown. We previously observed two forms of pol γ isolated from HeLa cells, the single catalytic subunit alone and in complex with the accessory subunit (23,25). We previously proposed that although DNA replication would require high processive synthesis characterized by the two subunit complex, the role of pol γ in base excision repair would only require the single catalytic subunit. The pol γ catalytic subunit has been observed throughout mitochondria (non-replicating and replicating) of the cell whereas active replication of mtDNA has only been observed in the perinuclear mitochondria (33,34). The non-replicating form of pol γ presumably functions in base excision repair, which does not require the accessory subunit in vitro (35). We observed that although the accessory subunit appears to protect much of the pol γ activity from oxidative damage, the activity of the single catalytic subunit of pol γ, possibly functioning alone during mtDNA repair, may be compromised by oxidative damage.
Although many studies have focused on the oxidation of DNA, oxidation of proteins is well documented (12,36–38). Oxidative stress in yeast has been shown to modify mitochondrial proteins, including Hsp60, pyruvate dehydrogenase, α-keto-glutarate dehydrogenase, aconitase, cytosolic fatty acid synthase and glyceraldehyde 3-phosphate dehydrogenase (20). Oxidative stress in yeast appears to arrest many of the mitochondrial functions, such as the tricarboxylic acid cycle, thereby shifting glucose metabolism to the pentose phosphate pathway, which generates the NADPH needed by antioxidant enzymes (20). Protein oxidation in mitochondria from Drosophila appears specific to a few proteins. Investigation of oxidized carbonyl groups from Drosophila mitochondrial proteins appears to be limited to the adenine translocator (ANT) in the membrane (39) and to aconitase in the mitochondrial matrix (40). The oxidation of ANT and aconitase in Drosophila appeared to occur as an age-related process with a decrease in the activity of these proteins. The age-specific oxidation of aconitase may be part of a larger biological mechanism by which oxidative stress causes an age-specific decline in mitochondrial function (41). In addition to protein oxidation, the age-associated accumulation of point mutations in mtDNA cannot be ignored as a parallel process (2). In human cells, the detection of carbonyls from the mitochondrial matrix, indicative of oxidized proteins, did not limit the oxidation to a single protein as observed in Drosophila. We observed at least 10 different polypeptides oxidized to varying degrees in the mitochondrial lysate. The pol γ catalytic subunit along with two other proteins represented one of the more abundant proteins oxidized. Given the fact that pol γ is a rare protein, representing ∼0.008% of mitochondrial proteins (23), the oxidation we observed may be specific for a subset of mitochondrial proteins including pol γ. Oxidative inactivation of pol γ could slow mtDNA replication and eventually lead to inhibition of oxidative phosphorylation. The specific inactivation of pol γ may be part of a more general mechanism in response to oxidative stress. This could also explain why a 60 min treatment of cells with hydrogen peroxide leads to less efficient repair of mtDNA (10).
In summary, we have determined that pol γ is a target for oxidative stress and that this oxidation can inactivate the catalytic subunit. The accessory subunit, however, offered significant protection from this oxidation, suggesting that a key oxidizable residue is masked by the binding of p55 to the catalytic subunit. Although much research has focused on the oxidation of mtDNA, the oxidation of mitochondrial proteins and, specifically, pol γ should now be considered as targets of ROS. Damage to pol γ and other mtDNA replication and repair proteins can have a consequence on the overall function of the mitochondria as well as the cell.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Ben Van Houten and Janine Santos for critical evaluation of the manuscript. We thank Drs Matthew Longley and Mikhail Ponamarev for providing purified pol γ enzyme.
REFERENCES
- 1.Cortopassi G.A., Shibata,D., Soong,N.W. and Arnheim,N. (1992) A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc. Natl Acad. Sci. USA, 89, 7370–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michikawa Y., Mazzucchelli,F., Bresolin,N., Scarlato,G. and Attardi,G. (1999) Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science, 286, 774–779. [DOI] [PubMed] [Google Scholar]
- 3.Chance B., Sies,H. and Boveris,A. (1979) Hydroperoxide metabolism in mammalian organs. Physiol. Rev., 59, 527–605. [DOI] [PubMed] [Google Scholar]
- 4.Ballinger S.W., Van Houten,B., Jin,G.F., Conklin,C.A. and Godley,B.F. (1999) Hydrogen peroxide causes significant mitochondrial DNA damage in human RPE cells. Exp. Eye Res., 68, 765–772. [DOI] [PubMed] [Google Scholar]
- 5.Salazar J.J. and Van Houten,B. (1997) Preferential mitochondrial DNA injury caused by glucose oxidase as a steady generator of hydrogen peroxide in human fibroblasts. Mutat. Res., 385, 139–149. [DOI] [PubMed] [Google Scholar]
- 6.Richter C., Park,J.W. and Ames,B.N. (1988) Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl Acad. Sci. USA, 85, 6465–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sohal R.S., Agarwal,S., Candas,M., Forster,M.J. and Lal,H. (1994) Effect of age and caloric restriction on DNA oxidative damage in different tissues of C57BL/6 mice. Mech. Ageing Dev., 76, 215–224. [DOI] [PubMed] [Google Scholar]
- 8.Brown W.M., Prager,E.M., Wang,A. and Wilson,A.C. (1982) Mitochondrial DNA sequences of primates: tempo and mode of evolution. J. Mol. Evol., 18, 225–239. [DOI] [PubMed] [Google Scholar]
- 9.Brown W.M., George,M.J. and Wilson,A.C. (1979) Rapid evolution of animal mitochondrial DNA. Proc. Natl Acad. Sci. USA, 76, 1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yakes F.M. and Van Houten,B. (1997) Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl Acad. Sci. USA, 94, 514–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reznick A.Z. and Packer,L. (1994) Oxidative damage to proteins: spectrophotometric method for carbonyl assay. Methods Enzymol., 233, 357–363. [DOI] [PubMed] [Google Scholar]
- 12.Stadtman E.R. (1993) Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal catalysed reactions. Annu. Rev. Biochem., 62, 797–821. [DOI] [PubMed] [Google Scholar]
- 13.Dean R.T., Fu,S., Stocker,R. and Davies,M.J. (1997) Biochemistry and pathology of radical-mediated protein oxidation. Biochem. J., 324, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Means G.E. and Feeney,R.E. (1971) Reducing and oxidizing agents. In Chemical Modifications of Proteins. Holden-Day, San Francisco, CA.
- 15.Simat T.J. and Steinhart,H. (1998) Oxidation of free tryptophan and tryptophan residues in peptides and proteins. J. Agric. Food Chem., 46, 490–498. [DOI] [PubMed] [Google Scholar]
- 16.Avila M.A., Corrales,F.J., Ruiz,F., Sanchez-Gongora,E., Mingorance,J., Carretero,M.V. and Mato,I.M. (1998) Specific interaction of methionine adenosyltransferase with free radicals. Biofactors, 8, 27–32. [DOI] [PubMed] [Google Scholar]
- 17.Denu J.M. and Tanner,K.G. (1998) Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry, 37, 5633–5642. [DOI] [PubMed] [Google Scholar]
- 18.Sok D.E. (1999) Oxidative inactivation of brain alkaline phosphatase responsible for hydrolysis of phosphocholine. J. Neurochem., 72, 355–362. [DOI] [PubMed] [Google Scholar]
- 19.de la Mata I., Ramon,F., Obregon,V.V., Castillon,M.P. and Acebal,C. (2000) Effect of hydrogen peroxide on d-amino acid oxidase from Rhodotorula gracilis. Enzyme Microb. Technol., 27, 234–239. [DOI] [PubMed] [Google Scholar]
- 20.Cabiscol E., Piulats,E., Echave,P., Herrero,E. and Ros,J. (2000) Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J. Biol. Chem., 275, 27393–27398. [DOI] [PubMed] [Google Scholar]
- 21.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- 22.Ropp P.A. and Copeland,W.C. (1996) Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics, 36, 449–458. [DOI] [PubMed] [Google Scholar]
- 23.Longley M.J., Ropp,P.A., Lim,S.E. and Copeland,W.C. (1998) Characterization of the native and recombinant catalytic subunit of human DNA polymerase gamma: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry, 37, 10529–10539. [DOI] [PubMed] [Google Scholar]
- 24.Graves S.W., Johnson,A.A. and Johnson,K.A. (1998) Expression, purification and initial kinetic characterization of the large subunit of the human mitochondrial DNA polymerase. Biochemistry, 37, 6050–6058. [DOI] [PubMed] [Google Scholar]
- 25.Lim S.E., Longley,M.J. and Copeland,W.C. (1999) The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis and confers N-ethylmaleimide resistance. J. Biol. Chem., 274, 38197–38203. [DOI] [PubMed] [Google Scholar]
- 26.Johnson A.A., Tsai,Y., Graves,S.W. and Johnson,K.A. (2000) Human mitochondrial DNA polymerase holoenzyme: reconstitution and characterization. Biochemistry, 39, 1702–1708. [DOI] [PubMed] [Google Scholar]
- 27.Carrodeguas J.A. and Bogenhagen,D.F. (2000) Protein sequences conserved in prokaryotic aminoacyl-tRNA synthetases are important for the activity of the processivity factor of human mitochondrial DNA polymerase. Nucleic Acids Res., 28, 1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrodeguas J.A., Theis,K., Bogenhagen,D.F. and Kisker,C. (2001) Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer. Mol. Cell, 7, 43–54. [DOI] [PubMed] [Google Scholar]
- 29.Copeland W.C. and Wang,T.S. (1991) Catalytic subunit of human DNA polymerase alpha overproduced from baculovirus-infected insect cells. Structural and enzymological characterization. J. Biol. Chem., 266, 22739–22748. [PubMed] [Google Scholar]
- 30.Copeland W.C. and Wang,T.S. (1993) Mutational analysis of the human DNA polymerase alpha. The most conserved region in alpha-like DNA polymerases is involved in metal-specific catalysis. J. Biol. Chem., 268, 11028–11040. [PubMed] [Google Scholar]
- 31.Shull S., Heintz,N.H., Periasamy,M., Manohar,M., Janssen,Y.M., Marsh,J.P. and Mossman,B.T. (1991) Differential regulation of antioxidant enzymes in response to oxidants. J. Biol. Chem., 266, 24398–24403. [PubMed] [Google Scholar]
- 32.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 33.Davis A.F. and Clayton,D.A. (1996) In situ localization of mitochondrial DNA replication in intact mammalian cells. J. Cell Biol., 135, 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis A.F., Ropp,P.A., Clayton,D.A. and Copeland,W.C. (1996) Mitochondrial DNA polymerase gamma is expressed and translated in the absence of mitochondrial DNA maintenance and replication. Nucleic Acids Res., 24, 2753–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longley M.J., Prasad,R., Srivastava,D.K., Wilson,S.H. and Copeland,W.C. (1998) Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc. Natl Acad. Sci. USA, 95, 12244–12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stadtman E.R. (1992) Protein oxidation and aging. Science, 257, 1220–1224. [DOI] [PubMed] [Google Scholar]
- 37.Van Remmen H. and Richardson,A. (2001) Oxidative damage to mitochondria and aging. Exp. Gerontol., 36, 957–968. [DOI] [PubMed] [Google Scholar]
- 38.Liu J., Killilea,D.W. and Ames,B.N. (2002) Age-associated mitochondrial oxidative decay: improvement of carnitine acetyltransferase substrate-binding affinity and activity in brain by feeding old rats acetyl-l-carnitine and/or R-alpha-lipoic acid. Proc. Natl Acad. Sci. USA, 99, 1876–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan L.J. and Sohal,R.S. (1998) Mitochondrial adenine nucleotide translocase is modified oxidatively during aging. Proc. Natl Acad. Sci. USA, 95, 12896–12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan L.J., Levine,R.L. and Sohal,R.S. (1997) Oxidative damage during aging targets mitochondrial aconitase. Proc. Natl Acad. Sci. USA, 94, 11168–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das N., Levine,R.L., Orr,W.C. and Sohal,R.S. (2001) Selectivity of protein oxidative damage during aging in Drosophila melanogaster. Biochem. J., 360, 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]