Abstract
The p120ctn-binding partner Kaiso is a new member of the POZ-zinc finger family of transcription factors implicated in development and cancer. To understand the role of Kaiso in gene regulation and p120ctn-mediated signaling and adhesion, we sought to identify Kaiso-specific DNA binding sequences and potential target genes. Here we demonstrate that Kaiso is a dual specificity DNA-binding protein that recognizes the specific consensus sequence TCCTGCNA as well as methyl-CpG dinucleotides. A minimal core sequence CTGCNA was identified as sufficient for Kaiso binding. Two copies of the Kaiso-binding site are present in the human and murine matrilysin promoters, implicating matrilysin as a candidate target gene for Kaiso. In electrophoretic mobility shift assays, matrilysin promoter-derived oligonucleotide probes formed a complex with GST–Kaiso fusion proteins possessing the zinc finger domain but not with fusion proteins lacking the zinc fingers. We further determined that only Kaiso zinc fingers 2 and 3 were necessary and sufficient for sequence-specific DNA binding. Interestingly, Kaiso also possesses a methyl-CpG-dependent DNA-binding activity distinct from its sequence-specific DNA binding. However, Kaiso has a higher affinity for the TCCTGCNA consensus than for the methyl-CpG sites. Furthermore, the DNA-binding ability of Kaiso with either recognition site was inhibited by p120ctn. Kaiso thus appears to have two modes of DNA binding and transcriptional repression, both of which may be modulated by its interaction with the adhesion cofactor p120ctn.
INTRODUCTION
Kaiso is a new member of the BTB/POZ (Broad Complex, Tramtrak, Bric à brac/pox virus and zinc finger) family of zinc finger transcription factors, hereafter POZ-ZF proteins, which are strongly implicated in development and cancer (reviewed in 1,2). The majority of these proteins function as transcriptional repressors and are characterized by an N-terminal protein–protein interaction POZ domain and a C-terminal DNA-binding domain that consists of one or more Kruppel-like C2H2 zinc fingers. Like other members of the POZ-ZF family, Kaiso homodimerizes through its highly conserved POZ domain and it localizes to the nucleus through a presently unknown mechanism that may involve the POZ domain (3). Currently, the only known binding partner for Kaiso is the cell adhesion cofactor p120ctn. Surprisingly, however, this interaction is not mediated by the protein–protein interaction POZ domain of Kaiso but rather is mediated via a C-terminal region of Kaiso that encompasses its three putative DNA-binding zinc fingers (3). We postulate that perhaps the interaction of p120ctn with Kaiso through its C-terminus serves to regulate Kaiso DNA binding or transcriptional activity.
p120ctn is an Armadillo repeat protein that was originally identified as a Src kinase substrate and subsequently found to be a component of the E-cadherin–catenin cell adhesion complex (4–6). Like the classical catenins β-catenin and plakoglobin, p120ctn binds E-cadherin directly via its Arm domain (7). However, whereas β-catenin and plakoglobin bind E-cadherin at a C-terminal catenin-binding site and anchor the adhesion complex to the actin cytoskeleton, p120ctn binds E-cadherin at a distinct juxtamembrane site that has been implicated in the regulation of cadherin adhesive strength and cell motility (7–11). Since the interaction of p120ctn with Kaiso is also mediated via its Arm domain, p120ctn forms mutually exclusive complexes with either E-cadherin or Kaiso (3). Interestingly, the p120ctn–Kaiso interaction appears restricted to epithelial cells (our unpublished data), hinting at an E-cadherin or epithelial-dependent phenomenon influencing the p120ctn–Kaiso interaction. Moreover, the fact that Kaiso does not interact with any other component of the cadherin–catenin complex (i.e. E-cadherin, β-catenin, plakoglobin or α-catenin) (3) suggests that Kaiso plays an important and unique role in p120ctn function and signaling.
The majority of POZ-ZF proteins are transcriptional repressors and the most studied family members, the oncoproteins BCL-6 and PLZF, recruit the macromolecular histone deacetylase co-repressor complex that includes histone deacetylase (HDAC) and the co-repressors SMRT, NCoR and Sin3A (12–16). Since the POZ domain mediated the interactions with the components of the HDAC complex, it was postulated that HDAC recruitment was a common mechanism for all POZ-ZF transcriptional repressors. However, studies of two other POZ-ZF transcriptional repressors, the tumor suppressor HIC-1 and its homolog HRG22, revealed that their mechanism of repression does not involve the HDAC complex (17,18). While current evidence indicates that Kaiso is a transcriptional repressor (19,20), its mechanism of action is unknown. However, preliminary data from our laboratory indicate that Kaiso, like HIC-1, does not associate with the HDAC complex. This suggests that (i) direct recruitment of the HDAC complex may not be a common mechanism of transcriptional repression by the POZ-ZF family of transcription factors and (ii) that Kaiso and HIC-1 may have a novel mechanism of transcriptional repression.
To fully understand the physiological role of Kaiso in signaling or cell adhesion and to gain further insight into its mechanism of transcriptional repression, this study focuses on the DNA-binding properties of Kaiso and the initial identification of potential target genes. We performed a cyclic amplification and selection of targets (CAST) analysis and identified TCCTGCNA (where N is any nucleotide) as a Kaiso-specific DNA binding consensus sequence. The presence of two copies of this optimal consensus site in both the human and mouse matrilysin promoters and the sequence-specific binding of Kaiso to matrilysin-derived oligonucleotides implicates matrilysin as a Kaiso target gene. Recently it was reported that Kaiso also binds CpG (cytidine–guanidine dinucleotide) methylated sites in the S100A4 gene promoter (20). This implicates S100A4 as another potential Kaiso target gene and suggests an alternative DNA recognition site for Kaiso. Consistent with those observations, we confirmed that Kaiso specifically binds S100A4-derived oligonucleotides containing methylated CpG sites. However, we found that Kaiso has a slightly higher affinity for the sequence-specific consensus site TCCTGCNA than for the methyl-CpG site. Interestingly, p120ctn can inhibit Kaiso DNA-binding ability to either recognition site.
MATERIALS AND METHODS
Plasmids
pGEX-Kaiso constructs were generated using pBluescript-Kaiso as a template for PCR amplification of various fragments spanning the C-terminal two-thirds of Kaiso that include the zinc finger domain. pGEX-KaisoΔPOZ was generated using the primers 5′-GCGCGAATTCCCAGACTCAGCAGTCAGT-3′ and 5′-GCGCCTCGAGCGTTGACTTGTTCTGAGG-3′ and spanned nt 1285–2166. The fragment was then subcloned in-frame into the EcoRI and XhoI sites of pGEX-4T. pGEX-KaisoΔPOZΔZF was constructed using the primers 5′-GCGCGAATTCGAGCTTGGTGTCCCACTG-3′ and 5′-GCGCCTCGAGTCTTCGCAAGCTTGTCAG-3′, which amplified a fragment spanning nt 598–1782 that was then subcloned into the EcoRI and XhoI sites of pGEX-4T. The full panel of pGEX-Kaiso constructs was likewise generated by PCR amplification of the desired fragments and subcloning into pGEX. The pGEX constructs were then used to transform DH5α bacteria. Bacterial cultures were grown for 90 min at 30°C before protein expression was induced by the addition of 100 mM IPTG to a final concentration of 0.1 mM. Cultures were grown for an additional 3 h and the bacteria pelleted by centrifugation and frozen at –80°C. The frozen bacterial pellets were thawed and resuspended in 10 ml cold phosphate-buffered saline pH 7.4, 0.1% NP-40, and lysed by sonication. The sonicates were centrifuged and the fusion proteins purified from the supernatants by incubation for 30 min with 500 µl glutathione–Sepharose beads/ml lysate. The protein–Sepharose slurry was washed twice with DNA-binding buffer and stored at 4°C or the glutathione S-transferase (GST)–Kaiso fusion proteins were eluted from the beads with 10 mM reduced glutathione in 50 mM Tris pH 8.0. The proteins were subjected to SDS–PAGE and visualized by Coomassie blue staining to confirm their size and purity.
DNA-binding site selection
Random oligonucleotide selection and amplification reactions were performed with bacterially expressed GST–Kaiso fusion proteins purified and bound on glutathione–agarose beads. A 50 µl slurry of protein-bound beads was incubated at 4°C with ∼100 ng oligonucleotide randomers, 5′-CGCGGATCCTGCAGCTCGAG-(30N)-GTCGACAAGCTTCTAGAGCA-3′, in 1× binding buffer (25 mM HEPES pH 7.5, 100 mM KCl, 1 mM EDTA, 10 mM MgCl2, 0.1% NP-40, 5% glycerol, 1 mM DTT) with 160 ng poly(dI–dC) and 160 ng BSA/ml. Unbound randomers were removed by four washes with 1× binding buffer. Selected oligomers were released from the protein-bound beads by boiling and an aliquot subjected to PCR amplification with the primers 5′-CGCGGATCCTGCAGCTCGAG-3′ and 5′-TGCTCTAGAAGCTTGTCGAC-3′. The amplified Kaiso-selected DNA was then incubated with a fresh aliquot of GST–Kaiso-bound agarose beads. This sequence of binding, boiling and PCR amplification represents one CAST cycle. Six sequential rounds of randomer selection by the GST–Kaiso-bound agarose beads followed by PCR amplification (94°C for 1 min, 52°C for 2 min and 72°C for 2 min for 10 cycles) were performed. The final PCR products were gel purified using the Mermaid kit (Bio 101, Carlsbad, CA), subcloned into pBluescript and sequenced by the dideoxynucleotide chain termination method with a Sequenase v.2.1 Sequencing Kit (USB, Cleveland, OH). A total of 56 clones were sequenced and the consensus DNA-binding site was determined by visual examination of the sequences.
Electrophoretic mobility shift assays (EMSA)
An aliquot of 50 ng double-stranded oligonucleotide was end-labeled with polynucleotide kinase and [γ-32P]ATP for 45 min at 37°C. Labeled probe was purified by centrifugation through a Clontech Chroma-spin TE-10 column according to the manufacturer’s instructions (Clontech, Palo Alto, CA). The two oligonucleotides used were human matrilysin Kaiso site oligo sequence 5′-GTGCTTCCTGCCAATAACG-3′ and murine matrilysin Kaiso site oligo sequence 5′-GTTCCTCCTGCCAATATAAAAAC-3′. Oligonucleotides were cytosine-methylated using SssI methylase according to the manufacturer’s instructions (NEB, Beverly, ME). The labeled probe was quantitated in a scintillation counter and 20 000–50 000 c.p.m. oligonucleotide was incubated with bacterially expressed GST–Kaiso in 1× DNA-binding buffer for 10 min at room temperature followed by a 30 min incubation on ice. Reactions were run on a 4% polyacrylamide/0.5× TBE gel containing 2.5% glycerol at 200 V for 3–4 h, dried and subjected to autoradiography. The cold competition assay utilized 20, 40 or 100 ng unlabeled probe and 1 ng purified, labeled probe, such that unlabeled probe was present in 20, 40 and 100 times excess, respectively. The p120ctn co-incubation assays were performed with the following modifications. Aliquots of 2.5 or 5 µg GST–p120ctn or 5 µg GST proteins were preincubated with GST–Kaiso at 4°C for 1.5 h to allow specific Kaiso–p120ctn protein–protein interactions. A sample of 50 000 c.p.m. oligonucleotide probe was then added and the reaction further incubated at room temperature for 25 min followed by 30 min on ice to allow Kaiso–DNA binding.
RESULTS
Kaiso is a sequence-specific DNA-binding protein
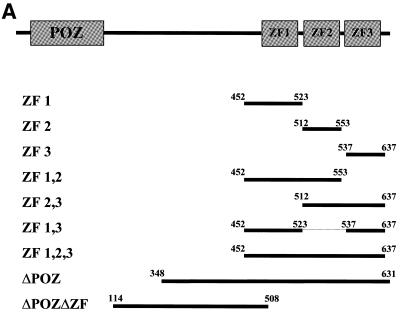
The structural similarity of Kaiso to the DNA-binding POZ-ZF transcription factors and the presence of three highly conserved Kruppel-like C2H2 zinc fingers at its C-terminus suggested that Kaiso bound DNA. However, the fact that the p120ctn-binding site of Kaiso encompasses its zinc finger domain raised the possibility that the zinc fingers were involved in protein–protein interactions as well as DNA binding. To resolve this issue, we performed a CAST analysis (21) using the bacterially expressed GST–KaisoΔPOZ fusion protein to determine if Kaiso had DNA-binding activity and if it did, to identify a Kaiso-specific DNA recognition site (Fig. 1A). This truncated KaisoΔPOZ fusion protein, containing the putative DNA-binding zinc finger domain but lacking the protein–protein interaction POZ domain, was used to circumvent the potential inhibitory DNA-binding action of the POZ domain (1). The GST fusion protein was affinity purified and bound to glutathione–agarose beads before incubation with double-stranded randomers containing 30 bp of random nucleotide sequence flanked at the N- and C-termini with 20 bp of known sequence containing a restriction site and a PCR priming sequence. The selected and amplified randomers were subcloned and 56 independent clones were sequenced. A candidate consensus DNA binding site, TCCTGCNA, with a highly conserved core sequence (CTGCNA) was deduced (Fig. 1B). Forty-eight of the 56 clones sequenced had at least one site and eight of the 48 clones had two perfect copies of the consensus. The remaining eight clones had no site, a partial site or unreadable sequence. Bacterially expressed GST and GST–KaisoΔPOZΔZF (no protein–protein interaction domain and no DNA-binding domain) were used as negative controls and did not bind DNA. These findings thus established that the Kaiso zinc finger domain does indeed mediate its DNA binding.
Figure 1.
Kaiso binds the consensus DNA sequence TCCTGCNA. (A) Schematic representation of Kaiso highlighting regions expressed as GST fusion proteins for EMSA studies. Amino acid residue positions are indicated above the bars. (B) Sequence analysis of randomers selected during CAST by GST–KaisoΔPOZ. The nucleotide sequences of 56 clones selected by Kaiso during CAST were examined for a consensus site. Thirty-two representative clones are shown with the derived consensus sequence TCCTGCNA highlighted. Sequences shown include the entire selected randomer and flanking primer sequence. Perfect matches (shaded boxes and bold font) and imperfect matches (shaded boxes and plain font) are indicated.
Kaiso binds specifically to a matrilysin-derived oligonucleotide
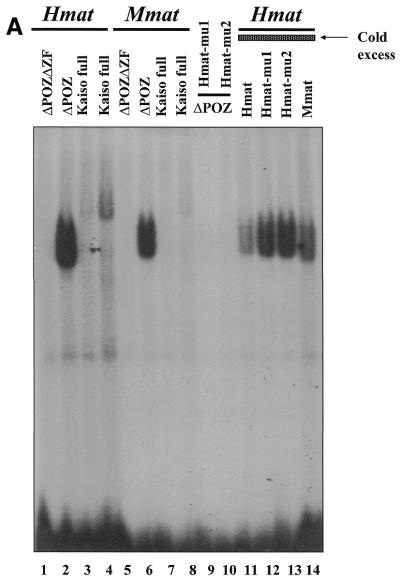
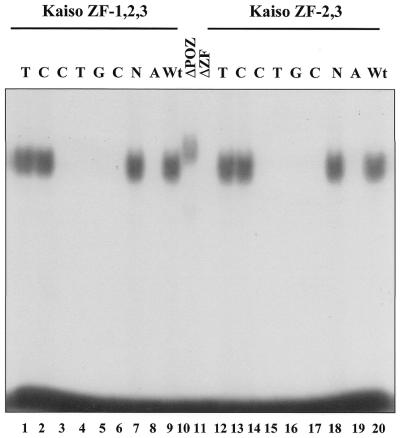
Having identified a Kaiso consensus sequence, we were next interested in determining whether any known genes possessed the Kaiso-binding site and were potential targets for transcriptional regulation by Kaiso. Since the p120ctn interaction with Kaiso was reminiscent of the interaction between the classical catenin, β-catenin, and the transcription factor TCF, we questioned whether p120ctn and Kaiso would target the same genes as β-catenin–TCF. Of the known β-catenin–TCF target genes, myc, cyclin D1 and matrilysin (22–25), we found that the human and mouse matrilysin promoters each had at least two perfectly conserved copies of the Kaiso-binding DNA consensus site. To confirm that the deduced Kaiso consensus sequence was physiologically relevant and that Kaiso could bind the matrilysin promoter through this recognition site, two double-stranded oligomers based on the human and mouse matrilysin promoter sequences were used in an EMSA with bacterially expressed GST–Kaiso. Both the human and murine matrilysin-derived oligomers formed a complex with GST–Kaiso containing the zinc fingers but not with GST–Kaiso lacking the zinc fingers (Fig. 2A, compare lanes 1 and 2, human matrilysin probe, and lanes 5 and 6, murine matrilysin probe). In addition, complex formation with the human matrilysin oligo (Hmat) could be competed with 100-fold excess unlabeled, wild-type Hmat oligo (Fig. 2A, lane 11) and to a lesser extent with unlabeled wild-type Mmat oligo (Fig. 2A, lane 14). However, complex formation was not competed with excess mutant Hmat oligos possessing a modified Kaiso consensus (Fig. 2, lanes 12 and 13). Like other POZ-ZF proteins, full-length Kaiso bound the oligo with less affinity than the Kaiso zinc finger domain alone (Fig. 2A, compare lanes 3, 4, 7 and 8 to lanes 2 and 6). This is believed to be due to the presence of the inhibitory POZ domain (1), which facilitates homodimerization and perhaps induces conformational changes that hinder DNA binding. The binding specificity of the Kaiso zinc finger domain for the matrilysin-derived oligo was also confirmed by use of EDTA as a zinc chelating agent to abrogate binding (Fig. 2B, lanes 7 and 8). Kaiso–DNA binding was restored upon addition of excess zinc ions (Fig. 2B, lanes 4 and 5). Mutational analysis of the matrilysin-derived oligonucleotide further validated the Kaiso consensus site. A panel of murine matrilysin-derived mutated oligos was incubated with GST–KaisoZF1,2,3 or GST–KaisoZF2,3 and EMSA revealed the core nucleotides of the Kaiso-binding site as CTGCNA (Fig. 3 and summarized in Table 1). Kaiso–DNA complex formation was completely abolished when any of the core nucleotides (CTGCNA) were changed. This is consistent with the core consensus nucleotides present in the selected randomer clones and further confirms the specificity of the Kaiso–DNA interaction.
Figure 2.
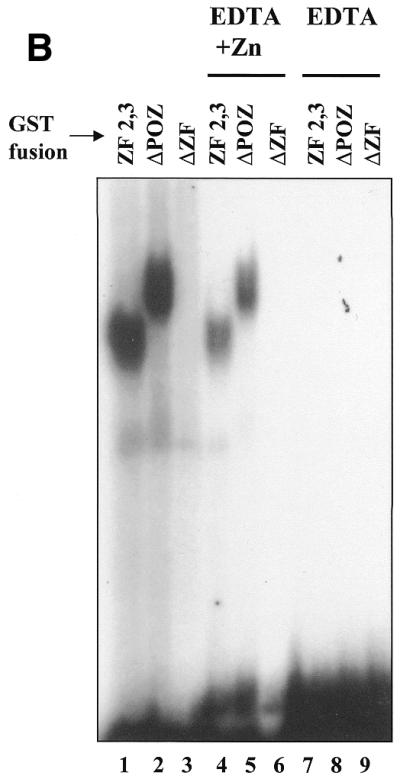
Kaiso binds a matrilysin-derived probe with a perfect Kaiso-binding site. (A) Gel shifts were performed using a radiolabeled 19 bp oligonucleotide containing the Kaiso-binding site based on the human matrilysin promoter or a 23 bp oligonucleotide based on the murine matrilysin promoter and bacterially expressed GST–KaisoΔPOZ (zinc fingers only), GST–KaisoΔPOZΔZF (no zinc fingers or POZ domain) or full-length GST–Kaiso. Competition assays were performed with 100-fold excess of cold unlabeled oligonucleotide as indicated. Only GST–KaisoΔPOZ and, to a lesser extent, full-length GST–Kaiso bound and shifted the labeled oligos (lanes 2, 4, 6 and 8). (B) GST–Kaiso fusion proteins were incubated with murine matrilysin probe in the presence or absence of 500 mM EDTA or in the presence of 500 mM EDTA and 2 M zinc sulfate. As predicted, Kaiso–DNA complex formation is dependent on the presence of zinc ions (compare lanes 7 and 8 to lanes 1, 2, 4 and 5).
Figure 3.
Mapping the core nucleotides of the Kaiso DNA binding site. Various oligonucleotides were synthesized with a 1 nt change each relative to the Kaiso consensus site (see Table 1 for exact sequences). Kaiso–DNA complex formation was completely abolished by single nucleotide changes of the core sequence CTGCNA but not by nucleotide changes outside the putative core.
Table 1. Kaiso binding to human and murine matrilysin DNA probes.
| Probe | Oligonucleotide sequence | Binding |
|---|---|---|
| Hmat | GTGCTTCCTGCCAATAACGa | ++++ |
| mu-Hmat | GTGCTTCCCGCCAATAACG | – |
| mu-Hmat-me | GTGCTTCCMGCCAATAACGb,c | ++++ |
| Mmat | GTTCCTCCTGCCAATATAAAAAC | ++++ |
| Mmat-mu1 | GTTCCTACTGCCAATATAAAAAC | ++ |
| Mmat-mu2 | GTTCCTCATGCCAATATAAAAAC | + |
| Mmat-mu3 | GTTCCTCCGGCCAATATAAAAAC | – |
| Mmat-mu4 | GTTCCTCCTTCCAATATAAAAAC | – |
| Mmat-mu5 | GTTCCTCCTGACAATATAAAAAC | – |
| Mmat-mu6 | GTTCCTCCTGCCCATATAAAAAC | – |
| Mmat-mu7 | GTTCCTCCTGCAAATATAAAAAC | + |
| Mmat-mu8 | GTTCCGCCTGCCAATATAAAAAC | ++++ |
aThe Kaiso DNA binding site is underlined.
bmu-Hmat-me is the methylated oligonucleotide.
cBold nucleotides represent changes from the conserved sequence.
Kaiso zinc fingers 2 and 3 mediate its sequence-specific DNA binding
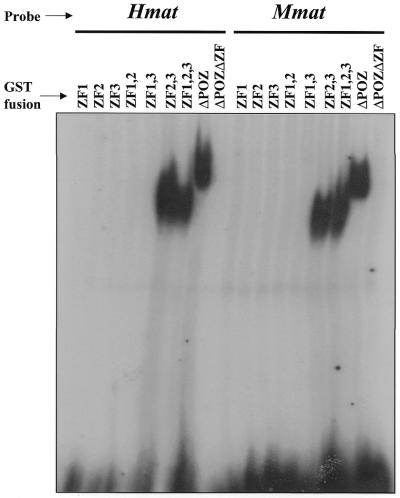
To map the specific region of the Kaiso zinc finger domain involved in DNA binding, we generated a panel of GST–Kaiso fusion protein deletion mutants (Fig. 1A) and analyzed their DNA-binding ability with both human and murine matrilysin-derived oligos (Fig. 4). Our gel shifts revealed that none of the individual zinc fingers were able to bind the human or murine matrilysin-derived oligos. However, GST–Kaiso fusion proteins containing all three zinc fingers or zinc fingers 2 and 3 formed a complex with the oligos (Fig. 4). GST–Kaiso fusion proteins containing zinc fingers 1 and 2 or zinc fingers 1 and 3 did not form protein–DNA complexes with the oligos (Fig. 4). This indicates that zinc fingers 2 and 3 are necessary and sufficient for binding to both the human and murine matrilysin-derived oligos.
Figure 4.
Kaiso zinc fingers 2 and 3 mediate DNA binding. Gel shifts were performed using a panel of GST–Kaiso deletion mutants incubated with human or murine matrilysin-derived oligonucleotides as indicated. None of the individual Kaiso zinc fingers bound DNA. The minimal region required for DNA binding encompassed zinc fingers 2 and 3.
Kaiso binds a methylated S100A4 promoter-derived oligonucleotide
A recent study reported that Kaiso is a DNA methylation-dependent transcriptional repressor and component of the MECP1 methyl-CpG-binding complex (20). In that study, sequence analysis of the methyl-CpG-bound proteins from nuclear extracts of the adenocarcinoma cell line CSML-0 identified Kaiso as a key component. Purified recombinant Kaiso was subsequently found to bind with high affinity to a methylated S100A4-derived probe with the sequence MGMGCCCAAMG, where M represents 5-methylcytosine. The S100A4 protein, also known as Metastasin, is a member of a family of calcium-binding proteins with roles in signal transduction, cell proliferation, cell adhesion and tumor metastasis (reviewed in 26). Since the CpG recognition sequence in that report differed significantly from our CAST-determined Kaiso binding site (CGCGCCCAACG versus TCCTGCNA), we wanted to determine whether Kaiso could indeed efficiently recognize and bind to two distinct DNA sites.
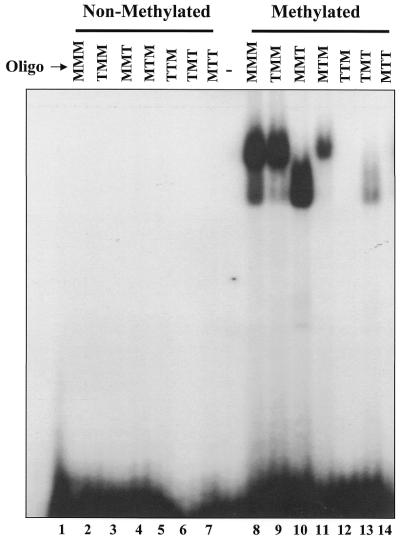
Like the human and murine-derived matrilysin probes, the S100A4/mts methyl-CpG probe formed complexes with GST–Kaiso containing the zinc fingers (i.e. ZF1,2,3, ZF2,3 and ΔPOZ) but not with GST–Kaiso lacking the zinc fingers (data not shown). To clarify the disparity in the DNA recognition sequences, we compared the binding ability of the minimal GST–Kaiso fusion (Kaiso-ZF2,3) to a panel of oligonucleotides based on the S100A4/mts methyl-CpG probe. We designed six oligonucleotides with the CpG sites mutated to give various combinations of one or two CpG sites compared with the wild-type consensus sequence with three CpG sites (Table 2). First, none of the non-methylated oligonucleotides formed protein–DNA complexes with Kaiso (Fig. 5, lanes 1–7). However, consistent with Prokhortchouk et al. (20), we found that the first two methylcytosines in the S100A4/mts probe were the most critical for Kaiso binding (Fig. 5 and Table 2). Of the three methyl-CpG sites, we found that only the second methylated CpG site displayed an individual affinity for Kaiso, although its affinity was significantly reduced relative to the probes containing two or three CpG sites (Fig. 5, compare lane 13 with lanes 8–11). It is also interesting to note that oligonucleotides mts-1 (with methylated CpG sites 2 and 3) and mts-3 (with methylated CpG sites 1 and 2) bound more efficiently than mts-2, where CpG sites 1 and 3 were methylated. The common feature between the mts-1 and mts-3 oligos is the methylated CpG site 2, and this further underscores the importance of the second CpG site in Kaiso recognition and binding. Unexpectedly, the methylated, Kaiso-bound oligo migrated as two complexes, the significance of which remains unknown. It is possible that these complexes represent oligonucleotides bound with either one (lower complex) or two (upper complex) Kaiso peptides.
Table 2. Binding of Kaiso to methylated S100A4/metastasin probes.
| Probe | Oligonucleotide sequence | Binding |
|---|---|---|
| mts | AGCAGCCGCGCCCAACGCTGGGAGa | – |
| mts-me | AGCAGCMGMGCCCAAMGCTGGGAGb,c | ++++ |
| mts-1 | AGCAGCTGMGCCCAAMGCTGGGAG | +++ |
| mts-2 | agcagcMGTGcccaaMGctgggag | ++ |
| mts-3 | AGCAGCMGMGCCCAATGCTGGGAG | +++ |
| mts-4 | AGCAGCTGTGCCCAAMGCTGGGAG | – |
| mts-5 | AGCAGCTGMGCCCAATGCTGGGAG | + |
| mts-6 | AGCAGCMGTGCCCAATGCTGGGAG | – |
aCpG sites are underlined.
bM represents cytosine-methylated nucleotide.
cBold nucleotides represent changes from the conserved sequence.
Figure 5.
Kaiso binds specifically to methylated CpG sites in S100A4-derived probes. A panel of S100A4 (mts) promoter-derived oligonucleotides with 1 or 2 NT changes in the CpG recognition site were used in EMSA experiments with GST–KaisoΔPOZ. MMM refers to methylation at all three CpG sites, MTM, methylation at sites 1 and 3, etc. None of the non-methylated probes bound GST–Kaiso (lanes 1–7). However, methylated wild-type and mutated oligos containing at least two of the three CpG sites formed complexes with GST–Kaiso (lanes 8–14). Kaiso also formed a complex with the mts-5 probe (TMT) in which only the second CpG site was methylated (lane 13).
Kaiso binds the sequence-specific consensus site with higher affinity
To further characterize the apparent dual specificity DNA binding activity of Kaiso, we performed EMSA competition experiments. GST–KaisoZF2,3 fusion proteins were incubated with labeled S100A4-derived methylated oligonucleotides and cold excess Hmat- or Mmat-derived oligos containing the consensus TCCTGCNA. As seen in Figure 6A, Kaiso–methyl-CpG DNA complex formation was significantly reduced (>90%) by a 40-fold excess of the Hmat and Mmat oligos containing the sequence-specific Kaiso-binding site and completely abolished by a 60 times excess. The specificity of the competition was further demonstrated by the failure of 60 times excess mutant Mmat oligo to affect Kaiso–CpG DNA complex formation. In contrast, when the reciprocal experiment was performed and Kaiso was incubated with Hmat- or Mmat-derived oligonucleotides containing the consensus Kaiso binding site and competed with excess unlabeled methylated S100A4-derived oligonucleotides, the Kaiso–DNA consensus complex was not affected until 60 times excess methylated S100A4 oligo was used (compare Fig. 6A and B). This suggests that although Kaiso may be a dual recognition DNA-binding transcription factor, it has a higher affinity for the sequence-specific consensus site TCCTGCNA identified by our CAST analysis than for methyl-CpG dinucleotides.
Figure 6.
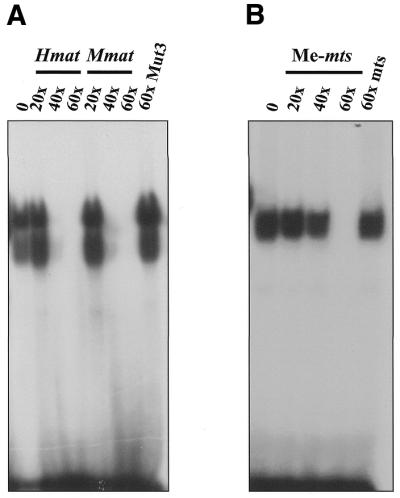
Kaiso binds with higher affinity to TCCTGCNA than to methylated CpG sites. GST–KaisoZF2,3 fusion proteins were incubated with labeled wild-type mts methylated oligonucleotides and cold excess Hmat or Mmat oligonucleotides containing the consensus TCCTGCNA (A) or incubated with Mmat oligonucleotides containing the consensus Kaiso-binding site and competed with excess unlabeled methylated S100A4-derived oligonucleotides (B). A 60 times excess of S100A4 was required to compete with the Mmat TCCTGCNA sequence, in contrast to 40 times excess Mmat to compete with S100A4.
p120ctn inhibits Kaiso DNA binding
In previous studies we found that the p120ctn-binding region of Kaiso mapped to the C-terminal third of the protein that encompassed the zinc finger domain (3). Interestingly, however, Kaiso constructs containing the ZF domain alone did not bind p120ctn, indicating that the ZF domain itself did not interact with p120ctn. This suggested that the p120ctn-binding site is a non-contiguous epitope flanking the DNA-binding ZF domain (3). These observations led us to question whether the p120ctn interaction with Kaiso could interfere with Kaiso DNA-binding ability. To address this question, we performed EMSA experiments and preincubated GST–p120ctn with the various GST–Kaiso fusion proteins for 1 h before addition of the radiolabeled oligonucleotide probe. As seen in Figure 7, preincubation of Kaiso with 10-fold excess p120ctn protein did indeed inhibit the GST–KaisoΔPOZ fusion protein from binding both the matrilysin (Hmat) and the methylated S100A4 (mts) oligonucleotides. Interestingly, however, preincubation with p120ctn had no effect on the DNA-binding ability of GST–KaisoZF1,2,3 or GST–KaisoZF2,3 fusion proteins.
Figure 7.
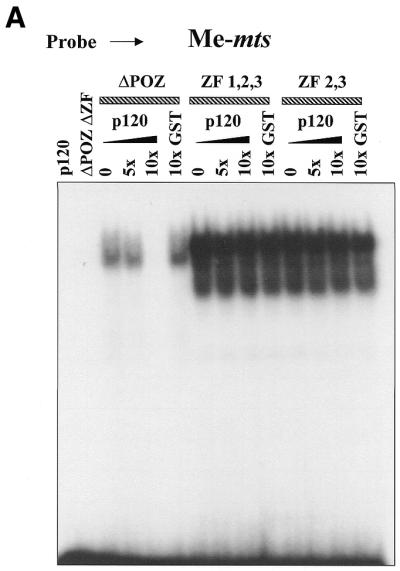
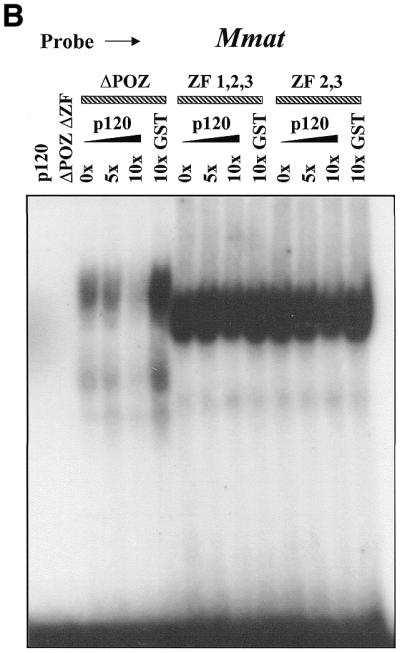
p120ctn inhibits Kaiso DNA binding. GST–KaisoΔPOZ, ZF1,2,3 and ZF2,3 fusion proteins were preincubated with the indicated amounts of GST–p120ctn before the addition of labeled Hmat-derived or labeled, methylated S100A4-derived oligonucleotides. GST–p120ctn specifically inhibited formation of the KaisoΔPOZ–DNA complexes with either recognition sequence. However, there was no effect on KaisoZF1,2,3–DNA or KaisoZF2,3–DNA complex formation.
DISCUSSION
Our recent discovery of a specific and direct interaction between the POZ-ZF transcription factor Kaiso and the cell adhesion cofactor p120ctn hints at their involvement in a signaling pathway that may link cell adhesion events to gene expression. However, although the p120ctn–Kaiso interaction is reminiscent of the much-studied β-catenin–TCF interaction in the Wnt signaling pathway (27,28), there is presently no known event or signaling molecule that triggers the nuclear translocation of p120ctn. Nonetheless, while the exact role of nuclear p120ctn remains to be determined, our findings suggest that its interaction with the C-terminal DNA-binding region of Kaiso may serve to regulate Kaiso–DNA binding and perhaps transcriptional activity.
As a first step towards identifying Kaiso target genes that may be part of a signaling pathway involving p120ctn and Kaiso, we performed a CAST analysis to empirically determine a consensus Kaiso-specific DNA-binding site. The elucidated consensus site, TCCTGCNA, was found at least once in 85% of the clones sequenced. This high frequency of occurrence and the presence of two perfect copies of the consensus in 14% of the clones further attest to the specificity of the Kaiso–DNA interaction. Mutational analysis of the site further defined the core sequence as CTGCNA. Deletion studies using truncated GST–Kaiso fusion proteins revealed that Kaiso zinc fingers 2 and 3 were necessary and sufficient for DNA binding. Since each zinc finger on average interacts with 3–4 nt (29,30), a consensus site of 8 nt is consistent with only two of Kaiso’s three zinc fingers mediating its DNA binding.
Intriguingly, the p120ctn-binding site of Kaiso encompasses the zinc finger domain (3) and may be a non-contiguous epitope flanking the ZF domain. One possibility is that zinc finger 1, which does not appear to be involved in DNA binding, is specifically involved in p120ctn binding. Although the C2H2 zinc fingers are typically associated with DNA binding, there are instances where they mediate protein binding, e.g. the closely related POZ protein ZF-5, the erythroid transcription factor GATA-1, the Myc-binding protein Miz-1 and the MDB-binding protein MIZF (31–34). The results of our p120ctn/Kaiso preincubation experiments (Fig. 7) neither confirm nor exclude this possibility, since p120ctn inhibited the binding of KaisoΔPOZ (residues 348–631, which include all three zinc fingers) but not the more truncated proteins Kaiso-ZF1,2,3 (residues 453–638) and Kaiso-ZF2,3 (residues 515–638). These results suggest that the key Kaiso residues for p120ctn binding lie within the 104 amino acid region (residues 348–453) upstream of its zinc finger domain and the seven amino acid region (residues 632–638) downstream of the zinc finger domain since, in our previous studies, we found that amino acid residues 3′ and 5′ of the ZF domain were necessary for p120ctn binding (3). However, the possibility still remains that zinc finger 1 is involved in p120ctn binding. This close physical juxtaposition of the p120ctn-binding site with the Kaiso DNA-binding motif could facilitate regulation of Kaiso transcriptional properties by p120ctn, either by direct steric hindrance of Kaiso DNA-binding ability or by inducing a conformational change in Kaiso.
Two perfect copies of the optimal Kaiso site were found in the human and murine matrilysin gene promoters and we demonstrated that Kaiso binds specifically to oligonucleotides derived from these promoters. This suggests that the Kaiso DNA consensus sequence in the matrilysin promoter may be physiologically relevant and implicates matrilysin as a potential Kaiso target gene. Matrilysin (MMP-7) is a unique member of the zinc-dependent family of matrix metalloproteinases (MMP) that degrade components of the extracellular matrix. It is distinguished from other MMPs by its preferential expression in epithelial cells, in contrast to the other MMPs that are more commonly expressed in connective tissue and fibroblasts (35). In addition, Matrilysin is implicated in invasion and metastasis (36) and is one of the target genes of the Wnt/β-catenin/TCF signaling pathway (22,37). Reports of a density-dependent regulation of matrilysin expression by E-cadherin (38) and the observation that E-cadherin is a substrate for cleavage by Matrilysin (39) provide additional links between Matrilysin and cadherin-mediated cell adhesion. Future experiments using wild-type matrilysin promoter sequences or matrilysin promoter sequences with a mutated Kaiso-binding site in Luciferase reporter assays should determine if matrilysin is a bona fide Kaiso target gene. In addition, the relationship, if any, between the β-catenin–TCF and Kaiso–p120ctn regulation of matrilysin would provide significant insights into these two distinct pathways.
The discovery that Kaiso also binds methyl-CpG sites in the S100A4 promoter was surprising because the methyl-CpG recognition sequence is quite distinct from our empirically determined Kaiso-binding site. CpG methylation is a hallmark of higher eukaryotic genomes and plays an important role in tissue-specific gene expression, X chromosome inactivation and genomic imprinting (40–42). Although this hypermethylated DNA is usually associated with inactive chromatin and gene silencing during embryogenesis, aberrant CpG methylation correlates with the etiology of various diseases. For example, in many cancers, tumor suppressor genes are hypermethylated and consequently inactive. While the molecular mechanism of this methyl-CpG gene silencing is unclear, it appears to involve a family of methyl-CpG-binding proteins (MeCPs or MBDs) that are characterized by a conserved methyl-binding domain (MBD) (43). If the methyl-CpG recognition site were indeed physiologically relevant to Kaiso function, then Kaiso would be the first methyl-CpG-binding protein without the canonical MBD, and it would be the first POZ-ZF transcription factor implicated in methylation-dependent gene silencing.
While several studies have linked transcriptional repression by the POZ-ZF proteins to recruitment of the histone deacetylase complex (12–16), the POZ-ZF proteins have never been linked to methylation-dependent transcriptional silencing. Interestingly, however, there is increasing evidence linking methylation-dependent gene silencing to the recruitment of the histone deacetylase complex (40,44–46). This raises the possibility that POZ-ZF transcription factors like Kaiso, HIC-1 and HRG22, which do not interact directly with the HDAC complex, may exert their repressive effects indirectly through a methylation-dependent mechanism.
While we have demonstrated that Kaiso binds two distinct DNA sequences, the physiological relevance of these sites and the biological consequences of Kaiso binding remain to be determined. For example, it is possible that one binding site is linked to p120ctn adhesion and signaling and the other is not. One paradox is that methylated DNA is usually found in condensed and transcriptionally inactive DNA, so why would a transcriptional repressor (Kaiso) need to bind and ‘repress’ an already inactive gene? One possibility is that during development and embryogenesis, the association of Kaiso with methylated DNA serves to prevent the demethylation and subsequent activation of its target genes until necessary. Alternatively, in tumor cells, Kaiso may bind hypermethylated tumor suppressor genes and maintain them in an inactive state. Another provocative idea is that the association of Kaiso with methyl-CpG DNA keeps Kaiso in an ‘inactive’ state until it is ‘activated’ by an appropriate signal or binding partner that causes a conformational change, releasing Kaiso from the methyl-CpG site and allowing it to bind and repress target genes bearing the sequence-specific Kaiso consensus site. Identifying the putative signaling trigger or event that modulates p120ctn nuclear translocation and/or Kaiso activity should provide valuable clues and allow us to examine these possibilities. This would significantly contribute to our understanding of Kaiso function and the relevance of its bi-modal DNA-binding activity.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Dr Egor Prokhortchouk for sharing his S100A4 CpG site sequence before publication. This work was supported by a CIHR grant (MOP-42405) to J.M.D. and a Center Grant from the National Cancer Institute (CA 68485) and NIH grant CA 55724 to A.B.R.
REFERENCES
- 1.Bardwell V.J. and Treisman,R. (1994) The POZ domain: a conserved protein-protein interaction motif. Genes Dev., 8, 1664–1677. [DOI] [PubMed] [Google Scholar]
- 2.Albagli O., Dhordain,P., Deweindt,C., Lecocq,G. and Leprince,D. (1995) The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ., 6, 1193–1198. [PubMed] [Google Scholar]
- 3.Daniel J.M. and Reynolds,A.B. (1999) The catenin p120ctn interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol. Cell. Biol., 19, 3614–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reynolds A.B. and Daniel,J.M. (1997) p120ctn: a Src-substrate turned catenin. In Cowin,P. and Klymkowsky,M. (eds), Cytoskeletal-Membrane Interactions and Signal Transduction. Landes Bioscience, Austin, TX, Chapter 3, pp. 31–48.
- 5.Reynolds A.B., Daniel,J., McCrea,P., Wheelock,M.M., Wu,J. and Zhang,Z. (1994) Identification of a new catenin: the tyrosine kinase substrate p120cas associates with E-cadherin complexes. Mol. Cell. Biol., 14, 8333–8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anastasiadis P.Z. and Reynolds,A.B. (2000) The p120 catenin family: complex roles in adhesion, signaling and cancer. J. Cell Sci., 113, 1319–1334. [DOI] [PubMed] [Google Scholar]
- 7.Daniel J.M. and Reynolds,A.B. (1995) The tyrosine kinase substrate p120cas binds directly to E-cadherin but not to the adenomatous polyposis coli protein or α-catenin. Mol. Cell. Biol., 15, 4819–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thoreson M.A., Anastasiadis,P.Z., Daniel,J.M., Ireton,R.C., Wheelock,M.J., Johnson,K.R., Hummingbird,D.K. and Reynolds,A.B. (2000) Selective uncoupling of p120ctn from E-cadherin disrupts strong adhesion. J. Cell Biol., 148, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozawa M. and Kemler,R. (1998) The membrane-proximal region of the E-cadherin cytoplasmic domain prevents dimerization and negatively regulates adhesion activity. J. Cell Biol., 142, 1605–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulson A.F., Mooney,E., Fang,X., Ji,H. and McCrea,P.D. (2000) Xarvcf, Xenopus member of the p120 catenin subfamily associating with cadherin juxtamembrane region. J. Biol. Chem., 275, 30124–30131. [DOI] [PubMed] [Google Scholar]
- 11.Yap A.S., Niessen,C.M. and Gumbiner,B.M. (1997) The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening and interaction with p120ctn. J. Cell Biol., 14, 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David G., Alland,L., Hong,S.-H., Wong,C.-W., DePinho,R.A. and Dejean,A. (1998) Histone deactylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene, 16, 2549–2556. [DOI] [PubMed] [Google Scholar]
- 13.Dhordain P., Albagli,O., Lin,R.J., Ansieau,S., Quief,S., Leutz,A., Kerckeart,J.-P., Evans,R.M. and Leprince,D. (1997) Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl Acad. Sci. USA, 94, 10762–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grignani F., De Matteis,S., Nervi,C., Tomassoni,L., Galmetti,V., Cioce,M., Fanello,M., Ruthardt,M., Ferrara,F.F., Zamir,I., Seiser,C., Grignani,F., Lazar,M.A., Minucci,S. and Pelicci,P.G. (1998) Fusion proteins of the retinoic acid receptor-α recruit histone deactylase in promyelocytic leukemia. Nature, 391, 815–818. [DOI] [PubMed] [Google Scholar]
- 15.Hong S.-H., David,G., Wong,C.-W., Dejean,A. and Privalsky,A.L. (1997) SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor α (RARα) and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc. Natl Acad. Sci. USA, 94, 9028–9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin R.J., Nagy,L., Inoue,S., Shao,W., Miller,W.H.,Jr and Evans,R.M. (1998) Role of the histone deacetylase complex in acute promyelocytic leukemia. Nature, 391, 811–814. [DOI] [PubMed] [Google Scholar]
- 17.Deltour S., Guerardel,C. and Leprince,D. (1999) Recruitment of SMRT/N-CoR-mSin3A-HDAC-repressing complexes is not a general mechanism for BTB/POZ transcriptional repressors: the case of HIC-1 and γFBP-B. Proc. Natl Acad. Sci. USA, 96, 14832–14836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deltour S., Pinte,S., Guerardel,C. and Leprince,D. (2001) Characterization of hrg22, a human homologue of the putative tumor suppressor gene hic1. Biochem. Biophys. Res. Commun., 287, 427–434. [DOI] [PubMed] [Google Scholar]
- 19.Kim S.W., Fang,X., Ji,H., Paulson,A.F., Daniel,J.M., Ciesiolka,M., van Roy,F. and McCrea,P.D. (2001) Isolation and characterization of XKaiso, a transcriptional repressor that associates with the catenin Xp120ctn in Xenopus laevis. J. Biol. Chem., 277, 8202–8208. [DOI] [PubMed] [Google Scholar]
- 20.Prokhortchouk A., Hendrich,B., Jorgensen,H., Ruzov,A., Wilm,M., Georgiev,G., Bird,A. and Prokhortchouk,E. (2001) The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev., 15, 1613–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blackwell T.K. and Weintraub,H. (1990) Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science, 250, 1104–1110. [DOI] [PubMed] [Google Scholar]
- 22.Crawford H.C., Fingleton,B.M., Rudolph-Owen,L.A., Goss,K.J.H., Rubinfeld,B., Polakis,P. and Matrisian,L.M. (1999) The metalloproteinase matrilysin is a target of β-catenin transactivation in intestinal tumors. Oncogene, 18, 2883–2891. [DOI] [PubMed] [Google Scholar]
- 23.He T.C., Sparks,A.B., Rago,C., Hermeking,H., Zawel,L., da Costa,L.T., Morin,P.J., Vogelstein,B. and Kinzler,K.W. (1998) Identification of c-MYC as a target of the APC pathway. Science, 281, 1509–1512. [DOI] [PubMed] [Google Scholar]
- 24.Shtutman M., Zhurinsky,J., Simcha,I., Albanese,C., D’Amico,M., Pestell,R. and Ben-Ze’ev,A. (1999) The cyclin D1 gene is a target of the β-catenin/Lef-1 pathway. Proc. Natl Acad. Sci. USA, 96, 5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tetsu O. and McCormick,F. (1999) β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature, 398, 422–426. [DOI] [PubMed] [Google Scholar]
- 26.Sherbet G.V. and Lakshmi,M.S. (1998) S100A4 (MTS1) calcium binding protein in cancer growth, invasion and metastasis. Anticancer Res., 18, 2415–2422. [PubMed] [Google Scholar]
- 27.Gumbiner B.M. (1997) Carcinogenesis: a balance between β-catenin and APC. Curr. Biol., 7, 443–446. [DOI] [PubMed] [Google Scholar]
- 28.Peifer M. (1997) β-Catenin as oncogene: the smoking gun. Science, 275, 1752–1753. [DOI] [PubMed] [Google Scholar]
- 29.Jacobs G.H. (1992) Determination of the base recognition positions of zinc fingers from sequence analysis. EMBO J., 11, 4507–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klevit R.E. (1991) Recognition of DNA by Cys2,His2 zinc fingers. Science, 253, 1367. [DOI] [PubMed] [Google Scholar]
- 31.Crossley M., Merika,M. and Orkin,S.H. (1995) Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol., 15, 2448–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Numoto M., Yokoro,K. and Koshi,J. (1999) ZF5, which is a Kruppel-type transcriptional repressor, requires the zinc finger domain for self-association. Biochem. Biophys. Res. Commun., 256, 573–578. [DOI] [PubMed] [Google Scholar]
- 33.Peukert K., Staller,P., Schneider,A., Carmichael,G., Hanel,F. and Eilers,M. (1997) An alternative pathway for gene regulation by Myc. EMBO J., 16, 5672–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekimata M., Takahashi,A., Murakami-Sekimata,A. and Homma,Y. (2001) Involvement of a novel zinc finger protein, MIZF, in transcriptional repression by interacting with a methyl-CpG binding protein, MBD2. J. Biol. Chem., 276, 42632–42638. [DOI] [PubMed] [Google Scholar]
- 35.Wilson C.L. and Matrisian,L.M. (1998) Matrilysin. In Parks,W.C. and Mecham,R.P. (eds), Matrix Metalloproteinases. Academic Press, San Diego, CA, pp. 149–184.
- 36.Powell W.C., Knox,J.D., Navre,M., Grogan,T.M., Kittelson,J., Nagle,R.B. and Bowden,G.T. (1993) Expression of the metalloproteinase matrilysin in DU-145 cells increases their invasive potential in severe combined immunodeficient mice. Cancer Res., 53, 417–422. [PubMed] [Google Scholar]
- 37.Brabletz T., Jung,A., Dag,S., Hlubek,F. and Kirchner,T. (1999) β-Catenin regulates the expression of the matrix metalloproteinase-7 in human colorectal cancer. Am. J. Pathol., 155, 1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borchers A.H., Sanders,L.A. and Bowden,G.T. (1997) Regulation of matrilysin expression in cells of squamous cell carcinoma by E-cadherin-mediated cell-cell contact. J. Cancer Res. Clin. Oncol., 123, 13–20. [DOI] [PubMed] [Google Scholar]
- 39.Noe V., Fingleton,B., Jacobs,K., Crawford,H.C., Vermeulen,S., Steelant,W., Bruyneel,E., Matrisian,L.M. and Mareel,M. (2001) Release of an invasion promoter E-cadherin fragment by matrilysin and stromelysin-1. J. Cell Sci., 114, 111–118. [DOI] [PubMed] [Google Scholar]
- 40.Bird A.P. and Wolffe,A.P. (1999) Methylation-induced repression–belts, braces and chromatin. Cell, 99, 451–454. [DOI] [PubMed] [Google Scholar]
- 41.Colot V. and Rossignol,J.L. (1999) Eukaryotic DNA methylation as an evolutionary device. Bioessays, 21, 402–411. [DOI] [PubMed] [Google Scholar]
- 42.Szyf M. (2001) Towards a pharmacology of DNA methylation. Trends Pharmacol. Sci., 22, 350–354. [DOI] [PubMed] [Google Scholar]
- 43.Wade P.A. (2001) Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene, 20, 3166–3173. [DOI] [PubMed] [Google Scholar]
- 44.Jones P.L., Veenstra,G.J., Wade,P.A., Vermaak,D., Kass,S.U., Landsberger,N., Strouboulis,J. and Wolffe,A.P. (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet., 19, 187–191. [DOI] [PubMed] [Google Scholar]
- 45.Nan X., Ng,H.H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenman,R.N. and Bird,A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- 46.Rountree M.R., Bachman,K.E., Herman,J.G. and Baylin,S.B. (2001) DNA methylation, chromatin inheritance and cancer. Oncogene, 20, 3156–3165. [DOI] [PubMed] [Google Scholar]