ABSTRACT
Adenylate cyclase family members have recently received attention as novel therapeutic targets. However, the significance of adenylate cyclase 9 (ADCY9) in breast cancer has not been elucidated. Here, we evaluated ADCY9 expression in breast cancer (BC) cell lines, and polymerase chain reaction array analysis was performed to determine the correlations between ADCY9 expression levels and 84 tumor-associated genes. The association of ADCY9 messenger RNA (mRNA) expression levels in clinical breast cancer specimens with patients’ clinicopathological factors and prognosis was evaluated. The database of cancer cell line showed that estrogen receptor-positive and progesterone receptor-positive cells expressed higher ADCY9 mRNA levels. ADCY9 expression showed positive correlations with several oncogenes, such as TGFB1, CDKN1A, and BAX in the polymerase chain reaction array analysis. We defined the ratio of ADCY9 mRNA expression levels in breast cancer and adjacent noncancerous tissues as the “C/N ratio”. Among 149 patients with BC, estrogen receptor-positive and progesterone receptor-positive patients exhibited higher C/N ratios than estrogen receptor-negative and progesterone receptor-negative patients, respectively. Patients in the lowest C/N ratio quartile experienced shorter prognosis periods. The C/N ratio of ADCY9 was found as an independent prognostic factor for disease-free survival. Thus, ADCY9 expression is high in hormone receptor-positive breast cancer, and its low expression indicates a poor prognosis in patients with breast cancer.
Key Words: breast cancer, ADCY9, estrogen receptor, prognostic marker
INTRODUCTION
Breast cancer (BC) is the most commonly diagnosed cancer among women.1 Medication therapies, including chemotherapy, endocrine therapy, and molecular targeting drugs, have recently been selected based on the expression status of the estrogen receptor (ER), progesterone receptor (PgR), and human epidermal growth factor receptor 2 (HER2) in primary tumors. Although the prognosis of patients with BC has improved owing to the development of novel therapeutic drugs, metastatic BC remains difficult to cure.2 An understanding of the oncological functions of unknown molecules and the development of prognostic biomarkers is essential to overcome intractable BC.
The adenylate cyclase (ADCY) superfamily is a group of glycoproteins that regulate intracellular signaling and catalyze the transformation of adenosine triphosphate (ATP) to cyclic adenosine monophosphate (cAMP).3 Recently, overactivation of ADCY has been implicated in modulating cancer progression and resistance to chemotherapy and immunotherapy.3 Among the members of the ADCY superfamily, the oncological roles of ADCY9 have been poorly studied. Although previous studies have reported the roles of ADCY9 in colon and lung cancer,4-6 it has been assigned contradictory functions depending on the carcinoma. Only few studies have evaluated the relationship of ADCY9 messenger RNA (mRNA) expression with patient clinicopathological factors and the prognosis in BC.
In this study, we focused on ADCY9 and investigated its expression in BC cell lines and patient tumor samples to elucidate its significance and determine whether it could be used as a prognostic marker in BC.
MATERIALS AND METHODS
Sample collection
In this study, a total of 13 BC cell lines (BT-20, BT-474, BT-549, HCC1419, HCC1954, Hs578T, MCF7, MDA-MB-231, MDA-MB-361, MDA-MB-415, MDA-MB-468, SK-BR-3, and ZR-75-1) and two noncancerous breast epithelial cell lines (MCF-10A and MCF-12A) were used. We purchased BT-549, HCC1419, HCC1954, and Hs578T cell lines from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan), and BT-474, MCF-7, and MCF-12A were provided from Johns Hopkins University (Baltimore, MD, USA) following the appropriate material transfer agreement. Other cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). These cells were cultured in RPMI 1640 (Sigma-Aldrich, St. Louis, MO, USA) with 10% fetal bovine serum in an atmosphere of 5% CO2 at 37 °C.7 The expression statuses of ER, PgR, and HER2 in each cell line were referred from previous studies that evaluated these expression.8,9
Clinical specimens were obtained from patients who had operations at Nagoya University Hospital between December 2002 and November 2009 and had available postoperative surveillance data. They were histologically diagnosed as BC and their cancerous specimens were resected with a diameter of approximately 1.5 mm and frozen to be stored in −80 °C freezer. The pathological stage of each patient was classified according to the Union for International Cancer Control staging system for BC (8th edition). The postoperative medication therapy was determined at the physician’s discretion based on each patient’s general condition, pathological features, and BC subtypes.
This study complied with the Declaration of Helsinki and was approved by the institutional review board (approval number: 2019-0028). All participants provided written informed consent for the use of their clinical samples and data, which have been stored in a locked locker in our office. The collected human specimens were anonymized so that it could be linked and have been stored in a −80 °C freezer in our office. Patient data was also anonymized and the consolidated forms have been stored in a password-locked hard disk and kept locked in our office.
Quantitative real-time reverse transcription polymerase chain reaction
ADCY9 mRNA expression levels in cell lines and clinical samples were evaluated using quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). RNA was extracted from the cell lines (8.0 × 106 cells per cell line) and 149 BC specimens. Complementary DNA (cDNA) was synthesized by the way previously described.10 To normalize ADCY9 mRNA expression levels, GAPDH mRNA levels were used as a house-keeping gene. The primers specific for each gene were as follows: ADCY9, forward 5′-TGGTTGCTGACCAGTTGAAA-3′ and reverse 5′-ACCTCAAAGCCAGAAAGCAA-3′, which generated a 106-bp product; GAPDH, forward 5′-GAAGGTGAAGGTCG-GAGTC-3′ and reverse 5′-GAAGATGGTGATGGGATTTC-3′, which generated a 226-bp product. SYBR Green PCR core reagent kit (Applied Biosystems) was used with these cycling conditions: one cycle at 95 °C for 10 min, followed by 40 cycles at 95 °C for 5 s and 60 °C for 60 s, using an ABI StepOnePlus real-time PCR System (Applied Bio-systems, Foster City, CA, USA).11 Each sample was conducted in triplicate.
Cancer Cell Line Encyclopedia
The mRNA expression levels of ADCY9 in 58 BC cell lines were obtained from the public database of Cancer Cell Line Encyclopedia (https://sites.broadinstitute.org/ccle/). We accessed it on January 12, 2022.
PCR array analysis
The correlation between the mRNA expression levels of ADCY9 and cancer-related genes was investigated with the kit of RT2 Profiler PCR Array Human for Oncogenes & Tumor Suppressor Genes (Qiagen, Hilden, Germany) that can evaluate the expression levels of 84 cancer-related genes. The assay was performed in accordance with the manufacturer’s protocol. The relative expression level of each gene was obtained by being normalized with the GAPDH level.11
Kaplan–Meier survival analysis
The public database of Kaplan–Meier Plotter online software (http://kmplot.com/analysis/index.php?p=background) was employed to analyze relapse-free survival in large number of patients with BC in relation to the expression of ADCY9 by classifying its expression levels by median.12 It was accessed on August 13, 2023.
Statistical analyses
Differences in ADCY9 mRNA levels between the two groups were compared with the Mann–Whitney U test. The correlation between the expression levels of the two genes were analyzed using the Spearman’s rank correlation test. The χ2 test was conducted to analyze the association between ADCY9 mRNA expression levels and clinicopathological factors. Prognosis, such as disease-free survival (DFS) and overall survival rates, was evaluated with the Kaplan–Meier method, and survival curves were compared using the log-rank test. To identify prognostic factors, multivariate regression analyses was performed using the Cox proportional-hazards model. These analyses were performed using JMP 12 software (SAS Institute, Inc, Cary, NC, USA). P values of < 0.05 were defined as statistically significant.
RESULTS
ADCY9 mRNA expression in BC cell lines
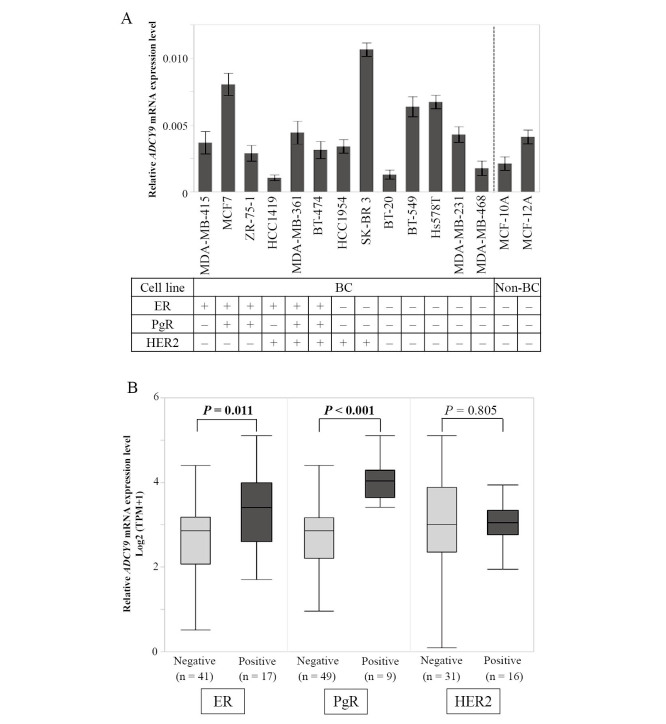
ADCY9 mRNA expression levels varied among the 13 BC cell lines and two non-cancerous mammary cell lines (Fig. 1A). No significant differences were observed in relation to positivity of ER (P = 0.568), PgR (P = 0.758), or HER2 (P = 0.884). To compensate for the small number of cell lines, the Cancer Cell Line Encyclopedia database was used to evaluate ADCY9 mRNA expression levels in 58 cell lines. ER-positive (n = 17) and PgR-positive (n = 9) cells expressed higher ADCY9 mRNA levels than ER-negative (n = 41) and PgR-negative (n = 49) cells, respectively (ER, P = 0.011; PgR, P < 0.001; Fig. 1B). No significant difference was observed between HER2-positive (n = 16) and HER2-negative (n = 31) cells (P = 0.805; Fig. 1B).
Fig. 1.
ADCY9 mRNA expression levels in BC cell lines
Fig. 1A:Analysis of ADCY9 mRNA expression. Bar graphs indicate relative ADCY9 mRNA levels, which were obtained from the ADCY9 value divided by GAPDH value. ADCY9 mRNA expression levels were heterogeneous among cell lines. The expression status of ER and HER2 was determined from previous studies.8,9
Fig. 1B:In the Cancer Cell Line Encyclopedia database, ER-positive and PgR-positive BC cell lines expressed higher ADCY9 mRNA levels, in comparison with the ER-negative and PgR-negative BC cell lines, respectively. No significant differences were observed between HER2-positive and HER2-negative BC cells.
ADCY9: adenylate cyclase 9
BC: breast cancer
ER: estrogen receptor
HER2: human epidermal growth factor receptor 2
PgR: progesterone receptor
mRNA: messenger RNA
The correlations between ADCY9 and the expression levels of 84 cancer-related genes in 13 BC cell lines were evaluated via PCR array analysis, and the expression levels of several oncogenes, including TGFB1, CDKN1A, BAX, and BRCA2, were positively correlated with that of ADCY9 (Table 1 and Supplementary Table).
Table 1.
Correlations between mRNA expression levels of ADCY9 and cancer-related genes
| Genes | Official full names | Correlation coefficients | P - values |
| TGFB1 | transforming growth factor beta 1 | 0.709 | 0.007 |
| CDKN1A | cyclin dependent kinase inhibitor 1A | 0.687 | 0.010 |
| BAX | BCL2 associated X, apoptosis regulator | 0.659 | 0.014 |
| SMAD4 | SMAD family member 4 | 0.659 | 0.014 |
| BRCA2 | BRCA2 DNA repair associated | 0.654 | 0.015 |
| NRAS | NRAS proto-oncogene, GTPase | 0.654 | 0.015 |
| NFKBIA | NFKB inhibitor alpha | 0.648 | 0.017 |
| RAF1 | Raf-1 proto-oncogene, serine/threonine kinase | 0.599 | 0.031 |
| MLH1 | mutL homolog 1 | 0.566 | 0.044 |
ADCY9: adenylate cyclase 9
Patient characteristics
This study included 149 BC patients. All patients were women with a median age of 52 years (range, 26–78 years). The median duration of patient follow-up was 121.6 months (range, 10–191 months). The characteristics of the patients are summarized in Table 2. Patients who showed positivity for ER, PgR, or HER2 were grouped as “non-triple-negative.” As four of the five patients whose HER2 statuses were unknown showed ER-positivity, these patients were categorized as “non-triple-negative”; thus, 15 patients were classified as triple-negative, 133 as non-triple-negative, and 1 as unknown.
Table 2.
Clinicopathological characteristics of 149 patients with breast cancer
| Clinicopathological parameter | |
| Age, median (range) | 52 (26–78) |
| ≤ 60 years | 97 (65.1%) |
| > 60 years | 52 (34.9%) |
| Histology | |
| DCIS | 6 (4.0%) |
| IDC | 133 (89.3%) |
| ILC | 4 (2.7%) |
| Others | 6 (4.0%) |
| UICC T factor a | |
| Tis | 6 (4.0%) |
| T1 | 60 (40.3%) |
| T2 | 70 (47.0%) |
| T3 | 8 (5.4%) |
| T4 | 5 (3.3%) |
| Node status | |
| Negative | 73 (49.0%) |
| Positive | 76 (51.0%) |
| UICC pathological stage a | |
| 0 | 6 (4.0%) |
| I | 41 (27.5%) |
| II | 70 (47.0%) |
| III | 31 (20.8%) |
| IV | 1 (0.7%) |
| ER status | |
| Positive | 113 (75.8%) |
| Negative | 36 (24.2%) |
| PgR status | |
| Positive | 102 (68.5%) |
| Negative | 47 (31.5%) |
| HER2 status | |
| Positive | 36 (24.2%) |
| Negative | 108 (72.5%) |
| Unknown | 5 (3.3%) |
| Triple-negative | |
| Yes | 15 (10.1%) |
| No | 133 (89.3%) |
| Unknown | 1 (0.6%) |
| Adjuvant therapy | |
| Endocrine therapy alone | 46 (30.9%) |
| Chemotherapy alone | 27 (18.1%) |
| Chemotherapy and endocrine therapy | 60 (40.3%) |
| None | 16 (10.7%) |
DCIS: ductal carcinoma in situ
ER: estrogen receptor
HER2: human epidermal growth factor 2
IDC: invasive ductal carcinoma
ILC: invasive lobular carcinoma
PgR: progesterone receptor
UICC: Union for International Cancer Control
a Pathological stages were classified using the UICC staging system for breast cancer (8th edition).
Clinical and prognostic significance of ADCY9 mRNA levels
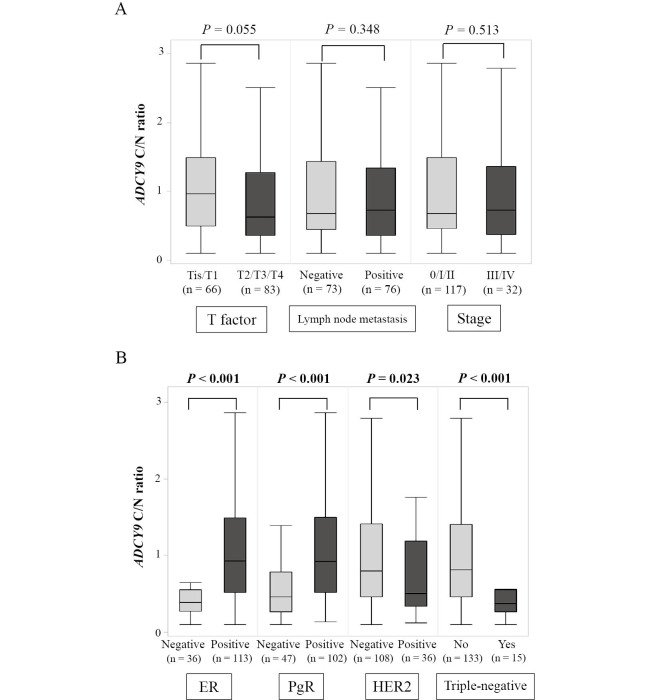
The ADCY9 mRNA levels in the BC tissues 89 (59.7%) of the 149 patients were lower than those in the adjacent normal tissues. We calculated the ratio of ADCY9 mRNA expression levels in the BC and adjacent noncancerous tissues and defined it as the “C/N ratio”. The C/N ratio of ADCY9 did not significantly differ between patients with Tis/T1 (n = 66), T2/T3/T4 (n = 83; P = 0.055), negative and positive lymph node metastases (n = 73 and 76, respectively; P = 0.348), Union for International Cancer Control (UICC) stage 0/I/II (n = 117), and stage III/IV (n = 32, P = 0.513; Fig. 2A) cancers.
Fig. 2.
ADCY9 C/N ratio in breast cancer tissues
Fig. 2A:Relationship between the ADCY9 C/N ratio and pathological factors. No significant differences were observed in relation to the T factor, lymph node metastasis, or stage.
Fig. 2B:Relationship between the ADCY9 C/N ratio and the conventional biomarkers. ADCY9 C/N ratio in the ER-positive and PgR-positive BC groups were significantly higher than those in the ER-negative and PgR-negative groups, respectively. The HER2-positive group showed a lower ADCY9 C/N ratio than the HER2-negative group. The triple-negative group showed a lower ADCY9 C/N ratio than the non-triple-negative group.
ADCY9: adenylate cyclase 9
C/N ratio: ratio of mRNA expression levels between cancerous and noncancerous tissues
ER: estrogen receptor
HER2: human epidermal growth factor receptor 2
PgR: progesterone receptor
In assessments of the relevance of conventional biomarkers (Fig. 2B), the ADCY9 C/N ratio in ER-positive specimens (n = 113) was significantly higher than that in ER-negative specimens (n = 36; P < 0.001); the ADCY9 C/N ratio in PgR-positive specimens’ (n = 102) was also higher than that in PgR-negative specimens (n = 47; P < 0.001); and HER2-positive specimens (n = 36) expressed a lower C/N ratio of ADCY9 than HER2-negative specimens (n = 108; P = 0.023; missing HER2 data for five patients). Additionally, the ADCY9 C/N ratio was lower in triple-negative BC (n = 15) than that in non-triple-negative BC (n = 133; P < 0.001; missing data for one patient; Fig. 2B).
In the subsequent analyses, the patients with the lowest quartile of ADCY9 C/N ratio were designated as the “low ADCY9 group” (n = 38), while the remaining patients were designated as the “high ADCY9 group” (n = 111). While these two groups showed no significant associations with the T factor (P = 0.065), node status (P = 0.324), and pathological stage (P = 0.703), the low ADCY9 group showed a significant association with the ER-negative (P < 0.001), PgR-negative (P < 0.001), HER2-positive (P = 0.014), and triple-negative status (P = 0.013; Table 3).
Table 3.
Associations between ADCY9 mRNA expression and clinicopathological characteristics of 149 patients with breast cancer
| Clinicopathological parameters | Low ADCY9 group (n = 38) | High ADCY9 group (n = 111) | P -values |
| Age | |||
| ≤ 60 years | 22 | 75 | 0.284 |
| > 60 years | 16 | 36 | |
| Histology | |||
| DCIS | 1 | 5 | |
| IDC | 35 | 98 | 0.785 |
| ILC | 0 | 4 | |
| Others | 2 | 4 | |
| UICC T factor a | |||
| Tis/T1 | 12 | 54 | 0.065 |
| T2/T3/T4 | 26 | 57 | |
| Node status | |||
| Negative | 16 | 57 | 0.324 |
| Positive | 22 | 54 | |
| UICC pathological stage a | |||
| 0/I/II | 29 | 88 | 0.703 |
| III/IV | 9 | 23 | |
| ER status | |||
| Positive | 19 | 94 | < 0.001 |
| Negative | 19 | 17 | |
| PgR status | |||
| Positive | 15 | 87 | < 0.001 |
| Negative | 23 | 24 | |
| HER2 status | |||
| Positive | 15 | 21 | 0.014 |
| Negative | 22 | 86 | |
| Triple-negative | |||
| Yes | 8 | 7 | 0.013 |
| No | 29 | 104 | |
| Adjuvant therapy | |||
| Endocrine therapy alone | 5 | 41 | |
| Chemotherapy alone | 13 | 14 | 0.003 |
| Chemotherapy and endocrine therapy | 14 | 46 | |
| None | 6 | 10 |
ADCY9: adenylate cyclase 9
DCIS: ductal carcinoma in situ
ER: estrogen receptor
HER2: human epidermal growth factor 2
IDC: invasive ductal carcinoma
ILC: invasive lobular carcinoma
PgR: progesterone receptor
UICC: Union for International Cancer Control
a Pathological stages were classified using the UICC staging system for breast cancer (8th edition).
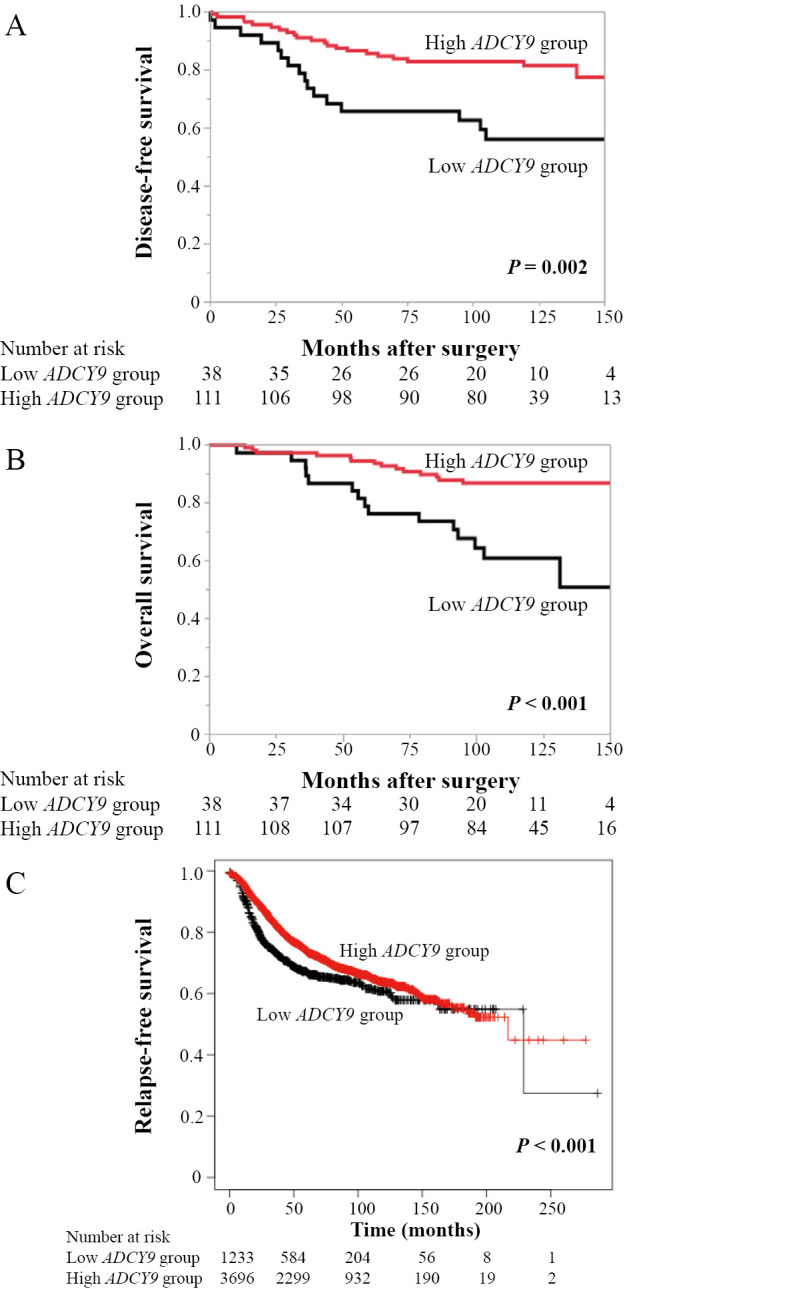
The low ADCY9 group showed a significantly shorter DFS than the high ADCY9 group (5-year DFS rates, low ADCY9 group: 65.8%, high ADCY9 group: 85.6%; P = 0.002; Fig. 3A). The overall survival of the low ADCY9 group was also shorter than that in the high ADCY9 group (5-year overall survival rates: low ADCY9 group, 76.3%; high ADCY9 group, 94.6%; P < 0.001; Fig. 3B). Multivariate analysis of DFS identified T factor T2/T3/T4 (hazard ratio: 2.24; 95% confidence interval: 1.02–5.65, P = 0.044), positive lymph node metastasis (hazard ratio: 4.02; 95% confidence interval: 1.79–9.99, P < 0.001), and the low ADCY9 group (hazard ratio: 2.00; 95% confidence interval: 1.01–3.88, P = 0.047) as independent prognostic factors (Table 4). To compensate for the small sample size of our cohort, the prognostic impact of ADCY9 expression was investigated using Kaplan–Meier Plotter. Similarly, when patients were assigned to either the lowest quartile (low ADCY9 group) or to other quartiles, the low ADCY9 group (n = 1233) exhibited worse relapse-free survival than the high ADCY9 group (n = 3696; hazard ratio = 0.77; P < 0.001; Fig. 3C).
Fig. 3.
Relationship between the ADCY9 C/N ratio and prognosis
Fig. 3A, B:In the cohort that evaluated our samples, the low ADCY9 group showed shorter disease-free survival (A) and overall survival (B) than the other patients.
Fig. 3C:According to the Kaplan–Meier Plotter, patients in the lowest-quartile ADCY9 group exhibited shorter relapse-free survival than the high ADCY9 group.
ADCY9: adenylate cyclase 9
C/N ratio: ratio of mRNA expression levels between cancerous and noncancerous tissues
Table 4.
Prognostic factors for disease-free survival in 149 patients with breast cancer
| Variables | n | Univariate | Multivariate | ||||
| Hazard ratios | 95% CI | P -values | Hazard ratios | 95% CI | P -values | ||
| Age (> 60 years) | 52 | 1.62 | 0.83 – 3.10 | 0.146 | |||
| UICC T factor (T2/T3/T4) | 83 | 3.81 | 1.77 – 9.46 | < 0.001 | 2.24 | 1.02 – 5.65 | 0.044 |
| Lymph node metastasis (positive) | 76 | 4.12 | 1.97 – 9.68 | < 0.001 | 4.02 | 1.79 – 9.99 | < 0.001 |
| ER status (negative) | 36 | 2.03 | 1.00 – 3.95 | 0.049 | 1.23 | 0.48 – 2.80 | 0.649 |
| PgR status (negative) | 47 | 1.84 | 0.95 – 3.52 | 0.071 | |||
| HER2 status (positive) | 36 | 1.68 | 0.81 – 3.30 | 0.157 | |||
| Triple-negative (yes) | 15 | 2.56 | 1.03 – 5.53 | 0.043 | 2.63 | 0.86 – 7.93 | 0.088 |
| Low ADCY9 expression | 38 | 2.64 | 1.36 – 5.05 | 0.005 | 2.00 | 1.01 – 3.88 | 0.047 |
ADCY9: adenylate cyclase 9
CI: confidence interval
ER: estrogen receptor
HER2: human epidermal growth factor 2
PgR: progesterone receptor
UICC: Union for International Cancer Control
DISCUSSION
In this study, ADCY9 mRNA expression was positively associated with ER and PgR expression in both BC cell lines and clinical specimens. Moreover, a low C/N ratio of ADCY9 was associated with poor survival and was found to be an independent prognostic factor. These results implied that activated ADCY9 plays certain roles in BC despite lacking the function analyses.
The activation of ADCY proteins drives the production of cAMP from ATP, leading to elevated intracellular cAMP levels that play important roles in cancer cell proliferation, apoptosis, invasion, angiogenesis, and immune escape.3 Several recent studies have shown that interventions targeting the steps upstream and downstream of the cAMP signaling pathway show anticancer effects and enhance the sensitivity of conventional therapeutic drugs3,13,14; thus, ADCY is considered a potential target of anticancer therapies. The ADCY family consists of 10 subtypes, each of which is encoded by an independent gene.3 The expression of ADCY subtypes varies in various cancers; some are highly expressed in tumors and participate in tumor formation and development as oncogenes, while others may show low or even no expression.3 Among them, ADCY4 mRNA expression is significantly downregulated in BC tissues in comparison with normal tissues, and high ADCY4 expression is correlated with improved survival in patients with BC.15 In contrast, downregulation and hypermethylation of ADCY6 has been associated with a better prognosis in patients with BC.16
Among the ADCY family members, the oncological roles of ADCY9 have been poorly studied, and several studies have reported inconsistent findings regarding its roles in various malignancies. A cohort study of ADCY9 polymorphisms in colon cancer showed that the ADCY9 expression level was higher in cancerous tissues than that in adjacent tissues, and low ADCY9 expression in cancer tissues was associated with a longer DFS.4 Immunohistochemical analyses showed that ADCY9 is highly expressed in advanced colon cancer and is a poor prognostic marker,5 which implies that ADCY9 performs a tumor-progressive role. In contrast, a recent study showed tumor-suppressive activities of ADCY9 in lung adenocarcinoma.6 Accordingly, ADCY9 expression was downregulated in cancerous tissues, and high ADCY9 expression led to a better prognosis and was an independent prognostic factor.6 In addition, overexpression of ADCY9 was shown to restrain the proliferation, invasion, and migration of lung adenocarcinoma cell lines via micro-RNA.6 The possible reason why the significance of ADCY9 expression differs among the cancer types is because the expression level of ADCY9 varies depending on the tissue,3 and the involved signaling-pathways and their weights on cellular function also differ. In BC, although computational analysis identified ADCY9 as a hub gene in the cisplatin-responsive network using the MCF-7 cell line,17 no studies have reported the relationship between ADCY9 expression and patients’ clinicopathological features and prognosis.
In this study, we found an association between the ADCY9 C/N ratio, hormone receptor status, and prognosis. As these results were based on observational data, the underlying mechanism was difficult to explain. Nevertheless, based on the results of our PCR array analysis in BC cells and those of previous studies, several possible explanations can be inferred. Regarding the relationship between ADCY9 and hormone receptors, ADCY9 was identified as the most differentially expressed gene in the ER signature based on the results of gene expression analysis in Chinese patients with BC, and it was reported to be located in the calcium- and gonadotropin-releasing hormone signaling pathways.18 Another study reported that ADCY9 is associated with the ER signaling pathway in non-small cell lung cancer, which may improve prognosis.19 Although ADCY9 levels in our cell lines were heterogenous even among same subtypes, the Cancer Cell Line Encyclopedia database showed the positive relation between ADCY9 levels and hormone receptor positive status, which was validated in clinical samples. The results of the PCR array analysis showed a positive correlation between the expression levels of ADCY9 and CDKN1A, which encodes p21. Expression of p21 has been reported to correlate with ER-positive status in BC.20 These results together imply that ADCY9 may be involved in the ER signaling pathway in BC. However, BRCA2, whose expression level positively correlated with that of ADCY9 in the PCR array analysis, was highly expressed in ER-negative BC patients.21 Therefore, further pathway analyses are required.
The most novel finding of this study was that patients with a low ADCY9 C/N ratio had a poor prognosis; thus, the ADCY9 C/N ratio was identified as a prognostic factor. Several possible explanations can be provided for this observation. First, the low ADCY9 group contained more patients with ER-negative status, who are more likely to show a worse prognosis than patients with ER-positive status.22 However, as the multivariate analysis of DFS identified the ADCY9 C/N ratio as a prognostic factor independent of hormone receptor status, this was not considered the only reason. The second possible reason can be determined from the PCR array results. The expression levels of TGFB1 and SMAD4, downstream targets of TGFB1, showed high correlation coefficients with those of ADCY9. TGF-β1, a multifunctional cytokine, performs contradictory roles in BC. Although it is linked to increased tumor progression in the late stages, it inhibits cellular proliferation and promotes apoptosis in the early stages.23 CDKN1A, which also showed high correlation with ADCY9, encodes p21, a downstream mediator of TGF-β1 and p53.20,24,25 Patients with BC and low p21 expression levels have a poor prognosis,20,24 supporting our results. Moreover, the expression level of BAX, which encodes a pro-apoptotic protein,26 was correlated with that of ADCY9. BAX is a key gene involved in apoptosis and is regulated by p53 and cAMP.3,26 Overexpression of ADCY9 in cancer cell lines leads to increased apoptosis in lung adenocarcinomas.6 On the basis of these observations, it is suggested that the low C/N ratio of ADCY9 may reflect the downregulation of these tumor-suppressive pathways, especially those related to pro-apoptosis, which creates a favorable environment for cancer cell survival.
This study had some limitations. The oncological functions of ADCY9 and its regulatory mechanisms in BC cells have not been evaluated. Therefore, basic functional analyses based on ADCY9 inhibition are needed. Although we have described a possible explanation based on the results of the PCR array analysis, further experiments can yield a better understanding of the pathways through which ADCY9 affects the survival of BC cells. Moreover, our clinical data were retrospective in nature. Adjuvant therapeutic interventions such as endocrine therapy, chemotherapy and molecular targeted therapy, may have affected patient prognosis. In addition, status of drug usage such as glucocorticoids and contraceptives, which can affect the expression status of hormone receptors,27-29 were unknown. However, because it is unlikely that many Japanese patients have taken these drugs, their impact on our results is considered limited.
Although we validated our data using a large public database to compensate for this limitation, validation using prospectively collected data would have been more desirable. Despite these limitations, this is the first study to demonstrate a relationship between ADCY9 mRNA expression and BC prognosis. Our findings can help clarify the molecular mechanisms underlying BC and may have possible clinical applications. For example, assessment of ADCY9 mRNA levels in resected samples may be helpful for evaluating each patient’s prognosis and selecting adjuvant medication therapy regimens. Further mechanistic studies are required to overcome these limitations.
CONCLUSION
This study suggests that ADCY9 mRNA expression is high in hormone receptor-positive BC, and its low expression in cancerous tissue indicates a poor prognosis in patients with BC.
AUTHOR CONTRIBUTIONS
Conceptualization: M.S. and M.K.; experiments: K.S., Y.A., I.S., T.I. (Takahiro Ichikawa), and T.I. (Takahiro Inaishi); data analysis: K.S., M.S., Y.A., I.S., and E.K.; resources (cell lines): M.H.; data and sample collection: K.S., M.S., Y.A., I.S., T.I. (Takahiro Ichikawa), and T.I. (Takahiro Inaishi); writing the manuscript: K.S., and M.S.; supervision: N.M. The final manuscript has been approved by all the authors. All the authors have read and agreed to the published version of the manuscript.
ACKNOWLEDGEMENT
We are grateful to David Sidransky, MD, director of the Otolaryngology Department of Johns Hopkins University School of Medicine (Baltimore, MD, USA) for providing the BT-474, MCF-7, and MCF-12A cell lines.
DISCLOSURE STATEMENT
All authors declare that we have no conflicts of interest.
FUNDING
This research received no external funding.
SUPPLEMENTARY INFORMATION
Correlations between mRNA expression levels of ADCY9 and 84 cancer-related genes
Abbreviations
- ADCY
adenylate cyclase
- BC
breast cancer
- C/N ratio
ratio of mRNA expression levels between cancerous and noncancerous tissues
- DFS
disease-free survival
- ER
estrogen receptor
- HER2
human epidermal growth factor receptor 2
- mRNA
messenger RNA
- PCR
polymerase chain reaction
- PgR
progesterone receptor
REFERENCES
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed]
- 2.Iwata H. Future treatment strategies for metastatic breast cancer: curable or incurable? Breast Cancer. 2012;19(3):200–205. doi: 10.1007/s12282-011-0267-4. [DOI] [PubMed]
- 3.Guo R, Liu T, Shasaltaneh MD, Wang X, Imani S, Wen Q. Targeting Adenylate Cyclase Family: New Concept of Targeted Cancer Therapy. Front Oncol. 2022;12:829212. doi: 10.3389/fonc.2022.829212. [DOI] [PMC free article] [PubMed]
- 4.Li H, Liu Y, Liu J, et al. Assessment of ADCY9 polymorphisms and colorectal cancer risk in the Chinese Han population. J Gene Med. 2021;23(2):e3298. doi: 10.1002/jgm.3298. [DOI] [PubMed]
- 5.Yi H, Wang K, Jin JF, et al. Elevated Adenylyl Cyclase 9 Expression Is a Potential Prognostic Biomarker for Patients with Colon Cancer. Med Sci Monit. 2018;24:19–25. doi: 10.12659/msm.906002. [DOI] [PMC free article] [PubMed]
- 6.Tang Y, Wang T, Zhang A, et al. ADCY9 functions as a novel cancer suppressor gene in lung adenocarcinoma. J Thorac Dis. 2023;15(3):1018–1035. doi: 10.21037/jtd-22-1027. [DOI] [PMC free article] [PubMed]
- 7.Inaishi T, Shibata M, Ichikawa T, et al. Platelet isoform of phosphofructokinase accelerates malignant features in breast cancer. Oncol Rep. 2022;47(1):9. doi: 10.3892/or.2021.8220. [DOI] [PubMed]
- 8.Finn RS, Dering J, Conklin D, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11(5):R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed]
- 9.Subik K, Lee JF, Baxter L, et al. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast Cancer (Auckl). 2010;4:35–41. doi: 10.1177/117822341000400004. [DOI] [PMC free article] [PubMed]
- 10.Shibata M, Kanda M, Shimizu D, et al. Expression of regulatory factor X1 can predict the prognosis of breast cancer. Oncol Lett. 2017;13(6):4334–4340. doi: 10.3892/ol.2017.6005. [DOI] [PMC free article] [PubMed]
- 11.Ichikawa T, Shibata M, Inaishi T, et al. Synaptotagmin 13 Is Highly Expressed in Estrogen Receptor-Positive Breast Cancer. Curr Oncol. 2021;28(5):4080–4092. doi: 10.3390/curroncol28050346. [DOI] [PMC free article] [PubMed]
- 12.Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed]
- 13.Follin-Arbelet V, Torgersen ML, Naderi EH, Misund K, Sundan A, Blomhoff HK. Death of multiple myeloma cells induced by cAMP-signaling involves downregulation of Mcl-1 via the JAK/STAT pathway. Cancer Lett. 2013;335(2):323–331. doi: 10.1016/j.canlet.2013.02.042. [DOI] [PubMed]
- 14.Zhang H, Kong Q, Wang J, Jiang Y, Hua H. Complex roles of cAMP-PKA-CREB signaling in cancer. Exp Hematol Oncol. 2020;9(1):32. doi: 10.1186/s40164-020-00191-1. [DOI] [PMC free article] [PubMed]
- 15.Fan Y, Mu J, Huang M, et al. Epigenetic identification of ADCY4 as a biomarker for breast cancer: an integrated analysis of adenylate cyclases. Epigenomics. 2019;11(14):1561–1579. doi: 10.2217/epi-2019-0207. [DOI] [PubMed]
- 16.Li W, Sang M, Hao X, Jia L, Wang Y, Shan B. Gene expression and DNA methylation analyses suggest that immune process-related ADCY6 is a prognostic factor of luminal-like breast cancer. J Cell Biochem. 2020;121(7):3537–3546. doi: 10.1002/jcb.29633. [DOI] [PubMed]
- 17.Fallahi H, Godini R. System-level responses to cisplatin in pro-apoptotic stages of breast cancer MCF-7 cell line. Comput Biol Chem. 2019;83:107155. doi: 10.1016/j.compbiolchem.2019.107155. [DOI] [PubMed]
- 18.Huang CC, Tu SH, Lien HH, et al. Concurrent gene signatures for han chinese breast cancers. PLoS One. 2013;8(10):e76421. doi: 10.1371/journal.pone.0076421. [DOI] [PMC free article] [PubMed]
- 19.Jia S, Li L, Xie L, Zhang W, Zhu T, Qian B. Transcriptome Based Estrogen Related Genes Biomarkers for Diagnosis and Prognosis in Non-small Cell Lung Cancer. Front Genet. 2021;12:666396. doi: 10.3389/fgene.2021.666396. [DOI] [PMC free article] [PubMed]
- 20.Wakasugi E, Kobayashi T, Tamaki Y, et al. p21(Waf1/Cip1) and p53 protein expression in breast cancer. Am J Clin Pathol. 1997;107(6):684–691. doi: 10.1093/ajcp/107.6.684. [DOI] [PubMed]
- 21.Egawa C, Miyoshi Y, Taguchi T, Tamaki Y, Noguchi S. Quantitative analysis of BRCA1 and BRCA2 mRNA expression in sporadic breast carcinomas and its relationship with clinicopathological characteristics. Jpn J Cancer Res. 2001;92(6):624–630. doi: 10.1111/j.1349-7006.2001.tb01140.x. [DOI] [PMC free article] [PubMed]
- 22.Kiba T, Inamoto T, Nishimura T, et al. The reversal of recurrence hazard rate between ER positive and negative breast cancer patients with axillary lymph node dissection (pathological stage I-III) 3 years after surgery. BMC Cancer. 2008;8:323. doi: 10.1186/1471-2407-8-323. [DOI] [PMC free article] [PubMed]
- 23.Zarzynska JM. Two faces of TGF-beta1 in breast cancer. Mediators Inflamm. 2014;2014:141747. doi: 10.1155/2014/141747. [DOI] [PMC free article] [PubMed]
- 24.Barbareschi M, Caffo O, Doglioni C, et al. p21WAF1 immunohistochemical expression in breast carcinoma: correlations with clinicopathological data, oestrogen receptor status, MIB1 expression, p53 gene and protein alterations and relapse-free survival. Br J Cancer. 1996;74(2):208–215. doi: 10.1038/bjc.1996.339. [DOI] [PMC free article] [PubMed]
- 25.Ryu B, Kern SE. The essential similarity of TGFbeta and activin receptor transcriptional responses in cancer cells. Cancer Biol Ther. 2003;2(2):164–170. doi: 10.4161/cbt.2.2.276. [DOI] [PubMed]
- 26.Thu HE, Hussain Z, Mohamed IN, Shuid AN. Eurycoma longifolia, A Potential Phytomedicine for the Treatment of Cancer: Evidence of p53-mediated Apoptosis in Cancerous Cells. Curr Drug Targets. 2018;19(10):1109–1126. doi: 10.2174/1389450118666170718151913. [DOI] [PubMed]
- 27.Cairat M, Al Rahmoun M, Gunter MJ, et al. Use of systemic glucocorticoids and risk of breast cancer in a prospective cohort of postmenopausal women. BMC Med. 2021;19(1):186. doi: 10.1186/s12916-021-02004-6. [DOI] [PMC free article] [PubMed]
- 28.Beaber EF, Malone KE, Tang MT, et al. Oral contraceptives and breast cancer risk overall and by molecular subtype among young women. Cancer Epidemiol Biomarkers Prev. 2014;23(5):755–764. doi: 10.1158/1055-9965.Epi-13-0944. [DOI] [PMC free article] [PubMed]
- 29.Lumachi F, Ermani M, Marino F, et al. Relationship between oral contraceptive therapy and estrogen receptor status in patients with breast cancer. Anticancer Res. 2008;28(1B):491–493. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlations between mRNA expression levels of ADCY9 and 84 cancer-related genes