Abstract
Background/Aim
The study examines whether DNA level mutations in the carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5) gene Pro-Glu-Leu-Pro-Lys (PELPK) motif differ between patients with appendiceal or colorectal adenocarcinoma. Significant differences between these two groups in correlation with development of metachronous liver metastases could help in the development of targeted therapies and preventative treatment approaches.
Patients and Methods
This retrospective comparative trial analysed 18 patients, 9 with appendiceal adenocarcinoma and 9 with colorectal adenocarcinoma. Genetic sequencing was conducted to detect mutations in the CEACAM5 gene PELPK motif. Data collection spanned from 2016 to 2022, with analysis completed in 2024 at a single tertiary care referral centre, where all participants underwent cytoreductive surgery with hyperthermic intraperitoneal chemotherapy.
Results
No DNA mutations were detected in the CEACAM5 gene PELPK motif in either the study groups. Despite this, significant (but not unexpected) differences were observed between the two groups regarding operative time, peritoneal cancer index, and length of hospital stay (p=0.031, 0.001, and 0.005, respectively). Patients with colorectal adenocarcinoma also had significantly more synchronous liver metastases present at time of their peritonectomies (p=0.029).
Conclusion
The absence of DNA level mutations in the CEACAM5 gene PELPK motif underscores the need for further research at the mRNA and protein levels to better understand the biological distinctions between these two groups. Future studies should focus on exploring alternative molecular pathways that may contribute to the differing clinical profiles of appendiceal and colorectal adenocarcinoma patients.
Keywords: Carcino-embryonic antigen, CEA, CEACAM5, CEA receptor, mutation, appendiceal adenocarcinoma, colorectal adenocarcinoma, PELPK motif, cytoreductive surgery
Recent findings published in an editorial Letter to the British Journal of Surgery propose a novel hypothesis to explain the disparity in liver metastases between appendiceal (AA) and colorectal adenocarcinoma (CA) (1). The hypothesis suggests that in appendiceal cancer, as the disease progresses, mechanical changes cause carcino-embryonic antigen (CEA) to bypass the portal vein, instead rerouting via the parietal peritoneum to the inferior vena cava. This diverges from the pathway described by Lee and Lee, where CEA from colorectal cancer enters the portal circulation, allowing binding to CEA receptor (CEAR) in hepatic Kupffer cells, which potentially triggers liver metastases (2). Further validation of this hypothesis is needed, particularly by replicating the methodology of Tabuchi et al. (3) in appendiceal cancer patients, specifically through comparative analysis of CEA levels in portal and peripheral venous blood. Ethics approval is currently being pursued for this study at our institution.
An additional explanation for the difference in liver metastasis rates between AA (2-5%) and CA (25%) could lie in mutations within the Pro-Glu-Leu-Pro-Lys (PELPK) binding region of the CEA glycoprotein. A study by Zimmer and Thomas in 2001 found that mutations in the CEA of colorectal cancer patients resulted in lower binding affinity to Kupffer cell receptors, potentially explaining the absence of liver metastases in these patients despite elevated serum CEA levels (4). We propose that appendiceal cancer patients may exhibit even higher rates of mutations in the PELPK region, potentially accounting for the lower incidence of liver metastases despite higher CEA levels, as noted in our earlier study.
To explore this hypothesis, archived tissue specimens were obtained from patients who underwent primary peritonectomies for AA or CA (n=9 per group). These specimens were processed at the Australian Genome Research Facility (AGRF) for gene sequencing and bioinformatics analysis to detect mutations in the CEACAM5 gene PELPK motif. With the primary objective being to determine whether there is a proportional difference in the CEACAM5 PELPK motif mutations between patients with AA or CA through the genomic sequencing analysis of bio-banked tissue specimens collected during cytoreductive surgery (CRS).
This study sought to address the knowledge gap regarding CEACAM5 PELPK motif mutations and their association with liver metastasis in AA and CA. By bridging this gap, there is hope that a better understanding of the role of CEA in liver metastasis development might prevail and ultimately there be a contribution to the advancement of targeted immunotherapies for CA, aiming to achieve improved patient outcomes and 5-year survival rates. Secondarily, this study also sought to assess differences in CEA immunohistochemistry (IHC) expression between patients with AA or CA. This study also represents the first genetic investigation into the association between CEACAM5 PELPK motif mutations and liver metastases rates in these distinct adenocarcinomas.
Patients and Methods
Ethical approval. This retrospective cohort study was reported in accordance with the STREGA 2009 guidelines (5). It focused on investigating DNA level mutations in the CEACAM5 gene PELPK motif in patients with AA or CA. Ethics approval was granted by the South Eastern Sydney Local Health District Human Research Ethics Committee (Approval number: 2023/PID00138) for the retrospective use of human tissue samples and the associated clinical data. This study adhered to the principles outlined in the Human Tissue Act 1983, ensuring that additional consent was obtained where necessary. All participants completed written consent forms as per the above protocol.
Study design. This retrospective comparative cohort study aimed to analyse DNA-level mutations in the CEACAM5 gene PELPK motif in patients diagnosed with AA or CA who underwent CRS at St George Hospital’s Peritonectomy Unit. Two cohorts, comprising patients with AA or CA, were compared in terms of their genetic profiles from bio-banked tumour tissue samples. Based on prior studies and statistical power calculations, a minimum of 6 patients per cohort was required, with an expected total of 20 eligible participants recruited over a six-year period from a single centre, St George Hospital, in Kogarah, Sydney, New South Wales, Australia. The study took place at a single site – St George Hospital, Sydney. Tissue samples were procured from the hospital’s biobank and analysed at the AGRF. Recruitment of archived tissue from eligible participants occurred over a six-year period. Patients with histologically confirmed AA or CA who underwent CRS and had bio-banked tissue samples available for genomic sequencing were included. Meanwhile, patients with incomplete clinical records, insufficient bio-banked tissue, or lack of consent for further genomic analysis were excluded.
Methods for follow-up. Patients were followed up for a total of five years post-CRS or until their death. Follow-up assessments included clinical reviews, radiological imaging, and laboratory tests to monitor for recurrence or metastasis, particularly focusing on metachronous liver metastases. Follow-up intervals were scheduled at three, six-, and twelve-months post-operation, and then annually for five years. Clinical data, including survival status, recurrence, and any new metastases, were recorded during each follow-up visit. Patients who did not complete the full five-year follow-up due to early death had their data censored at the time of their last assessment.
Variables. The main outcome variable was the categorical presence of mutations in the CEACAM5 gene PELPK motif. The secondary variables included IHC expression of CEA in tumour tissues from both cohorts. Potential confounders such as patient demographics (age, sex, comorbidities) and tumour characteristics (e.g., grade, stage) were considered.
DNA extraction and sequencing. Tissue samples were retrieved from bio-banked specimens, and genomic DNA were extracted following standard protocols. Sequencing was performed at the AGRF using high-throughput sequencing platforms to assess mutations in the CEACAM5 gene PELPK motif. Genotyping followed standardised allele calling algorithms, and sequencing quality was monitored by calculating error rates and call rates.
Immunohistochemistry (IHC). IHC was conducted to compare CEA expression levels across both groups using established protocols. CEA rabbit polyclonal antibody (Catalog Number: 236A-1; Sigma-Aldrich, St. Louis, MO, USA) were used in routinely processed, neutral-buffered formalin-fixed paraffin-embedded (FFPE) human colonic tissue section tissues (7,8). Tissue sections were approximately 4 μm in thickness and placed on positively charged slides. Deparaffinised tissues were pre-treated with heat-induced epitope retrieval. Staining intensity and distribution were manually qualitatively assessed and interpreted using light microscopy by a licensed pathologist experienced in IHC procedures.
Statistical analysis. The study size of 18 participants (9 AA, 9 CA) were determined based on prior statistical calculations and were designed to ensure adequate power to detect differences in mutation rates and CEA expression. A two-sample independent design were used based on the Tabuchi et al. (3) study, with an enrolment ratio of 1:1, alpha set to 0.05, and power at 90%, the total estimated sample size required is six patients per group. Mutation proportions in the CEACAM5 PELPK motif were compared between the AA and CA groups using the N-1 Chi-squared test. Descriptive statistics [mean±standard deviation (SD) or median and interquartile range (IQR), depending on distribution] were used for demographic and clinical data (6). Multivariate logistic regression models were controlled for potential confounders. Statistical significance was set at p<0.05. No patients were lost to follow-up. Variables where greater than 20% of the data were missing were excluded from any further multivariate logistic regression model analyses. CEA levels and mutation frequencies were treated as continuous variables, while categorical variables such as the presence or absence of mutations were analysed using appropriate statistical tests (N-1 chi-squared).
Subgroup analyses were conducted based on CEA expression levels and the use of HIPEC during surgery. Interactions between variables such as CEACAM5 PELPK mutation status and clinical outcomes (e.g., development of liver metastases) were also explored. No sensitivity analyses were required as missing data was handled by excluding those variables where greater than 20% of the data were missing from any further multivariate logistic regression model analyses. Regarding the Hardy-Weinberg equilibrium (HWE), the allelic distributions at the CEACAM5 gene PELPK motif in both the AA and CA cohorts were tested for HWE using a chi-squared test. Genotypes for the CEACAM5 PELPK motif were inferred through high-throughput sequencing data, processed using standard genotype-calling algorithms from the AGRF. Haplotypes were reconstructed using the PHASE software (Version: 2.1.1, Stephens Lab, Chicago, IL, USA) (9,10). Variant-calling filters were applied to minimise genotyping errors and ensure the reliability of inferred haplotypes. Principal component analysis on the genotype data were used to detect genetic substructure within the study population. The Bonferroni correction was applied for conservative significance estimation, and the Benjamini-Hochberg procedure was used to control the false discovery rate. Relatedness among participants was evaluated using KING software (Version: 2.3.1, Wei-Min Chen, Charlottesville, VA, USA), which calculates pairwise kinship coefficients (11).
Efforts to minimise bias included strict adherence to standardised tissue handling protocols and consistent laboratory methods for DNA extraction and sequencing. Potential biases due to pharmacotherapy, particularly the use of HIPEC during CRS, were controlled for through subgroup analyses.
Results
A total of 686 potentially eligible patients who underwent peritonectomy since January 1996 were examined for eligibility from a prospectively collated database. Of these, nineteen patients met the eligibility criteria, however, due to lack of provision of consent by one of the patient’s next-of-kin, only eighteen patients were included in the study and analysed. All eighteen patients had their primary tumour specimens successfully complete DNA extraction, quality control, and library preparation for next-generation sequencing. Twelve have completed their follow-up (i.e., either five years post-index operation or until their demise), while the remaining six patients are still under routine clinical and radiological follow-up. Of the above twelve, five had completed their five years of follow-up (three of those with CA and two of those with AA), with a further seven patients (two with CA and five with AA) having passed away.
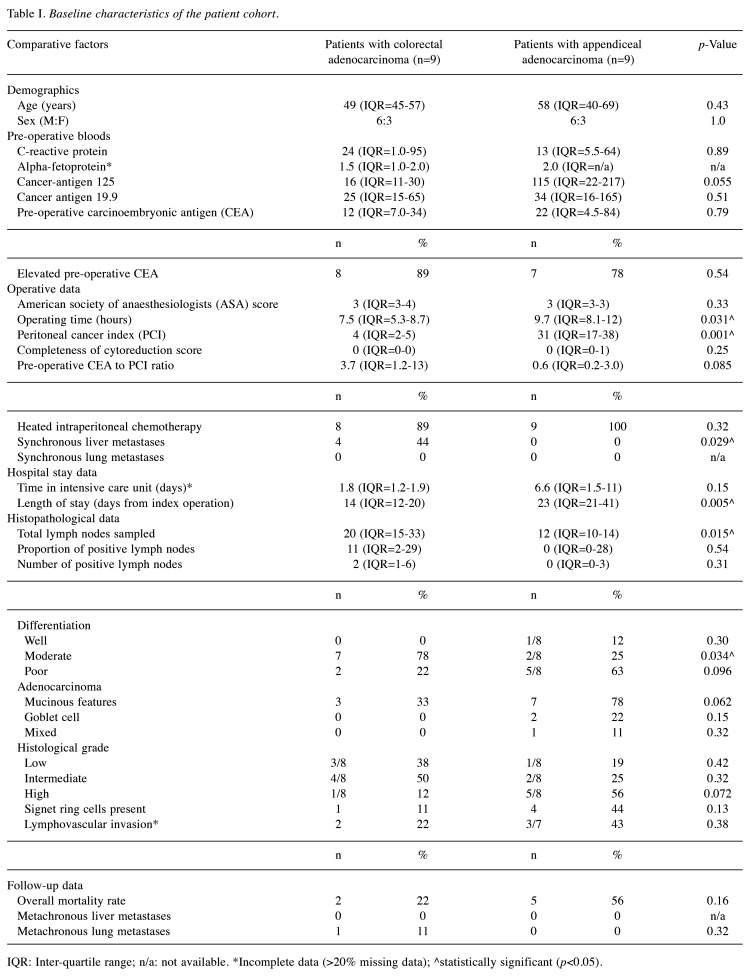
The study included nine patients with CA, and nine patients with AA. The median age of patients with CA were 49 years (IQR=45-57), while the median age of patients with AA was 58 years (IQR=40-69; p=0.43). Six males and three females formed part of both the CA and AA groups, respectively. Missing data were noted from patient’s alpha-fetoprotein (n=9; six with CA and three with AA; NB: this was not tested for on their screening pre-operative bloods). Four (44%) patients with CA had synchronous liver metastases at the time of their index operation. However, no patients developed metachronous liver metastases during their follow-up time, thus far. One patient with CA did develop a metachronous lung metastasis during their follow-up time. Two (22%) patients with CA (mean time to death: data not available) and five (56%) patients with AA (mean time to death: 13.3±5.4 months) died during their follow-up. Baseline characteristics and other detailed outcome data, including survival rates and recurrence patterns, co-morbidities, tumour characteristics, and pre-operative blood results are provided in Table I.
Table I. Baseline characteristics of the patient cohort.
IQR: Inter-quartile range; n/a: not available. *Incomplete data (>20% missing data); ^statistically significant (p<0.05).
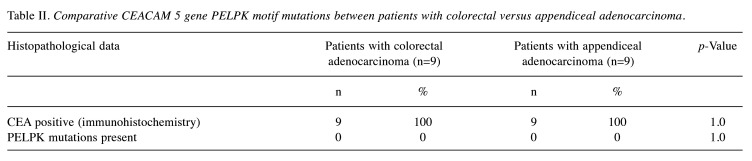
No variations from the reference DNA were detected in the CEACAM5 gene responsible for the PELPK binding motif in any of the 18 patients, however, IHC analysis for CEA in the primary tumour tissue specimens was positive in all 18 patients (Table II). Close familial relationships were not identified, and sensitivity analyses confirm results were not biased by related individuals.
Table II. Comparative CEACAM 5 gene PELPK motif mutations between patients with colorectal versus appendiceal adenocarcinoma.
Discussion
This study is the first to investigate potential genetic differences in the CEACAM5 gene PELPK motif between patients with AA and CA and builds off our institution’s previous research in this field (12,13). The main finding being that no mutations were detected in this gene region across both patient groups. Despite differences in clinical outcomes, including the incidence of liver metastases, this suggests that the PELPK motif is highly conserved at the DNA level in these cancers. However, IHC analysis confirmed the presence of CEA expression across both CA and AA groups, indicating CEA expression despite the absence of genetic mutations in the PELPK motif, reinforcing the role of CEA in tumour biology but suggesting that factors other than PELPK motif mutations might drive metastatic patterns in these cancers.
The strengths of this novel study reigns through as it is the first of its kind to compare the proportions of genetic mutations in the CEACAM5 gene in the region responsible for the PELPK binding motif for the CEA receptor between patients with CCA and AA. The primary limitation of this study is the small sample size, with only eighteen patients enrolled. While this cohort is reflective of a rare cancer population, larger studies are needed to confirm these findings. Despite this limitation, the findings provide insight into the molecular landscape of CEA expression in these cancers, supporting the hypothesis that mechanisms other than PELPK motif mutations may contribute to the observed differences in metastatic behaviour between AA and CA. The authors also believe that similar findings would have been encountered even if more patients had been enrolled. No significant sources of bias were identified, and all genotyping was performed under stringent quality controls.
Our results are consistent with prior studies that have found limited mutational variability in the CEACAM5 gene among colorectal cancer patients. For instance, Zimmer and Thomas in 2001 similarly found minimal variation in CEA genes in patients with CA with elevated CEA levels (4). The absence of mutations in our study suggests that post-transcriptional or post-translational modifications of CEA or other microenvironmental factors may contribute to differences in metastatic spread between CA and AA. Additionally, as noted in studies such as Tabuchi et al. (3) portal CEA levels may not entirely correlate with metastatic potential, indicating the need for further mechanistic research. Saraji et al. (14), on the other hand, demonstrated that CEACAM6 enhances metastasis in metastatic prostate cancer by promoting cell proliferation, migration, and suppressing apoptosis, suggesting CEACAM6 as a potential therapeutic target. Alongside Raghav et al.’s (15) findings on the unique molecular profile of AA, our study further supports that the CEACAM family may play varied roles in cancer progression across different tumour types.
This study opens new avenues for future research. The absence of mutations in the PELPK motif points to other molecular mechanisms being responsible for the differences in liver metastasis rates between AA and CA patients. This could lead to the development of alternative diagnostic tests, such as IHC assays, to identify at-risk patients for liver metastasis based on CEA PELPK expression rather than genetic mutations.
Availability of Data and Materials
The data that support the findings of this study are available from the corresponding author, A.C., upon reasonable request.
Conflicts of Interest
The Authors declare no potential conflicts of interest.
Authors’ Contributions
The first author (A.C.) participated in all aspects of the research study including writing of the manuscript. The second author (S.B.) participated in sample collection and distribution. The third and fourth authors (N.A. & D.L.M.) participated in the study supervision.
Acknowledgements
The authors would like to acknowledge the patients who consented to participate in this study and the members of the St George Peritonectomy Unit for their involvement. Funding was kindly provided by the Royal Australasian College of Surgeons in the form of a Small Project Grant which was used to fund the work performed by the Australian Genome Research Fund.
References
- 1.Cristaudo AT, Morris DL. Hypothesis to explain the disparity in the proportion of liver metastases between appendiceal and colorectal cancer. Br J Surg. 2022;109(4):e63–e64. doi: 10.1093/bjs/znab472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JH, Lee SW. The roles of carcinoembryonic antigen in liver metastasis and therapeutic approaches. Gastroenterol Res Pract. 2017;2017:7521987. doi: 10.1155/2017/7521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tabuchi Y, Deguchi H, Imanishi K, Saitoh Y. Comparison of carcinoembryonic antigen levels between portal and peripheral blood in patients with colorectal cancer correlation with histopathologic variables. Cancer. 1987;59(7):1283–1288. doi: 10.1002/1097-0142(19870401)59:7<1283::aid-cncr2820590709>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 4.Zimmer R, Thomas P. Mutations in the carcinoembryonic antigen gene in colorectal cancer patients: Implications on liver metastasis. Cancer Res. 2001;61(7):2822–2826. [PubMed] [Google Scholar]
- 5.Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, Khoury MJ, Cohen B, Davey-Smith G, Grimshaw J, Scheet P, Gwinn M, Williamson RE, Zou GY, Hutchings K, Johnson CY, Tait V, Wiens M, Golding J, van Duijn C, McLaughlin J, Paterson A, Wells G, Fortier I, Freedman M, Zecevic M, King R, Infante-Rivard C, Stewart A, Birkett N, STrengthening the REporting of Genetic Association Studies STrengthening the REporting of Genetic Association Studies (STREGA): an extension of the STROBE statement. PLoS Med. 2009;6(2):e22. doi: 10.1371/journal.pmed.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furey E. Descriptive Statistics Calculator. Available at https://www.calculatorsoup.com/calculators/statistics/descriptivestatistics.php. [Last accessed on November 23, 2023]
- 7.Sheahan K, O’Brien MJ, Burke B, Dervan PA, O’Keane JC, Gottlieb LS, Zamcheck N. Differential reactivities of carcinoembryonic antigen (CEA) and CEA-related monoclonal and polyclonal antibodies in common epithelial malignancies. Am J Clin Pathol. 1990;94(2):157–164. doi: 10.1093/ajcp/94.2.157. [DOI] [PubMed] [Google Scholar]
- 8.Alkushi A, Irving J, Hsu F, Dupuis B, Liu CL, Van De Rijn M, Gilks CB. Immunoprofile of cervical and endometrial adenocarcinomas using a tissue microarray. Virchows Arch. 2003;442(3):271–277. doi: 10.1007/s00428-002-0752-4. [DOI] [PubMed] [Google Scholar]
- 9.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76(3):449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cristaudo AT, Jennings SB, Morris DL. Comparison of proportion of elevated carcinoembryonic antigen levels in patients with appendiceal and colorectal adenocarcinoma: a systematic review and meta-analysis. Anticancer Res. 2022;42(9):4217–4235. doi: 10.21873/anticanres.15922. [DOI] [PubMed] [Google Scholar]
- 13.Garrett C, Cristaudo A, Barat S, Morris DL. Increased incidence of liver metastases in colorectal versus appendiceal adenocarcinoma peritonectomy patients despite equivocal survival. Anticancer Res. 2023;43(10):4657–4662. doi: 10.21873/anticanres.16661. [DOI] [PubMed] [Google Scholar]
- 14.Saraji A, Wulf K, Stegmann-Frehse J, Kang D, Offermann A, Shaghoyan G, Jonigk D, Kühnel MP, Perner S, Kirfel J, Sailer V. CEACAM6 promotes lung metastasis via enhancing proliferation, migration and suppressing apoptosis of prostate cancer cells. Cancer Genomics Proteomics. 2024;21(4):405–413. doi: 10.21873/cgp.20459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raghav K, Shen JP, Jácome AA, Guerra JL, Scally CP, Taggart MW, Foo WC, Matamoros A, Shaw KR, Fournier K, Overman MJ, Eng C. Integrated clinico-molecular profiling of appendiceal adenocarcinoma reveals a unique grade-driven entity distinct from colorectal cancer. Br J Cancer. 2020;123(8):1262–1270. doi: 10.1038/s41416-020-1015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, A.C., upon reasonable request.