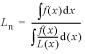Abstract
Bi-stranded abasic clusters, an abasic (AP) site on one DNA strand and another nearby AP site or strand break on the other, have been quantified using Nfo protein from Escherichia coli to produce a double-strand break at cluster sites. Since recent data suggest that Nfo protein cleaves inefficiently at some clusters, we tested whether polyamines, which also cut at AP sites, would cleave abasic clusters at higher efficiency. The data show that Nfo protein cleaves poorly at clusters containing immediately opposed AP sites and those separated by 1 or 3 bp. Putrescine (PUTR) cleaved more efficiently than spermidine or spermine, and did not cleave undamaged DNA. It cleaved abasic clusters in oligonucleotide duplexes more effectively than Nfo protein, including immediately opposed or closely spaced clusters. PUTR cleaved more efficiently than Nfo protein by a factor of ∼1.7 or ∼2 for DNA that had been γ-irradiated in moderate or non-radioquenching conditions, respectively. This suggests that the DNA environment during irradiation affects the spectrum of cluster configurations. Further comparison of PUTR and Nfo protein cleavage may provide useful information on abasic cluster levels and configurations induced by ionizing radiation.
INTRODUCTION
Abasic sites (apurinic and apyrimidinic sites, AP) and oxidized bases constitute common forms of cellular DNA damage induced by ionizing radiation and radiomimetic drugs (1,2). It was predicted (3,4), and subsequently shown (5–7) that ionizing radiation produces clustered damages (locally multiply damaged sites) in addition to isolated lesions. While isolated lesions are generally repaired efficiently, clustered DNA damages have been suggested to be more difficult or impossible for cells to repair (8–11). Studies of the efficiency of different repair enzymes in cleaving oligonucleotide duplexes containing closely spaced DNA lesions showed that closely spaced AP sites, oxidized bases or single-strand breaks (SSBs) reduced or eliminated recognition and/or cleavage (9–11).
AP sites are recognized by AP endonucleases, which incise DNA at the modified site and initiate repair (12). One of the most conserved and well-characterized AP endonucleases is Escherichia coli endonuclease IV (Nfo protein) (13). Endonuclease IV, like the majority of AP endonucleases, cleaves DNA at the 5′ side of the AP site, generating 3′-hydroxyl and 5′-phosphate termini. This enzyme can recognize regular as well as oxidized AP sites arising by radiation-induced hydroxyl radical abstraction of a proton from the C1′, C2′ and, more frequently, C4′ positions of the sugar moiety (2). Recent data indicate that Nfo protein may also have a DNA glycosylase-like activity incising at some oxidized pyrimidines (14), but enzyme cleavage of irradiated DNA containing multiple modified bases may differ from that in oligonucleotides containing a single modified base (15).
AP sites are also recognized and cleaved by polyamines such as spermine+4 (SP), spermidine+3 (SPD) and putrescine+2 (PUTR). It has been suggested that polyamines can cleave both regular and oxidized AP sites (8,16). In contrast to AP endonucleases, polyamines cleave at the 3′ side of the AP site (17). They also cleave AP sites opposing a SSB, which are resistant to cleavage by some AP endonucleases (8,16,18).
In this study, we assessed the efficiency of different polyamines in detecting and cleaving at abasic sites introduced in T7 DNA by ionizing radiation (γ-rays). To compare directly the abilities of Nfo protein and PUTR to recognize and cleave at abasic sites, we used oligonucleotide constructs containing bi-stranded abasic clusters containing AP sites at positions ranging from directly opposing to 5 bp apart in either the 5′ or 3′ direction. The influence of different cluster configurations on incision by the two different agents was determined. We then compared the cleavage by PUTR and Nfo protein of T7 DNA irradiated with γ-rays in different radioquenching conditions to determine the levels of abasic clusters introduced by ionizing radiation under these conditions and to assess the fraction of abasic clusters detected by Nfo protein (7).
MATERIALS AND METHODS
Preparation of T7 DNA substrates
Bacteriophage T7 DNA, a linear double-stranded 40 kb duplex, was prepared by dialyzing the phage against T7 diluent (50 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 500 mM NaCl), diluted to an OD260 of ∼12 (∼4.8 × 1012 phage/ml). Phage was then transferred to sterile, silicone-stoppered tubes and treated with an equal volume of phenol (Mallinckrodt, Georgetown, KY). The phenol was reagent grade, which had been purified further by redistilling while bubbling with N2, collected under double-distilled H2O that had been boiled for 20 min and stored under boiled, deionized, double-distilled H2O under N2 in foil-wrapped bottles at –20°C; just before use, it was saturated with 40 mM NaCl, 30 mM Tris, pH 8.0, 1.0 mM EDTA. The tube was rolled slowly in a near horizontal position for 5 min. The mixture was centrifuged (11 950 g for 15 min) and the upper solution was removed to a fresh tube using a wide-bore pipette to prevent shearing of DNA. The DNA solution was treated three times with phenol as above (until the interface after centrifugation was clear), extracted twice with water-saturated s-butanol, dialyzed repeatedly against Super DNA Dialysis Buffer (0.5 M NaCl, 10 mM EDTA, 10 mM Tris–HCl, pH 7.0), then against Standard DNA Dialysis Buffer (1 mM EDTA, 10 mM Tris–HCl, pH 7.0, 50 mM NaCl) until the A270 of the dialysis solution was no greater than that of fresh buffer. The A260/A280 ratio of the resulting DNA solution was >1.95. The T7 DNA was finally extensively dialyzed against 20 mM potassium phosphate buffer, pH 7.4, or 10 mM Tris–HCl, pH 8.0, producing different radioquenching conditions.
Irradiation
DNA (50 µg/ml) was irradiated in plastic tubes on ice with 137Cs γ-rays at dose rates from 0.16 to 1.6 Gy/min. DNA in 20 mM potassium phosphate buffer (pH 7.4) was irradiated with 0–10 Gy and DNA in 10 mM Tris–HCl (pH 8.0) was irradiated with 0–500 Gy. Irradiations were carried out at the Brookhaven Controlled Environment Radiation Facility; dosimetry was performed by the certified operator of the facility using thermoluminescent lithium fluoride chips.
Endonuclease IV and polyamine treatment
After irradiation, the DNA samples were brought to final concentrations of 70 mM HEPES–KOH, pH 7.6, 100 mM KCl, 0.25 mM EDTA, 1 mM DTT, 50 ng/µl bovine serum albumin (BSA). Escherichia coli Nfo protein (endonuclease IV) was purified to homogeneity according to Levin et al. (19). Its specific activity (1.2 × 1014 AP sites/µg/min) was determined by cleavage of supercoiled pUC18 DNA (2.68 kb; Bayou Biolabs, Harahan, LA) containing an average of 0.7 AP sites/molecule. The non-specific activity of this enzyme stock was assessed on undamaged pUC18 DNA substrate and found to be negligible.
The amount of Nfo protein required to give complete cleavage of abasic clusters was determined by titrating T7 DNA irradiated in phosphate with increasing amounts of enzyme (see inset in Fig. 5); a quantity of 10 ng was chosen per 150 ng DNA. T7 DNA was incubated with Nfo protein or without enzyme for 30 min at 37°C in 70 mM HEPES–KOH, pH 7.6, 100 mM KCl, 0.25 mM EDTA, 1 mM DTT, 50 ng/µl BSA in a total reaction volume of 10 µl. After digestion was complete, enzymes were removed by addition of proteinase K and EDTA to 1.33 mg/ml and 0.1 M, respectively, and incubation at 37°C for 16 h. A neutral stop mixture (0.125% bromophenol blue, 0.5% sodium lauryl sulfate in 50% glycerol) was then added to ensure dissociation of any persistent DNA–protein complexes.
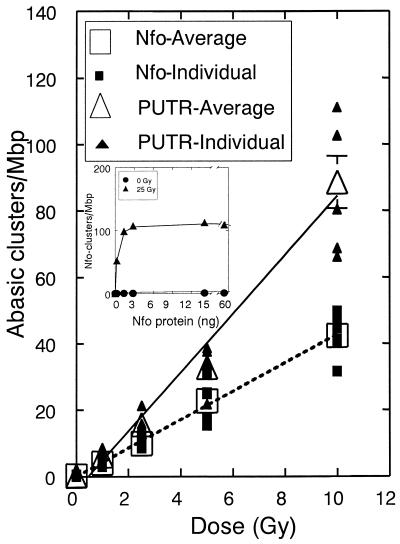
Figure 5.
Frequencies of abasic clusters detected by 1 mM PUTR and Nfo protein for T7 DNA irradiated with γ-rays in 20 mM phosphate buffer. Lines, least squares linear fit of the average points (large open symbols); error bars, standard errors of the means (for fitting parameters see Table 2). In some cases the errors are smaller than the symbols showing the average values. (Inset) Titration of irradiated (25 Gy) and unirradiated (0 Gy) T7 DNA with different amounts of Nfo protein.
Polyamine hydrochlorides (SP, SPD and PUTR; Sigma, St Louis, MO) were dissolved in double-distilled, sterile water at 1, 10 and 100 mM for SP, SPD and PUTR, respectively. DNA samples intended for polyamine treatment were dialyzed against low ionic strength polyamine buffer (buffer P, 5 mM HEPES–KOH, 0.1 mM EDTA, 1 mM KCl, pH 7.25) by placing 20 µl of DNA solution on a sterile Millipore microdialysis membrane (Millipore Co., Bedford, MA), then floating it on buffer P for 1 h on ice. The samples were then placed in plastic tubes and stored at 4°C. DNAs diluted ∼1:10 in the same buffer were equilibrated at 28°C for 10 min, then incubated at 37°C for 30 min with increasing polyamine concentrations. The reaction was stopped and polyamines were dissociated from DNA by the addition of ice-cold 5 M NaCl to a final concentration of 0.5 M NaCl, and incubation continued at 28°C for 30 min. Samples were placed in wells of a 0.4% agarose gel, in Tris–acetate buffer (40 mM Tris, 20 mM acetate, 0.15 mM EDTA, 18 mM NaCl, pH 8) and equilibrated for 1 h.
T7 abasic cluster measurement
Frequencies of abasic clusters were determined as previously described (20,21). Briefly, samples and double-stranded molecular length standards were electrophoresed using static field electrophoresis (30 V, 6°C, with buffer recirculation) for frequencies >∼30 sites/Mb or unidirectional pulsed field electrophoresis [15 V/cm, 0.3 s pulse, 10 s interpulse period, 16 h, 10°C, with buffer recirculation (22)] for lower damage frequencies. Gels were stained, destained and imaged as previously described (21). From the molecular length standards, a DNA dispersion curve (correlating electrophoretic migration with DNA length) was calculated, and from the dispersion curve and the experimental DNA profiles, the number average molecular length (Ln) of each sample DNA population was calculated from equation 1,
where L(x) is the length of the DNA molecules that migrated to position x and f(x)dx is the intensity of ethidium fluorescence from DNA molecules at that position. From the profiles of irradiated and unirradiated (or enzyme-treated, polyamine-treated and untreated DNA populations), the number average length (Ln) of each DNA distribution was calculated. From these Ln values, the frequencies of double-strand breaks (DSBs) (φDSB) and of abasic clusters (φCL) were calculated by equations 2–4,
φDSB = [1/Ln(+rad)] – [1/Ln(–rad)] 2
φCL = [1/Ln(+rad, +enzyme)] – [1/Ln(+rad, –enzyme) 3
φCL = [1/Ln(+rad, +polyamine)] – [1/Ln(+rad, –polyamine) 4
where 1/Ln(+rad), 1/Ln(–rad), 1/Ln(+rad, +enzyme or polyamine) and 1/Ln(+rad, –enzyme or polyamine) are the reciprocals of the Ln values of samples that were irradiated, unirradiated, irradiated and treated with enzyme or polyamine or irradiated and not treated with enzyme or polyamine, respectively.
Preparation of oligonucleotides substrates
Oligonucleotides (see Table 1) (Operon, Alameda, CA) were annealed by mixing the single-stranded oligonucleotides (B–H) in equimolar concentrations with a complementary strand (either A, which contains uracil at position 12, or Ac, with no uracil) in STE (10 mM Tris–HCl, 1 mM EDTA, 50 mM NaCl, pH 7.8), heated to 95°C for 3 min and then allowed to cool slowly to room temperature overnight. The stock concentration of the annealed oligonucleotide samples was measured spectrophotometrically and adjusted to 150 µg/ml. The annealing efficiency was checked by electrophoresis (60 V, 3 h) on a non-denaturing 18% polyacrylamide gel, which was stained using a DNA PhastGel silver staining kit (Pharmacia Biotech, Peapack, NJ) as indicated in the manufacturer’s instructions. An electronic image was obtained and the double- and single-stranded species were quantified. To quantitate cleavage of the oligonucleotides, we needed a sensitive method that would produce a linear signal with DNA mass. Since PhastGel silver staining was much more sensitive than ethidium bromide, we tested whether the signal obtained by electronic imaging of a silver-stained gel was directly proportional to the quantity of DNA in a band. By measuring the signals from samples of increasing quantities of oligonucleotides electrophoresed on the same gel (silver-stained), we determined empirically that the absolute value of the ‘area’ under each band on the image was directly proportional to the mass of DNA in the band. The inset in Figure 4 shows this proportionality between signal and amount of DNA. The level of single-stranded molecules was <5%.
Table 1. Sequences of oligonucleotide duplexes containing uracil sites (X) at various bi-stranded positions.
| Duplex | Sequence | Position |
|---|---|---|
| E | 5′-AGAGGAXATGTATGTATGGAGAG-3′ | –5 |
| A | 3′-TCTCCTATACAXACATACCTCTC-5′ | |
| D | 5′-AGAGGATAXGTATGTATGGAGAG-3′ | –3 |
| A | 3′-TCTCCTATACAXACATACCTCTC-5′ | |
| C | 5′-AGAGGATATGXATGTATGGAGAG-3′ | –1 |
| A | 3′-TCTCCTATACAXACATACCTCTC-5′ | |
| B | 5′-AGAGGATATGTXTGTATGGAGAG-3′ | 0 |
| A | 3′-TCTCCTATACAXACATACCTCTC-5′ | |
| F | 5′-AGAGGATATGTAXGTATGGAGAG-3′ | +1 |
| A | 3′-TCTCCTATACAXACATACCTCTC-5′ | |
| G | 5′-AGAGGATATGTATGXATGGAGAG-3′ | +3 |
| A | 3′-TCTCCTATACAXACATACCTCTC-5′ | |
| H | 5′-AGAGGATATGTATGTAXGGAGAG-3′ | +5 |
| A | 3′-TCTCCTATACAXACATACCTCTC-5′ | |
| B | 5′-AGAGGATATGTXTGTATGGAGAG-3′ | Control (C) |
| AC | 3′-TCTCCTATACATACATACCTCTC-5′ |
The uracil residues were converted to AP sites as described in Materials and Methods and the cluster configurations are those of David-Cordonnier et al. (37), in which negative and positive numbers indicate that the lesions (here AP sites) lie 3′ and 5′ to each other, respectively, while 0 indicates that the AP sites are directly opposed (29).
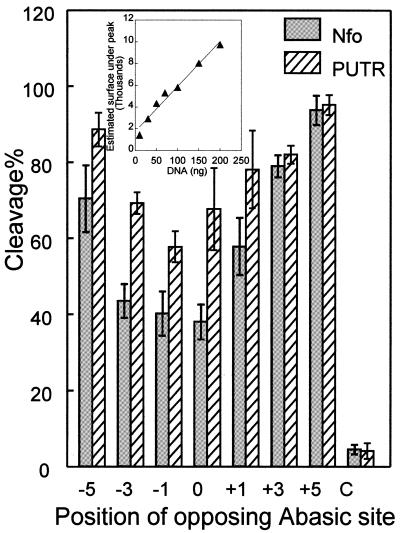
Figure 4.
Cleavage efficiency by Nfo protein and PUTR of the oligonucleotide substrates shown in Table 1 (see also Fig. 3), including the BAC control double-stranded oligo labeled C. Points, averages of three independent experiments; error bars, standard errors of the means. Cleavage efficiency is given as a percentage of conversion of intact oligonucleotides of 23 bp to smaller sized DNA fragments for the oligonucleotides shown in Table 1. (Inset) Linearity test for quantitation of silver stained oligonucleotide (CA, shown in Table 1). A least squares line (r = 0.98) is shown.
To prepare oligonucleotide substrates containing AP sites at specific positions (Table 1), the double-stranded oligonucleotides containing uracil (U) in one or both strands were incubated at 37°C with 1 U uracil-DNA glycosylase (UDG) (New England Biolabs, Beverly, MA) in 10 µl of STE for 30 min (23). The efficiency of formation of an AP site was >98% as determined from the strand breaks introduced by heating the duplexes to 95°C for 10 min in 0.5 M NaOH and PAGE as described above.
Treatment of oligonucleotides with Nfo protein or PUTR
The quantity of Nfo protein required for maximal cleavage of an abasic cluster (oligonucleotide HA in Table 1, +5) that is efficiently cut by this enzyme (24) was determined by titrating this oligonucleotide (150 ng) with increasing amounts of Nfo protein. More than 95% of the 23mer was cleaved by 7 ng enzyme in a 30 min incubation at 37°C in STE buffer containing 1 mM DTT, and this ratio of enzyme to oligonucleotide was chosen for all further experiments. Samples were then treated with proteinase K as described above.
For polyamine treatment, the UDG-treated DNA duplexes were treated with proteinase K and precipitated by addition of an equal volume of isopropanol, incubation at room temperature for 20 min and centrifugation at 14 000 g for 30 min. After centrifugation, the supernatant was removed and the precipitated DNA was resuspended in buffer P and equilibrated for 10 min. The DNA was then precipitated, the supernatant removed and the precipitated DNA chilled on ice, air dried for 5 h and resuspended in buffer P. Oligonucleotides were then incubated with polyamine at 37°C for 30 min. The reaction was stopped by addition of ice-cold 5 M NaCl to a final concentration of 0.5 M and subsequent incubation at 28°C for 30 min.
RESULTS
Abasic clusters and their measurement
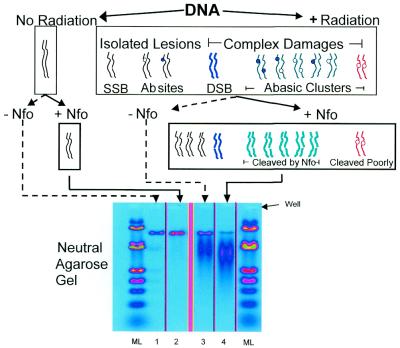
Ionizing radiation induces isolated lesions, shown here as strand breaks and abasic sites, and complex damages (e.g. DSBs and abasic clusters) in DNA. DSBs and bi-stranded clustered damages that can be converted to DSBs through cleavage by a lesion class-specific enzyme can be quantified by measuring the change in the average size of the DNA molecules in a sample (25). Figure 1 shows the principles of cluster recognition and quantitation using neutral gel electrophoresis and number average length analysis, in this case for DSBs and abasic clusters. Treatment of irradiated DNA with Nfo protein converts many of the abasic clusters to de novo DSBs (in addition to the DSBs produced directly by radiation). The figure also shows that Nfo protein does not cleave all cluster conformations at the same rate (24). Lane 4 shows that Nfo protein treatment of the DNA results in the appearance of additional, smaller molecules, the signature of clustered abasic damages. Abasic clusters recognized by Nfo protein are termed Nfo-‘abasic’ clusters to designate the agent recognizing them and its principal substrate (26). Sample DNAs and molecular length standards were electrophoresed in neutral agarose, stained with ethidium bromide and a quantitative image obtained. The yield of abasic clusters for the Nfo-treated irradiated DNA samples (lane 4) was calculated as described above (see equations 1–4 in Materials and Methods).
Figure 1.
Principles of identification and measurement of abasic clustered damages. DNA was exposed to ionizing radiation and a control sample was unirradiated. Radiation induces isolated lesions, shown here as regular abasic sites (open circles), oxidized abasic sites (filled circles) and SSBs, as well as complex bi-stranded damages containing closely spaced multiple lesions on opposing strands. Among the complex damages are DSBs and abasic clusters (one abasic site with another abasic site or an abasic site with a SSB). Many radiation-induced abasic clusters (green molecules) are converted to additional DSBs by Nfo protein cleavage; however, some configurations of abasic clusters are resistant to Nfo action (red molecule). Cleavage at isolated sites does not reduce the DNA size under neutral conditions. Since unirradiated DNA contains few if any clusters, Nfo protein treatment does not reduce the DNA size upon electrophoresis under neutral conditions (cf. lane 1 with lane 2). Radiation-induced DSBs (blue molecule) reduce the size of the DNA population (lane 3); Nfo protein cleaves at Nfo-‘abasic’ clusters, producing additional smaller daughter DNA molecules (lane 4). The panels in the gel are taken from an electronic image of a single gel; extraneous lanes were removed for clarity. The intensity of fluorescence of ethidium bromide bound to DNA is coded by a pseudo-color rainbow transformation, in which blue represents the lowest quantity of DNA and red the highest.
Polyamine cleavage of unirradiated and γ-irradiated T7 DNA
Polyamines can also cleave DNA at abasic sites (8,16), and we suspected that they might be able to cleave abasic clusters shown to be refractory to Nfo protein cleavage (24,27). To determine whether polyamines might provide improved recognition and cleavage at abasic clusters, we first tested the efficiency and specificity of cleavage of irradiated and unirradiated T7 DNA by SP, SPD and PUTR.
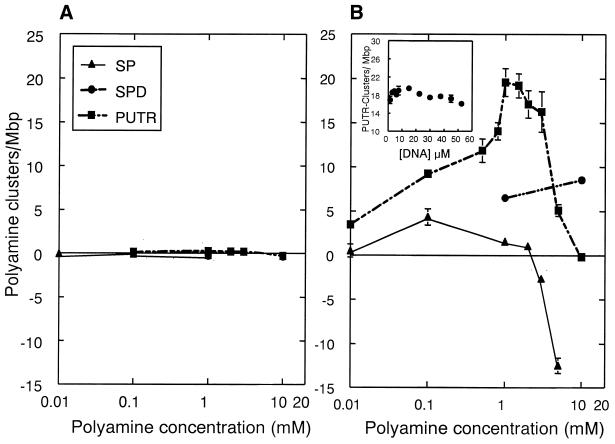
Figure 2 shows the results of such cleavage experiments using unirradiated T7 DNA (Fig. 2A) as well as DNA irradiated in 20 mM potassium phosphate buffer (2.5 Gy) (Fig. 2B). Figure 2A shows that these polyamines had little or no activity on unirradiated DNA. Figure 2B shows that SP recognized few bi-stranded AP sites, presumably abasic clusters (SP-‘abasic’ clusters). Further, at concentrations greater than ∼0.1 mM, it interfered with measurement of cleavage, giving apparently negative frequencies. This reflects formation of SP–DNA complexes with lower migration on the neutral gel that would (artifactually) be recorded as larger molecules. SPD revealed more abasic cluster sites, but only at relatively high concentrations.
Figure 2.
Cleavage of γ-irradiated DNA by polyamines. T7 DNA was unirradiated (A) or irradiated in 20 mM phosphate buffer with 2.5 Gy (B). DNA was incubated with increasing concentrations of SP, SPD or PUTR and the frequencies of cleavage at bi-stranded abasic clusters determined by gel electrophoresis and number average length analysis. Points are the averages of at least three independent experiments; error bars, standard errors of the means. In some cases, the errors are smaller than the data points. The lines were fitted to the points by eye. Negative values result from association of SP and PUTR with DNA, retarding its migration in the gel. (Inset) Cleavage by 1 mM PUTR of T7 DNA (irradiated with 2.5 Gy) as a function of DNA concentration (in base pairs).
PUTR was more effective in bi-stranded cluster cleavage (PUTR-‘abasic’ clusters), and at 1 mM produced ∼19 PUTR clusters/Mb for DNA irradiated with 2.5 Gy. The apparent decreased cleavage at high PUTR concentrations is probably the result of PUTR–DNA complexes, similar to those observed with SP. We also tested cleavage by 1 mM PUTR of irradiated T7 DNA at concentrations from 1 to 60 µM. The inset in Figure 2B shows that the level of cleavage by 1 mM PUTR is approximately constant over this range of DNA concentrations. We thus chose PUTR at 1 mM in buffer P to evaluate further as a potential abasic cluster-cleaving agent.
Cleavage efficiency of Nfo protein and putrescine at closely spaced bi-stranded abasic sites
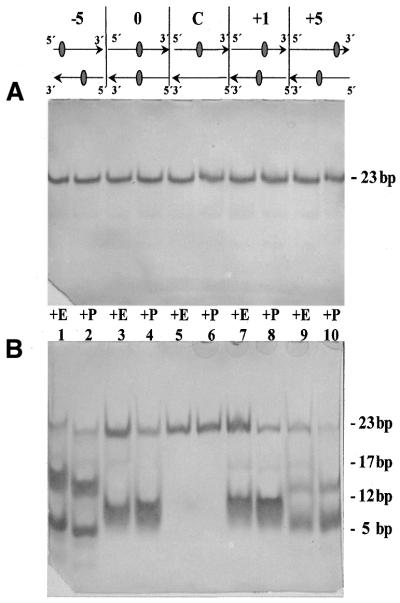
To test whether PUTR was cutting at abasic clusters, and to compare the cleavage efficiencies of PUTR and Nfo protein, we designed a series of oligonucleotides containing uracil residues at specific sites, plus one unsubstituted oligonucleotide. We annealed these oligonucleotides, then treated them with UDG to produce double-stranded species containing abasic clusters of different configurations, as well as a control containing a single abasic site on one strand. Table 1 shows the sequences of the resulting double-stranded oligonucleotides.
Figure 3 shows a diagram of five such oligonucleotides, plus electronic images of polyacrylamide gels containing oligonucleotides with U residues in specific bi-stranded cluster conformations (Fig. 3A) or abasic sites in the same positions (Fig. 3B). These oligonucleotides were treated with Nfo protein (E) or PUTR (P), then electrophoresed under non-denaturing conditions to measure the production of DSBs at abasic cluster sites. Figure 3A shows that neither Nfo protein nor PUTR produces DSBs in any of the uracil-containing oligonucleotides, whether they contain clustered (–5, 0, +1 or +5) or isolated (C) residues. Further, lanes 5 and 6 of Figure 3B show that neither Nfo protein nor PUTR produces a DSB in DNA containing only a single abasic site, evidence for the absence of any non-specific cleavage.
Figure 3.
Action of Nfo protein (E) or PUTR (P) on double-stranded oligonucleotides, containing one (C, lanes 5 and 6) or two (lanes 1–4 and 7–10) altered sites at the positions shown (see Table 1). In (A) the sites are uracil (U) residues; in (B) the sites are AP sites.
Cleavage by Nfo protein or PUTR at the bi-stranded abasic sites produces a DSB, and thus two smaller fragments. For the oligonucleotides containing +5 and –5 clusters, the resulting daughter fragments are sufficiently different in size that they are resolved on the 18% polyacrylamide gel, producing two readily distinguishable bands (Fig. 3B, lanes 1, 2, 9 and 10). In the very closely opposed clusters, the smaller fragments are not well resolved (lanes 3, 4, 7 and 8); however, cleavage results in the disappearance of the intact 23 bp species and production of a smaller product band.
We then quantified cleavage by Nfo and PUTR of the abasic clusters. Figure 4 shows that Nfo protein cleaves the –5, +3 and +5 abasic clusters efficiently, with lower activity for the –3, –1, 0 and +1 clusters. The cluster 0 (directly opposing AP sites) is the least efficiently incised, while the –1 and –3 clusters were incised approximately equally. PUTR cleavage of all the clusters was generally very efficient, approximately equalling that of Nfo protein for the +5 cluster and providing higher cleavage efficiency at all other cluster configurations. Even though it cleaved the 0 and –1 clusters somewhat less efficiently, the corresponding decrease in cleavage by Nfo protein was much greater. These results indicate that PUTR is a more effective cleavage agent for abasic clusters than is E.coli Nfo protein.
Abasic clusters in γ-irradiated T7 DNA
Using the conditions we developed for PUTR cleavage at abasic clusters, we tested cleavage by PUTR or Nfo protein of T7 DNA irradiated in phosphate buffer with 0–10 Gy of γ-rays. Figure 5 shows that the levels of abasic clusters for T7 DNA irradiated in phosphate buffer detected by the two agents increased linearly with dose. PUTR cleavage indicates the presence of about twice as many abasic clusters as did Nfo protein. The ratio (RP) of the slopes for the two agents for DNA irradiated in phosphate indicates an average of about 2 times higher cleavage for PUTR (Table 2).
Table 2. Yields (F) for cluster induction derived from linear fitting of the data in Figures 5 and 6.
| Cleavage agent | F (clusters/Mb/Gy) phosphate (P) | F (clusters/Mb/Gy) Tris–HCl (T) |
|---|---|---|
| Nfo protein | 4.27 ± 0.12 (FP, Nfo) | 0.032 ± 0.003 (FT, Nfo) |
| PUTR | 8.85 ± 0.72 (FP, PUTR) | 0.056 ± 0.002 (FT, PUTR) |
| Ratios Ri = Fi, putr/Fi, endo i = P or T | 2.07 ± 0.06 (RP = FP, PUTR/FP, Nfo) | 1.65 ± 0.17 (RT = FT, PUTR/FT, Nfo) |
The number of clusters/Mb (y) were fitted to dose (x) according to the equation y = a + F·x (least mean squares fit). The ratios (Ri) of the frequencies for cluster induction by γ-rays in T7 DNA, detected by Nfo protein or PUTR, irradiated in phosphate (RP) or 10 mM Tris–HCl (RT), respectively, are also shown. The ratios were found to be statistically different at the 10% level (Fisher’s z-test).
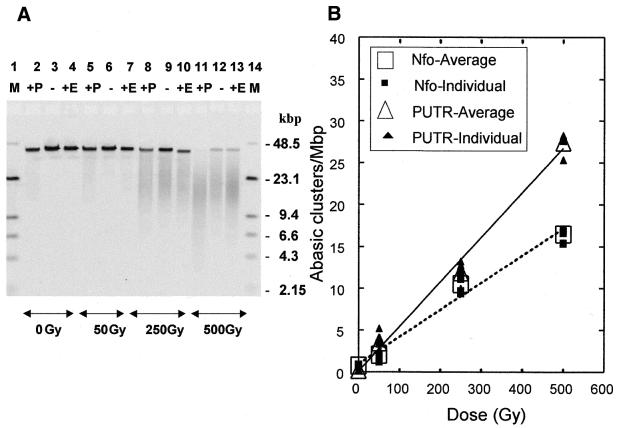
To test the impact of the environment of the DNA during irradiation on cluster production, we irradiated T7 DNA with γ-rays under moderate radical scavenging conditions, i.e. 10 mM Tris–HCl, pH 8.0 (1.5 × 107/s •OH scavenging capacity) and determined the levels of abasic clusters. An electronic image of a typical gel containing such DNA samples is shown in Figure 6A. The DNA profiles show that the size of the DNA molecules decreases with radiation dose (resulting from radiation-induced DSBs) and decreases further upon cleavage by PUTR (P) or Nfo protein (E). The higher level of fragmentation produced by PUTR compared with Nfo protein is especially evident in the 250 and 500 Gy samples. Figure 6B shows the results of quantitation of gels such as the one shown in Figure 6A. PUTR gives higher cleavage of the T7 DNA irradiated in the presence of Tris. The ratio (RT) of the slopes for the two agents for DNA irradiated in Tris indicates an average of about 1.7 times higher cleavage for PUTR (Table 2).
Figure 6.
Detection of abasic clusters by PUTR (P) and Nfo protein (E) in T7 DNA irradiated with γ-rays in 10 mM Tris. (A) Electronic image of a neutral electrophoretic gel used for analyzing abasic clusters induced in T7 DNA by γ-rays. Samples are in groups of three (+P, PUTR-treated; –, untreated; +E, Nfo-treated) for 0, 50, 250 and 500 Gy. Molecular length standard DNAs (M, lanes 1 and 14), λ and HindIII-digested λ DNA (48.5 and 23.1, 9.4, 6.6, 4.3, 2.15 Kb, respectively). Lanes are taken from an electronic image of a single gel and are rearranged in sequence for clarity. (B) Frequencies of abasic clusters detected by 1 mM PUTR and Nfo protein for T7 DNA irradiated with γ-rays in phosphate buffer. Lines, least squares linear fit of the average points (large open symbols); error bars, standard errors of the means (for fitting parameters see Table 2).
DISCUSSION
Theoretical and experimental studies indicate that ionizing radiation can produce complex DNA lesions, i.e. several base damages, AP sites or SSBs on opposing DNA strands within a few helical turns (25,28). Bi-stranded DNA damage clusters can be measured by cleavage at constituent lesion sites of a cluster by enzymes or other agents, producing a de novo DSB. Nfo protein, which recognizes and cleaves oxidized and regular AP sites, has provided a means of recognizing and quantifying abasic clusters, specifically Nfo-‘abasic’ clusters. However, recent data suggest that Nfo protein may not cleave efficiently at all abasic clusters, recognizing immediately opposing, –1 and –3 clusters particularly poorly (24). The data in Figures 3 and 4 show that Nfo protein cleaves +5 (>90% cleavage), +3 (∼80%), –5 (∼70%) and +1 (∼60%) clusters reasonably well, but fails to cleave more than half of the 0, –1 and –3 clusters. These results are in agreement with previous data suggesting that AP endonucleases (E.coli exonuclease III, E.coli endonuclease IV and human AP endonuclease 1, APE1) may be less efficient in the cleavage of some closely spaced lesions, e.g. an AP site opposite an AP site or a nucleotide gap (for a review see 29).
NMR data indicate that closely spaced AP sites on opposing strands (directly opposing or 1–3 bp apart) can create significant distortions in the structure of DNA near the AP sites (30), possibly leading to reduced recognition and/or cleavage by AP endonucleases. In the oligonucleotides shown in Table 1, the AP site opposes a purine, producing an intrahelical conformation of the orphan base, which may also contribute to decreased recognition by the enzyme (31). The asymmetrical inhibition for the 0, –1 and –3 clusters may be related to a decreased ability of Nfo protein to recognize and/or bind to the specific cluster. Crystallographic data on endonuclease IV recognition and cleavage 5′ to AP sites suggests that initial detection of AP sites is mediated by direct contacts between specific amino acid residues and the DNA minor groove immediately 3′ to the AP site (13). The fact that –1 and –3 clusters are also cleaved less efficiently may be related to this initial recognition and binding process in which the enzyme binds to DNA immediately 3′ of the AP site. In the case of the –1 and –3 clusters, the opposing AP site in strand A (or gap, if the AP site was already nicked) could distort the DNA and interfere with this interaction. Finally, the great difficulty encountered by the enzyme in cleaving the 0 cluster may be related to partial local denaturation around the cluster (32) and to the fact that endonuclease IV requires a double-stranded DNA substrate (33).
Since several polyamines have been shown to cleave AP sites, it seemed possible that such molecules might provide more complete cleavage of abasic clusters. Indeed, we found that PUTR provided the highest cleavage of irradiated DNA (Fig. 2A), while showing virtually no scission of unirradiated DNA (Fig. 2B). Examination of its cleavage of oligonucleotide duplexes containing specific abasic cluster configurations confirmed that it produced higher cleavage of all clusters (Fig. 4), especially those most resistant to Nfo protein. Neither Nfo protein nor PUTR, however, cleaved all clusters completely. Polyamine molecules bind preferentially to double-stranded DNA molecules (34) and it has been shown that the cleavage rate of PUTR at AP sites is reduced to about one-half when the DNA substrate is single stranded (35). The relatively low cleavage by PUTR of the 0, –1 and +1 clusters might reflect either a local helix distortion at these clusters and/or a type of steric hindrance.
The PUTR-‘abasic’ cluster data allow us to revise the previous estimates of the relative proportions of specific cluster frequencies (20). PUTR recognized approximately twice as many abasic clusters in DNA irradiated in phosphate as did Nfo protein (compare slopes FP,PUTR and FP,Nfo of the regression lines in Table 2). Thus, the revised estimates of abasic clusters as revealed by PUTR gives the ratio of complex damages of 1 DSB:2 oxyPur clusters:3 abasic clusters:0.5 oxyPyr cluster, with abasic clusters comprising some 46% of the complex damages and DSBs ∼15%.
In DNA irradiated in radioquenching Tris buffer, PUTR reveals about 1.7 times as many abasic clusters as does Nfo protein (Table 2). Previous values on abasic cluster detection by Nfo protein cleavage indicated an ∼136-fold decrease in T7 DNA irradiated in 10 mM Tris–HCl relative to that in phosphate (20). Similarly, our data show an ∼133-fold decrease. The DNA environment thus seems to affect both the absolute frequency and the ratios of the damage types. Using the improved estimates of abasic clusters, we calculate that the apparent ratio of complex damages is 1 DSB:1.4 oxyPur clusters:∼0.7 abasic cluster:0.75 oxyPyr cluster. Abasic clusters would thus comprise some 17% of the complex damages and DSBs ∼25%. The cellular milieu provides a highly radioquenching environment. Therefore, compared with previous measurements using Nfo protein to detect abasic clusters in human cells (26), we speculate that PUTR would also reveal a similarly higher level of abasic clusters, resulting in 1 DSB:1 oxyPur cluster:1.2 abasic clusters:0.9 oxyPyr cluster. In this case, abasic clusters would account for almost 30% of the radiation-induced complex damages and DSBs ∼25%.
Figure 4 shows that PUTR and Nfo protein cleavage differs principally at 0, –1, –3 and +1 clusters and to a lesser extent at –3 and +3 clusters. This suggests that the decrease in the differences in abasic cluster detection found for the two agents (see ratios RP for phosphate and RT for Tris) cleaving T7 DNA irradiated in the two environments reflects different cluster populations. More specifically, at low scavenging conditions, a higher level of more closely spaced clusters (0, 1 and 3 bp apart) or more closely spaced lesions within a cluster are formed, contributing to the observed differences between PUTR and Nfo protein (see Fig. 5). The closely spaced clusters may arise from a higher number and denser radical attacks on the DNA molecule compared with the Tris environment.
The major inhibition of Nfo protein activity by the presence of an opposing AP site (or AP site opposite a gap) within a very short base pair region (0–3 bp apart) may play a critical role in cell survival during the repair of clustered damages. Since similar inhibition was also observed with other AP endonucleases, e.g. E.coli exonuclease III and human APE1 (11,36), we suggest that this clustered damage is repaired by sequential processing of its constituent lesions, if repaired at all by this pathway. In this way, the cell minimizes the conversion of abasic clustered damages to DSBs, which may be lethal for the cell.
In this study we tested the hypothesis that closely spaced bi-stranded abasic clusters in DNA may be resistant to Nfo protein cleavage at the lesions. We also tested the ability of polyamines to cleave abasic clusters. By using PUTR, the most efficient polyamine in our assays, we detected significant differences between PUTR and Nfo abasic cluster cleavage at different cluster configurations. We then used this simple and more efficient technique to measure abasic clusters in irradiated DNA. Comparison of Nfo protein and PUTR cleavage may prove to be a powerful tool in the detection of damage clusters and identification of their configurations in cells exposed to ionizing radiation or radiomimetic drugs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Peter Guida for T7 DNA and Drs R. Setlow, J. C. Sutherland and C. de los Santos for helpful discussions and critical reading of the manuscript. We also thank Dr K. Thompson for valuable suggestions on the statistical evaluation of the data, Ms D. Monteleone for her help in gel imaging and quantitative analysis and R. Sautkulis for his help in dosimetry and irradiations. This research was supported by the Low Dose Program of the Office of Biological and Environmental Research of the US Department of Energy and by a grant from the National Institutes of Health (R01 CA 86897) to B.M.S.
REFERENCES
- 1.von Sonntag C. (1987) The Chemical Basis of Radiation Biology. Taylor & Francis, London.
- 2.Haring M., Rudiger,H., Demple,B., Boiteux,S. and Epe,B. (1994) Recognition of oxidized abasic sites by repair endonucleases. Nucleic Acids Res., 22, 2010–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward J.F. (1994) The complexity of DNA damage: relevance to biological consequences. Int. J. Radiat. Biol., 66, 427–432. [DOI] [PubMed] [Google Scholar]
- 4.Goodhead D.T. (1994) Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int. J. Radiat. Biol., 65, 7–17. [DOI] [PubMed] [Google Scholar]
- 5.Prise K.M., Pullar,C.H. and Michael,B.D. (1999) A study of endonuclease III-sensitive sites in irradiated DNA: detection of alpha-particle-induced oxidative damage. Carcinogenesis, 20, 905–909. [DOI] [PubMed] [Google Scholar]
- 6.Milligan J.R., Aguilera,J.A., Nguyen,T.T., Paglinawan,R.A. and Ward,J.F. (2000) DNA strand-break yields after post-irradiation incubation with base excision repair endonucleases implicate hydroxyl radical pairs in double-strand break formation. Int. J. Radiat. Biol., 76, 1475–1483. [DOI] [PubMed] [Google Scholar]
- 7.Sutherland B., Bennett,P.V., Sidorkina,O. and Laval,J. (2000) DNA damage clusters induced by ionizing radiation in isolated DNA and in human cells. Proc. Natl Acad. Sci. USA, 97, 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Povirk L.F. and Houlgrave,C.W. (1988) Effect of apurinic/apyrimidinic endonucleases and polyamines on DNA treated with bleomycin and neocarzinostatin: specific formation and cleavage at closely opposed lesions in complementary strands. Biochemistry, 27, 3850–3857. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhry M.A. and Weinfeld,M. (1997) Reactivity of human apurinic/apyrimidinic endonuclease and Escherichia coli exonuclease III with bistranded abasic sites in DNA. J. Biol. Chem., 272, 15650–15655. [DOI] [PubMed] [Google Scholar]
- 10.Harrison L., Hatahet,Z. and Wallace,S. (1999) In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J. Mol. Biol., 290, 667–684. [DOI] [PubMed] [Google Scholar]
- 11.David-Cordonnier M.H., Cunniffe,S.M.T., Hickson,I.D. and O’Neill,P. (2002) Efficiency of incision of an AP site within clustered DNA damage by the major human AP endonuclease. Biochemistry, 41, 634–642. [DOI] [PubMed] [Google Scholar]
- 12.Demple B., Johnson,A. and Fung,D. (1986) Exonuclease III and endonuclease IV remove 3′ blocks from DNA synthesis primers in H2O2-damaged Escherichia coli. Proc. Natl Acad. Sci. USA, 83, 7731–7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosfield D.J., Guan,Y., Haas,B.J., Cunningham,R.P. and Tainer,J.A. (1999) Structure of the DNA repair enzyme Endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell, 98, 397–408. [DOI] [PubMed] [Google Scholar]
- 14.Ischenko A.A. and Saparbaev,M.K. (2002) Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature, 415, 183–187. [DOI] [PubMed] [Google Scholar]
- 15.Dizdaroglu M., Burgess,S.M., Jaruga,P., Hazra,T.K., Rodriguez,H. and Lloyd,R.S. (2001) Substrate specificity and excision kinetics of Escherichia coli endonuclease VIII (Nei) for modified bases in DNA damaged by free radicals. Biochemistry, 40, 12150–12156. [DOI] [PubMed] [Google Scholar]
- 16.Lindahl T. and Andersson,A. (1972) Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry, 11, 3618–3623. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura J. and Swenberg,A.J. (1999) Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res., 59, 2522–2526. [PubMed] [Google Scholar]
- 18.Steigher J.R. and Povirk,L.F. (1990) Effect of in vitro cleavage of apurinic/apyrimidinic sites on bleomycin-induced mutagenesis of repackaged lambda phage. Mutat. Res., 240, 93–100. [DOI] [PubMed] [Google Scholar]
- 19.Levin J.D., Johnson,A.W. and Demple,B. (1988) Homogeneous Escherichia coli Endonuclease IV. Characterization of an enzyme that recognizes oxidative damage in DNA. J. Biol. Chem., 263, 8066–8071. [PubMed] [Google Scholar]
- 20.Sutherland B.M., Bennett,P.V., Weinert,E., Sidorkina,O. and Laval,J. (2001) Frequencies and relative levels of clustered damages in DNA exposed to gamma rays in radioquenching vs. nonradioquenching conditions. Environ. Mol. Mutagen., 38, 159–165. [DOI] [PubMed] [Google Scholar]
- 21.Sutherland J.C., Monteleone,D.C., Trunk,J.G., Bennett,P.V. and Sutherland,B.M. (2001) Quantifying DNA damage by gel electrophoresis, electronic imaging and number average length analysis. Electrophoresis, 22, 843–854. [DOI] [PubMed] [Google Scholar]
- 22.Sutherland J.C., Monteleone,D.C., Mugavero,J.H. and Trunk,J. (1987) Unidirectional pulsed-field electrophoresis of single- and double-stranded DNA in agarose gels: analytical expression relating mobility and molecular length and their application in the measurement of strand breaks. Anal. Biochem., 162, 511–520. [DOI] [PubMed] [Google Scholar]
- 23.David-Cordonnier M.H., Boiteux,S. and and O’Neill,P. (2001) Efficiency of excision of 8-oxo-guanine within DNA clustered damage by XRS5 nuclear extracts and purified human OGG1 protein. Biochemistry, 40, 11811–11818. [DOI] [PubMed] [Google Scholar]
- 24.Rasouli-Nia A., Chaudhry,M.A. and Weinfeld,M. (2001) 13th Symposium on Microdosimetry. Stresa, Italy.
- 25.Sutherland B.M., Bennett,P.V., Sidorkina,O. and Laval,J. (2000) Clustered damages and total lesions induced in DNA by ionizing radiation: oxidized bases and strand breaks. Biochemistry, 39, 8026–8031. [DOI] [PubMed] [Google Scholar]
- 26.Sutherland B.M., Bennett,P.V., Sutherland,J.C. and Laval,J. (2002) Clustered DNA damages induced by X-rays in human cells. Radiat. Res. , 157,611–616. [DOI] [PubMed] [Google Scholar]
- 27.Steighner R.J. and Povirk,L.F. (1990) Bleomycin-induced DNA lesions at mutational hot spots: implications for the mechanism of double-strand cleavage. Proc. Natl Acad. Sci. USA, 87, 8350–8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikjoo H., O’Neill,P., Wilson,E.W. and Goodhead,D. (2001) Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat. Res., 156, 577–583. [DOI] [PubMed] [Google Scholar]
- 29.Weinfeld M., Rasouli-Nia,A., Chaudhry,M.A. and Britten,R.A. (2001) Response of base excision repair enzymes to complex DNA lesions. Radiat. Res., 156, 584–589. [DOI] [PubMed] [Google Scholar]
- 30.Lin Z. and de los Santos,C. (2001) NMR characterization of clustered bistrand abasic site lesions: effect of orientation on their solution structure. J. Mol. Biol., 308, 341–352. [DOI] [PubMed] [Google Scholar]
- 31.Lhomme J., Constant,J.-F. and Demeunynck,M. (2000) Abasic DNA structure, reactivity and recognition. Biopolymers, 52, 65–83. [DOI] [PubMed] [Google Scholar]
- 32.Gelfand C.A., Plum,G.E., Grollman,A.P., Johnson,F. and Breslauer,K.J. (1996) The impact of a bistrand abasic lesion on DNA duplex properties. Biopolymers, 38, 439–445. [DOI] [PubMed] [Google Scholar]
- 33.Lindahl T. (1982) DNA repair enzymes. Annu. Rev. Biochem., 51, 61–87. [DOI] [PubMed] [Google Scholar]
- 34.Morgan E.J., Blankenship,W.J. and Mathews,R.H. (1986) Association constants for the interaction of double-stranded and single-stranded DNA with spermine, spermidine, putrescine, diaminopropane, N1- and N8-acetylspermidine and magnesium: determination from analysis of the broadening of thermal denaturation curves. Arch. Biochem. Biophys., 246, 225–232. [DOI] [PubMed] [Google Scholar]
- 35.Male R., Fosse,V.M. and Kleppe,K. (1982) Polyamine-induced hydrolysis of apurinic sites in DNA and nucleosomes. Nucleic Acids Res., 10, 6305–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrison L., Hatahet,Z., Purmal,A.A. and Wallace,S.S. (1998) Multiply damaged sites in DNA: interactions with Escherichia coli endonucleases III and VIII. Nucleic Acids Res., 26, 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David-Cordonnier M.H., Laval,J. and O’Neill,P. (2000) Clustered DNA damage: influence on damage excision by XRS5 nuclear extracts and Escherichia coli Nth and Fpg proteins. J. Biol. Chem., 275, 11865–11873. [DOI] [PubMed] [Google Scholar]