Abstract
A first step in Cre-mediated site-specific DNA recombination is binding to the two 13 bp repeats of the 34 bp site loxP. Several nucleotides within loxP do not directly contact the bound enzyme, yet mutation at two of these base pairs, at positions 11 and 12 in each repeat, results in a 100 000-fold reduction in recombination. To understand better how Cre selects DNA sequences for recombination, we combined DNA shuffling mutagenesis and a forward selection strategy to obtain Cre mutants that recombine at 100% efficiency a mutant loxK2 site carrying these dinucleotide changes. The role of the several mutations found in these Cre isolates was analyzed both in vivo and biochemically with purified enzymes. A single mutation at E262 accounts for most but not all of the enhanced activity at loxK2. Secondary mutations act in one or more of three ways: enhancement of loxK2 binding, accelerated synthesis of Cre in vivo or faster DNA recombination at the alternative spacer region present in loxK2. Systematic analysis of all 20 natural amino acids at position E262 shows that the naturally occurring glutamate residue at this position provides the optimal balance of efficiency of recombination at loxP and maximal discrimination against loxK2.
INTRODUCTION
Site-specific DNA recombinases are the engine for a host of powerful new genome manipulation strategies that have become standard techniques in determining gene function in mammals and in facilitating the genetic engineering of both plants and animals (1–4). These strategies include temporally and spatially conditional gene ablation or activation, targeted gene insertion and the generation of novel chromosomal rearrangements, including reciprocal translocations, inversions and segmental aneuploidies. Of particular importance is the Cre recombinase of bacteriophage P1, now used extensively for conditional gene knockout in mice. A key factor in the adoption of Cre for genome engineering has been the alacrity and high degree of selectivity with which Cre catalyzes conservative site-specific DNA recombination in vivo at a specific site, the 34 bp loxP site from bacteriophage P1.
Although the selectivity with which Cre chooses DNA sites to recombine is quite stringent, high level expression of Cre in Saccharomyces cerevisiae leads to recombination at endogenous lox-like DNA sequences in the yeast genome at a low, but appreciable, frequency (5). Even more striking are the numerous Cre-dependent chromosome rearrangements that have been observed in spermatids of transgenic mice expressing Cre from the strong protamine 1 promoter (6). To examine in greater detail the fidelity with which Cre selects a target site for DNA recombination we have isolated Cre mutants that have gained the ability to efficiently recognize and catalyze DNA recombination at a synthetic lox-like sequence called loxK2 that shares only 56% identity with loxP.
Consideration of the cryptic lox sequences identified previously in yeast indicates that sites with changes at positions 11 and 12 may be particularly susceptible to recognition by Cre: >80% of the independent isolates obtained carried changes at these positions (5). These nucleotides lie near the middle of the two 13 bp inverted repeats of loxP to which Cre binds. Inspection of the co-crystal structure of Cre with its substrate DNA, however, indicates that Cre makes no contact with these residues (7). Here we show that mutant lox sites, such as loxK2, that incorporate changes at these positions are very poorly recombined by Cre. Mutant recombinases with an altered selectivity can, however, be generated using DNA shuffling mutagenesis (8) coupled with a powerful genetic selection. The variant Cre recombinases we obtained recombine loxK2 at nearly 100% efficiency with no loss of loxP activity. To analyze these mutants in detail we employed both gain-of-function and loss-of-function in vivo recombination assays. In addition, to facilitate understanding of how particular amino acid changes influence the interaction of Cre with its target site, several mutant Cre proteins were purified to homogeneity for analysis. Remarkably, a single amino acid change E262→G contributes to most, but not all, of the >100 000-fold increase in recombinase activity at loxK2. Additional mutations in these mutants augment loxK2 DNA recombination in several ways, including tighter Cre binding to loxK2.
MATERIALS AND METHODS
Bacterial strains and plasmids
Plasmids were constructed and propagated using Escherichia coli DH5α (Invitrogen). The constitutive lacZ gene from pMB1041 (9) was flanked by directly repeated loxP sites and inserted into the unique XhoI site of λD69 (10). Lysogenization of DH5ΔlacU169 (11) with the resulting phage generated the loxP2 lacZ indicator strain BS583. Transformation of DH5α and BS583 with the loxK22 plasmid pBS584 yielded the indicator/selector strains BS1491 and BS1494, respectively. To construct pBS584 the small HindIII–BglII fragment between the RSV promoter and the neo structural gene of RSVneo (12) was replaced by two directly repeated loxK2 sites (5′-GAT ACA ACG TAT ATA CCT TTC TAT ACG TTG TAT T-3′) flanking an EGFP stuffer gene (Clontech) and the transcriptional terminator rrnBT1T2 from pBAD33 (13).
Protein purification
Wild-type (wt) and mutated Cre proteins were expressed to high levels in E.coli BL21(DE3) using the T7 expression vector pRH200 (14) carrying the corresponding cre gene. The Cre purification procedure (15) was modified by adding a Superose sizing column after P11 ion exchange chromatography. Typical yields were 0.5–2 mg from 800 ml of cultured cells with a protein purity of >99% as determined by SDS–PAGE. Concentrations were determined by spectrophotometry at 280 nm using an ɛ280 for wt Cre of 1.17e5 M–1 cm–1 (14). Western blot analysis using preadsorbed rabbit anti-Cre polyclonal antibody was as previously described (14). [125I]protein A from AP Biotech was used for detection.
Mutagenesis and mutant selection
The DNA shuffling procedure (8) was employed for random mutagenesis of cre. Primers for amplification of the cre gene were 5′-BSB436 (5′-AAATAATCTAGACTGAGTGTGAAATGTCC-3′) and 3′-BSB376 (5′-ATATATAAGCTTATCATTTACGCGTTAATGG-3′), introducing an XbaI and HindIII cloning site, respectively (underlined). The PCR product from the shuffling reaction was ligated into the expression vector pBAD33 (13) and electroporated into the selector/indicator strain BS1494. Immediately after electroporation cells were cultured in 1 ml of SOB medium supplemented with 20 mM MgCl2 and also 20 mM l-arabinose to induce cre expression. After the indicated time (2 h or 30 min) glucose was added to a final concentration of 30 mM to quench cre expression. Cells were grown for another 2 h to ensure adequate expression of the neo gene and then plated on LB plates containing the appropriate antibiotics for selection (ampicillin for pBS584, chloramphenicol for the cre expression plasmid and kanamycin to select for loxK22 recombination). After overnight incubation at 37°C candidate colonies in which loxK22 recombination had presumably occurred were pooled and plasmid DNA was purified for the next round of mutagenesis.
Specific single amino acid substitutions in Cre were obtained by site-specific mutagenesis with the Quick-Change Kit (Stratagene). Plasmid constructs and cre mutants were cycle sequenced using a PE ABI PRISM 310 Genetic Analyzer (Perkin Elmer).
In vitro DNA recombination
Cre-mediated DNA recombination in vitro was at a 2:1 molar ratio of Cre protein to binding site (lox half-site) or 66.7 nM Cre and 8.3 nM lox2 substrate in a volume of 12 µl (50 mM Tris–HCl pH 7.8, 200 mM NaCl, 5 mM spermidine and 3 mM EDTA). The reaction was incubated at 30°C for 30 min followed by heat inactivation of Cre at 70°C for 10 min. The reaction mix was digested with an appropriate restriction enzyme to facilitate identification of recombination products, extracted once with CHCl3 and analyzed by agarose gel electrophoresis. Recombination frequencies were calculated after quantitation of recombinant products and unreacted substrate using AlphaImager 2000 software (Alpha Innotech).
Electrophoretic mobility shift assay
Gel shift experiments were performed under identical conditions to those for DNA recombination except that (i) incubation was for 20 min at 30°C and (ii) the substrate for Cre binding was the gel-purified 270 bp PvuII polylinker-containing fragment of pUC19 into which the indicated lox site had been cloned. After incubation, a 10 ng heparin challenge was added to the reaction mix to eliminate non-specific DNA binding (16). DNA was electrophoresed on a 6% polyacrylamide (TBE) gel and visualized by staining with SYBR Gold (Molecular Probes).
Structure representation
Visualization of the structure of Cre, as well as the fitting of mutated side chains, was performed using SwissProt pdb Viewer (17). Altered side chains were modeled into the crystal structure (7) to a local energetic minimum keeping DNA coordinates fixed.
RESULTS
Mutant isolation
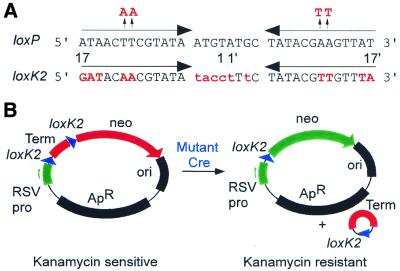
To investigate the role of the thymidine residues at positions 11 and 12 (and adenines at 11′ and 12′) in the 13 bp inverted repeats of the loxP site, we substituted adenines (thymidines) at these positions (Fig. 1A). Preliminary experiments both in vivo and in vitro with purified Cre recombinase (see below) indicated that these alterations reduced Cre-mediated DNA recombination by at least several orders of magnitude. To devise a genetic selection for the isolation of Cre mutants able to overcome the block to DNA recombination, we incorporated these four substitutions (two in each 13 bp inverted repeat) into a lox-like DNA sequence, loxK2, that differs in total at 15 positions from the 34 bp loxP site (Fig. 1A). In addition to the adenine (and thymidine) substitutions the loxK2 site carries two other types of base pair changes: (i) the recombinationally proficient 8 bp loxFAS1 spacer (18) was substituted for the loxP spacer to prevent any potentially complicating loxK2 recombination with loxP; (ii) several alterations were made at the outer few positions of the repeats (positions 15–17) to facilitate plasmid construction. Such alterations have previously been observed to have relatively little effect on Cre-mediated recombination (5,19,20).
Figure 1.
Selection for expanded Cre substrate recognition. (A) The 34 bp nucleotide sequences of the wt loxP and mutant loxK2 sites are shown. The two 13 bp inverted repeats of the sites are indicated by horizontal arrows and are separated by 8 bp, traditionally called the spacer region. Since the scissile phosphates on the upper and lower strands of the loxP site are set 1 bp in from the two inverted repeats, the actual crossover region in loxP is 6 bp. Vertical arrows at positions 11 and 12 indicate mutations that block Cre-mediated recombination. Mutated residues of the loxK2 site are shown in red: alterations in the inverted repeats involved in Cre binding and in the FAS1 spacer (18) of loxK2 are shown in upper and lower case, respectively. Nucleotide positions are numbered using the previously established convention (27). (B) The loxK22 selection plasmid carries a loxK2-flanked transcription terminator cassette inserted between the neo structural gene and the RSV promoter. This renders the neo gene inactive. Excisive recombination by a mutant Cre results in eviction of the transcription terminator and activates expression of the neo gene.
Recombination assays both in vitro and in vivo confirmed that Cre was completely inactive on loxK2 sites. To select for cre mutants that had acquired the ability to recognize and recombine loxK2 we designed a gain-of-function selection plasmid for recombination (Fig. 1B). Selection relies on activation of a quiescent neo gene by Cre-mediated excision of a loxK2-flanked transcription terminator interposed between the neo structural gene and an RSV promoter which is naturally active in E.coli (21). Thus, gain of resistance to kanamycin signals that recombination has occurred between two loxK2 sites. To simultaneously monitor for loxP recombination the loxK22 reporter plasmid was introduced into an E.coli strain carrying a single copy chromosomal lacZ gene flanked by two directly repeated loxP sites.
Random mutagenesis of the entire cre gene by DNA shuffling (8) gave mutant cre pools that were subjected to a kanamycin selection for loxK22 recombination. To better ensure that very active cre mutants were obtained, selection was imposed after only brief expression of cre by using the arabinose-inducible ara promoter on pBAD (13). Under these conditions wt Cre-mediated recombination at loxK2 was not detected, occurring at a frequency of <1 × 10–5 (Table 1). With a similar loxP2 reporter plasmid under the same induction conditions the frequency of recombination was 100% (data not shown). Four consecutive rounds of mutagenesis were carried out, the selected candidates from the previous round serving as template mix for the next. With each round the cre gene pools contained an increased number of cre mutants having enhanced proficiency for loxK22 recombination (Table 1). To further hone the selection stringency for loxK2 proficiency, we shortened the window of time for arabinose-induced cre expression from 2 h in rounds 1 and 2 to only 30 min for rounds 3 and 4. Even with these very stringent selection conditions, numerous KanR colonies were obtained, suggesting that the mutagenesis and selection procedure had given rise to an abundance of potent Cre mutants proficient for loxK22 recombination.
Table 1. Enrichment of cre mutants that recombine loxK2.
| Round | Induction (h) | KanR colonies (%) | KanR colonies (no.) | Colora |
|---|---|---|---|---|
| wt Cre | 2 | <0.001 | 0 | White |
| 1 | 2 | 0.16 | 6 | White |
| 2 | 2 | 0.56 | 47 | White |
| 3 | 0.5 | 0.2 | 36 | White |
| 4 | 0.5 | 4.6 | 102 | White |
aScored on X-gal + kanamycin selection plates following transfection of E.coli BS1494.
Because the bacterial strain used for selection carried a single copy chromosomal loxP2[lacZ] cassette, we simultaneously monitored for Cre-mediated recombination at loxP. Cre mutants that had completely lost the ability to catalyze DNA recombination at loxP appeared blue on X-gal. However, no blue KanR colonies were obtained (Table 1), suggesting that all of the mutants recovered by selection for recombination at loxK2 retained significant recombinational activity at loxP.
Characterization of cre mutants
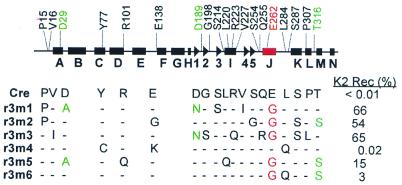
Sequencing of six randomly chosen cre mutants from the third round of mutagenesis revealed three to eight mutations per isolate (Fig. 2). Re-assay for loxK22 DNA recombination in the selector strain showed that five of the six had considerably enhanced recombinational activity on loxK2 compared with the wt enzyme, with three of the mutants showing >50% loxK2 recombination compared with no recombination with wt Cre. Strikingly, all five carry the same E262→G amino acid change. This result implicated the glutamate to glycine exchange at position 262 (E262G) of Cre protein as critical for recognition of the loxK2 site. Nevertheless, there is an ∼10-fold variation in recombinational proficiency at loxK2 amongst these five isolates, indicating that several of the other mutations found in the various isolates must play an ancillary role in augmenting loxK22 recombination. Identical amino acid changes in Cre re-occurring in more than one loxK2-proficient Cre mutant were at three other positions: D29A, D189N and T316S (Fig. 2).
Figure 2.
Distribution of Cre mutations obtained from DNA shuffling. The secondary structure of Cre recombinase (7) is represented by rectangles (α-helices) and triangles (β-strands). Aligned with the sequence of wt Cre are DNA sequences of six Cre mutants obtained after the third round of mutagenesis (r3m1–6, i.e. round 3, mutants 1–6). Recombinational activity at the loxK2 site was determined by electroporation of the tester strain BS1491 with pBAD33 carrying the indicated mutant cre gene, induction for 0.5 h and selection for kanamycin resistance after an additional outgrowth for 2 h to allow neo expression. The percentage of kanamycin-resistant colonies was determined by comparison with cells plated non-selectively. The E262G mutation common to all mutants with enhanced loxK2 activity is shown in red, as is the J helix within which it resides. Amino acid changes occurring in two or more mutant isolates with enhanced loxK2 activity are shown in green.
To confirm the central role of the E262G mutation in conferring loxK2 recognition, singly mutated E262G cre and several double mutants were synthesized and evaluated for excisive DNA recombination at both loxP and loxK2. To accurately compare loxP2 and loxK22 recombination in vivo at an equal copy number of substrate per cell, bacterial strains with a resident wt or mutant cre on an l-arabinose-inducible plasmid were electroporated with either a loxP2 neo or a loxK22 neo multicopy reporter plasmid in which the neo gene is flanked by wt or mutant lox sites. After brief cre induction cells were plated with selection for the reporter plasmid backbone marker (AmpR) and the resulting transformants were scored for presence or loss of the lox-flanked neo marker. Only colonies arising from cells that had excised the neo gene on all copies of the incoming reporter plasmid score as kanamycin sensitive, i.e. as recombination positive. Figure 3 shows that although wt Cre is completely blocked for loxK2 recombination, the E262G mutant exhibits 28% recombination on loxK2 with no diminution of activity on loxP. In contrast, the multiply mutated original isolate Cre-r3m3 is nearly as proficient on loxK2 as wt Cre is on loxP. Again, the multiple mutant does not appear to be reduced for loxP recombination. Combining E262G with any of the three most commonly occurring secondary amino acid changes indicated that these secondary mutations provide a small boost to recombination but are not sufficient by themselves when combined with E262G to give the full activity shown by Cre-r3m3.
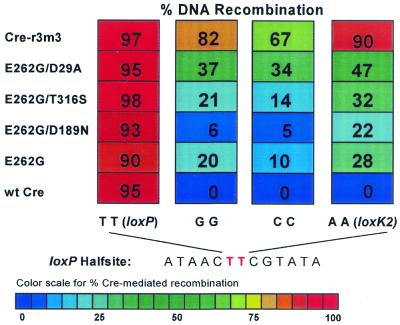
Figure 3.
Modulation of Cre-E262G recombination at mutant lox sites. Shown are recombination frequencies for wt and the indicated mutant Cre recombinases at loxP and at mutant lox sites that differ from loxP at positions 11 and 12. Escherichia coli DH5α carrying a resident pBAD construct expressing the indicated cre mutant was transformed with the indicated lox2 neo plasmids and cre expression induced for 1 h before plating non-selectively to yield about 200 colonies/experiment. Sensitivity to kanamycin, signifying excisive recombination, was determined by replica plating. The frequencies shown are calculated from at least two independent experiments.
Evaluation of mutant Cre activity on two additional mutant loxK2-like sites gave the same general result (Fig. 3). Similar to loxK2, the thymidines at positions 11 and 12 were changed to either guanosines or cytosines. In both cases wt Cre activity was blocked and the E262G mutation restored recombinational proficiency, with the multiple Cre-r3m3 mutant displaying significantly more activity than the E262G single mutant. Two of the secondary mutations, D29A and T316S, modestly enhanced E262G-mediated recombination, whereas the third, D189N, appeared to be mildly inhibitory, especially on the GG and CC lox site substitutions. These results confirm a critical role for positions 11 and 12 of the lox site in restricting Cre-mediated recombination to sites having thymidines at these positions, and implicate E262 as being a key sensor for the ‘correct’ lox sequence.
Secondary mutations
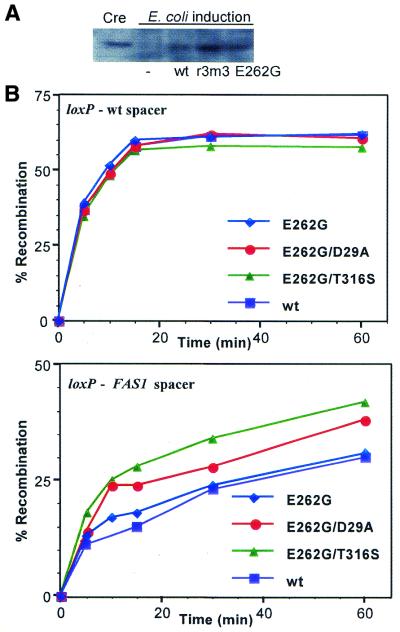
In both types of in vivo excision assays (neo activation and neo deletion) secondary mutations modestly enhance the ability of E262G to recombine loxK2. How might this occur? Because cre expression was limited to only a short period of time to ensure recovery of very active cre mutants, one indirect way to enhance recombination would be to increase the rate of Cre production during induction in the cell. Western blot analysis with specific anti-Cre antibody shows that this is the case for the multiply mutated Cre-r3m3 isolate (Fig. 4A). After only 1 h of arabinose induction a greater amount of Cre is detected with the multiple mutant than with either wt or singly mutated E262G Cre. Mutations that increase Cre synthesis and thereby hasten loxK2 recombination during a short induction time window may explain the recurrence in several isolates of silent mutations at positions P15, S214 and S287 (Fig. 2), but this was not examined in detail.
Figure 4.
Role of secondary mutations in enhanced recombination at loxK2. (A) Accelerated Cre synthesis in vivo. Escherichia coli DH5α strains expressing the indicated cre variants in pBAD33 were grown to an OD600 of 0.5 and cre expression induced for 1 h with 0.2% l-arabinose. Cells from 1 ml of culture were harvested, lysed and 30 µl of the sonicated crude extracts subjected to SDS–PAGE, followed by western analysis with polyclonal anti-Cre serum. (B) Enhanced recombination with the FAS1 spacer. Purified mutant and wt Cre were assayed for activity in vitro on two lox2 constructs, differing only by their 8 bp spacers. Reactions were as described in Materials and Methods, with termination at the indicated time points by heat inactivation of Cre. Recombination frequencies were determined by quantitation of recombination products and unprocessed substrate after gel electrophoresis.
Subsequent to DNA binding and synapsis of lox sites by Cre the spacer regions of the synapsed lox sites are non-base paired during recombination. Although there is no strict requirement for specific DNA sequences in the 8 bp spacer region, different sequences in the spacer could affect the ease with which unpairing occurs. To detect kinetic changes in recombination specific to the spacer rather than to the 13 bp binding sites, we purified both wt Cre protein and the E262G mutant, as well as double mutant proteins having E262G combined with either D29A or T316S, the two secondary mutations in Figure 3 that enhanced E262G activity in vivo. Recombination in vitro with these purified proteins showed that on an authentic loxP2 substrate the kinetics of excisive recombination were indistinguishable for all of these proteins (Fig. 4B). In contrast, with replacement of the 8 bp loxP spacer with the FAS1 spacer in the loxP sites of the recombination substrate, both the E262G T316S double mutant and the E262G D29A double mutant showed distinct enhancement of recombination compared with either wt Cre or the single E262G mutant. This indicates that these secondary mutations probably conferred a slight selective advantage in recombining the loxK2 site during initial mutant selection. Similar in vitro recombination experiments showed that wt Cre, Cre E262G and Cre-r3m3 all recombined loxP sites carrying the FAS1 spacer with nearly identical efficiencies, consistent with the absence in Cre-r3m3 of both the D29A and T316S mutations. It is likely that effects of the spacer region on DNA recombination occur subsequent to Cre binding to the lox site. Gel shift analysis and surface plasmon resonance DNA binding experiments revealed no differences in binding of any of these proteins to lox sites with the FAS1 spacer (A.W.Rüfer and B.Sauer, manuscript in preparation), in accord with previous work that the specific DNA-binding determinants of the loxP site lie in the 13 bp inverted repeat elements. Thus at least some of the mutations recovered after selection for loxK2 recombination can act to modestly enhance recombination of lox sites carrying the FAS1 spacer.
Mutant site recombination and DNA binding in vitro
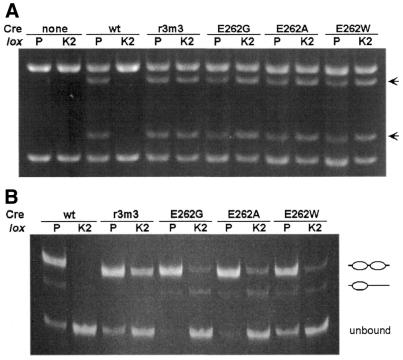
To address the role of E262G in loxK2 recognition and recombination we constructed several different E262 single amino acid substitution mutants and then overexpressed each of them in E.coli. Each mutant protein was purified to 99% homogeneity as was also wt Cre and the multiple Cre-r3m3 mutant. In addition to the single E262G mutant Cre we constructed two other single amino acid substitution mutants: E262A and E262W. We suspected that the substitution of alanine with its small side chain would behave similarly to E262G on loxK2 but that substitution with tryptophan with its bulky side chain would not and might even block recombination with loxP. Recombination in vitro on both loxP2 and loxK22 substrates with each of these purified Cre proteins showed that each of the mutant proteins efficiently recombined loxK2 including, surprisingly, E262W, whereas wt Cre was completely inactive on the loxK2 site (Fig. 5A). None of the mutant enzymes exhibited any reduction in loxP2 recombination compared with wt Cre.
Figure 5.
Recognition of loxK2 in vitro. (A) DNA recombination was assayed in a 30 min reaction at 30°C using either wt or the indicated mutant Cre protein on both a loxP2 and loxK22 substrate as described in Materials and Methods. Recombination products (arrows) as well as unrecombined substrate were detected by electrophoresis on a 0.8% agarose gel after cleavage with the restriction enzyme AlwN1. This enzyme cuts each recombination product once and the parental plasmid twice. (B) Electrophoretic mobility shift experiments were performed as described in Materials and Methods using a 270 bp DNA fragment carrying either a loxP or a loxK2 site. Indicated are shifted fragments with either one (lower band) or two (upper band) Cre molecules per lox site (14).
Presumably wt Cre fails to recombine loxK2 sites because the protein is unable to recognize and bind to this site, whereas the mutant Cre proteins are now able to do so. To examine mutant Cre activity more closely in vitro, we conducted DNA binding assays under conditions identical to those of the recombination assays. Binding of wt Cre to loxP resulted in two gel-shifted complexes (Fig. 5B), the slowest mobility band corresponding to loxP with two molecules of Cre, one bound to each 13 bp inverted repeat, and the faster band corresponding to a complex with a single Cre molecule bound to one of the two inverted repeats (14). All of the mutant Cre proteins bind to loxP at least as well as the wt enzyme. In addition, all of the mutant Cre proteins bind to the loxK2 site, whereas wt Cre is completely unable to bind loxK2. Strikingly, Cre-r3m3 shows distinctly better binding to loxK2 than any of the single E262 substitution mutants. In particular, there is a more fully occupied loxK2 complex with Cre-r3m3: the single amino acid substitution mutants show less overall binding and there is a proportionally greater amount of the faster complex having only a single Cre molecule bound. This suggests that one or more of the additional mutations present in the Cre-r3m3 protein confer greater affinity and/or cooperativity in binding to the loxK2 site. Detailed kinetic analysis of DNA binding to loxK2 by these mutant proteins shows that this is indeed the case (A.W.Rüfer and B.Sauer, manuscript in preparation).
Recombinational selectivity by residue 262
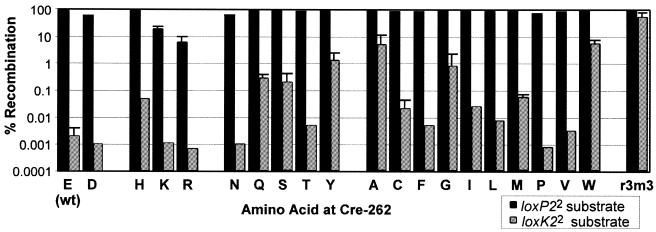
For a more complete determination of the role of residue 262 in the discrimination of loxK2 versus loxP recognition and recombination we systematically replaced the wt glutamate residue at position 262 with each of the 19 other amino acids by site-directed mutagenesis. Each single amino acid substitution mutant was tested in E.coli for both loxK22 and loxP2 recombination (Fig. 6), using reporter strains to monitor loss of a single copy loxP-flanked lacZ gene or activation of neo on the multicopy loxK22 selection plasmid originally used for Cre mutant isolation. Substitution of E262 with tyrosine, glycine, alanine, glutamine, serine or tryptophan markedly enhanced DNA recombination at loxK2 with no diminution of recombination at loxP. Both the alanine and tryptophan mutants, for example, increased recombination at loxK2 several thousand-fold, confirming the results seen in vitro. Still, neither mutant was as proficient as one of the original mutant isolates, cre-r3m3. Other amino acid substitutions conferred only a modest (5–100-fold) increase in loxK2 recombination with little effect on loxP recombination. In contrast, several amino acid substitutions (aspartate, asparagine, proline, lysine and arginine) had a negative effect, concomitantly depressing both loxP2 and loxK22 recombination. Thus, E262 plays a critical role in effectively discriminating against loxK2 recombination while allowing optimal DNA recombination at loxP.
Figure 6.
Contribution of amino acid residue 262 to Cre Activity. All 19 possible amino acid substitutions at Cre E262 were assayed for DNA recombination in vivo at a single copy loxP2 substrate and at a multiple copy loxK22 substrate using the indicator strain BS583 and the selector strain BS1494, respectively. Recombination was scored as described in Figure 1. Induction conditions were as described in Figure 2.
Although the multicopy loxK22 selection plasmid and the single copy loxP2 chromosomal reporters provide a relative ranking of recombination for either loxK2 or loxP, they do not directly compare loxK2 versus loxP recombination for the singly mutated Cre because of the difference in copy number of the recombination substrates. We therefore used the more stringent neo excision plasmid assay from Figure 3 to compare directly the relative efficiencies of loxP and loxK2 recombination for several of the ‘best’ single amino acid mutations obtained by site-directed mutagenesis (Table 2). All of the single amino acid substitution mutants examined recombined loxK2 at least 6000-fold better than wt Cre. In fact, they were only slightly less efficient (4–30%) on loxK2 than on loxP. Again, the cre-r3m3 initial isolate was more proficient at loxK22 recombination than any of the single E262 mutants, recombining loxK2 and loxP at roughly identical efficiencies. None of the mutants examined were diminished for recombinational activity on loxP. Thus, specific mutations of E262 dramatically improve loxK22 recombination by abrogating the stringency of Cre target site selection imposed by the glutamate at position 262.
Table 2. Excisive recombination from a plasmid substrate in E.coli.
| Substitution at Cre E262 | neo gene excision (%)a | |
|---|---|---|
| loxP2 | loxK22 | |
| wt (Glu) | 100 | <0.01 |
| Tyr | 100 | 67.3 |
| Gln | 100 | 75.5 |
| Gly | 100 | 85.6 |
| Ala | 100 | 87.6 |
| Trp | 100 | 95.5 |
| Cre-r3m3 | 100 | 99.8 |
aEscherichia coli DH5α carrying pBAD33 expressing the indicated cre mutant was transformed with either pBS632 (loxP2 neo) or pBS633 (loxK22 neo), induced for 2.5 h with 0.2% l-arabinose and then plated non-selectively to yield at least 1000 colonies. Resistance to kanamycin was determined by replica plating. Frequencies are the average of three independent experiments.
DISCUSSION
Both the in vivo and in vitro assays show clearly that the 100 000-fold block to wt Cre-mediated recombination at loxK2 can be completely eliminated by mutation of the Cre protein. Importantly, these mutants are less selective in choosing a target recombination site but are undiminished in DNA binding and in their ability to catalyze DNA recombination, as shown by DNA recombination in vitro with purified enzyme and in E.coli by carefully limiting expression of Cre to only a small window of time prior to selection for recombination. The primary alteration in Cre permitting this dramatic expansion of site selectivity is the mutation of glutamate at position 262 to any of several amino acids, with glycine, alanine and tryptophan being particularly favorable to loxK2 binding and DNA recombination. In addition, mutations at other positions in Cre can enhance loxK2 DNA recombination in vitro by further increasing binding affinity for the loxK2 site and also by enhancing synthesis of Cre protein in vivo.
One other means for improved recombination at loxK2 uncovered by genetic selection concerns the spacer region. Although the spacer does not specify binding of the lox site by Cre we show here that the rapidity with which Cre recombines lox sites having the FAS1 spacer, as is present in loxK2, is decreased compared with those having the normal loxP spacer. As shown by the mutant Cre-r3m2 protein, mutation of Cre can permit recombination of lox sites with the FAS1 spacer at a rate similar to that seen with loxP. Because the single amino acid substitutions at E262 do not have enhanced recombination with lox sites having the FAS1 spacer, other secondary mutations in Cre-r3m2, or combinations of these, are likely to allow more rapid recombination at the alternative FAS1 spacer.
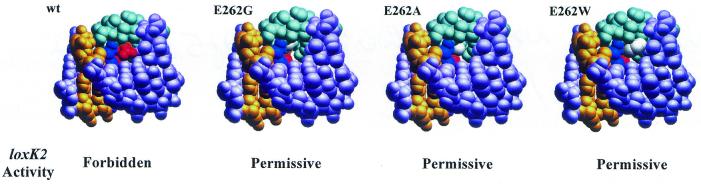
How does a single amino acid change, that at E262 in Cre, cause such drastic changes in target site selectivity of the enzyme? From the crystal structure (7), mutation of the glutamate at position 262 results in the loss of a DNA backbone contact at the adenine at position 8 of the lox site. Since the mutants isolated here are not diminished in activity on loxP this contact cannot be critical for binding or recombination. The region of Cre that includes E262 quite nicely fills the major groove in this region of the loxP inverted repeat element (Fig. 7). Presumably mutation of positions 11 and 12 (yellow) of the lox site disturbs this cosy arrangement, thereby preventing Cre binding. In contrast, the three mutations that confer high level loxK2 recognition and recombination, E262G, E262A and E262W, when modeled to fit into the crystal structure, all increase the accessible surface within this protein–DNA interface comprised of the N-terminus of the J helix in Cre and positions 6–12 of loxP. We speculate that although located distal to positions 11 and 12 that are altered in loxK2, mutation of E262 may enhance flexibility within Cre and thus nullify discrimination against loxK2 recognition. Both R259 (blue) and E266 (red) reside in this critical protein–DNA interface and are likely to play a role here. In particular, R259 inserts deep into the major groove to establish two hydrogen bonds with guanosine 10 of loxP. E266 establishes one hydrogen bond with the R259 side chain as well and may therefore contribute to the orientation of R259 within this interface. Alteration of the T-A base pairs at positions 11 and 12 in loxP may cause clashes with this R259–E266 arrangement and result in loss of Cre binding, especially since E262 constrains the side chain of R259. Substitution of the E262 side chain with that of glycine, alanine and even tryptophan (Fig. 7) is predicted by modeling to allow a higher degree of freedom for both the R259 and the E266 side chains, thus inducing tolerance for the nucleotide alterations present at positions 11 and 12 of loxK2. Although in the absence of structural information this scenario is highly speculative, it does suggest that replacement of E262 by large, non-planar positively charged residues like lysine or arginine might inhibit even loxP recombination by disturbing this cosy protein–DNA interface, and this is precisely what we observed (Fig. 6).
Figure 7.
Substitutions in Cre E262 that allow loxK2 recombination. Using the structure of the non-cleaving Cre monomer bound to the loxP inverted repeat (7) the glutamate at position 262 was replaced with glycine, alanine or tryptophan, keeping the surrounding residues fixed in space and fitting the substitutions to an energetic minimum and avoiding steric clashes. Shown in light plum are positions 6–13 of the lox site (see Fig. 1) with positions 11 and 12 (mutated in loxK2) highlighted in yellow. Residues of Cre shown (in cyan) are located within a 7 Å radius from the Cα of E262. The three residues of the J helix of Cre that fill the DNA major groove, R259, E262 and E266, are colored by type, as are the three substitutions at E262: non-polar residues in gray, acidic residues in red and basic residues in blue.
These results provide insight into how Cre selects a target DNA for site-specific DNA recombination. The two thymidines at position 12′ and 11′ (Fig. 1A) do not contact Cre directly. They more likely impose selectivity on Cre because substitutions at these positions would prevent formation of a critical DNA contact by R259 (22). Remarkably, the glutamate at position 262 appears to be the optimal choice for imposing this selectivity while allowing undiminished recombination at loxP. Although several studies have described Cre mutants that display improved recombination at non-loxP DNA sequences (23–25), such a major effect on site discrimination by a single amino acid change is unusual. The Cre mutants described by one of these studies (24) were also obtained using a DNA shuffling mutagenesis procedure, and several of these display much better activity on a mutant lox site than on loxP. Although none of the more than a dozen different amino acid changes in each of those mutants were analyzed in detail and the mutants were not assayed in vitro, in several of the mutants the glutamate at position 262 was replaced, suggesting that some relaxation of site selectivity accompanied the evolution of a new specificity. Similarly, in a second study (25) Cre mutants recognizing yet another mutant lox site (mutated at positions 9–11) again displayed multiple amino acid substitutions and again included mutation of E262.
The design of recombinases that can recognize new sites is appealing both because the resulting enzymes are likely to be useful for genetic engineering and because their analysis will provide a deeper appreciation of how proteins select target DNA sequences with high specificity. Because it is likely that the change in specificity of a protein by evolution proceeds through a stage of relaxed specificity (26), the mutant cre genes that we describe here may represent a useful starting point for the directed development of recombinases that recognize distinctly non-lox-like DNA targets. However, for such recombinases with altered specificity to be useful in vivo they must also display a very high degree of site selectivity to avoid recognition of unintended genomic sequences that may lead to unexpected genomic aberrations. The observation here that seemingly neutral mutations at E262 can result in a slight but appreciable increase in recognition of non-loxP sequences underscores the importance in such strategies of maintaining or enhancing the very high degree of site selectivity naturally exhibited by wt Cre recombinase.
Acknowledgments
ACKNOWLEDGEMENTS
We thank N. Dominguez for expert technical assistance with protein purification and the German National Scholarship Foundation (Bonn, Germany) for financial support to A.W.R. in the early stages of this work.
REFERENCES
- 1.Kilby N.J., Snaith,M.R. and Murray,J.A. (1993) Site-specific recombinases: tools for genome engineering. Trends Genet., 9, 413–421. [DOI] [PubMed] [Google Scholar]
- 2.Kühn R., Schwenk,F., Aguet,M. and Rajewsky,K. (1995) Inducible gene targeting in mice. Science, 269, 1427–1429. [DOI] [PubMed] [Google Scholar]
- 3.Sauer B. (1993) Manipulation of the transgene by site-specific recombination: use of the cre recombinase. Methods Enzymol., 225, 890–900. [DOI] [PubMed] [Google Scholar]
- 4.Sauer B. (1998) Inducible gene targeting in mice using the Cre/lox system. Methods, 14, 381–392. [DOI] [PubMed] [Google Scholar]
- 5.Sauer B. (1992) Identification of cryptic lox sites in the yeast genome by selection for chromosome translocations that confer multiple drug resistance. J. Mol. Biol., 223, 911–928. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt E.E., Taylor,D.S., Prigge,J.R., Barnett,S. and Capecchi,M.R. (2000) Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc. Natl Acad. Sci. USA, 97, 13702–13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo F., Gopaul,D.N. and Van Duyne,G.D. (1997) Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature, 389, 40–46. [DOI] [PubMed] [Google Scholar]
- 8.Stemmer W.P.C. (1994) Rapid evolution of a protein in vitro by DNA shuffling. Nature, 370, 389–391. [DOI] [PubMed] [Google Scholar]
- 9.Berman M.L. and Beckwith,J. (1979) Use of gene fusions to isolate promoter mutants in the transfer RNA gene TyrT of Escherichia coli. J. Mol. Biol., 130, 303–315. [DOI] [PubMed] [Google Scholar]
- 10.Silhavy T.J., Berman,M.L. and Enquist,L.W. (1984) Experiments with Gene Fusions. Cold Sring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 11.Sauer B. and Henderson,N. (1988) The cyclization of linear DNA in Escherichia coli by site-specific recombination. Gene, 70, 331–341. [DOI] [PubMed] [Google Scholar]
- 12.Gorman C., Padmanabhan,R. and Howard,B.H. (1983) High efficiency DNA-mediated transformation of primate cells. Science, 221, 551–553. [DOI] [PubMed] [Google Scholar]
- 13.Guzman L.-M., Belin,D., Carson,M.J. and Beckwith,J. (1995) Tight regulation, modulation and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol., 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mack A., Sauer,B., Abremski,K. and Hoess,R. (1992) Stoichiometry of the Cre recombinase bound to the lox recombining site. Nucleic Acids Res., 20, 4451–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abremski K. and Hoess,R. (1984) Bacteriophage P1 site-specific recombination: purification and properties of the Cre recombinase protein. J. Biol. Chem., 259, 1509–1514. [PubMed] [Google Scholar]
- 16.Wierzbicki A., Kendall,M., Abremski,K. and Hoess,R. (1987) A mutational analysis of the bacteriophage P1 recombinase Cre. J. Mol. Biol., 195, 785–794. [DOI] [PubMed] [Google Scholar]
- 17.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 18.Sauer B. (1996) Multiplex Cre/lox recombination permits selective site-specific DNA targeting to both a natural and an engineered site in the yeast genome. Nucleic Acids Res., 24, 4608–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sternberg N., Hoess,R. and Abremski,K. (1983) The P1 lox-Cre site-specific recombination system: properties of lox sites and biochemistry of lox-Cre interactions. In Cozzarelli,N.R. (ed.), Mechanisms of DNA Replication and Recombination, UCLA Symposia on Molecular and Cellular Biology. Alan R. Liss, New York, NY, Vol. 10, pp. 671–684.
- 20.Sauer B., Whealy,M., Robbins,A. and Enquist,L. (1987) Site-specific insertion of DNA into a pseudorabies virus vector. Proc. Natl Acad. Sci. USA, 84, 9108–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antonucci T.K., Wen,P. and Rutter,W.J. (1989) Eukaryotic promoters drive gene expression in Escherichia coli. J. Biol. Chem., 264, 17656–17659. [PubMed] [Google Scholar]
- 22.Kim S., Kim,G., Lee,Y. and Park,J. (2001) Characterization of Cre-loxP interaction in the major groove: hint for structural distortion of mutant Cre and possible strategy for HIV-1 therapy. J. Cell. Biochem., 80, 321–327. [PubMed] [Google Scholar]
- 23.Hartung M. and Kisters-Woike,B. (1998) Cre mutants with altered DNA binding properties. J. Biol. Chem., 273, 22884–22891. [DOI] [PubMed] [Google Scholar]
- 24.Buchholz F. and Stewart,A.F. (2001) Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat. Biotechnol., 19, 1047–1052. [DOI] [PubMed] [Google Scholar]
- 25.Santoro S.W. and Schultz,P.G. (2002) Directed evolution of the site specificity of Cre recombinase. Proc. Natl Acad. Sci. USA, 99, 4185–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumura I. and Ellington,A.D. (2001) In vitro evolution of beta-glucoronidase into a beta-galactosidase proceeds through non-specific intermediates. J. Mol. Biol., 305, 331–339. [DOI] [PubMed] [Google Scholar]
- 27.Hoess R.H., Ziese,M. and Sternberg,N. (1982) P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc. Natl Acad. Sci. USA, 79, 3398–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]