Abstract
Synthetic single-stranded DNA vectors have been used to correct point and frameshift mutations in episomal or chromosomal targets in the yeast Saccharomyces cerevisiae. Certain parameters, such as the length of the vector and the genetic background of the organism, have a significant impact on the process of targeted gene repair, and point mutations are corrected at a higher frequency than frameshift mutations. Genetic analyses reveal that expression levels of the recombination/repair genes RAD51, RAD52 and RAD54 can affect the frequency of gene repair. Overexpression of RAD51 enhances the frequency 4-fold for correction of an episomal target and 5-fold for correction of a chromosomal target; overexpression of RAD54 is also effective in stimulating gene repair, to the same extent as RAD51 in the chromosomal target. In sharp contrast, RAD52 gene expression serves to reduce gene repair activity in rescue experiments and in experiments where RAD52 is overexpressed in a wild-type strain. This may suggest an antagonist role for Rad52p. Consistent with this notion, the highest level of targeted repair occurs when the RAD51 gene is overexpressed in a strain of yeast deficient in RAD52 gene function.
INTRODUCTION
Single base changes in genomic DNA can be directed by synthetic oligonucleotides. These vectors are used either to correct genetic mutations or to introduce stop codons in genes, which terminate protein expression or produce truncated versions of the protein. One class of vectors, known as chimeric oligonucleotides, or chimeras, are self-complementary duplex structures that fold spontaneously into a double-hairpin configuration, a molecule consisting of a double-stranded stem connected by hairpin-capped ends (for a review see 1). One strand of the stem is chimeric, consisting of a DNA and 2′-O-methyl RNA backbone. The double-stranded stem contains the duplex targeting region that is homologous to the target sequence, with the exception of a single base embedded in the middle of this region (2,3). Once bound, the chimera acts as a template for nucleotide exchange and the complementary base is inserted into the target helix directed by the chimera. This reaction is governed by the normal cellular processes of DNA recombination and repair.
Initial reports indicated that the chimera could act at the episomal (4) and the chromosomal level (5). Chimera-directed nucleotide exchange was demonstrated in the alkaline phosphatase gene of cultured human HuH-7 cells (6) and in a factor IX gene of rat hepatocytes in vivo (7), among many others. Single base correction has also been demonstrated in animal models, including the Gunn rat model for Crigler-Najjar syndrome (8), a rat model for carbonic anhydrase deficiency (9) and in the canine (10) and murine (11) models of muscular dystrophy. The chimeric oligonucleotide also proved useful in targeted gene conversion of the tyrosinase gene in albino mouse melanocytes and in the appropriate mouse model, a system providing the means for facile phenotypic readout (12,13). Recently, Li et al. (14) demonstrated targeted conversion of a mutant β-globin gene in MZL cells at a correction efficiency of 1.9%, confirming the initial reports of Cole-Strauss et al. (5). Tagalakis and colleagues then employed this technique to repair a mutation in the APO2E gene and achieved correction frequencies exceeding 30% (15).
A systematic dissection of the functional domains of the chimera (16,17) revealed that the contiguous DNA region is the domain that directs the majority of nucleotide exchange and the RNA region confers an enhanced level of stability onto the conjoined complex of vector and target (18). Single-stranded molecules containing RNA were almost devoid of correction activity, while the DNA construct with the same central mismatch was only one-fifth as active as the chimera in a cell-free extract assay. In the latter case, the lower rate of conversion is likely attributable to rapid degradation of single-stranded DNA. An immediate and reproducible enhancement of activity was observed in mammalian and plant cells using a novel single-stranded DNA targeting vector containing a specific number of modified terminal linkages. Hence, an important step in stabilizing and elevating frequencies of gene repair has involved the simplification of vector design. This new generation of vectors is synthesized more efficiently, as compared to the chimera, with a higher percentage of full-length molecules in the final product.
To begin to understand the mechanism of targeted gene repair in vivo, we chose to examine the role of DNA-associated enzymes, particularly those involved in recombination and/or repair, in the model organism Saccharomyces cerevisiae. Yeast has been used previously by us in studies of DNA repair directed by synthetic oligonucleotides (19–21), and many recombination/repair genes in yeast are conserved in higher eukaryotes.
RAD51, RAD52 and RAD54, members of the RAD52 epistasis group, code for proteins that are central to the initial steps of recombination. Other proteins, such as Rad55p and Rad57p, have been shown to participate in DNA pairing (22), but their roles appear auxiliary to those of Rad51p, Rad52p and Rad54p, respectively. Rad51p and Rad54p have been shown to be required for efficient D-loop formation (23,24). Rad51p acts as a DNA strand transferase capable of assimilating an oligonucleotide into a circular plasmid at regions of complementarity. Rad54p is a Swi2/Snf2-like factor, which promotes D-loop formation by Rad51p (23–25), presumably by transient helix destabilization induced by Rad54p-promoted chromatin remodeling. The joint molecule (D-loop) is the central intermediate in the process of targeted gene repair (16), and the proteins that catalyze its formation are likely to control the rate-limiting factors of the reaction. Rad52p is known to aggregate at DNA lesions and form foci that may represent active sites of DNA repair (26). Rad52p also stimulates DNA annealing and helps ‘load’ Rad51p onto single-stranded DNA prior to strand transfer (27), with the subsequent displacement of RPA. Recent mutational analyses reveal that Rad51p–Rad52p and Rad51p–Rad54p interactions occur on partially overlapping interfaces (28). All of these DNA-associated reactions could impact on the gene repair process and modulate its frequency.
We demonstrated previously that targeted gene repair of a plasmid using modified single-stranded DNA vectors occurs readily in S.cerevisiae (29). Based on these results, and the fact that the D-loop is a critical intermediate in the pairing phase of gene repair, it seemed reasonable to predict that proteins involved in DNA strand assimilation would be good candidates for enzymes that regulate the frequency of repair. As described above, central players in D-loop formation in yeast are Rad51p, Rad54p and, perhaps, Rad52p. Herein, we demonstrate that the activity of all three genes impact on the gene repair reaction, albeit in different ways. Rad51p and Rad54p appear to be important for gene repair activity, while Rad52p acts in an antagonistic fashion. Overexpression of RAD51 or RAD54, but not RAD52, results in both elevated frequencies of targeted gene repair in episomal and chromosomal targets.
MATERIALS AND METHODS
Plasmid DNA constructs and yeast strains
Plasmids pAURHyg(rep)eGFP and pAURHyg(ins)eGFP were constructed by inserting a cassette containing a mutant hygromycin gene and a fused eGFP gene into the pAUR123 shuttle vector, thus producing a stable episomal plasmid (29). This fusion cassette was inserted into pAUR101 to form the integrative plasmid pAUR101Hyg(rep)eGFP. Plasmids pYNRad51, pYNRad52 and pYNRad54 were constructed by inserting the S.cerevisiae genes RAD51, RAD52 and RAD54, respectively, into plasmid pYN132, a modification of pYX132 (R&D Systems Europe, UK) containing a NdeI site in place of the NcoI site (a kind gift from Dr W. K. Holloman, Cornell University School of Medicine). Briefly, the RAD51, RAD52 and RAD54 genes were amplified from yeast strain LSY678 genomic DNA using the following sets of primers: Rad51F (5′-CGGAATTCTCATATGTCTCAAGTTCAAGAACAAC) and Rad51B (5′-GTGCTCGAGCCTACTCGTCTTCTTCTCTGG); Rad52F (5′-CGGAATTCATGGCGTTTTTAAGCTATTTTGC) and Rad52B (5′-GCTCTCGAGTCAAGTAGGCTTGCGTGCATGCAG); Rad54F (5′-CAGAATTCCTGATGGCAAGACGCAGATTACCAG) and Rad54B (5′-TGACTGCTCGAGGTAATGACCCCCCGACGATCGA). The PCR products were cut with EcoRI and XhoI (New England BioLabs) and ligated into pYN132; proper insertion and orientation were confirmed by DNA sequencing. The following yeast strains were a kind gift of Dr Lorraine Symington (Columbia University): LSY678 (MATa leu 2-3, 112 trp 1-1 ura 3-1 his 3-11, 15 ade 2-1 can 1-100); LSY402 (LSY678 rad51::LEU2); LSY386 (LSY678 rad52::LEU2); LSY403 (LSY678 rad54::LEU2.
Oligonucleotides
Single-stranded oligonucleotide vectors Hyg3S/25NT, Hyg3S/40NT, Hyg3S/60NT, Hyg3S/74NT, Hyg3S/74T, Hyg3S/100NT and Kan3S/70T were synthesized as described previously (29). Hyg3S/74NT was designed to target the non-transcribed strand of the hygromycin gene and Hyg3S/74T is directed against the transcribed strand. Kan3S/70T is a non-specific oligonucleotide bearing no complementarity to the hygromycin gene (29).
Transformation of S.cerevisiae
Plasmids and oligonucleotides were transformed into yeast cells by electroporation. Yeast cells were grown in yeast-peptone-dextrose (YPD) or selective media at 30°C to a density of ∼2 × 107 cells/ml, then harvested and washed with dH2O (twice) and 1 M sorbitol. The cells were resuspended in 120 µl of 1 M sorbitol and aliquots (40 µl) of yeast suspension (1 × 108 cell) were electroporated using a Gene Pulser Apparatus (Bio-Rad, Gaithersberg, MD) at the following settings: 1.5 kV, 25 µF, 200 Ω, 1 pulse, 5 s pulse length. Cells were then allowed to recover in 3 ml of YPD medium supplemented with 1 M sorbitol for 16 h and were plated (200 µl) on YPD + hygromycin (300 µg/ml) plates, YPD + aureobasidin A (0.5 µg/ml) plates or on plates lacking tryptophan (SC–trp). Colony counts were determined using an AccuCount 1000 (Biologics) and tabulated from five independent experiments done in triplicate. For the uptake studies, strains LSY678, LSY402, LSY386 and LSY403 were grown at 30°C to a density of ∼107 cells/ml in 40 ml of YPD medium, washed twice with 25 ml of sterile H2O and once with 1 ml of 1 M sorbitol, and then resuspended in 120 µl of 1 M sorbitol. Aliquots (40 µl, 108 cells) were electroporated with 5 µl of 4 µM [32P]Hyg3S/74NT oligonucleotide and recovered in 1 ml of YPD for 30 min. Cells were washed twice with 1 M sorbitol and the radioactivity was detected with a LS6500 scintillation counter (Beckman, Fullerton, CA).
Complementation by RAD51, RAD52 or RAD54
Plasmids pYN132, pYNRad51, pYNRad52 and pYNRad54 were electroporated into LSY678, LSY402(Δrad51), LSY386(Δrad52) and LSY403(Δrad54), respectively. Cells were spread on plates lacking tryptophan, and the presence of the plasmids was confirmed by colony PCR. The overexpression of functional Rad51p, Rad52p and Rad54p in yeast was confirmed by complementation of the sensitivity to methyl methanesulfonate (MMS) seen in the deletion strains. Yeast were cultured in SC–trp medium until the OD600 reached 0.1, then a 10 µl aliquot of a series of 10-fold diluted cell suspensions was plated on SC–trp plates or on 0.020% (v/v) freshly made MMS plates. Cells were cultured at 30°C for 3 days and the growth remaining was visualized and photographed.
Overexpression of RAD51, RAD52 and RAD54 in the episomal yeast strains
Yeast strains LSY678, LSY402(Δrad51), LSY386(Δrad52) and LSY403(Δrad54), bearing either episomal plasmid pAURHyg(rep)eGFP or pAURHyg(ins)eGFP, were transformed with one of four plasmids, pYN132, pYNRad51, pYNRad52 or pYNRad54, then spread on SC–trp medium plates containing aureobasidin A. The presence of the plasmids was confirmed by colony PCR. All targeting experiments were carried out in trp– medium, selective for the RAD expression constructs.
Creation of the integrated hygromycin gene target and overexpression of Rad51p, Rad52p and Rad54p
Integrative plasmid pAUR101Hyg(rep)eGFP was linearized with StuI (New England Biolabs) and electroporated into LSY678 following the protocol described above. Resistant colonies were selected on YPD + aureobasidin A plates, then confirmed by colony PCR and Southern blotting. The integrated hygromycin gene was targeted with vector Hyg3S/25NT, Hyg3S/40NT, Hyg3S/60NT, Hyg3S/74NT, Hyg3S/74T or Hyg3S/100NT and selected as described above. LSY678 bearing the integrated hygromycin cassette was transformed with plasmid pYN132, pYNRad51, pYNRad52 or pYNRad54 and confirmed by colony PCR. These strains were cultured in SC–trp medium to a cell density of 107/ml. Hyg3S/74NT was electroporated and the cells were plated on YPD + hygromycin plates (200 µl of undiluted yeast cells) or on YPD + aureobasidin A plates (200 µl of 105 diluted yeast cells).
RESULTS
Assay system and electrocompetency of wild-type and mutant yeast cell lines
Two mutant plasmids were used as substrates for gene repair with modified single-stranded DNA vectors (29). Each plasmid contains a mutated hygromycin gene (Fig. 1): plasmid pAURHyg(rep)eGFP harbors a point mutation creating a stop codon, while pAURHyg(ins)eGFP contains a single base insertion at the same codon site. These plasmids also contain CEN4 and ARS1 elements for maintenance at low copy number. Correction of either mutation by the oligonucleotide vector Hyg3S/74NT or Hyg3S/74T (also shown in Fig. 1) results in resistance to hygromycin. The plasmids were introduced into the indicated yeast strain by electroporation and maintained stably under selection (29). Subsequently, the single-stranded vectors were electroporated and the cells were allowed to recover for 16 h under aureobasidin selection (0.25 µg/ml). Cells were analyzed for correction of the hygromycin mutation by spreading on agar plates containing hygromycin and by diluting a second sample of cells onto plates containing aureobasidin A, ensuring that the vast majority of cells targeted contained targets of the oligonucleotide vector. The correction efficiency is determined by dividing the number of Hygr colonies by the number of Aurr colonies.
Figure 1.
Genetic assay systems. Gene repair assays utilize plasmids pAURHyg(rep)eGFP and pAURHyg(ins)eGFP and oligonucleotides. Plasmid pAURHyg(rep/ins)eGFP has a synthetic expression cassette, which contains a hygromycin B gene and an eGFP gene fused to the alcohol dehydrogenase (ADH1) constitutive promoter and a selective marker of AUR1-C. Plasmid pAURHyg(rep/ins)eGFP contains a point mutation at nt 137 of the hygromycin B coding sequence, which makes a replacement mutation of TA(T) (tyrosine) to TA(G) (stop codon, indicated by * in frame) or makes an insertion frameshift mutation of TA(T) to TA(AT). Correction of pAURHyg(rep)eGFP requires the replacement of the mutant G residue with a C residue and pAURHyg(ins)eGFP requires the removal of A or T and replacement with a C residue. Hyg3S/74NT and Hyg3S/74T are single-stranded oligonucleotides with a length of 74 bases. NT refers to the oligonucleotides targeting the non-transcribed strand of the hygromycin gene and T those targeting the transcribed strand. *, phosphorothiate linkages within the oligonucleotide and located at both 3′ and 5′ termini as described previously (29).
Intrinsic to this assay is the electrocompetency of the yeast cell and, in particular, mutant cell lines, which vary at only one locus. The experimental protocol relies on electroporation of the oligonucleotide vector and, in some cases, the expression vector for episomal gene repair. Similarly, targeting experiments carried out at the chromosomal level involve the same two elements. Thus, a fundamental aspect of gene repair is to measure yeast transformation. To analyze for plasmid electroporation efficiencies, plasmid pAURHyg(wt)eGFP containing the wild-type hygromycin gene was introduced into strains LSY678, LSY402(Δrad51), LSY386(Δrad52) and LSY403(Δrad54). The number of hygromycin-resistant colonies was approximately equal for all three strains (data not shown), indicating that plasmid transformation is equally proficient. To test for the electrocompetency of the oligonucleotide vector, Hyg3S/74NT was radiolabeled at the 5′ end and the purified molecule introduced into each of the strains listed above. As shown in Table 1, LSY402 and LSY386 have comparable electrocompetencies when compared with wild-type (LSY678), while LSY403(Δrad54) exhibits 60% of the wild-type activity. This difference should be taken into consideration when comparing correction efficiencies. In addition, Danhash et al. (30) demonstrated that introduction of plasmid DNA into yeast by electroporation can result in a slow growth phenotype. And, while our growth curves did not exhibit obvious differences, this phenomenon should be taken into consideration when interpreting the data.
Table 1. Electrocompetency of Δrad51, Δrad52 and Δrad54 with 32P-labeled oligonucleotides.
| Strains | 32P (c.p.m.) | Cell density (107/ml) | 32P (c.p.m./108 cells) | Relative radioactivity |
|---|---|---|---|---|
| LSY678 | 580758.8 | 1.35 | 430191.7 | 1 |
| LSY402 | 577503.4 | 1.35 | 427780.3 | 0.99 |
| LSY386 | 600765.9 | 1.38 | 435337.6 | 1.01 |
| LSY403 | 372718.1 | 1.52 | 245209.3 | 0.57 |
Strains LSY678, LSY402, LSY386 and LSY403 were electroporated with 4 µM [32P]74mer oligonucleotide (5 µl) and the radioactivity incorporated into the respective cell line determined (see Materials and Methods).
Targeted gene repair in yeast recombination/repair mutants with or without complementation
Table 2 outlines the results of targeting experiments conducted in the three mutant strains and a single wild-type strain. Strain LSY678 does not contain any deleted genes from the Rad52p epistasis group and is, therefore, essentially the wild-type control for these experiments. The target plasmid pAURHyg(rep)eGFP was introduced into each strain previously and is maintained by growing the cells under aureobasidin A selection. The indicated plasmid construct plus 5 µg Hyg3S/74NT was electroporated into each of the four yeast strains. Plasmid pYN132 is the empty expression vector. The correction efficiency was again determined by dividing the number of hygromycin-resistant colonies appearing by the number of aureobasidin A-resistant colonies. As depicted in Table 2, strains containing the indicated deletion, rad51, rad52 or rad54, demonstrate a reduced capacity to support the correction of pAURHyg(rep)eGFP, with LSY402(Δrad51) having the most severe impact. In contrast, gene repair activity in LSY386(Δrad52) is elevated or at least unaffected when standard deviation extremes are considered. A similar profile of activity is observed when pAURHyg(ins)eGFP is the target (Table 3). In both cases, RAD51 and RAD54 appear to be required for repair, while the RAD52 gene appears less important for the correction of a point mutation because, in this case, only a modest reduction in repair activity was observed.
Table 2. Complementation of Δrad51, Δrad52 and Δrad54 in episomal gene repair of a point mutation.
| Strain | Construct | Hygr (mean ± SD) | CE (105) |
|---|---|---|---|
| LSY678 | pYN132 | 2632.25 ± 389.48 | 12.64 ± 3.66 |
| pYNRad51 | 5008.33 ± 401.44 | 49.41 ± 13.68 | |
| pYNRad52 | 276.13 ± 49.78 | 1.52 ± 0.31 | |
| pYNRad54 | 4156.57 ± 897.58 | 34.76 ± 5.56 | |
| LSY402(Δrad51) | pYN132 | 232.56 ± 42.11 | 1.01 ± 0.21 |
| pYNRad51 | 7189.32 ± 688.74 | 45.91 ± 12.37 | |
| pYNRad52 | 202.14 ± 30.31 | 0.94 ± 0.35 | |
| pYNRad54 | 507.41 ± 96.34 | 1.70 ± 0.27 | |
| LSY386(Δrad52) | pYN132 | 2315.14 ± 578.75 | 17.63 ± 7.95 |
| pYNRad51 | 12400.57 ± 1356.16 | 299.51 ± 41.76 | |
| pYNRad52 | 908.51 ± 127.12 | 8.28 ± 1.32 | |
| pYNRad54 | 9180.32 ± 1774.58 | 201.97 ± 33.15 | |
| LSY403(Δrad54) | pYN132 | 614.79 ± 79.92 | 3.29 ± 0.53 |
| pYNRad51 | 1224.27 ± 125.88 | 6.58 ± 0.92 | |
| pYNRad52 | 291.40 ± 67.02 | 1.64 ± 0.26 | |
| pYNRad54 | 1846.51 ± 430.68 | 10.86 ± 2.28 |
Strains of LSY678, LSY402, LSY386 and LSY403 harboring plasmid pAURHyg(rep)eGFP and either pYN132, pYNRad51, pYNRad52 or pYNRad54 were electroporated with 5 µg Hyg3S/74NT and then spread on YPD + hygromycin plates. Average hygromycin-resistant colony counts from these experiments are presented, as well as the correction efficiency (CE) calculated as hygromycin-resistant colonies divided by aureobasidin A-resistant colonies. Means ± SD are average counts from five experiments plus or minus standard deviation.
Table 3. Complementation of Δrad51, Δrad52 and Δrad54 in episomal gene repair of a frameshift mutation.
| Strain | Construct | Hygr (mean ± SD) | CE (105) |
|---|---|---|---|
| LSY678 | pYN132 | 1811.25 ± 162.76 | 8.26 ± 1.46 |
| pYNRad51 | 2882.75 ± 248.16 | 16.67 ± 6.70 | |
| pYNRad52 | 143.75 ± 61.45 | 0.54 ± 0.22 | |
| pYNRad54 | 1952.25 ± 599.96 | 12.11 ± 5.27 | |
| LSY402(Δrad51) | pYN132 | 72.14 ± 19.37 | 0.56 ± 0.14 |
| pYNRad51 | 6164.56 ± 560.03 | 17.60 ± 4.56 | |
| pYNRad52 | 206.35 ± 65.68 | 0.60 ± 0.36 | |
| pYNRad54 | 359.51 ± 65.80 | 1.63 ± 0.32 | |
| LSY386(Δrad52) | pYN132 | 339.91 ± 108.99 | 3.65 ± 1.6 |
| pYNRad51 | 7646.78 ± 1073.12 | 139.82 ± 49.17 | |
| pYNRad52 | 45.53 ± 12.07 | 0.16 ± 0.06 | |
| pYNRad54 | 2502.67 ± 933.95 | 17.81 ± 6.68 | |
| LSY403(Δrad54) | pYN132 | 262.37 ± 86.74 | 2.16 ± 0.70 |
| pYNRad51 | 408.39 ± 55.27 | 2.65 ± 0.85 | |
| pYNRad52 | 416.47 ± 170.29 | 0.83 ± 0.17 | |
| pYNRad54 | 840.35 ± 41.65 | 6.04 ± 1.90 |
Strains of LSY678, LSY402, LSY386 and LSY403 bearing plasmid pAURHyg(ins)eGFP, pYN132, pYNRad51, pYNRad52 or pYNRad54 were electroporated with 5 µg Hyg3S/74NT. Average hygromycin-resistant colonies numbers and correction efficiency are presented, as well as standard deviation; n = 5 experiments.
Next, we attempted to rescue the gene repair in the mutant strains and to see if overexpression of each of these genes can boost activity in the wild-type strain. To begin, RAD51, RAD52 and RAD54 were cloned into a yeast expression vector pYN132 under the control of a constitutive promoter, TPI. Since plasmid targets pAURHyg(rep)eGFP and pAURHyg(ins)eGFP are already present in the strains (maintained under aureobasidin A selection), the pYN expression cassette was chosen because it could be maintained under a different, trp, selection. To begin testing the effect of these genes directly, cells with pAURHyg(rep)eGFP or pAURHyg(ins)eGFP were electroporated with Hyg3S/74NT and gene repair was ascertained as a function of the appearance of Hygr colonies. Overexpression of RAD51, RAD52 and RAD54, respectively, was tested first in LSY678 and, as also shown in Table 2, RAD51 and RAD54 facilitated an increase in gene repair, while overexpression of RAD52 decreased repair efficiency. Specifically, the expression construct containing RAD51 produced a 4-fold increase in correction of pAURHyg(rep)eGFP, while RAD54 was less effective. When mutant strains lacking one of these genes served as the host for these experiments, the overexpression of RAD51 rescued the activity in the Δrad51 deletion mutant, producing a comparable level of enhancement observed when pYNRAD51 was overexpressed in LSY678. However, neither RAD52 nor RAD54 had much impact on repair activity in LSY402(Δrad51). Similarly, overexpression of RAD54 rescued the Δrad54 mutant and restored gene repair activity to near wild-type levels, but to only one-third the level of activity seen when RAD54 was overexpressed in LSY678. Overexpression of RAD51 in the Δrad54 strain also led to the re-establishment of activity in this mutant. Interestingly, the overexpression of RAD52 in LSY386 did not restore activity and, in fact, a significant reduction in gene repair activity was observed, suggesting further that Rad52p may act in a suppressive fashion. Overexpression of RAD51 not only restored activity in the LSY386(Δrad52) strain, but also produced activity that exceeded wild-type levels, even under conditions in which RAD51 was overexpressed in the wild-type background. In particular, overexpression of RAD51 in a Δrad52 mutant strain increased targeted gene repair ∼17-fold over the mutant strain and >23-fold above wild-type levels. A similar level of enhancement was also observed when RAD54 was overexpressed in LSY386, reinforcing the belief that the activity of Rad54p in this reaction is reduced in the presence of functional Rad52p.
As previously reported (1,29,31), the level of gene repair is reduced when a frameshift mutation is targeted for repair. The results presented in Table 3 confirm and extend this observation. Plasmid pAURHyg(ins)eGFP served as the target in strains LSY678, LSY402, LSY386 and LSY403, respectively. In each strain, RAD51, RAD52 or RAD54 was overexpressed and the gene repair activity analyzed once again. The pattern of enhancement was very similar to that seen in previous targeting experiments using pAURHyg(rep)eGFP (Table 2). Overexpression of RAD51 or RAD54 increased the repair activity in LSY678, while RAD52 overexpression reduced gene repair activity. Again, the overexpression of RAD51 in LSY386(Δrad52) exhibited the highest level of enhancement (nearly 17-fold above wild-type levels), while overexpression of RAD54 in LSY386 was lower here than its enhancement of point mutation repair. It is noteworthy that overexpression of PMS1, RAD57 or RAD10 in the pYN132 vector did not affect the targeted repair reaction (data not shown). These control experiments indicate that the type of gene inserted in pYN132 is important.
The results observed on RAD51 overexpression in the Δrad52 strain may initially seem paradoxical, but these data suggest that Rad51p and Rad52p operate in pathways that may share an essential common factor or process the single-stranded vector differently. For example, the Rad51p-driven reaction may be the favored pathway and effective in initiating complexes that mature more productively into repair events. In contrast, Rad52p-driven pathways may be less effective in creating the initial reaction intermediate for gene repair. The consistent reduction in repair activity observed with overexpression of RAD52 in all strains strongly suggests a suppressive or antagonistic role. Thus, the absence of RAD52 gene activity eliminates this ‘sink effect’ and enables a higher level of repair activity, perhaps directed by Rad51p with or without Rad54p.
Overexpression of the RAD51, RAD52 and RAD54 genes produces functional proteins
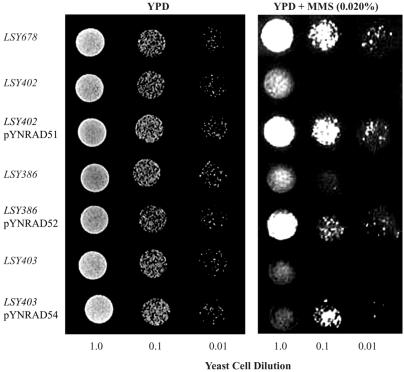
To confirm that overexpression from the pYN vector produced the desired protein in functional form, an MMS sensitivity assay was performed. We tested for the presence of functional protein by its capacity to protect against the DNA-damaging agent MMS, an established functional assay for the activity of this set of recombination/repair genes. In this assay, the growth of the mutant cells is dependent on the capacity of the overexpressed protein to function in double-stranded break repair. As seen in Figures 2 and 3, Δrad51, Δrad52 and Δrad54 mutants (the same strains used for the gene repair assays) are unable to grow on 0.02% MMS, while the wild-type strain grows well. Overexpression of RAD51, RAD52 or RAD54 restored resistance to MMS in the appropriate mutant strains, as evidenced by the presence of cell growth in the strains bearing the overexpression constructs. These results confirm that functional Rad51p, Rad52p and Rad54p are produced from their respective overexpression constructs. In addition, RT–PCR analyses performed on all expression construct/yeast strain combinations revealed that a similar level of each gene is being expressed (data not shown).
Figure 2.
Resistance to MMS conferred by overexpression. Aliquots of 10 µl of a series of 10-fold diluted yeast cells were loaded on YPD and YPD containing 0.020% (v/v) MMS plates and cultured at 30°C for 3 days. LSY678 yeast wild-type strain bearing pYN132 plasmids; LSY402 yeast bearing pYN132; LSY402 yeast bearing pYNRad51; LSY386 yeast bearing pYN132; LSY386 yeast bearing pYNRad52; LSY403 yeast bearing pYN132; LSY403 yeast bearing pYNRad54.
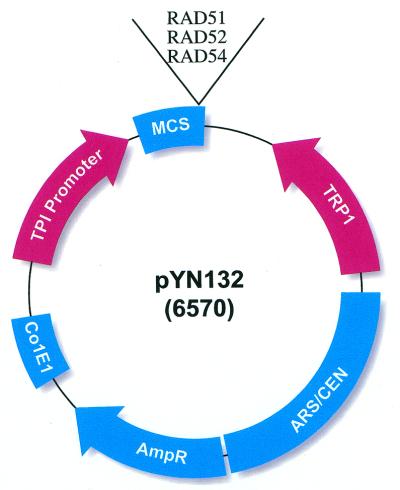
Figure 3.
Structure of gene expression plasmid pYN132. Plasmid pYN132 contains a constitutive triosephosphate isomerase (TPI) promoter, a tryptophan (TRP1)-selective marker and an ARS/CEN sequence, which maintain the plasmid in low copy numbers in yeast. ScRAD51, RAD52 and RAD54 cDNA were PCR amplified and inserted into pYN132 at EcoRI/XhoI sites.
Gene repair of an integrated chromosomal target
The results of episomal targeting indicate that Rad51p, Rad52p and Rad54p are factors in regulating the frequency of gene repair positively and negatively. To translate these observations to the level of the chromosome, pAURHyg(rep)eGFP was integrated into LSY678 at the AUR1 locus in chromosome XI. Clones containing the integrated plasmid were selected by growth in aureobasidin A and a new strain, LSY678(int)Hyg, was isolated and confirmed by Southern blotting. Thus, to begin the next phase of the experiments, we examined chromosomal gene repair at a basal level by simply introducing either Hyg3S/74NT or Hyg3S/74T into LSY678(int)Hyg. Previously, we found that an oligonucleotide vector designed to direct correction on the non-transcribed (or non-template) strand produced a higher level of gene repair than the corresponding vector targeted to the transcribed strand (29). The same two vectors can be used to direct the repair of the point mutation integrated in a yeast chromosome. As shown in Table 4, repair of the chromosomal mutation was, in fact, observed, and increasing the amount of each Hyg3S vector produced correspondingly more hygromycin-resistant colonies. Surprisingly, the level of gene repair of the chromosomal target with these single-stranded vectors is approximately two-thirds the level of episomal repair. Consistent with the episomal gene targeting reported in Liu et al. (29), the vector targeting the non-transcribed strand produced a higher level of gene repair at each dosage tested, with a maximum at 7.5 µg and declining slightly at 10.0 µg. DNA sequence analyses confirmed that the repair activity was precise and the targeted base was indeed changed (data not shown).
Table 4. Strand bias for gene repair of a chromosomal point mutation in LSY678 [strain LSY678(Int)Hyg].
| Vector | Vector concentration (µg) | Hygr (mean ± SD) | CE (105) |
|---|---|---|---|
| Hyg3S/74NT | 0 | 0 | 0 |
| 2.5 | 612.88 ± 153.22 | 4.64 ± 0.88 | |
| 5.0 | 1213.75 ± 178.99 | 8.39 ± 1.76 | |
| 7.5 | 2125.34 ± 246.25 | 19.67 ± 4.37 | |
| 10.0 | 2011.25 ± 552.86 | 18.59 ± 5.58 | |
| Hyg3S/74T | 0 | 0 | 0 |
| 2.5 | 165.40 ± 26.23 | 1.25 ± 0.32 | |
| 5.0 | 356.76 ± 47.03 | 2.47 ± 0.64 | |
| 7.5 | 758.93 ± 97.32 | 7.03 ± 0.76 | |
| 10.0 | 718.21 ± 206.46 | 6.51 ± 1.42 |
LSY678(Int)Hyg containing an integrated copy of pAUR(Int)Hyg(rep)eGFP was electroporated with varying amounts of the indicated vector. Average hygromycin resistance and the correction efficiency of five experiments plus standard deviation are presented.
We also reported earlier that the length of the targeting vector was a parameter in the frequency of gene repair (1,29,31). In general, as the oligonucleotide was lengthened, the level of gene repair of an episomal target was increased until a 100mer was used, and then activity was seen to diminish. The same length dependence experiment was carried out in LSY678(Int)Hyg in order to determine if the optimal vector length for episomal gene repair held for a chromosomal mutation. Using the same Hyg3S vectors with lengths varying from 25 to 100 nt, the repair of the integrated hygromycin point mutation was evaluated. As shown in Table 5, the frequency of gene repair increased as a function of the length of the vector, with a 74mer showing maximal correction, and decreasing when a 100mer was used as the correction vector. These results parallel those found when repair of an episomal mutation was analyzed as a function of length (29).
Table 5. Vector length influences the repair of a chromosomal mutation [strain LSY678(Int)Hyg].
| Vector | Hygr (mean ± SD) | CE (105) |
|---|---|---|
| Hyg3S/25NT | 21.23 ± 6.51 | 0.13 ± 0.05 |
| Hyg3S/40NT | 529.52 ± 64.15 | 4.49 ± 0.39 |
| Hyg3S/60NT | 1051.25 ± 104.86 | 6.73 ± 0.79 |
| Hyg3S/74NT | 1374.22 ± 143.56 | 8.85 ± 1.30 |
| Hyg3S/100NT | 116.51 ± 39.31 | 0.91 ± 0.27 |
LSY678(Int)Hyg containing an integrated copy of pAUR(Int)Hyg(rep)eGFP was electroporated with 220 pmol indicated vector. Average hygromycin-resistant colonies and the correction efficiency of five experiments with standard deviation are presented.
Enhancement of gene repair by overexpression of RAD51, RAD52 and RAD54
The results presented in Tables 2 and 3 indicate that the frequency of targeted gene repair can be increased by the overexpression of RAD51 or RAD54 and that overexpression of RAD52 decreases activity in LSY678. The same experiment could now be carried out in LSY678(Int)Hyg by introducing the overexpression constructs and monitoring the corrective activity of Hyg3S/74NT on chromosomal repair. As shown in Table 6, repair of the integrated hygromycin mutation was less than episomal repair, but the addition of expression constructs containing either RAD51 or RAD54 elevated the targeting frequency ∼5-fold. Overexpression of RAD52, however, did not increase the level of targeted gene repair and, in fact, decreased activity. Overall, these results are quite similar to those obtained when an episome served as the template for repair. Taken together, these results suggest that repair of a chromosomal target can be enhanced by overexpression of RAD51 or RAD54 and, perhaps, suppressed by overexpression of RAD52.
Table 6. Overexpression of RAD51, RAD52 and RAD54 in LSY678(Int)Hyg.
| Expression construct | Hygr (mean ± SD) | CE (105) |
|---|---|---|
| pYN132 | 278.74 ± 71.75 | 7.71 ± 2.23 |
| pYNRad51 | 1683.37 ± 321.73 | 38.17 ± 4.50 |
| pYNRad52 | 286.36 ± 68.73 | 4.95 ± 1.68 |
| pYNRad54 | 1693.21 ± 257.17 | 37.14 ± 3.51 |
LSY678(Int)Hyg strains containing an integrated copy of pAUR(Int)Hyg(rep)eGFP and either pYN132, pYNRad51, pYNRad52 or pYNRad54 were electroporated with 5 µg Hyg3S/74NT. Average hygromycin-resistant colonies and the correction efficiency of five experiments with standard deviation are shown.
DISCUSSION
Targeted gene repair directed by oligonucleotides may become an effective technique for altering single nucleotides at specific sites if the molecular mechanism is elucidated. Previous studies have demonstrated its versatility by successful application in mammalian cells (12,13,15,32–34 and references therein), animal models (6–8,10), plants (35–37) and yeast (1,29,31). Within some of these systems, however, a range of correction frequencies has been observed, frustrating and impeding subsequent efforts. To this end, we established a yeast model system for defining the reaction parameters of targeted repair in the hope of isolating and eventually manipulating the factors that regulate the frequency.
Earlier studies suggest that the repair reaction likely consists of two fundamental phases: DNA pairing and DNA repair (18). Biochemical evidence supports the notion that the oligonucleotide vector (here a single-stranded moiety) conjoins with the target helix to form a joint molecule known as a D-loop (16,17). Thus, we chose to study prominent eukaryotic proteins (recombinases) known to promote DNA pairing and analyze their impact on the frequency of targeted gene repair.
This study focused on three genes from the RAD52 epistasis group: RAD51, RAD52 and RAD54. These three genes are believed to be central players in the initial steps of homologous recombination. As shown in Table 2, RAD51, RAD52 and RAD54 appear to regulate the episomal gene repair pathway, as deletion of any of these genes alters activity. Deletion of RAD51 or RAD54 had a severe impact on gene repair, while the absence of RAD52 gene function resulted in either no reduction or a modest increase in activity. The results are similar whether the target is a point or frameshift mutation. Since evidence exists to support the coordinated activity of Rad51p and Rad54p in D-loop formation (24), these data are consistent with the notion that this type of joint molecule may, in fact, be a reaction intermediate in targeted gene repair and perhaps that its construction is rate limiting. In addition, these data suggest that the same factors influencing the DNA pairing phase are important to the overall reaction, regardless of what type of single base mutation is present at the target site.
Complementation of LSY402(Δrad51) or LSY403(Δrad54) with the corresponding gene led to nearly complete rescue of episomal gene repair activity. In contrast, overexpression of RAD52 in LSY386(Δrad52) led to a reduction in gene activity. In fact, Rad52p production suppressed gene repair in all cell lines tested herein. But, overexpression of RAD51 or RAD54 in strain LSY386(Δrad52) resulted not only in the total recovery of gene repair activity, but also in a significant enhancement above the Δrad52 baseline activity, as well as wild-type levels. Previous cell-free extract data (31) suggested that yeast strains containing a deletion in the RAD52 gene exhibited a higher level of gene repair activity, and in vivo data presented herein align with the notion that functional RAD52 may act to inhibit the reaction. Its deletion apparently sets the stage for an expanded enhancement of gene repair when RAD51 or RAD54 is overexpressed. When the electrocompetency data are taken into consideration (see Table 1), the level of enhancement directed by RAD54 overexpression approximates that of RAD51.
Many of the observations obtained in studies of episomal repair extend to the level of the chromosome. First, gene repair appears to be equally efficient (or inefficient) whether the target is episomal or chromosomal (integrated). Second, vectors designed to hybridize to the non-transcribed strand are more effective in directing repair than those hybridizing to the transcribed strand, confirming our earlier observations (29). Third, there is an enhancement of repair activity as a function of vector length, up to 74 bases, but a reduction in activity when a 100mer is used. Finally, overexpression of either RAD51 or RAD54 leads to significant enhancement of chromosomal repair and overexpression of RAD52 reduced activity.
The current model of DNA pairing in eukaryotes during double-strand break repair centers around the activity of Rad51p. Initially, single-stranded DNA is bound by RPA and then processed by Rad52p for loading with Rad51p, with subsequent displacement of RPA. The nucleoprotein filament containing DNA and Rad51p searches for homology with the assistance of Rad54p, acting as a chromatin remodeler (23,25,38), destabilizing nucleosomal DNA and eventually enabling stable base pairing between the single-stranded vector and the target.
Our overall observations can be explained by this model with one exception. Based on the above scenario, the enhancement of gene overexpression of the recombinase Rad51p would be predicted to enhance gene repair, because Rad51p is likely to catalyze DNA strand transfer, assimilating the vector onto the target. And any activity, such as that of Rad54p, which promotes nucleosome repositioning or DNA unwinding, thereby producing a more accessible DNA target, would also be predicted to elevate the frequency of gene repair. In fact, our data confirm this prediction. However, according to the paradigm of double-strand break repair, a role of Rad52p is to load Rad51p onto a single-stranded DNA molecule (here the vector). But, in every reaction tested herein, the overexpression of RAD52 led to either no enhancement or a significant reduction in activity. We also observed that in the absence of this gene, the overexpression of RAD51 or RAD54 led to the highest level of targeted gene repair. D-loop formation, which is likely to happen in both episomal and chromosomal targeting, likely requires the activity of the DNA recombinases, such as Rad51p (with or without Rad54p), which can promote strand invasion and strand assimilation. Rad52p, in contrast, does not promote D-loop formation, but does catalyze DNA annealing. Thus, Rad52p could be competing directly with Rad51p or Rad54p by binding to the single-stranded vector in an attempt to find a complementary partner. The need for the ‘loading activity’ of Rad52p may be substantially reduced when either Rad51p or Rad54p is present in abundance. Hence, by simple equilibrium shifts, the need for Rad52p-associated Rad51p-promoted reactions may be diminished.
The data provided in this paper reveal that Rad51p, Rad52p and Rad54p impact on, and perhaps regulate, the process of targeted gene repair directed by modified single-stranded DNA molecules. Thus, it is clear that by understanding the proteins involved in the targeted gene repair mechanism and by carefully manipulating the content of these proteins within cells and the genetic background of the cell, we may increase the frequency and bring the potential of therapeutic gene repair closer to reality.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Lorraine Symington for many of the yeast strains in this study. We thank Ms Elizabeth Feather for manuscript preparation and Mr Eric Roberts for graphics and illustrations. This work was supported by a grant from the National Institutes of Health (1R01CA89325-01A) and from NaPro Biotherapeutics.
REFERENCES
- 1.Rice M.C., Czymmek,K. and Kmiec,E.B. (2001) The potential of nucleic acid repair in functional genomics. Nat. Biotechnol., 19, 321–326. [DOI] [PubMed] [Google Scholar]
- 2.Kmiec E.B. (1999) Targeted gene repair. Gene Ther., 6, 1–3. [DOI] [PubMed] [Google Scholar]
- 3.Brachman E.E. and Kmiec,E.B. (2002) The biased evolution of targeted gene repair. Curr. Opin. Mol. Ther., 4, 171–176. [PubMed] [Google Scholar]
- 4.Yoon K., Cole-Strauss,A. and Kmiec,E.B. (1996) Targeted gene correction of episomal DNA in mammalian cells mediated by a chimeric RNA.DNA oligonucleotide. Proc. Natl Acad. Sci. USA, 93, 2071–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole-Strauss A., Yoon,K., Xiang,Y., Byrne,B.C., Rice,M.C., Gryn,J., Holloman,W.K. and Kmiec,E.B. (1996) Correction of the mutation responsible for sickle cell anemia by an RNA-DNA oligonucleotide. Science, 273, 1386–1389. [DOI] [PubMed] [Google Scholar]
- 6.Kren B.T., Cole-Strauss,A., Kmiec,E.B. and Steer,C.J. (1997) Targeted nucleotide exchange in the alkaline phosphatase gene of HuH-7 cells mediated by a chimeric RNA/DNA oligonucleotide. Hepatology, 25, 1462–1468. [DOI] [PubMed] [Google Scholar]
- 7.Kren B.T., Bandyopadhyay,P. and Steer,C.J. (1998) In vivo site-directed mutagenesis of the factor IX gene by chimeric RNA/DNA oligonucleotides. Nature Med., 4, 285–290. [DOI] [PubMed] [Google Scholar]
- 8.Kren B.T., Parashar,B., Bandyopadhyay,P., Chowdhury,N.R., Chowdhury,J.R. and Steer,C.J. (1999) Correction of the UDP-glucuronosyltransferase gene defect in the Gunn rat model of Crigler-Najjar syndrome type I with a chimeric oligonucleotide. Proc. Natl Acad. Sci. USA, 96, 10349–10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai L.-W. and Lien,Y.H. (1999) Homologous recombination based gene therapy. Exp. Nephrol., 7, 11–14. [DOI] [PubMed] [Google Scholar]
- 10.Bartlett R.J., Stockinger,S., Denis,M.M., Bartlett,W.T., Inverardi,L., Le,T.T., thi Man N., Morris,G.E., Bogan,D.J., Metcalf-Bogan,J. and Kornegay,J.N. (2000) In vivo targeted repair of a point mutation in the canine dystrophin gene by a chimeric RNA/DNA oligonucleotide. Nat. Biotechnol., 18, 615–622. [DOI] [PubMed] [Google Scholar]
- 11.Rando T.A., Disatnik,M.H. and Zhou,L.Z. (2000) Rescue of dystrophin expression in mdx mouse muscle by RNA/DNA oligonucleotides. Proc. Natl Acad. Sci. USA, 97, 5363–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexeev V. and Yoon,K. (1998) Stable and inheritable changes in genotype and phenotype of albino melanocytes induced by an RNA-DNA oligonucleotide. Nat. Biotechnol., 16, 1343–1346. [DOI] [PubMed] [Google Scholar]
- 13.Alexeev V., Igoucheva,O., Domashenko,A., Cotsarelis,G. and Yoon,K. (2000) Localized in vivo genotypic and phenotypic correction of the albino mutation in skin by RNA-DNA oligonucleotide. Nat. Biotechnol., 18, 43–47. [DOI] [PubMed] [Google Scholar]
- 14.Li Z.H., Liu,D.P., Yin,W.X., Guo,Z.C. and Liang,C.C. (2001) Targeted correction of the point mutations of beta-thalassemia and targeted mutagenesis of the nucleotide associated with HPFH by RNA/DNA oligonucleotides: potential for beta-thalassemia gene therapy. Blood Cells Mol. Dis., 27, 530–538. [DOI] [PubMed] [Google Scholar]
- 15.Tagalakis A.D., Graham,I.R., Riddell,D.R., Dickson,J.G. and Owen,J.S. (2001) Gene correction of the apolipoprotein (Apo) E2 phenotype to wild-type ApoE3 by in situ chimeraplasty. J. Biol. Chem., 276, 13226–13230. [DOI] [PubMed] [Google Scholar]
- 16.Gamper H.B., Cole-Strauss,A., Metz,R., Parekh,H., Kumar,R. and Kmiec,E.B. (2000) A plausible mechanism for gene correction by chimeric oligonucleotides. Biochemistry, 39, 5808–5816. [DOI] [PubMed] [Google Scholar]
- 17.Gamper H.B., Parekh,H., Rice,M.C., Bruner,M., Youkey,H. and Kmiec,E.B. (2000) The DNA strand of chimeric RNA/DNA oligonucleotides can direct gene repair/conversion activity in mammalian and plant cell-free extracts. Nucleic Acids Res., 28, 4332–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamper H.B., Hou,Y.M. and Kmiec,E.B. (2000) Evidence for a four-strand exchange catalyzed by the RecA protein. Biochemistry, 39, 15272–15281. [DOI] [PubMed] [Google Scholar]
- 19.Moerschell R.P., Tsunasawa,S. and Sherman,F. (1988) Transformation of yeast with synthetic oligonucleotides. Proc. Natl Acad. Sci. USA, 85, 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto T., Moerschell,R.P., Wakem,L.P., Komar-Panicucci,S. and Sherman,F. (1992) Strand-specificity in the transformation of yeast with synthetic oligonucleotides. Genetics, 131, 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto T., Moerschell,R.P., Wakem,L.P., Ferguson,D. and Sherman,F. (1992) Parameters affecting the frequencies of transformation and co-transformation with synthetic oligonucleotides in yeast. Yeast, 8, 935–948. [DOI] [PubMed] [Google Scholar]
- 22.Sung P. (1997) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev., 11, 1111–1121. [DOI] [PubMed] [Google Scholar]
- 23.Petukhova G., Stratton,S. and Sung,P. (1998) Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature, 393, 91–94. [DOI] [PubMed] [Google Scholar]
- 24.Van Komen S., Petukhova,G., Sigurdsson,S., Stratton,S. and Sung,P. (2000) Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell, 6, 563–572. [DOI] [PubMed] [Google Scholar]
- 25.Mazin A.V., Bornarth,C.J., Solinger,J.A., Heyer,W.D. and Kowalczykowski,S.C. (2000) Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell, 6, 583–592. [DOI] [PubMed] [Google Scholar]
- 26.Lisby M., Rothstein,R. and Mortensen,U.H. (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl Acad. Sci. USA, 98, 8276–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortensen U.H., Bendixen,C., Sunjevaric,I. and Rothstein,R. (1996) DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl Acad. Sci. USA, 93, 10729–10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krejci L., Damborsky,J., Thomsen,B., Duno,M. and Bendixen,C. (2001) Molecular dissection of interactions between Rad51 and members of the recombination-repair group. Mol. Cell. Biol., 21, 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Liu L., Rice,M.C. and Kmiec,E.B. (2001) In vivo gene repair of point and frameshift mutations directed by chimeric RNA/DNA oligonucleotides and modified single-stranded oligonucleotides. Nucleic Acids Res., 29, 4238–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danhash N., Gardner,D.C. and Oliver,S.G. (1991) Heritable damage to yeast caused by transformation. Biotechnology, 9, 179–182. [DOI] [PubMed] [Google Scholar]
- 31.Rice M.C., Bruner,M., Czymmek,K. and Kmiec,E.B. (2001) In vitro and in vivo nucleotide exchange directed by chimeric RNA/DNA oligonucleotides in Saccharomyces cerevisae. Mol. Microbiol., 40, 857–868. [DOI] [PubMed] [Google Scholar]
- 32.Igoucheva O., Alexeev,V. and Yoon,K. (2001) Targeted gene correction by small single-stranded oligonucleotides in mammalian cells. Gene Ther., 8, 391–399. [DOI] [PubMed] [Google Scholar]
- 33.Bandyopadhyay P., Kren,B.T., Ma,X. and Steer,C.J. (1998) Enhanced gene transfer into HuH-7 cells and primary rat hepatocytes using targeted liposomes and polyethylenimine. Biotechniques, 25, 282–292. [DOI] [PubMed] [Google Scholar]
- 34.Bandyopadhyay P., Ma,X., Linehan-Stieers,C., Kren,B.T. and Steer,C.J. (1999) Nucleotide exchange in genomic DNA of rat hepatocytes using RNA/DNA oligonucleotides. Targeted delivery of liposomes and polyethyleneimine to the asialoglycoprotein receptor. J. Biol. Chem., 274, 10163–10172. [DOI] [PubMed] [Google Scholar]
- 35.Beetham P.R., Kipp,P.B., Sawycky,X.L., Arntzen,C.J. and May,G.D. (1999) A tool for functional plant genomics: chimeric RNA/DNA oligonucleotides cause in vivo gene-specific mutations. Proc. Natl Acad. Sci. USA, 96, 8774–8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu T., Peterson,D.J., Tagliani,L., St Clair,G., Baszczynski,C.L. and Bowen,B. (1999) Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proc. Natl Acad. Sci. USA, 96, 8768–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu T., Mettenburg,K., Peterson,D.J., Tagliani,L. and Baszczynski,C.L. (2000) Engineering herbicide-resistant maize using chimeric RNA/DNA oligonucleotides. Nat. Biotechnol., 18, 555–558. [DOI] [PubMed] [Google Scholar]
- 38.Eisen J.A., Sweder,K.S. and Hanawalt,P.C. (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res., 23, 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]