Abstract
Riboswitches are RNAs that recognize ligands and regulate gene expression. They are typically located in the untranslated region of bacterial messenger RNA and consist of an aptamer and an expression platform. In this study, we examine the folding pathway of the Vc2 (Vibrio cholerae) riboswitch aptamer domain, which targets the bacterial secondary messenger cyclic-di-GMP. We demonstrated by nuclear magnetic resonance (NMR) and isothermal titration calorimetry that the stable folding of the Vc2 riboswitch requires an adequate supply of Mg2+, Na+ and K+ ions. We found that Mg2+ has a crucial role in the pre-folding of the aptamer, while K+ is essential for establishing the long-range G-C interactions and stabilizing the ligand binding pocket. Precise imino proton assignments revealed the progressive folding of the aptamer. The results indicate that the P2 helix consists of weaker and more dynamic base pairs compared to the P1b helix, allowing the rearrangement of the base pairs in the P2 helix during the folding process required for effective ligand recognition. This study provides a profound understanding riboswitch architecture and dynamics at the atomic level under physiological conditions as well as structural information on apo-state RNA.
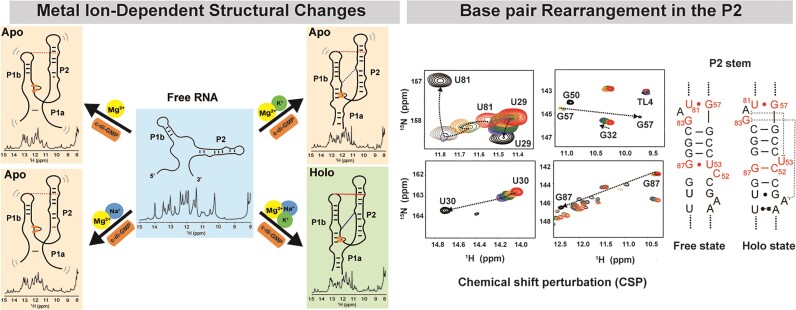
Graphical Abstract
Graphical Abstract.
Introduction
RNAs perform various biological functions. Many biologically active RNAs can adopt complex 3D structures under the influence of cofactors, such as ions and small molecules, to function in the cellular environment (1). Despite being composed of four chemically similar building blocks, RNAs can fold into complex structures and recognize specific small molecules to perform diverse functions, as evidenced by the naturally occurring metabolite-sensing riboswitches (2).
A riboswitch is a structural RNA motif that is commonly found in the 5′-untranslated region (UTR) of messenger RNA. It regulates protein expression upon binding to small molecules. More than 56 distinct riboswitch classes have been experimentally validated, and the ligands they sense include a wide variety of biologically relevant elemental ions and fundamental metabolites, many of which are derived from RNA nucleotides or their precursors (3–6). A riboswitch consists of two essential elements: the aptamer domain and the expression platform. The aptamer domain specifically binds to a ligand, whereas the expression platform is responsible for controlling gene expression through its ability to switch conformation between two different secondary structures in response to ligand binding at the aptamer (3,7,8). Understanding the structure of ligand-bound aptamers is crucial, but equally important is comprehending how the conformational changes in the aptamer and expression platform can facilitate gene regulation. Therefore, it is essential to examine both the ligand-bound and ligand-free states of riboswitches (9–11).
While many studies have focused on the structures of the aptamer-ligand complexes of riboswitches, there have been only a few studies on the structure of the apo state of the riboswitch, which is the ligand sensing phase (12). One reason is that the dynamics and flexibility make it difficult to capture its structure. In addition, the structure of the apo state of riboswitches may be less stable compared with that of the ligand-bound state. Mg2+ ions are shown to be crucial for the pre-organization of aptamer structures in SAM-II and other riboswitches, as they alter RNA structure to form primitive ligand binding pockets and promote long-range helical interactions (13–15). This process, crucial for efficient ligand binding, suggesting the importance of investigation of both apo and holo states for a comprehensive understanding of riboswitch folding dynamics.
Cyclic di-GMP [bis-(3′-5′)-cyclic dimeric guanosine monophosphate], class I riboswitches specifically bind c-di-GMP (16). These riboswitches are present in pathogens, such as Clostridium difficile, Vibrio cholerae and Bacillus anthracis. Vibrio cholerae encodes two riboswitches (Vc1 and Vc2) that directly bind and respond to changes in the intracellular concentration of c-di-GMP (17). Vc1 is encoded by the 5′-UTR of the putative adhesin, gbpA. The binding of c-di-GMP to Vc1 enhances the production of GbpA, making this riboswitch an ‘on’ switch. Vc2, which has extensive sequence identity with Vc1, is encoded by the 5′-UTR of the tfoY gene and functions as an ‘off’ switch, because it represses TfoY production (16–18). In 2009, two crystal structures of the Vc2 c-di-GMP-I riboswitch aptamer domain were solved, and functional and structural analyses were conducted (19,20). The ligand-bound formation of the P1 helix in the aptamer domain induces the repression of translation, and Mg2+ ions play an important role in promoting ligand binding. Molecular dynamic (MD) simulation studies have shown that Mg2+ ion binding increases the size of the c‐di‐GMP binding site pocket, which promotes subsequent c‐di‐GMP molecule binding (21). Gel shift assay, single molecule fluorescence resonance energy transfer (smFRET) and isothermal titration calorimetry (ITC) analyses of site-specific mutants that disrupt interhelical long-range interactions through base pairing and/or stacking tertiary interactions revealed that these tertiary interactions are not only required for binding of c-di-GMP, but affect the structure of RNA in the absence of ligand (22,23). However, these results provide limited information and are still insufficient for describing the RNA conformational changes at the atomic level. Furthermore, only a few X-ray structures of ligand-free riboswitches have been successfully determined, which is important for understanding the ligand recognition pathway; however, crystal packing stabilizes only one conformation from the ensemble of conformers accessible to the ligand-free aptamer in solution (24,25). Solution NMR spectroscopy is a powerful tool for the direct and unambiguous elucidation of hydrogen bonding patterns in nucleic acids and readily resolves canonical and non-Watson–Crick base pairing patterns associated with RNA folding (26). In addition, NMR has the advantage of studying RNA molecules in a biologically relevant context under near-physiological conditions, such as temperature, pH and ionic and metabolite concentrations (27,28). There have been combined NMR and smFRET studies on adenine riboswitch folding, in which both methods play complementary roles in providing different information on RNA conformational changes (29). Here, we provide accurate RNA conformational states by comparing our NMR results with those previously reported for c-di-GMP riboswitches.
In this study, we conducted NMR experiments on the full-length 85-nucleotide (nt) long c-di-GMP Vc2 riboswitch from V. cholerae to probe the conformational changes of its three states, ‘free’, ‘apo’ and ‘holo’ states, at varying concentrations of Na+, K+, Mg2+ and c-di-GMP. By observing changes in secondary structures while monitoring base pair formation, the folding process of the riboswitch aptamer was accurately chased at the atomic level. In addition, the role of tertiary interactions in the stability and folding of RNA was further characterized using site-specific mutants by NMR (30) and ITC experiments (31). Our results indicate that the aptamer RNA was folded into a preorganized structure by Mg2+-induced long-range interactions and further stabilized by forming a compact and rigid tertiary structure upon binding with the c-di-GMP ligand. The conformational dynamics and detailed aptamer folding pathway are described. Our NMR experiments have enabled us to observe the folding process of large-sized riboswitches in detail. This method allowed us to follow the rearrangement of base pairs and to quantify multiple conformations of the aptamer in its apo state.
Materials and methods
RNA preparation
For NMR studies, non-labeled and 13C, 15N-labeled full-length aptamer RNA (85 mer, nts 10–99), truncated P1b (nts 21–46) and P2 (nts 50–89) were designed. A single GC base pair was added at the terminus of both truncated P1b and P2, respectively. RNAs <82 nucleotides were prepared by in vitro transcription using a His6-tagged P266L T7 RNA polymerase mutant (32) with synthetic DNA templates (IDT). The RNA transcription reaction was performed in 40 mM Tris-HCl (pH 8.0), 1 mM spermidine, 0.01% Triton X-100, 2.5 mM dithiothreitol (DTT), 4 mM of each ribonucleotide triphosphate (rNTP), T7 RNA polymerase and 25 mM MgCl2 at 37 °C for 5 h (33,34). The synthesized RNAs were precipitated with ethanol, purified by 15% denaturing polyacrylamidegel electrophoresis, electroeluted (Elutrap, Whatman) and subjected to anion exchange chromatography (GE Healthcare). Fractions containing purified RNAs were dialyzed against water using an Amicon filtration system (Millipore). The RNAs were diluted in 10 mM sodium phosphate (NaPi) buffer (pH 6.5), heated to 95°C, cooled on ice and concentrated to 0.5–1 mM. For full-length aptamer RNA synthesis, a DNA template was prepared using the Giga Kit (Qiagen), and RNA was transcribed as described above. Non-labeled and 13C- and 15N-labeled c-di-GMP ligands were purchased (Biolog).
NMR spectroscopy
The NMR experiments were performed at 288 K or 298 K on a Bruker 800 MHz spectrometer (Avance III HD) equipped with a cryogenic Z-gradient triple resonance TCI probe. NMR samples of ∼500 μM were prepared in 10 mM NaPi, pH 6.5, and 10% D2O. The 1D proton, 2D 1H-1H NOESY, 2D 1H-15N HSQC and 2D 1H15N15N COSY experiments were conducted (35–37). The NMR spectra were processed using TopSpin 3.5pl7 software (Bruker), and the resulting data were analyzed using NMRFAM-SPARKY (38). The hydrogen exchange rates (kex) of the imino protons of free, apo and holo states of the RNA were measured by a water magnetization transfer experiment (39,40). The apparent longitudinal relaxation rate constants (R1a = 1/T1a) of the imino protons were determined by semi-selective inversion recovery 1D NMR experiments. Peaks were identified using Mnova software (Mestrelab), and signal intensities were normalized and fitted using the equation, 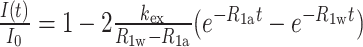 where I0 and I(t) are the peak intensities of the imino proton in the water magnetization transfer experiments at times 0 and t, respectively, and R1w is the apparent longitudinal relaxation rate constants for the water. To obtain chemical shift perturbation (CSP) values, 2D 1H-15N HSQC experiments were run on 0.5 mM 13C, 15N-labeled c-di-GMP riboswitch aptamer RNA at varying concentrations of MgCl2 and c-di-GMP. CSPs of imino protons were calculated using the equation,
where I0 and I(t) are the peak intensities of the imino proton in the water magnetization transfer experiments at times 0 and t, respectively, and R1w is the apparent longitudinal relaxation rate constants for the water. To obtain chemical shift perturbation (CSP) values, 2D 1H-15N HSQC experiments were run on 0.5 mM 13C, 15N-labeled c-di-GMP riboswitch aptamer RNA at varying concentrations of MgCl2 and c-di-GMP. CSPs of imino protons were calculated using the equation,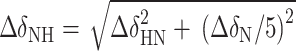 , where
, where 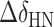 and
and 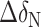 are the chemical shift differences in the 1H and 15N dimensions, respectively (41). The chemical shift values for 15N and 1H of the wil-type aptamer and the c-di-GMP ligand were listed in Supplementary Table S1.
are the chemical shift differences in the 1H and 15N dimensions, respectively (41). The chemical shift values for 15N and 1H of the wil-type aptamer and the c-di-GMP ligand were listed in Supplementary Table S1.
ITC experiments
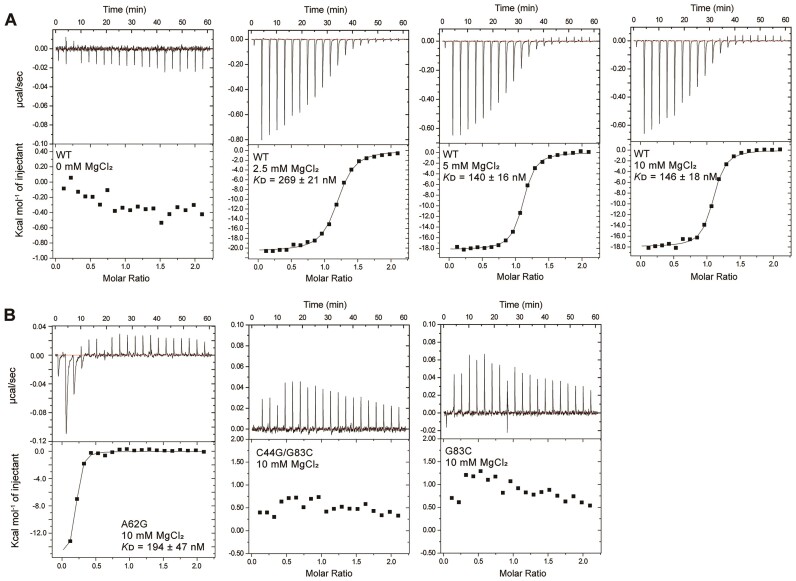
ITC experiments were performed using an ITC 200 instrument (MicroCal, Inc.), and the data were analyzed using ORIGIN 7.0. ITC experiments of wild-type (wt) aptamer RNA or its mutants and c-di-GMP were performed in 10 mM NaPi, pH 6.5, containing 30 mM NaCl, 30 mM KCl and 0–10 mM MgCl2 at 298 K. RNA (20 μM) was placed in the cell and c-di-GMP (200 μM) in the syringe. To prevent oligomerization of c-di-GMP induced by K+, c-di-GMP was incubated for 2 h at 60°C and cooled to room temperature before the experiments. Titrations were performed using 19 injections of 2 μL each with 180-s intervals. The experimental raw data were fitted using a one-site binding model. Titration data were analyzed using a nonlinear least-square curve-fitting algorithm with the following three floating variables: binding stoichiometry, binding constant (KD) and enthalpy change during the interaction. For the mutants (G83C, C44G/G83C, A62G), experiments were performed only under the condition of 10 mM MgCl2. Specifically, for the A62G mutant, 10 mM MgCl2 was added, and ligand binding experiments were conducted immediately afterward. The RNA was then stored at room temperature for 24 h, with experiments repeated over several days at intervals.
Results
Secondary structure of the aptamer domain of the Vc2 riboswitch
Aptamer construct design
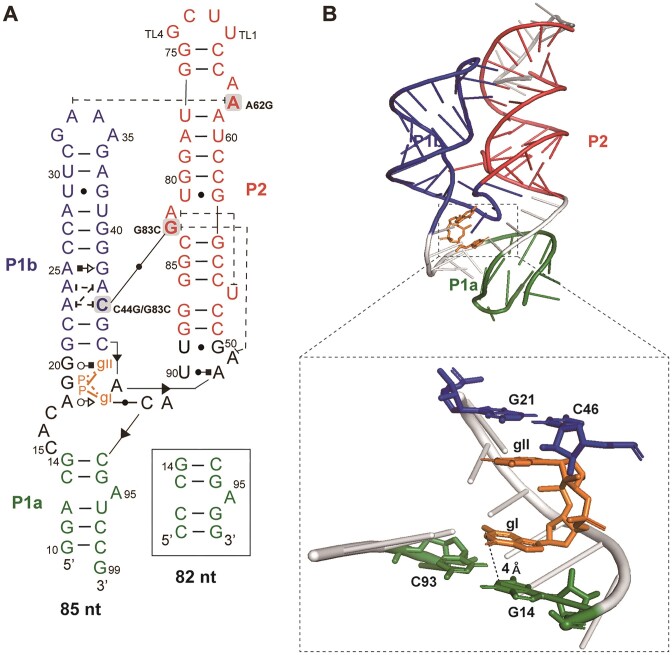
The c-di-GMP riboswitch aptamer construct employed in the NMR study included the full sequence (nts 10–99) (Figure 1A). To increase stability and in vitro transcription efficiency, two G-C base pairs and a single G were added to the terminus of the P1a stem, with a stable UUCG tetraloop (42,43) used to seal the P2 apical stem in the construct. Because the size of the 85-nt long aptamer and its intricate tertiary structure poses a formidable challenge for NMR resonance assignment, truncated P1b and P2 constructs were strategically designed (Supplementary Figure S1). The assignments obtained from these constructs were used for the full-length aptamer.
Figure 1.
(A) Secondary structure of a fully folded c-di-GMP riboswitch aptamer from V. cholera. The structure of 85-nt RNA is shown, and for the shorter 82 nt RNA, the P1a region (green) is replaced as indicated in the box. Nucleotides were added to the terminus of the P1a stem to increase the stability and transcription efficiency, and a stable UUCG tetraloop was added to P2 in the NMR construct. The point mutations of C44G/G83C, G83C and A62G are indicated. (B) The tertiary structure of the c-di-GMP riboswitch aptamer (PDB ID: 3MXH). The ligand binding pocket is zoomed in and the residues involved in ligand binding are labeled. The residues in P1a, P1b, P2 and the ligand are colored in green, blue, red and orange, respectively.
Free and holo state RNA secondary structures
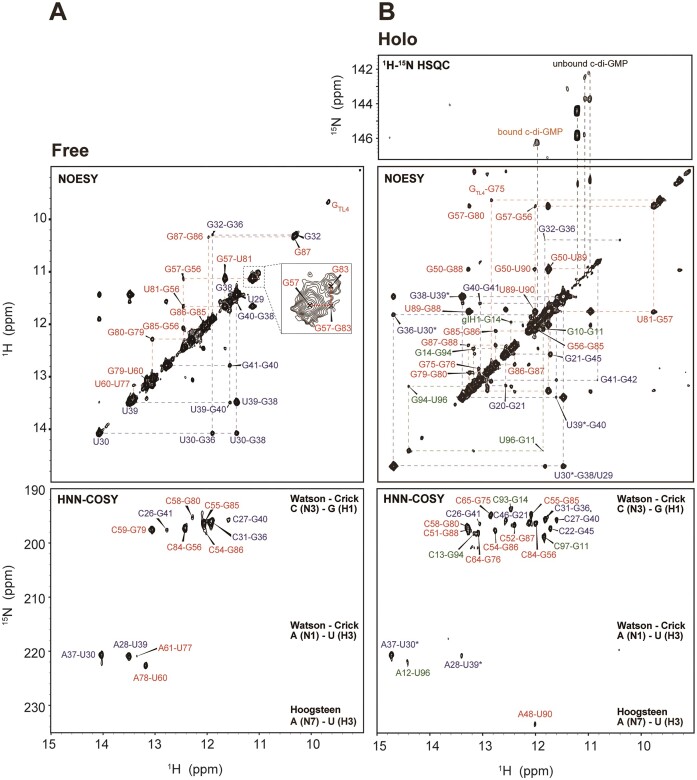
The structural changes within the aptamer were scrutinized by monitoring the signals from the imino protons of G and U, which were observed during the formation of base pairs by hydrogen bonding. Therefore, NMR assignments were required for all detectable imino protons under diverse conditions, such as varying concentrations of Na+, K+, Mg2+ and ligands. Typically, the Watson–Crick and G•U wobble base pairs were identified through the NOE cross peaks occurring between neighboring imino protons of the Us and/or Gs in the 2D 1H-1H NOESY spectra, with assignments being confirmed through an analysis of the 2D 1H15N15N COSY spectra (Figure 2). These experiments indicated that the ‘truncated’ P1b stem exhibits sequential imino-to-imino cross peaks arising from the C31-G36 to the C26-G41 base pairs. Likewise, the ‘truncated’ P2 stem also showed sequential imino-to-imino cross peaks from the C65-G75 to the G50-U89 base pairs (Supplementary Figure S1A and B). Based on these assignments, the base pairs and bulge nucleotides in the secondary structures of the truncated P1b and P2 were identified. A particularly noteworthy base pair to highlight is U53•G86 in the ‘truncated’ P2, which undergoes dramatic base pair alterations to U53•G87 and C52-G87 during ligand recognition (Supplementary Figure S1C). This is discussed in further detail below.
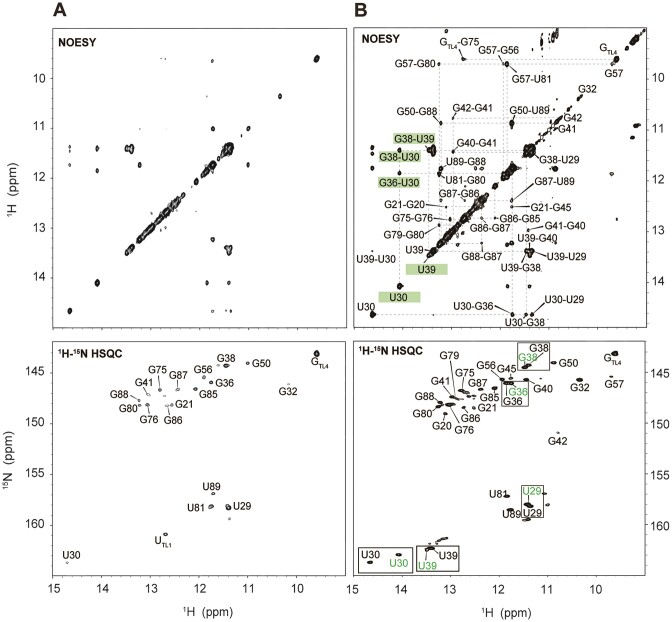
Figure 2.
2D 1H-1H NOESY and 2D 1H15N15N COSY spectra of (A) free and (B) holo state of c-di-GMP riboswitch aptamer with full assignments. Assignments of bound and unbound c-di-GMP were performed using 15N-HSQC with 13C/15N-labeled c-di-GMP. The free state spectra were measured in 10 mM sodium phosphate buffer (NaPi) at pH 6.5, while the holo state spectra were measured in conditions of 30 mM Na+, 30 mM K+, 2.5 mM Mg2+ (saturated) and ligand present. All experiments were conducted at 288 K. The residues in P1a, P1b, P2 and the ligand are colored in green, blue, red and orange, respectively.
To determine the folding process of the full-length aptamer under various conditions of Na+, K+, Mg2+ and ligands through changes in the NMR resonances of imino protons (i.e. base pair alterations), resonance assignments for the aptamer were initially conducted in two states: the ‘free’ state, which contains only a phosphate buffer and lacks additional Na+, K+, Mg2+ and ligand, and the ‘holo’ state, in which all the components are present, resulting in a fully folded structure. In the free state, only 19 imino protons from the Us and Gs were detectable in the NMR spectrum (Figure 2A and Supplementary Figure S2). The structural configuration of P1b in the full-length aptamer closely resembled that of truncated P1b, as supported by the identification of base pairs ranging from C31-G36 to C26-G41, which shared almost identical imino proton resonances with the truncated P1b. However, P2 from the full-length aptamer forms a secondary structure different from that of truncated P2. Base pairs from U77-A61 to G86-C54 were observed, but the terminal base pairs of P2 (G75-C65, G76-C64, C51-G88 and G50•U89), all present in truncated P2, were not formed. Notably, the G87 resonance exhibited an up-field shift to the non-canonical G•U wobble pair (∼10 ppm) region, indicating base pairing with neighboring U53. Because of the different secondary structures, the spectra for truncated P2 and P2 of the full-length RNA did not align and resulted in distinct chemical shift values, although they showed a similar number of assigned imino protons (Figure 2A and Supplementary Figure S1B). No imino protons in the P1a stem were observed in the free state. In summary, the riboswitch aptamer in its free state only forms partially folded P1b and P2 structures, with the core ligand binding site remaining unstructured, which precludes the formation of the P1a helix.
In the presence of 30 mM Na+, 30 mM K+, 2.5 mM Mg2+ and one equivalent of the ligand required to form a completely folded aptamer (19,44), the remaining imino protons of aptamer were assigned. In the fully folded aptamer, as confirmed by its crystal structure, 23 Watson–Crick base pairs, four G•U wobble pairs, one Hoogsteen base pair and various non-canonical base pairs were observed. The 2D 1H-1H NOESY spectrum revealed assignments for 33 out of 38 G or U imino protons, excluding TL1 and TL2 in the UUCG tetraloop of P2, U53, U60 and U77 in the P2 stem, and G19 of P1b. In the 2D 1H15N15N COSY spectrum, a Hoogsteen base pair between A48 and U90 was evident (Figure 2B). In the crystal structure (19,20) A25 and G42 engage in Hoogsteen–sugar-edge hydrogen bonding (45). A distinct resonance observed at approximately 151 ppm in the 2D 1H-15N HSQC spectrum was notably downfield shifted compared with the typical resonances of Watson–Crick or wobble base pairs. This resonance is attributed to the imino proton of G42 (G42N1H1), which likely results from hydrogen bonding with the phosphate oxygen of A24 (Supplementary Figure S2B). Compared with the free state, in the holo state in solution, the aptamer appears to form most of the base pairs observed in the crystal structure (Figure 1B), which suggests a nearly identical secondary structure. This includes base pairs near the ligand binding site in P1b (G21-C46, G45-C22), in P2 (G50•U89, G57•U81, G79-C59, G80-C58, G88-C51, U90-A48) and in P1a (G11-C97, G14-C93, G94-C13, U96-A12) (Figure 1A). The absence of imino protons corresponding to the A61-U77 and U60-A78 base pairs seems to be influenced by the interhelical base stacking between A62 and A33.
Resonance assignment of c-di-GMP
RNA-bound and free c-di-GMP resonances in 2D 1H-1H NOESY spectra were identified. In the presence of K+, c-di-GMP assembles into complex structures, such as quadruplexes (27). Therefore, we examined K+-induced structural changes in c-di-GMP. Without K+, NMR signals for H8, NH2 and H1′ of c-di-GMP were detected at approximately 8, 6.1 and 5.8 ppm, respectively. The absence of any imino proton signal would suggest a monomeric state of c-di-GMP. In contrast, in the presence of K+, additional H1 imino protons were observed, indicating the formation of ligand oligomers (Supplementary Figure S3). Next, to identify the resonances specific to RNA-bound c-di-GMP, we compared the 2D 1H-1H NOESY (Nuclear Overhauser Effect Spectroscopy) spectrum of ligand-bound, non-labeled RNA with the 2D 1H-15N HSQC (Heteronuclear Single Quantum Coherence) spectrum of 13C,15N fully labeled c-di-GMP-bound non-labeled RNA. The imino proton of RNA-bound c-di-GMP appeared as a new peak at 12 ppm, which showed a NOE (Nuclear Overhauser Effect) correlation with G14H1 (Figure 2B). Within the crystal structure, G14H1 and gI-H1 were separated by ∼4 Å, a proximity sufficient for observing NOE (Figure 1B). This unique signal was attributed to gI-H1 from c-di-GMP, which formed a base pair with C92, as evident in the crystal structure. Notably, the gII-H1 imino signal was not detected, so this proton does not participate in H-bonding interactions as predicted by its crystal structure (19,20). The sharp diagonal peaks appearing at ∼11 ppm in the 2D 1H-1H NOESY were assigned as imino protons from the ligand in its quadruplex states.
Metal- and ligand-dependent aptamer folding
Na+, K+ and Mg2 are critical factors for the pre-folding of the aptamer, and ligand completes RNA folding. After achieving comprehensive assignments of imino proton resonances at preliminary ‘free’ and final ‘holo’ conformational states, we evaluated the aptamer's conformational states across diverse microenvironments. Systematically varying the concentrations of Na+, K+ and Mg2+ ions as well as the c-di-GMP ligand, we examined the resulting RNA conformational changes using 1D 1H NMR titration assays (Figure 3). We systematically examined the conformational states of RNA in its free state by sequentially adding Na+, K+, Mg2+, and ligands and monitoring the resulting spectral variations. Overall, the NMR spectrum of the aptamer showed more pronounced changes upon addition of Mg2+ ions compared with the presence of Na+ and K+ alone. Moreover, the introduction of a ligand alongside Mg2+ triggered further spectral changes, including the emergence of new peaks. In contrast, when only Na+ and K+ together, or Na+ and K+ individually, were present, the addition of the ligand resulted in only minimal detectable changes in the NMR spectrum. A detailed description of these structural transitions is provided below.
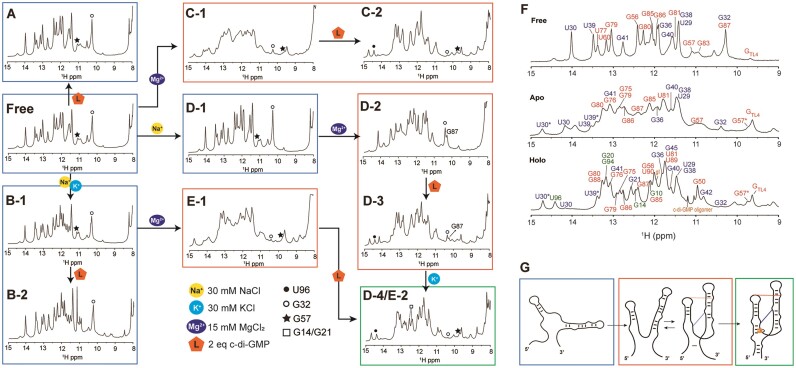
Figure 3.
(A–E) Imino proton area of the 1D 1H NMR spectra of c-di-GMP riboswitch aptamer at 288 K. The aptamer's folding process is influenced by the presence of Na+, K+ and Mg2+. The folding state of the riboswitch is indicated by color-coded boxes: blue for the unfolded state of P1a, orange for the partially folded state of P1a and green for the fully folded holo state of the aptamer. The U96, G32 and G57 imino resonance are shown as a solid circle, open circle and star, respectively and G14 and G21 are represented by a square. (F) 1D imino proton NMR spectra for the aptamer in the free, apo and holo states at 288 K. (G) Predicted aptamer structures during the folding process following the addition of Na+, K+, Mg2+ and c-di-GMP ligand.
We initially added only the ligand to the RNA without Na+, K+ or Mg2+, which resulted in no spectral changes. This suggests a lack of ligand interaction and no structural alterations in the RNA (Figure 3A). Subsequently, when Na+ and K+ were introduced before the ligand, a sharpening and separation of the overlapped peaks were observed (Figure 3B); however, these did not correspond to new peaks, as evidenced by the 2D 1H-15N HSQC spectra (Supplementary Figure S4A and B). Under these conditions, two imino protons near the ligand binding pocket in the holo state, G14H1 (12.45 ppm) and G21H1 (12.60 ppm), were absent. This indicates that while Na+ and K+ ions reduce repulsion among the RNA phosphate groups, leading to a more stable RNA structure, only portions of the P1b and P2 helix form, with the ligand binding pocket and P1a helix remaining unformed as no ligand binding occurs.
The addition of Mg2+ alone to RNA resulted in dramatic spectral changes, and subsequent ligand addition resulted in a spectrum resembling that of the holo state RNA (Figure 3C-1 and C-2). This suggests a complex structural transition toward a completely folded structure. The broader appearance of the spectrum compared with the holo state indicates that the RNA remains incompletely folded. The U96H3 peak emerged with the formation of the P1a helix. While residues in P1a, such as G14H1, produced imino protons that overlapped in the 1D NMR spectrum and were difficult to discern, U96H3 appeared as a distinct downfield peak at 14.5 ppm, serving as an excellent indicator of the formation of the lower stem of the P1a helix. In the presence of ligand and Mg2+, U96H3 was observed (Figure 3C-2, solid circle), yet G21H1 and G14H1 remained undetectable, which suggests partial formation of P1a and an incomplete ligand binding site.
Next, we examined the formation of long-range interactions in the 1D spectrum in the presence of Mg2+. As shown in the crystal structure, when this aptamer adopts a completely folded structure, it establishes two long-range interactions between P1b and P2. One is a ‘GAAA Tetraloop’ (GT)/‘Tetraloop Receptor’ (TR) (46,47) base-stacking interaction between A33 in P1b and A62 in P2, and the other is C44-G83 Watson–Crick base pair. The GT/TR interaction involves no hydrogen bonding, and the second, one base pair between two helices, does not protect imino protons from solvent exchange. Therefore, these long-range interactions could not be directly measured by imino proton observation. However, alterations in the imino protons of residues near the long-range interactions enabled indirect detection. Specifically, the GT/TR interaction led to line broadening at G32H1 adjacent to A33 in P1b and the disappearance of imino peaks corresponding to the U77-A61 and A78-U60 base pairs near A62 in P2. In addition, the formation of the C44-G83 long-range interaction in the holo state may be indirectly inferred from changes in the chemical shifts of the surrounding residues. Therefore, the sequential connectivity between G57H1 and G83H1 (Figure 2A), which is only visible when the RNA bases are oriented toward the inside of the helix in the free state, disappears in the holo state. Additionally, the significant up-field shift of G57H1 from 11 to 9.7 ppm next to G83 indicates that the G83 base is positioned outward from the helix, allowing interaction with C44. These results suggest the occurrence of a C44-G83 long-range base pair interaction.
Significant line broadening of G32H1 was observed even with Mg2+ alone, and upon subsequent ligand addition, this peak nearly vanished (Figure 3C-1 and 2, open circle; and Supplementary Figure S5C). An up-field shift in the peak for G57H1 at 9.75 ppm was also observed, which indicates that Mg2+ alone is sufficient to induce the two long-range interactions. In the ligand-free ‘apo’ state, in which only Na+, K+ and Mg2+ are present, the imino proton of G57 (11 ppm) becomes obscured by the emerging G50 imino peak upon ligand addition; however, in the presence of excess Mg2+, both the original and shifted peaks coexist at 11 ppm (Figure 3F, Apo), suggesting the co-existence of undocked and docked structures.
Next, we examined the spectral changes upon the sequential addition of Mg2+ and ligand in the presence of Na+ and K+. In the presence of Mg2+, the spectrum resembled that observed without Na+ and K+ (Figure 3C-1 and E-1), but upon further addition of the ligand, the aptamer adopted a fully folded form (Figure 3E-2). This indicates that complete folding of the aptamer requires the presence of Na+, K+, Mg2+ and the ligand.
To elucidate the role of Na+ and K+ in detail, we replicated the above experiment with the sequential addition of Na+ and K+. With Na+ alone, no structural change was observed (Figure 3D-1). The addition of Mg2+ in the presence of Na+ resulted in a spectrum similar to that without Mg2+ (Figure 3D-1,2) and did not produce any new significant resonances. Following ligand addition, however, spectral changes similar to those previously described were observed (Figure 3D-3 and C-2), but not peaks corresponding to G14H1 and G21H1, both at ∼12.5 ppm, which indicates the formation of the P1a helix and binding pocket, respectively (Figure 3F, Holo). When K+ was introduced, peaks for G14H1 and G21H1 finally appeared (Figure 3D-4). This confirms that the presence of K+ is crucial for the formation of the ligand binding site structure and the P1a helix. These 1D 1H NMR experiments enabled the prediction of secondary structural changes and the folding process of the aptamer, which are dependent on the presence of Na+, K+, Mg2+ and the ligand (Figure 3G).
We conducted additional experiments under complete Na+- free conditions, adjusting the pH using potassium phosphate while maintaining all other experimental parameters constant. In the presence of K+, Mg2+ and ligand, we observed the imino peaks for G21 and G14, indicating the formation of the ligand-binding pocket (Supplementary Figure S5E, rectangle). The resulting spectrum closely resembled that of the holo state spectrum; however, peaks for U30 and U39, associated with the unfolded RNA, were still observed. These results suggest that both Na+ and K+ are necessary to achieve a more stable folding, ultimately leading to a single RNA conformation.
Influence of P1a helix length on ligand binding
To investigate the importance of P1a helix length on ligand binding and aptamer stability, we conducted NMR experiments. The results showed that the helix length is crucial for achieving a stable holo-state aptamer structure. For the 85-nt aptamer, over 90% of the RNA adopted a folded structure with the addition of two equivalents of ligand. In contrast, for the 82-nt aptamer, only about 50% of the RNA folded, even with four equivalents of ligand under the same conditions (Figure 4). Specifically, in the 82-nt aptamer, imino signal from G20 interacting with gII were detected, but NOE signals between gI and G14, as well as peaks from U90 and G94, were absent, unlike in 85-nt aptamer where these signals are present when gI binds to RNA stably. This suggests partial ligand binding and incomplete folding in the shorter construct. These results highlight the necessity of an optimal P1a helix length for stable ligand binding and maintaining the functional aptamer structure.
Figure 4.
Imino proton region of the 2D 1H-1H NOESY and 2D 1H-15N HSQC spectrum of the 82 mer short aptamer in (A) 30 mM Na+, 30 mM K+ and 5 mM Mg2+, and in (B) 30 mM Na+, 30 mM K+, 5 mM Mg2+ and four equivalents of ligand, measured at 288 K. Only ∼50% of the completely folded aptamer was present according to the comparison of the two U30 peak intensities as well as the existence of double resonances observed for many residues, such as U29, G36, G38 and U39 as shown in the boxes. The residues in free state are colored in green.
Base pair dynamics in the P2 stem
In the free state, G87 of P2 resonates at 10.25 ppm, a characteristic frequency, in which a wobble base-paired with a G imino proton typically appears. Therefore, G87H1 seems to form a wobble base pair with U53 (Figure 2A and Supplementary Figure S1C). In the holo state, however, it forms a Watson–Crick base pair with C52, resulting in a downfield shift to 12.35 ppm (Figure 2B). This indicates that during the folding process, the base pairs interchange. Monitoring the G87 imino peak under varying conditions of Na+, K+, Mg2+ and ligands revealed that G87H1 exists as a wobble pair (G87H1wobble) in the free state and the presence of Na+, K+ and ligands (Figure 3A, B and D-1). However, in the absence of Na+ and K+, but in the presence of Mg2+ and ligands, the G87H1wobble peak disappears, suggesting that G87 will form a base pair with C52 (Figure 3C-1 and 2). Interestingly, in the presence of Na+, even the addition of Mg2+ retains the G87H1wobble (Figure 3D-2). Subsequently, adding ligands decreases its intensity, but this peak remains (Figure 3D-3), suggesting that in this state, G87H1 coexists in both GC and GU forms. U53, which formed a base pair with G87 within the helix in the free state, should flip out of the helix during the folding process induced by Mg2+, leading to a change in base pairing.
To elucidate the dynamics of RNA base pairs, we determined the hydrogen exchange rate constants (kex) of imino protons for the free, apo and holo states of the RNA at 298 K. The kex values of base-paired residues in stable helices were observed to be around 2–4 s-1. In the free state, U30 and G32 in the P1b helix exhibited high kex values of 8.50 ± 0.46 s-1 and 36.87 ± 3.12 s-1, respectively. Similarly, residues in the P2 helix, including G57, U77, U81, G83 and G87, showed large kex values, ranging from 8.84 ± 0.23 s-1 to 36.87 ± 3.12 s-1, indicating significant flexibility in the free state. In the apo state, the kex values for U30 and G32 in the P1b helix decreased, with G32 still retaining a relatively high value of 10.98 ± 0.60 s-1. The P2 helix residues, which had large kex values in the free state, also exhibited a marked reduction, with values ranging from 4.24 ± 0.16 s-1 to 5.43 ± 0.41 s-1. In the holo state, where the ligand is bound, the NMR signal for G32 in the P1b helix became very weak, making it difficult to measure its kex value. Notably, ligand binding promoted the formation of the P1a helix, allowing for the measurement of kex values for the corresponding residues with the values of ∼2 s-1. (Supplementary Figures S6 and S7, and Supplementary Table S2). These data suggest that ligand binding significantly stabilizes the overall RNA structure, particularly in the P1b and P2 helices, as reflected by the reduced exchange rates in the holo state.
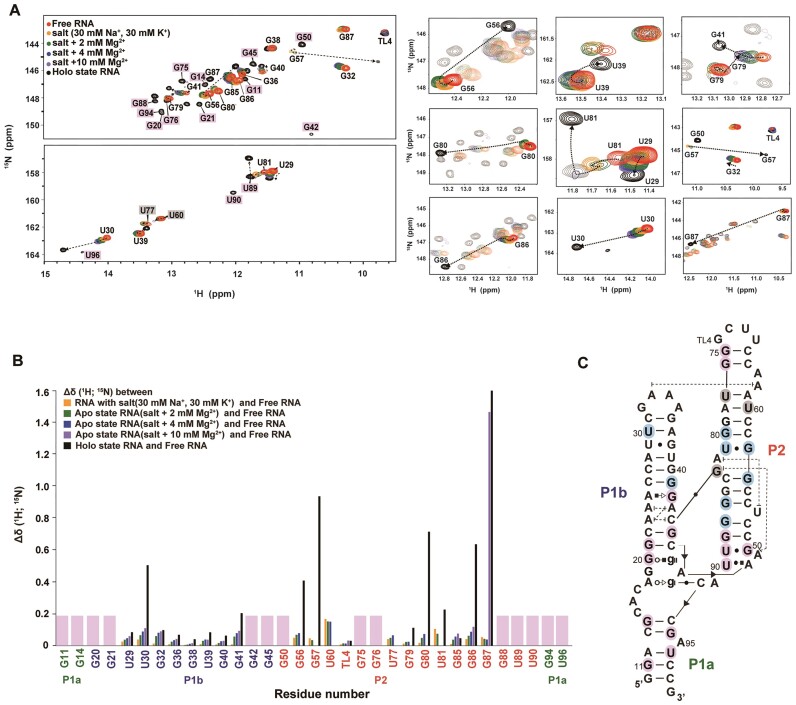
In 2D 1H-15N HSQC experiments, chemical shift changes for individual residues can be traced, which enables us to predict structural changes in RNA that were not visible in the 1D spectra. Unlike 1D 1H NMR experiments, in which various conditions were tested, we primarily observed aptamer folding by sequentially adding Na+, K+, Mg2+ and ligands (Figure 5 and Supplementary Figure S4). Similar to the 1D 1H NMR, there were very minor chemical shift changes for all residues by only adding Na+ and K+. The addition of Mg2+ caused spectral changes in the P1b and P2 regions. Peaks for G56, U60, U77, G79, G80, U81/G57 and G86 in P2 exhibited line broadening and eventually disappeared with increasing Mg2+ concentrations, indicating that these residues are in an intermediate exchange state between free and partially folded structures. Interestingly, under these conditions, U29, U30, G32, G36, G40 and G41 in P1b remained detectable, which indicated that these residues underwent fast exchange (Figure 5A). The results suggest that significant structural changes occurred in P2, including the rearrangement of many base pairs during interhelical docking during the folding process, whereas P1b mostly retained its secondary structure. In the presence of sufficient Mg2+, the addition of ligand caused huge chemical shift changes in U30 and G41 of P1b, and G56, G57, G80, U81, G86 and G87 of P2 (Figure 5B). As expected from the added Mg2+ results, major changes occurred in the P2 helix, which provides indirect evidence of the occurrence of a base pair rearrangement from G87•U53 to G87-C52, which in turn, induced A82-U53 base stacking. For P1b, notable chemical shift changes in U30 and G41 were observed. The large chemical shift change in U30 was linked to structural rearrangements in the GAAA tetraloop driven by stacking between A58 and A33. In contrast, the shift in G41 resulted from G83-C44 long-range interactions that stabilize the P1b bottom helix through the formation of base pairs near the ligand binding site. The formation of a stable-folded structure is supported by the presence of new imino resonances for G20, G21, G42 and G45 in P1b. In addition, the formation of the cross peaks, G11, G14, G94 and U96, confirmed the completion of the P1a helix.
Figure 5.
Characterization of c-di-GMP riboswitch folding pathway. (A) Chemical shift changes for the c-di-GMP riboswitch in 2D 1H-15N-HSQC spectra were tracked from free to holo states. The residues with significant or complex chemical shift changes are presented on the right. (B) CSP analysis calculated using the equation, 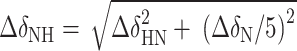 , which displays the comparative shifts between the free RNA, the four apo states, and the holo state RNA, denoted by different colors. (C) The RNA secondary structure is annotated to indicate these significant changes in cyan, whereas the residues that manifest exclusively in the holo state, and thus lack of CSP data, are consistently marked with pink throughout.
, which displays the comparative shifts between the free RNA, the four apo states, and the holo state RNA, denoted by different colors. (C) The RNA secondary structure is annotated to indicate these significant changes in cyan, whereas the residues that manifest exclusively in the holo state, and thus lack of CSP data, are consistently marked with pink throughout.
Quantitative analysis of the RNA folding equilibrium
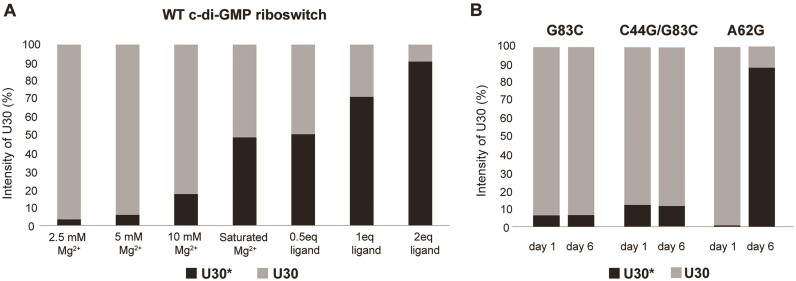
As briefly mentioned for the G87 case, during the gradual addition of Na+, K+, Mg2+, and ligand, some residues exhibited imino peaks at two distinct resonances. This indicates that two RNA structures before and after complete folding are in equilibrium and undergo a slow exchange. To determine the proportion of these conformers under each ionic condition, the relative intensity of the two imino peaks in the 1D spectrum was measured and the ratios of each conformer were quantitatively analyzed. Of the various residues, we selected U30 to analyze because the resonance of its imino peak was distinguishable in the apo (U30, 14.14 ppm) and holo (U30*, 14.7 ppm) states, unlike other residues (Figure 3F). At 2.5, 5 and 10 mM Mg2+, the RNA in the apo state consisted of 3%, 5% and 20%, respectively, and reached 50% under saturated Mg2+ conditions of 15 mM (Figure 6A). At this Mg2+ concentration, U30 imino peak picking was difficult because of line broadening and spectral overlap, so the RNA was washed in 2.5 mM Mg2+ buffer and the peak intensities were measured. This buffer exchange did not affect RNA folding as confirmed by comparison of the NMR spectra between before and after washing (data not shown). Subsequently, as the ligand was added, the percentage of U30* increased, reaching approximately 90% with a two-equivalent addition of the ligand (Figure 6A). These results indicate that RNA exists in an equilibrium between folded and unfolded states under various concentrations of Mg2+ and ligand.
Figure 6.
(A) The proportion of the two distinct equilibrium states of the c-di-GMP riboswitch determined by calculating the relative intensities of U30 (at 14.14 ppm) and U30* (at 14.7 ppm) in the presence of varying concentrations of Mg2+ and the ligand for the wt RNA. (B) Time-dependent structural transitions in the mutant aptamer by tracking the U30 and U30* signal intensity. Experiments were conducted under conditions containing 30 mM Na+, 30 mM K+, 15 mM Mg2+ and two equivalents of ligand, which are sufficient for aptamer folding. All experiments were performed at 288 K. The apo state is represented by the lighter shaded bars at a U30 peak intensity, and the darker shaded bars mark a U30* peak intensity, which signify the holo state.
Role of long-range interactions
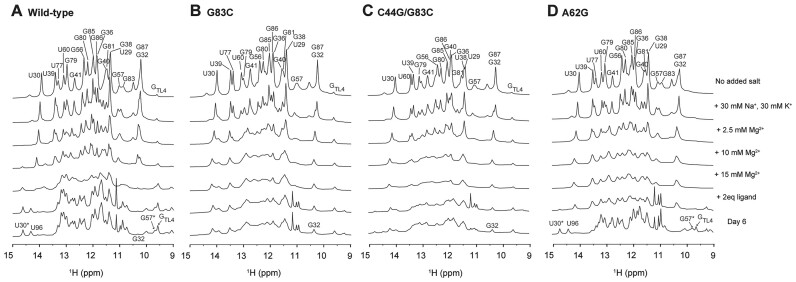
To characterize the role of the GT/TR (A33-A62) and C44-G83 tertiary interactions in the formation and stability of the ligand-docked structure, we synthesized three site-specific mutated RNAs, A62G, G83C and G83C/C44G, which were designed to disrupt these interactions (Figure 1A). Imino proton 1D 1H NMR experiments were conducted under the same conditions as the wt RNA NMR titration experiments as described above. RNA folding was also observed 6 days after ligand addition to investigate any potential occurrence of very slow RNA folding process (Figure 7).
Figure 7.
1D 1H NMR spectra recorded at 288 K for the riboswitch mutants: (A) Wild-type, (B) G83C, (C) C44G/G83C and (D) A62G. The specific buffer conditions used for each are detailed on the right.
For G83C mutant RNA, which was designed to disrupt long-range Watson–Crick base pairing, the spectrum was broadened compared with that of wt RNA. RNA did not fold into the holo state, with only a slight change even after 6 days (Figure 7B). A previous FRET study using the G83C mutant under similar conditions with sufficient Mg2+ and ligand reported that approximately 80% of the RNA remained undocked and only 20% formed the docked conformation (22). In our NMR experiment, when comparing the imino proton intensity of U30 and U30*, <10% of the RNA adopted a folded structure, with the majority remaining in an unfolded structure (Figure 6B). Thus, our results are consistent with that of the FRET results. Next, we examined the C44G/G83C mutant, which was designed to recover the loss of the G-C base pair from the G83C substitution. After adding two equivalents of ligand, small peaks for U30* and U96 were observed at 14.7 and 14.4 ppm, respectively, but no significant spectral changes were observed, similar to the results with G83C. These two peaks, which indicated complete folding, slightly increased in intensity after 6 days, but showed no further significant changes in the spectrum (Figures 6B and 7C). Finally, we tested the A62G mutant, which was designed to disrupt base-stacking interactions between A62 and A33 (Figures 6B and 7D). Interestingly, following the addition of Mg2+ and two equivalents of ligand, a spectrum similar to that of wt RNA in 10 mM Mg2+ was observed; however, U30* and U96 were almost absent. These peaks gradually reappeared over time, resulting in a spectrum similar to the holo state wt RNA (Figure 7D and Supplementary Figure S8A). This suggests that, over time, the A62G mutant, even without AA stacking, can still achieve a folding structure similar to that of wt RNA with ligand binding and the completion of P1a base pairs. The formation of U30* associated with base stacking suggests the possibility of AG stacking, instead of AA stacking. Thus, AA stacking may stabilize the tertiary structure of the holo state aptamer, which is otherwise unstable because of P2 base pair dynamics and an insufficient G83-C44 long-range interaction. AA stacking may act as a kinetic factor facilitating RNA folding and long-range GC base pairs are essential for stable docking of RNA.
To further understand their roles, ITC experiments were conducted under various Mg2+ concentrations (0, 2.5, 5, 10 mM) (Figure 8 and Table 1). For wt RNA, no ligand binding occurred without Mg2+, but 1:1 binding with N = 1 was formed in its presence, with KD values of 269 ± 21, 140 ± 16 and 146 ± 18 nM, showing tight binding as the Mg2+ concentration increased (Figure 8A). This was consistent with the NMR findings, in which increased Mg2+ facilitated ligand binding. For the G83C and C44G/G83C mutants, no ligand binding was observed (Figure 8B). For the A62G mutant, the KD value is 194 ± 47 nM, similar to that of wt RNA (146 ± 18 nM) (Figure 8B). However, with an N value of 0.17, this indicates that most RNA exists in a misfolded form, with only a small fraction binding to the ligand. Consistent with the NMR results above, after 3 days, the N value increases to 0.98, suggesting that most RNA adopts a holo state similar to that of wt RNA, achieving a 1:1 binding with the ligand (Supplementary Table S3 and Supplementary Figure S9). These findings suggest that the A62G mutant is initially unable to fully bind the ligand due to structural alteration caused by the mutation. However, over time, in the presence of Na+, K+ and Mg2+ ions, the RNA transition to a pre-folded conformation that promotes ligand binding. To confirm whether the A62G RNA was kinetically trapped in a misfolded state during folding, heating and cooling experiments were performed, which immediately resulted in an almost properly folded structure (Supplementary Figure S8B). This suggests that A62G mutation disrupt stacking between A33 and A62, leading to misfolding or kinetic trapping during early folding stages. Thus, stacking interactions appear to act as a kinetic facilitator, accelerating the formation of a stable tertiary structure. These results indicate that disrupting critical interactions like stacking can significantly delay riboswitch folding, potentially impairing its ability to rapidly respond to ligands and regulate gene expression in the cell.
Figure 8.
ITC results for the interaction of c-di-GMP with aptamer, which was performed in a buffer containing 10 mM NaPi, pH 6.5, with 30 mM NaCl and 30 mM KCl at 298 K. The results for c-di-GMP binding with (A) wt, and (B) mutant riboswitch aptamer at 10 mM Mg2+ concentrations are shown. The specific Mg2+ concentrations and the corresponding KD values are shown.
Table 1.
ITC results for the c-di-GMP ligand binding with wt and mutant aptamers at various Mg2+ concentrations
| Riboswitch/c-di-GMP | N | K D (nM) | ΔG (Kcal/mol) | TΔS (Kcal/mol) | ΔH (Kcal/mol) |
|---|---|---|---|---|---|
| WT (0 mM MgCl2) | N.D.a | N.D. | N.D. | N.D. | N.D. |
| WT (2.5 mM MgCl2) | 1.16 | 269 ± 21 | −8.9 ± 0.03 | −11.65 ± 0.09 | −20.62 ± 0.13 |
| WT (5 mM MgCl2) | 1.07 | 140 ± 16 | −9.4 ± 0.04 | −8.88 ± 0.10 | −18.24 ± 0.09 |
| WT (10 mM MgCl2) | 1.05 | 146 ± 18 | −9.3 ± 0.07 | −8.61 ± 0.17 | −17.94 ± 0.15 |
| A62G (10 mM MgCl2) | 0.17 | 194 ± 47 | −9.1 ± 0.14 | −6.71 ± 0.61 | −15.32 ± 0.59 |
| G83C (10 mM MgCl2) | N.D. | N.D. | N.D. | N.D. | N.D. |
| C44G/G83C (10 mM MgCl2) | N.D. | N.D. | N.D. | N.D. | N.D. |
aN.D.: Not detected.
Discussion
RNA alters its structure under appropriate conditions to function within the cell. Particularly, riboswitches, which are known for changing their structure upon ligand recognition to regulate gene expression, have become a subject of interest (2,7,48). In the present study, we examined the conformational states and binding affinity of the V. cholerae c-di-GMP riboswitch aptamer under various Na+, K+ and Mg2+ conditions, and upon ligand recognition using NMR and ITC. We tracked the imino signals from the NMR spectra during the formation of base pairs during the RNA folding process to identify the structural changes in the aptamer. Initially, we observed the imino signals generated by two homogeneous and stable structures: the free (buffer only) and holo (ligand-bound) states. Previous studies have elucidated the structure and ligand binding characteristics of the aptamer through crystal structures (19,20), and overall conformational changes of the c-di-GMP aptamer and the docking of its two helices were obtained from FRET studies (22). The FRET analyses have identified four distinct conformational states of the riboswitch in the absence of its ligand, c-di-GMP, demonstrating its structural diversity and the possibility of ligand-induced conformational transitions. Furthermore, the riboswitch is significantly triggered toward a ligand-binding optimized conformation by the presence of Mg2+ ions. The observed transitions in the NMR spectra can be related to the FRET populations, providing insight into the dynamic states of the riboswitch. For instance, in the absence of Mg2+ and ligand or in the presence of ligand alone (Figure 3B-1 and B-2), the aptamer remains largely unfolded, corresponding to the static undocked state observed in FRET. Upon introducing Mg2+, the aptamer adopts a more folded conformation, transitioning to the dynamic docked state (Figure 3E-1), where both dynamic docked and undocked states coexist. Finally, with the addition of c-di-GMP ligand, the fully folded conformation is stabilized, resulting in a higher population of the static docked state than other states (Figure 3E-2). Similarly, Figure 6 demonstrates the effect of increasing Mg2+ and ligand concentrations on aptamer folding in the presence of Na+ and K+. At lower Mg2+ concentrations, the aptamer predominantly resides in a static undocked state, shifting toward a more docked conformation with increasing Mg2+ concentration. The fully folded conformation is not stabilized until the ligand is introduced, as reflected by the increase in U30* peak intensity, which corresponds to the static docked state in FRET under saturating Mg2+ and ligand conditions. Additionally, the conformation of the riboswitch is profoundly affected by tertiary interactions, even those distant from the ligand binding site, stabilizing the riboswitch in a pre-organized state conducive to rapid and efficient ligand binding. Our study took this a step further through a detailed exploration of the intermediate structures, including the base pair changes that the aptamer undergoes in the apo (ligand-free) state, and how these changes contribute to ligand recognition.
Long-range interactions are critical for ligand binding in various riboswitches (49), with metal ions playing a significant role in riboswitch-mediated gene regulation (50,51). In this study, we demonstrated that both K+ and Mg2+ ions are essential for the pre-folding of the aptamer in the c-di-GMP riboswitch, driven by two key long-range interactions between the P1b and P2 helices. Specifically, Mg2+ promotes base-stacking interactions between A33 and A62 even without the ligand, while the C44-G83 interaction does not form with Na+ and Mg2+ alone but is stabilized in the apo state with Na+, K+ and Mg2+, as evidenced by changes in G83 connectivity and significant chemical shifts in G57H1. These findings indicate that while Mg2+ is crucial for GT/TR interactions, K+ is equally important for stabilizing the aptamer's pre-folded state, facilitating ligand binding. Previous studies support the role of K+ in RNA folding. For instance, K+ binding to the AA platform in the P4-P6 domain of the Tetrahymena Group I intron enhances GT/TR interactions (52), while the Guanidine-I riboswitch requires both Mg2+ and K+ for efficient ligand binding (53). Although K+ was not detected in the crystal structure of the c-di-GMP riboswitch, the TR domain forms a structural motif similar to the AA platform, suggesting a potential K+ binding site. However, since our results showed that GT/TR interactions can occur solely in the presence of Mg2+, we propose that K+ primarily contributes to stabilizing the conformation of the binding pocket and potentially driving GT/TR interactions. The C44G/G83C mutant was designed to restore the original GC base-pair, but ligand binding was not recovered, likely due to unintended structural disruptions introduced by the mutation. The swapping C44 and G83 may have altered nearby interactions, such as the stacking between G83 and A49, leading to changes in the RNA’s tertiary structure that disrupted the binding pocket. These structural changes likely prevented proper folding of the RNA and reduced its ligand affinity.
As indicated by the NMR assignments and CSP, there was little change in the chemical shifts between the free and holo states in the P1b helix, but significant changes were observed in the P2 helix. These findings align with the water magnetization transfer experiments, which revealed that base pairs in the P2 helix, unlike those in the P1b helix, exhibit dynamic imino proton exchange in the free state, as evidenced by high hydrogen exchange rate constants (kex). This dynamic flexibility in the P2 helix likely facilitates base pair rearrangements, enabling the RNA aptamer to efficiently transition into a ligand-bound structure in the presence of metal ions. Furthermore, the ligand-induced stabilization observed in the kex measurements supports the hypothesis that these structural dynamics play a critical role in ligand recognition. In particular, the decreased kex values in the apo and holo states suggest a progressive stabilization of the RNA upon ligand binding, with the P2 helix showing significant structural rigidity in the holo state. This stabilization is likely essential for precise ligand recognition and subsequent structural rearrangements. This may be the result of the evolutionary adaptation of the c-di-GMP riboswitch, which selects a nucleotide sequence in a manner that favors RNA structural changes for gene regulation.
As evidenced by various riboswitches studied so far, many of which bind ligands to form a three-way junction structure (54). Among these, the purine riboswitch shares a highly similar structure with the c-di-GMP-I riboswitch used in this study, and the formation of the P1 helix is crucial for gene expression (55). Another example is the SAM-VI riboswitch, in which the ligand (SAM) binding disrupts the P0 helix and triggers the formation of the P1 helix (56). This helix formation in the holo state is vital for stabilizing the structure and function of the riboswitch, thus preventing the binding of ribosomes to the ribosome binding site and inhibiting translation (57). This indicates that the formation of specific helices in response to ligand binding is a key mechanism for the structural and functional modulation of riboswitches. In the present study, the structure of the aptamer induced by metal ions forms part of P1a, even before ligand binding, but it completes the structure of the binding pocket and forms the full P1a helix only after ligand binding. As previously shown (22,58), the formation of P1a disrupts riboswitch terminator formation, forms an anti-terminator hairpin structure, and promotes translation. It is important to note that a sufficient length of the P1a helix is directly related to ligand binding as demonstrated by gel shift assays (20). Our NMR studies with the 82-nt aptamer confirmed the critical role of the P1a helix length. In this shorter construct, incomplete ligand binding and RNA folding were observed, emphasizing that a sufficient P1a helix length is essential for forming a stable holo-state aptamer structure. These similarities suggest a common structural mechanism among different riboswitches, in which the formation or restructuring of specific helical regions upon ligand binding is required for their regulatory function.
This study uncovers a significant insight in RNA biology, demonstrating the essential role of dynamic base pair rearrangements within the P2 helix for stable folding and specific ligand recognition by the Vc2 c-di-GMP riboswitch. Our results emphasize the intricate role of ion interactions in RNA molecule functionality, where Mg2+ ions facilitate the initial folding stages and K+ ions are instrumental in forming essential long-range interactions for structural integrity. We believe that this work not only deepens our understanding of riboswitch mechanisms at the atomic level but also establishes a new benchmark for investigating RNA folding dynamics in physiological conditions. The implications of our findings can be extended to the development of new antibacterial strategies, as highlighted in recent reviews (59,60). Furthermore, as shown in other studies (61) our findings underscore the utility of NMR spectroscopy in detailed structural analysis, enhancing our capacity to explore the action of RNA elements and their interactions with small molecules, which could lead to the identification of new drug targets. Additionally, this study provides valuable insights for synthetic biology, suggesting approaches for designing RNA molecules with specific functions, leveraging the critical role of ion-mediated folding and structural rearrangement.
Supplementary Material
Contributor Information
Ji-Yeon Shin, Advanced Analysis Data Center, Korea Institute of Science and Technology, Hwarang-ro 14-5, Seongbuk-gu, Seoul 02792, Republic of Korea; Department of Life Sciences, Korea University, 145, Anam-ro, Seongbuk-gu, Seoul 02841, Republic of Korea.
Seo-Ree Choi, Advanced Analysis Data Center, Korea Institute of Science and Technology, Hwarang-ro 14-5, Seongbuk-gu, Seoul 02792, Republic of Korea.
So Young An, Department of Agriculture Biotechnology, Seoul National University, 1 Gwanak-ro, Gwanak-gu, Seoul 08826, Republic of Korea.
Kyeong-Mi Bang, Advanced Analysis Data Center, Korea Institute of Science and Technology, Hwarang-ro 14-5, Seongbuk-gu, Seoul 02792, Republic of Korea; Department of Life Sciences, Korea University, 145, Anam-ro, Seongbuk-gu, Seoul 02841, Republic of Korea.
Hyun Kyu Song, Department of Life Sciences, Korea University, 145, Anam-ro, Seongbuk-gu, Seoul 02841, Republic of Korea.
Jeong-Yong Suh, Department of Agriculture Biotechnology, Seoul National University, 1 Gwanak-ro, Gwanak-gu, Seoul 08826, Republic of Korea; Research Institute of Agriculture and Life Sciences, Seoul National University, 1 Gwanak-ro, Gwanak-gu, Seoul 08826, Republic of Korea.
Nak-Kyoon Kim, Advanced Analysis Data Center, Korea Institute of Science and Technology, Hwarang-ro 14-5, Seongbuk-gu, Seoul 02792, Republic of Korea.
Data availability
The chemical shifts for the c-di-GMP riboswitch aptamer RNA in its holo- and free- states have been deposited in the Biological Magnetic Resonance Bank (https://bmrb.io/) with the accession numbers of 52 463 and 52 464, respectively.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Institutional Research Program of Korea Institute of Science and Technology [2V10021 to N.-K.K]; Rural Development Administration [PJ01495901]; National Research Foundation of Korea [2021R1A2C1003917 to J.-Y.S.]. Funding for open access charge: Korea Institute of Science and Technology.
Conflict of interest statement. None declared.
References
- 1. Al-Hashimi H.M., Walter N.G. RNA dynamics: it is about time. Curr. Opin. Struct. Biol. 2008; 18:321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Breaker R.R. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 2012; 4:a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Henkin T.M. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev. 2008; 22:3383–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Breaker R.R. The biochemical landscape of riboswitch ligands. Biochemistry. 2022; 61:137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Salvail H., Breaker R.R. Riboswitches. Curr. Biol. 2023; 33:R343–R348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bu F., Lin X., Liao W., Lu Z., He Y., Luo Y., Peng X., Li M., Huang Y., Chen X. et al. Ribocentre-switch: a database of riboswitches. Nucleic Acids Res. 2024; 52:D265–D272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Montange R.K., Batey R.T. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 2008; 37:117–133. [DOI] [PubMed] [Google Scholar]
- 8. Winkler W.C., Breaker R.R. Genetic control by metabolite-binding riboswitches. ChemBioChem. 2003; 4:1024–1032. [DOI] [PubMed] [Google Scholar]
- 9. Liberman J.A., Wedekind J.E. Riboswitch structure in the ligand-free state. Wiley Interdiscip. Rev. RNA. 2012; 3:369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ganser L.R., Kelly M.L., Herschlag D., Al-Hashimi H.M. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell. Biol. 2019; 20:474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee H.K., Lee Y.T., Fan L., Wilt H.M., Conrad C.E., Yu P., Zhang J., Shi G., Ji X., Wang Y.X. et al. Crystal structure of Escherichia coli thiamine pyrophosphate-sensing riboswitch in the apo state. Structure. 2023; 31:848–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haller A., Souliere M.F., Micura R. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc. Chem. Res. 2011; 44:1339–1348. [DOI] [PubMed] [Google Scholar]
- 13. Haller A., Altman R.B., Soulière M.F., Blanchard S.C., Micura R. Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proc. Natl Acad. Sci. U.S.A. 2013; 110:4188–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roy S., Lammert H., Hayes R.L., Chen B., LeBlanc R., Dayie T.K., Onuchic J.N., Sanbonmatsu K.Y. A magnesium-induced triplex pre-organizes the SAM-II riboswitch. PLoS Comput. Biol. 2017; 13:e1005406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rode A.B., Endoh T., Sugimoto N. Crowding shifts the FMN recognition mechanism of riboswitch aptamer from conformational selection to induced fit. Angew. Chem. Int. Ed. Engl. 2018; 57:6868–6872. [DOI] [PubMed] [Google Scholar]
- 16. Sudarsan N., Lee E.R., Weinberg Z., Moy R.H., Kim J.N., Link K.H., Breaker R.R. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008; 321:411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pursley B.R., Maiden M.M., Hsieh M.L., Fernandez N.L., Severin G.B., Waters C.M. Cyclic di-GMP regulates TfoY in Vibrio cholerae to control motility by both transcriptional and posttranscriptional mechanisms. J. Bacteriol. 2018; 200:e00578–e00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pursley B.R., Fernandez N.L., Severin G.B., Waters C.M. The Vc2 cyclic di-GMP-dependent riboswitch of Vibrio cholerae regulates expression of an upstream putative small RNA by controlling RNA stability. J. Bacteriol. 2019; 201:e00293-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kulshina N., Baird N.J., Ferre-D’Amare A.R. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat. Struct. Mol. Biol. 2009; 16:1212–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith K.D., Lipchock S.V., Ames T.D., Wang J., Breaker R.R., Strobel S.A. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat. Struct. Mol. Biol. 2009; 16:1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li C., Zhao X., Xie P., Hu J., Bi H. Molecular dynamics simulation on the allosteric analysis of the c-di-GMP class I riboswitch induced by ligand binding. J. Mol. Recognit. 2019; 32:e2756. [DOI] [PubMed] [Google Scholar]
- 22. Wood S., Ferre-D’Amare A.R., Rueda D. Allosteric tertiary interactions preorganize the c-di-GMP riboswitch and accelerate ligand binding. ACS Chem. Biol. 2012; 7:920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith K.D., Lipchock S.V., Strobel S.A. Structural and biochemical characterization of linear dinucleotide analogues bound to the c-di-GMP-I aptamer. Biochemistry. 2012; 51:425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stoddard C.D., Montange R.K., Hennelly S.P., Rambo R.P., Sanbonmatsu K.Y., Batey R.T. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010; 18:787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Serganov A., Huang L.L., Patel D.J. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008; 455:1263–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marusic M., Toplishek M., Plavec J. NMR of RNA- structure and interactions. Curr. Opin. Struct. Biol. 2023; 79:102532. [DOI] [PubMed] [Google Scholar]
- 27. Keller H., Weickhmann A.K., Bock T., Wohnert J. Adenine protonation enables cyclic-di-GMP binding to cyclic-GAMP sensing riboswitches. RNA. 2018; 24:1390–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bains J.K., Blechar J., de Jesus V., Meiser N., Zetzsche H., Furtig B., Schwalbe H., Hengesbach M. Combined smFRET and NMR analysis of riboswitch structural dynamics. Methods. 2019; 153:22–34. [DOI] [PubMed] [Google Scholar]
- 29. Warhaut S., Mertinkus K.R., Hollthaler P., Furtig B., Heilemann M., Hengesbach M., Schwalbe H. Ligand-modulated folding of the full-length adenine riboswitch probed by NMR and single-molecule FRET spectroscopy. Nucleic Acids Res. 2017; 45:5512–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Furtig B., Richter C., Wohnert J., Schwalbe H. NMR spectroscopy of RNA. ChemBioChem. 2003; 4:936–962. [DOI] [PubMed] [Google Scholar]
- 31. Kumar G.S., Basu A. The use of calorimetry in the biophysical characterization of small molecule alkaloids binding to RNA structures. Biochim. Biophys. Acta. 2016; 1860:930–944. [DOI] [PubMed] [Google Scholar]
- 32. Guillerez J., Lopez P.J., Proux F., Launay H., Dreyfus M. A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc. Natl Acad. Sci. U.S.A. 2005; 102:5958–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Milligan J.F., Uhlenbeck O.C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989; 180:51–62. [DOI] [PubMed] [Google Scholar]
- 34. Lorsch J. Methods in enzymology. Laboratory methods in enzymology: RNA. Preface. Methods Enzymol. 2013; 530:xxi. [DOI] [PubMed] [Google Scholar]
- 35. Dingley A.J., Nisius L., Cordier F., Grzesiek S. Direct detection of N-H•••N hydrogen bonds in biomolecules by NMR spectroscopy. Nat. Protoc. 2008; 3:242–248. [DOI] [PubMed] [Google Scholar]
- 36. Cromsigt J., van Buuren B., Schleucher J., Wijmenga S. Resonance assignment and structure determination for RNA. Methods Enzymol. 2001; 338:371–399. [DOI] [PubMed] [Google Scholar]
- 37. Dieckmann T., Feigon J. Assignment methodology for larger RNA oligonucleotides: application to an ATP-binding RNA aptamer. J. Biomol. NMR. 1997; 9:259–272. [DOI] [PubMed] [Google Scholar]
- 38. Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015; 31:1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee J.H., Pardi A. Thermodynamics and kinetics for base-pair opening in the P1 duplex of the Tetrahymena group I ribozyme. Nucleic Acids Res. 2007; 35:2965–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee J.H., Jucker F., Pardi A. Imino proton exchange rates imply an induced-fit binding mechanism for the VEGF165-targeting aptamer, Macugen. FEBS. Lett. 2008; 582:1835–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Williamson M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013; 73:1–16. [DOI] [PubMed] [Google Scholar]
- 42. Ennifar E., Nikulin A., Tishchenko S., Serganov A., Nevskaya N., Garber M., Ehresmann B., Ehresmann C., Nikonov S., Dumas P. The crystal structure of UUCG tetraloop. J. Mol. Biol. 2000; 304:35–42. [DOI] [PubMed] [Google Scholar]
- 43. Nozinovic S., Furtig B., Jonker H.R., Richter C., Schwalbe H. High-resolution NMR structure of an RNA model system: the 14-mer cUUCGg tetraloop hairpin RNA. Nucleic Acids Res. 2010; 38:683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith K.D., Lipchock S.V., Livingston A.L., Shanahan C.A., Strobel S.A. Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch. Biochemistry. 2010; 49:7351–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mladek A., Sharma P., Mitra A., Bhattacharyya D., Sponer J., Sponer J.E. Trans hoogsteen/sugar edge base pairing in RNA. Structures, energies, and stabilities from quantum chemical calculations. J. Phys. Chem. B. 2009; 113:1743–1755. [DOI] [PubMed] [Google Scholar]
- 46. Butcher S.E., Dieckmann T., Feigon J. Solution structure of a GAAA tetraloop receptor RNA. EMBO J. 1997; 16:7490–7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fiore J.L., Nesbitt D.J. An RNA folding motif: GNRA tetraloop-receptor interactions. Q. Rev. Biophys. 2013; 46:223–264. [DOI] [PubMed] [Google Scholar]
- 48. Overbeck J.H., Vögele J., Nussbaumer F., Duchardt-Ferner E., Kreutz C., Wöhnert J., Sprangers R. Multi-site conformational exchange in the synthetic neomycin-sensing riboswitch studied by F NMR. Angew. Chem. Int. Ed. Engl. 2023; 62:e202218064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jones C.P., Ferre-D’Amare A.R. Long-range interactions in riboswitch control of gene expression. Annu. Rev. Biophys. 2017; 46:455–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Woodson S.A. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005; 9:104–109. [DOI] [PubMed] [Google Scholar]
- 51. Ferre-D’Amare A.R., Winkler W.C. The roles of metal ions in regulation by riboswitches. Met. Ions Life Sci. 2011; 9:141–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Basu S., Rambo R.P., Strauss-Soukup J., Cate J.H., Ferre-D’Amare A.R., Strobel S.A., Doudna J.A. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat. Struct. Biol. 1998; 5:986–992. [DOI] [PubMed] [Google Scholar]
- 53. Trachman R.J. 3rd, Ferre-D’Amare A.R. An uncommon [K(+)(Mg(2+))(2)] metal ion triad imparts stability and selectivity to the Guanidine-I riboswitch. RNA. 2021; 27:1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Batey R.T. Structure and mechanism of purine-binding riboswitches. Q. Rev. Biophys. 2012; 45:345–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stagno J.R., Wang Y.X. Riboswitch mechanisms for regulation of P1 helix stability. Int. J. Mol. Sci. 2024; 25:10682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xue Y., Li J., Chen D., Zhao X., Hong L., Liu Y. Observation of structural switch in nascent SAM-VI riboswitch during transcription at single-nucleotide and single-molecule resolution. Nat. Commun. 2023; 14:2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Landgraf T., Volklein A.E., Furtig B., Schwalbe H. The cotranscriptional folding landscape for two cyclic di-nucleotide-sensing riboswitches with highly homologous aptamer domains acting either as ON- or OFF-switches. Nucleic Acids Res. 2022; 50:6639–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Inuzuka S., Kakizawa H., Nishimura K.I., Naito T., Miyazaki K., Furuta H., Matsumura S., Ikawa Y. Recognition of cyclic-di-GMP by a riboswitch conducts translational repression through masking the ribosome-binding site distant from the aptamer domain. Genes Cells. 2018; 23:435–447. [DOI] [PubMed] [Google Scholar]
- 59. Giarimoglou N., Kouvela A., Maniatis A., Papakyriakou A., Zhang J., Stamatopoulou V., Stathopoulos C. A riboswitch-driven era of new antibacterials. Antibiotics (Basel). 2022; 11:1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ellinger E., Chauvier A., Romero R.A., Liu Y.C., Ray S., Walter N.G. Riboswitches as therapeutic targets: promise of a new era of antibiotics. Expert Opin. Ther. Targets. 2023; 27:433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Camara M.B., Lange B., Yesselman J.D., Eichhorn C.D. Visualizing a two-state conformational ensemble in stem-loop 3 of the transcriptional regulator 7SK RNA. Nucleic Acids Res. 2024; 52:940–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The chemical shifts for the c-di-GMP riboswitch aptamer RNA in its holo- and free- states have been deposited in the Biological Magnetic Resonance Bank (https://bmrb.io/) with the accession numbers of 52 463 and 52 464, respectively.