Abstract
KLF4 (Krüppel-like factor 4 or gut-enriched Krüppel-like factor, GKLF) and KLF5 (Krüppel-like factor 5 or intestinal-enriched Krüppel-like factor, IKLF) are two closely related members of the zinc finger-containing Krüppel-like factor family of transcription factors. Although both genes are expressed in the intestinal epithelium, their distributions are different: Klf4 is primarily expressed in the terminally differentiated villus cells while Klf5 is primarily in the proliferating crypt cells. Previous studies show that Klf4 is a negative regulator of cell proliferation and Klf5 is a positive regulator of cell proliferation. In this study, we demonstrate that Klf5 binds to a number of cis-DNA elements that have previously been shown to bind to Klf4. However, while Klf4 activates the promoter of its own gene, Klf5 suppresses the Klf4 promoter. Moreover, Klf5 abrogates the activating effect of Klf4 on the Klf4 promoter and Klf4 abrogates the inhibitory effect of Klf5 on the same promoter. An explanation of this competing effect is due to physical competition of the two proteins for binding to cognate DNA sequence. The complementary tissue localization of expression of Klf4 and Klf5 and the opposing effect of the two Klfs on the Klf4 promoter activity may provide a basis for the coordinated regulation of expression of the Klf4 gene in the intestinal epithelium.
INTRODUCTION
Krüppel-like factors (KLFs) are zinc finger-containing transcription factors that exhibit homology to the Drosophila melanogaster segmentation gene product Krüppel (1). A subfamily of mammalian KLFs highly related to the erythroid Krüppel-like factor (EKLF/KLF1) has recently been described (2). This rapidly expanding subfamily currently has 12 members, which have been given numerical designations by the Human Gene Nomenclature Committee (3). KLFs can be transcriptional activators or repressors (4) and they bind to a similar DNA sequence that has either a CACCC homology or is rich in GC content (5). It is therefore not surprising that KLFs may interact with the same cis-element in the same gene. For example, both KLF12/AP2-rep and KLF9/BTEB1 bind to the CACCC element in the AP2α promoter (6). But, while KLF12 acts as a repressor, KLF9 acts as a strong activator of the AP2α promoter (6). In another example, both KLF6/Zf9 and KLF4/GKLF activate the human keratin 4 promoter through the CACCC motif and the two factors physically interact (7). Finally, both KLF4/GKLF and KLF5/IKLF bind to the CACCC element in the α-smooth muscle and SM22α promoters (8). In this case, KLF4 represses while KLF5 activates the induction of these two promoters by TGFβ (8).
The reciprocal effect of KLF4 and KLF5 in regulating gene expression merits additional consideration. Aside from their opposing biochemical effects on several smooth muscle gene promoters as described above, the cellular distribution and regulatory functions of KLF4 and KLF5 also exhibit an opposite pattern. For example, while Klf4 is mainly expressed in the post-mitotic differentiated villus epithelial cells of the intestinal tract (9), Klf5 is found primarily in the proliferating crypt cells (10). The cellular distribution of the two genes in the epidermis of the skin also mirrors that in the intestinal epithelium (11–13). In vitro, expression of Klf4 is associated with a process of growth arrest (9), while that of Klf5 mainly accompanies cellular proliferation (14). Moreover, forced expression of Klf4 leads to a G1/S cell cycle arrest (15,16) but that of Klf5 causes a transformed phenotype (14).
Because of the relatively restricted pattern of tissue expression of Klf4, we have been characterizing factors that regulate its expression. One such factor, Cdx2, previously shown to drive differentiation of intestinal epithelial cells (17), is shown to be a transactivator of the Klf4 promoter (18,19). In addition, Klf4 is capable of transactivating the promoter of its own gene through three closely spaced GC-boxes within the promoter (18). The present study demonstrates that Klf5 binds to the same DNA cis-elements as Klf4, including those within the Klf4 promoter. However, while Klf4 activates, Klf5 represses Klf4 promoter activity. Furthermore, the two factors can abrogate each other’s effect on the Klf4 promoter. Lastly, the two factors compete with each other for binding to the same cis-element. The results of these studies suggest that Klf4 and Klf5 have opposing biochemical functions in regulating expression of the Klf4 gene and may potentially be involved in a coordinated effort to orchestrate the proliferative and differentiated phenotype of the intestinal epithelium.
MATERIALS AND METHODS
Plasmid constructs
The eukaryotic expression construct containing full-length Klf4 (PMT3–Klf4) and the luciferase reporter construct containing 1.0 kb of the 5′ flanking promoter region of the Klf4 gene (Klf4–pGL2–Luciferase) were previously described (18). The expression construct containing full-length Klf5, pBK CMV–Klf5, was generously provided by J. Lingrel (University of Cincinnati, College of Medicine) (10). An EcoRI–KpnI fragment of the full-length Klf5 cDNA from pBK CMV–Klf5 was subcloned into the PMT3 vector to create PMT3–Klf5. The two PMT3 constructs were digested with appropriate restriction endonucleases to release the zinc finger portions of Klf4 and Klf5, which were then subcloned back into the PMT3 vector to create PMT3–Klf4–ZF and PMT3–Klf5–ZF, respectively. The internal control for transfection, pRL–CMV, was purchased from Promega Corporation (Madison, WI).
The prokaryotic expression vector pET–16b containing the zinc finger portion of Klf4 (amino acids 350–483) has previously been described (20). The C-terminal portion of Klf5 between amino acids 242 and 446 was cloned into the prokaryotic expression vector pET101/D. Both recombinant proteins contained a 10-histidine tag at the N-terminus and were purified by nickel affinity chromatography as described before (20). The apparent molecular weight for the resultant recombinant protein was 18 and 26 kDa, respectively, for Klf4 and Klf5.
Preparation of nuclear extracts
Nuclear extracts containing full-length Klf4 or full-length Klf5 were prepared from COS-1 cells transfected with PMT3–Klf4 or PMT3–Klf5. Nuclear extracts from cells transfected with PMT3 alone were used as controls. Extracts from transfected cells were prepared as previously described (18). Briefly, transfected cells were rinsed with ice-cold phospate-buffered saline, scraped, harvested and pelleted. The pellets were washed with 4 pack cell volume (p.c.v.) of solution containing 10 mM Tris–HCl pH 7.8, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT and a Complete Protease Inhibitor Cocktail Tablet (Roche, Indianapolis, IN). Following 10 min incubation on ice, the cells were lysed by 10 strokes of a Dounce homogenizer. Nuclei were collected by centrifugation (Sorvall Microspin) and resuspended in 2 p.c.v. of a lysis solution containing 420 mM KCl, 20 mM Tris–HCl pH 7.8, 1.5 mM MgCl2, 0.5 mM DTT, 20% glycerol and a Complete Protease Inhibitor Tablet (Roche). After incubation for 1 h at 4°C with gentle agitation, the solution was centrifuged and the supernatant dialyzed twice against 500 ml of 20 mM Tris–HCl pH 7.8, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 20% glycerol and a Complete Protease Inhibitor Tablet (Roche). Extracts were divided into small aliquots and immediately cryopreserved.
Electrophoretic mobility shift assays (EMSAs)
EMSAs were performed according to previously published protocols (9,20). The basic transcription element (BTE) probe contained a sequence in the promoter of the cytochrome P4501A1 gene that had previously been shown to bind to Klf4 (21). The target detection assay (TDA) probe was a previously selected consensus-binding site for KLF4 (20) and closely resembled the three GC-boxes in the promoter of the Klf4 gene (18). The GC-2 probe refers to the GC-box 2 sequence in the Klf4 promoter between nucleotide –108 and –86 that has previously been shown to bind Klf4 (18). Probes were labeled with [γ-32P]ATP using T4 polynucleotide kinase. In each reaction, 0.5 pmol of labeled probe was used. Nuclear extracts from PMT3-, PMT3–Klf4- or PMT3–Klf5-transfected cells, or purified recombinant Klf4 or Klf5 were incubated in a volume of 30 µl containing 20 mM Tris–HCl pH 7.2, 10 mM MgCl2, 150 mM NaCl, 20 mM KCl, 5 µM ZnCl2, 0.5 mM DTT, 5% glycerol and 2 µg poly(dI•dC) on ice for 30 min. The labeled probe was then added to the reaction and the incubation continued for another 15 min at room temperature. The DNA and DNA–protein complexes were resolved from one another by native 6.0% polyacrylamide gel electrophoresis and visualized by autoradiography.
Transfection and reporter assays
Transient transfection by lipofection of Chinese hamster ovary (CHO) cells with various DNA constructs (Klf4–pGL2–Luciferase, PMT3–Klf4, PMT3–Klf5, PMT3–Klf4–ZF and PMT3–Klf5–ZF) were performed as previously described (9,18,20). Transfections were performed in 6-well tissue culture dishes at 37°C for 16 h. All transfections included the internal standard pRL–CMV (Promega) to normalize for transfection efficiency. Luciferase and Renilla assays were performed using the Dual-Luciferase Assay (Promega).
RESULTS
Klf4 and Klf5 bind to similar cis-DNA elements
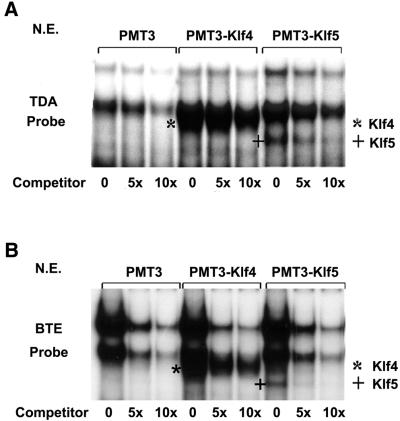
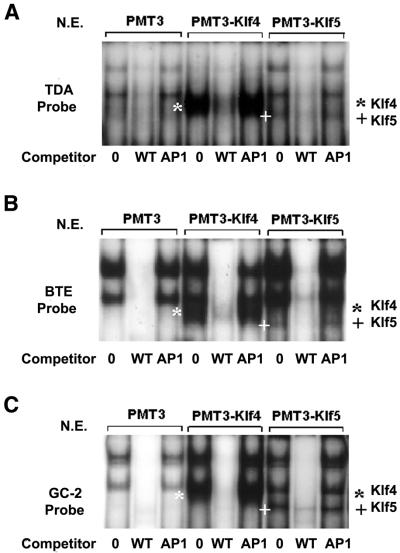
We first examined whether Klf4 and Klf5 bound to similar sequences by EMSAs using radiolabeled oligonucleotides containing either an established consensus sequence for Klf4 (TDA) (20) or the BTE (21). As shown in Figure 1A, two common DNA–protein complexes were formed when the TDA probe was incubated with nuclear extracts from PMT3-, PMT3–Klf4- and PMT3–Klf5-transfected cells. However, an additional complex was formed in the reaction that contained either PMT3–Klf4- or PMT3–Klf5-transfected cells. The Klf4–TDA complex is identified by * and the Klf5–TDA complex is identified by +. The specificities of all the DNA–protein complexes were demonstrated by their gradual disappearance upon addition of increasing amounts of unlabeled probe (Fig. 1A). A similar pattern was observed when BTE was used as a probe except that the intensities of the two common bands were stronger when compared with the TDA probe (Fig. 1B). The results suggest that Klf4 and Klf5 bind to similar DNA elements. Furthermore, both Klf4 and Klf5 bound to the GC-2 probe (Fig. 2C) which represents one of three GC-box sequences in the Klf4 promoter, all of which have been shown to bind Klf4 (18). The specificities of DNA–protein interaction with all three probes were further demonstrated by the failure of an unrelated AP1 probe to compete for the formation of the complexes (Fig. 2A–C).
Figure 1.
EMSAs of Klf4 and Klf5 on two cis-elements, TDA and BTE. EMSAs were performed using 10 µg nuclear extracts prepared from COS-1 cells transfected with an expression construct containing vector alone (PMT3), full-length Klf4 (PMT3–Klf4), or full-length Klf5 (PMT3–Klf5) and two radiolabeled oligonucleotide probes containing established cis-elements for Klf4: TDA (A) and BTE (B). Where indicated, an unlabeled cognate oligonucleotide was used as a competitor at 5× and 10× molar excess of the labeled probe. * represents the shifted Klf4–probe complex and + represents the shifted Klf5–probe complex. N.E., nuclear extracts.
Figure 2.
EMSAs of Klf4 and Klf5 on an additional cis-element, GC-2, that is present in the Klf4 promoter. EMSAs were performed with 10 µg nuclear extracts (N.E.) prepared from cells transfected with PMT3, PMT3–Klf4 or PMT3–Klf5 in the absence of any competitors (0), or in the presence of 25× molar excess of a cognate competitor (WT) or an unrelated competitor (AP1). Probes used include TDA (A), BTE (B) and GC-2 (C). * represents the shifted Klf4–probe complex and + represents the shifted Klf5–probe complex.
Klf4 activates and Klf5 represses the Klf4 promoter
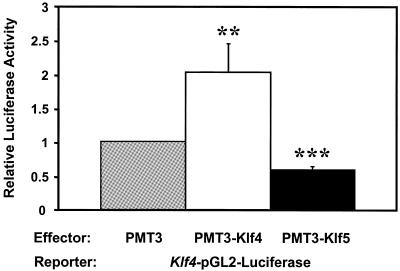
Recent work from our laboratory showed that Klf4 activates the promoter of its own gene by interacting with the GC-boxes in the proximal promoter (18). To determine whether Klf5 also influences the promoter activity of the Klf4 gene, we performed co-transfection studies using the Klf4–pGL2–Luciferase reporter and a eukaryotic expression construct containing either Klf4 or Klf5. As depicted in Figure 3, PMT3–Klf4 significantly activated the Klf4 promoter when compared with PMT3 alone, confirming previous findings. In contrast, PMT3–Klf5 significantly suppressed Klf4 promoter activity when compared with PMT3 alone. Thus, Klf4 and Klf5 have opposite effects on the activity of the Klf4 promoter.
Figure 3.
The opposite effects of Klf4 or Klf5 on the Klf4 promoter. Five micrograms each of the Klf4–pGL2–Luciferase reporter and PMT3, PMT3–Klf4 or PMT3–Klf5 and 2 µg pCMV–RL were co-transfected into CHO cells using Lipofectamine reagent (Promega). Firefly and Renilla luciferase assays were determined 24 h after transfection. Shown are the means of three independent experiments (n = 3) performed in triplicate. Relative luciferase activity was standardized with the internal control Renilla luciferase activity. Bars represent standard deviations. ** = P < 0.01, *** = P < 0.001 by Student’s t-test compared with PMT3 vector control.
Klf4 and Klf5 compete to regulate the Klf4 promoter
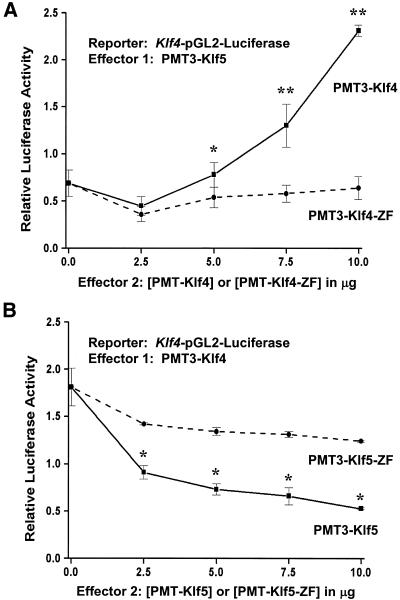
To further determine a possible interaction between Klf4 and Klf5 on the Klf4 promoter, we performed co-transfection experiments using the Klf4–pGL2–Luciferase reporter, a constant amount of PMT3–Klf5 and increasing amounts of PMT3–Klf4 or PMT3–Klf4–ZF. As seen in Figure 4A, in the absence of any competing Klf4, Klf5 suppressed the reporter activity, as in Figure 3. The addition of Klf4 removed this inhibitory effect in a dose-dependent fashion (solid line). In contrast, a construct including only the zinc finger portion of Klf4 failed to reverse the inhibitory effect on the Klf4 promoter (dashed line). Similarly, in a converse experiment with PMT3–Klf4 as the primary effector and Klf5 as the competing effector (Fig. 4B), Klf5 (solid line) but not Klf5–ZF (dashed line) abrogated the stimulatory effect of Klf4 on the Klf4 promoter activity. These results indicate that Klf4 and Klf5 not only exhibit an opposite effect on the Klf4 promoter, they actively compete with each other. Moreover, this competitive effect requires the full-length protein. The zinc fingers alone are not sufficient.
Figure 4.
The competing effects of Klf4 and Klf5 on the Klf4 promoter. (A) Five micrograms each of Klf4–pGL2–Luciferase reporter and effector 1 (PMT3–Klf5) were co-transfected into CHO cells in presence of increasing amounts of effector 2: either PMT3–Klf4 (solid line) or PMT–Klf4–ZF (dashed line). (B) Five micrograms each of Klf4–pGL2–Luciferase reporter and effector 1 (PMT3–Klf4) were co-transfected into CHO cells with increasing amounts of effector 2: either PMT3–Klf5 (solid line) or PMT3–Klf5–ZF (dashed line). The total amount of DNA transfected was kept constant at 25 µg by the addition of PMT3 DNA. Firefly and Renilla luciferase assays were determined 24 h after transfection. Shown are the means of four independent experiments (n = 4) performed in triplicate. The relative activity was determined from dividing the reporter activity of effector 1-transfected cells by that of PMT3-transfected cells. Bars represent standard deviations. * = P < 0.05, ** = P < 0.01 by Student’s t-test comparing full-length with zinc finger only-containing constructs.
Klf4 and Klf5 compete with each other for DNA binding
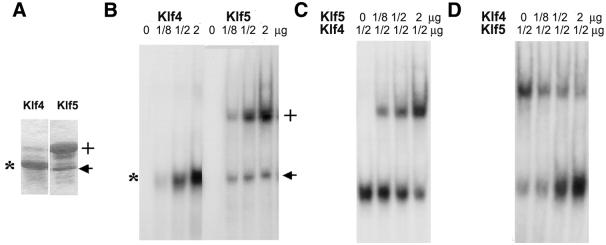
One possibility that may explain the competing effects of Klf4 and Klf5 on the Klf4 promoter is that the two proteins may compete with each other for binding to DNA. To address this possibility, we produced purified partial-length recombinant proteins that contained the zinc finger region of Klf4 and Klf5 (Fig. 5A). When incubated individually with a labeled GC-2 probe, each protein interacted avidly with the probe (Fig. 5B). We next performed EMSA of GC-2 using a constant amount of recombinant Klf4 and increasing amounts of Klf5. As seen, Klf5 reduced the binding of Klf4 to the probe in a dose-dependent fashion (Fig. 5C). Similarly, if an increasing amount of Klf4 was incubated with a constant amount of Klf5, the binding of Klf5 to the probe also gradually diminished (Fig. 5D). These results indicate that Klf4 and Klf5 compete with each other in binding to DNA.
Figure 5.
The competing effects of Klf4 and Klf5 on binding to DNA. (A) Purified recombinant proteins containing the zinc finger portion of Klf4 (*) and Klf5 (+) are shown in this Commassie blue-stained gel. The apparent molecular weight for Klf4 is 18 kDa and that for Klf5 is 26 kDa. The arrow points to a degradation product of Klf5, which apparently binds to DNA (B). (B) EMSAs of recombinant Klf4 and Klf5 on the GC-2 probes. Increasing amounts of purified recombinant Klf4 or Klf5 were used in the experiment. (C) EMSAs were performed using a constant amount of recombinant Klf4 and increasing amounts of Klf5. (D) EMSAs were performed with a constant amount of recombinant Klf5 and increasing amounts of Klf4.
DISCUSSION
The present study demonstrates how two closely related Klfs exert their effects on transcription of a target gene in an opposing manner. We show that Klf5 binds to several cis-elements previously established to be the binding sites for Klf4, including TDA, BTE and GC-2, the last a native cis-element in the Klf4 promoter. However, while Klf4 activates the promoter activity of its own gene, Klf5 has exactly the opposite effect. Moreover, when present together, the two Klfs interfere with each other in modulating the Klf4 promoter activity. We also show that a possible explanation for this interfering effect is due to competition of the two proteins for binding to DNA.
Opposing effects of other KLFs on the promoters of several other genes have previously been described. For example, Klf4 suppresses the activity of the cytochrome p4501A1 (CYP1A1) promoter while Sp1 activates it in a BTE-dependent fashion (21). Also, similar to the present study, Klf4 abrogates the stimulatory effect of Sp1 on the CYP1A1 gene in a dose-dependent manner. In this case, it was shown that the zinc fingers of Klf4 compete for binding to BTE with Sp1 (21). It is of interest to note that KLF5, also known as BTEB2 (10), is a strong activator of the CYP1A1 promoter (22). The findings on the CYP1A1 promoter would then be the first example in which KLF4 and KLF5 antagonize each other in regulating promoter activity. In another example, KLF4 and KLF5 bind to the same CACCC element present in the promoters of two smooth muscle genes, α-smooth muscle actin and SM22α (8). Here, KLF4 inhibits TGFβ-dependent stimulation of the SM22α promoter while KLF5 further activates TGFβ-dependent stimulation of the same promoter (8). Thus, evidence from literature to date indicates that KLF4 and KLF5 exert opposite effects on the promoters of the genes studied.
It should be noted that the biochemical behavior of KLF4 and KLF5 described above is a reflection of their biological behavior. For example, the cellular distribution of the two Klfs in several epithelial tissues is complementary, rather than redundant, to each other. Thus, Klf4 is mainly expressed in the post-mitotic villus epithelial cells of the intestine (9) and the suprabasal epidermal cells of the skin (11), while Klf5 is expressed mostly in the proliferating cells of the crypt in the intestine (10) and the basal epidermal cells of the skin (13). Moreover, the pattern of expression of the two genes in vitro contrasts each other. Klf4 is mainly expressed in the growth-arrested cells due to serum deprivation (9), contact inhibition (9) or DNA damage (21). Expression of Klf5, on the contrary, is associated with a proliferative state as seen during serum or mitogenic stimulation (14). Based on the results of current and past studies, one may speculate that the lack of crypt expression of Klf4 in the intestinal crypt epithelium may, in part, be due to the presence of Klf5 in this compartment.
A potential mechanism that may explain how Klf4 and Klf5 compete with each other to regulate the activity of the Klf4 promoter appears to be exerted at the level of DNA–protein interaction. As demonstrated in Figure 5, Klf5 actively inhibits the binding of Klf4 to the GC-2 probe, and vice versa. However, the zinc finger portion of one Klf is not sufficient in reversing the effect of the other Klf on the activity of the Klf4 promoter (Fig. 4). These results indicate that amino acid sequence outside the zinc fingers of both proteins is required for the full regulation of the Klf4 promoter. Klf4 has previously been shown to be a pleiotropic transcription factor, with activating and repressing effects (23). Mutational studies have also located the activating and repressive domains to regions outside the zinc fingers in the Klf4 polypeptide (24). KLF5, although known to be a transcription activator in most studies (10,22), can act as a repressor (25). One might therefore expect that some of these domains involved in transcriptional regulation by the two proteins are responsible for the interplay noted in the current study.
The opposing effects of Klf4 and Klf5 are reminiscent of the opposing effects of Cdx1 and Cdx2 along the crypt–villus axis of the intestinal epithelium. CDX1 and CDX2 are homeodomain transcription factors with sequence homology to the caudal gene of D.melanogaster, which in mammals are specifically expressed in the epithelium of small intestine and colon (26–29). Cdx1 is expressed in the proliferating crypt cells (27,30,31) while Cdx2 is expressed predominantly in the differentiated villus cells (28,32). Several studies have shown that CDX1 regulates cell proliferation (33,34) and CDX2 regulates cell differentiation and growth arrest (17,35).
Of interest, we have recently reported that the intestinal gatekeeper adenomatous polyposis coli (APC) induces Klf4 expression via Cdx2 (19). APC is a cytoplasmic protein that modulates the oncogenic Wnt signal transduction cascade by binding to β-catenin and promoting its phosphorylation by glycogen synthase kinase 3 β (GSK3β), for subsequent degradation (36). Our previous data, which showed that APC activation of Klf4 via Cdx2 occurred through a β-catenin independent pathway, have led to our proposal that these three proteins form a part of an enterocyte-specific tumor suppressor pathway in the sequence of APC → CDX2 → KLF4 → growth arrest (19). This pathway may become active as epithelial cells migrate up the intestinal villus and undergo differentiation with the simultaneous loss of proliferative potential. Interestingly, Wnt, which is the product of a proto-oncogene with opposing effects to APC, can up-regulate CDX1 but not CDX2 expression (37). Moreover, Wnt has recently been reported to induce mouse KLF5/BTEB2 through a β-catenin independent, protein kinase C dependent pathway (38). A search of the mouse Klf5 promoter (DDBJ/EMBL/GenBank accession no. AF285184) reveals several potential CDX1 binding sites (39). We may postulate that KLF5 is regulated by Wnt via CDX1 in the proliferating intestinal crypt cells while KLF4 is regulated by APC via CDX2 in the differentiating intestinal villus cells. Crosstalk and negative regulation between these opposing pathways, at the level of KLF4 and KLF5 as shown in this paper, as well as possibly higher levels involving CDX1, CDX2, Wnt and APC, tightly coordinate the intestinal epithelial cells as they proliferate, migrate and differentiate. Current studies are underway to address these interactions.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Jerry Lingrel for providing reagents. This work was supported in part by grants from the National Institutes of Health (DK52230 and CA84197 to V.W.Y. and DK59970 to D.T.D.). D.T.D. is a recipient of an American Digestive Health Foundation/American Gastroenterological Association Research Scholar Award and a Glaxo Institute for Digestive Health Basic Research Award.
REFERENCES
- 1.Schuh R., Aicher,W., Gaul,U., Cote,S., Preiss,A., Maier,D., Seifert,E., Nauber,U., Schroder,C. and Kemler,R. (1986) A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Krüppel, a Drosophila segmentation gene. Cell, 47, 1025–1032. [DOI] [PubMed] [Google Scholar]
- 2.Turner J. and Crossley,M. (1999) Mammalian Krüppel-like transcription factors: more than just a pretty finger. Trends Biochem. Sci., 24, 236–240. [DOI] [PubMed] [Google Scholar]
- 3.White J.A., McAlpine,P.J., Antonarakis,S., Cann,H., Eppig,J.T., Frazer,K., Frezal,J., Lancet,D., Nahmias,J., Pearson,P., Peters,J., Scott,A., Scott,H., Spurr,N., Talbot,C.,Jr. and Povey,S. (1997) Guidelines for human gene nomenclature. HUGO nomenclature committee. Genomics, 45, 468–471. [DOI] [PubMed] [Google Scholar]
- 4.Bieker J.J. (2001) Krüppel-like factors: three fingers in many pies. J. Biol. Chem., 276, 34355–34358. [DOI] [PubMed] [Google Scholar]
- 5.Dang D.T., Pevsner,J. and Yang,V.W. (2000) The biology of mammalian Krüppel-like factors. Int. J. Biochem. Cell Biol., 32, 1103–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imhof A., Schuierer,M., Werner,O., Moser,M., Roth,C., Bauer,R. and Buettner,R. (1999) Transcriptional regulation of the AP-2alpha promoter by BTEB-1 and AP-2rep, a novel wt-1/egr-related zinc finger repressor. Mol. Cell. Biol., 19, 194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okano J., Opitz,O.G., Nakagawa,H., Jenkins,T.D., Friedman,S.L. and Rustgi,A.K. (2000) The Krüppel-like transcriptional factors Zf9 and GKLF coactivate the human keratin 4 promoter and physically interact. FEBS Lett., 473, 95–100. [DOI] [PubMed] [Google Scholar]
- 8.Adam P.J., Regan,C.P., Hautmann,M.B. and Owens,G.K. (2000) Positive- and negative-acting Krüppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22αin vivo. J. Biol. Chem., 275, 37798–37806. [DOI] [PubMed] [Google Scholar]
- 9.Shields J.M., Christy,R.J. and Yang,V.W. (1997) Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J. Biol. Chem., 271, 20009–20017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conkright M.D., Wani,M.A., Anderson,K.P. and Lingrel,J.B. (1999) A gene encoding an intestinal-enriched member of the Krüppel-like factor family expressed in intestinal epithelial cells. Nucleic Acids Res., 27, 1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrett-Sinha L.A., Eberspaecher,H., Seldin,M.F. and de Crombrugghe,B. (1996) A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J. Biol. Chem., 271, 31384–31390. [DOI] [PubMed] [Google Scholar]
- 12.Segre J.A., Bauer,C. and Fuchs,E. (1999) Klf4 is a transcription factor required for establishing the barrier function of the skin. Nature Genet., 22, 356–360. [DOI] [PubMed] [Google Scholar]
- 13.Ohnishi S., Laub,F., Matsumoto,N., Asaka,M., Ramirez,F., Yoshida,T. and Terada,M. (2000) Developmental expression of the mouse gene coding for the Krüppel-like transcription factor KLF5. Dev. Dyn., 217, 421–429. [DOI] [PubMed] [Google Scholar]
- 14.Sun R., Chen,X.M. and Yang,V.W. (2001) Intestinal-enriched Krüppel-like factor (Krüppel-like factor 5) is a positive regulator of cellular proliferation. J. Biol. Chem., 276, 6897–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X.M., Johns,D.C., Geiman,D.E., Marban,E., Dang,D.T., Hamlin,G., Sun,R. and Yang,V.W. (2001) Gut-enriched Krüppel-like factor (Krüppel-like factor 4) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J. Biol. Chem., 276, 30423–30428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shie J.L., Chen,Z.Y., O’Brien,M.J., Pestell,R.G., Lee,M.E. and Tseng,C.C. (2000) Role of gut-enriched Krüppel-like factor in colonic cell growth and differentiation. Am. J. Physiol. Gastrointest. Liver Physiol., 279, G806–G814. [DOI] [PubMed] [Google Scholar]
- 17.Suh E. and Traber,P.G. (1996) An intestine-specific homeobox gene regulates proliferation and differentiation. Mol. Cell. Biol., 16, 619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahatan C.S., Kaestner,K.H., Geiman,D.E. and Yang,V.W. (1999) Characterization of the structure and regulation of the murine gene encoding gut-enriched Krüppel-like factor (Krüppel-like factor 4). Nucleic Acids Res., 27, 4562–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang D.T., Mahatan,C.S., Dang,L.H., Agboola,I.A. and Yang,V.W. (2001) Expression of the gut-enriched Krüppel-like factor (Krüppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on CDX2. Oncogene, 20, 4884–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shields J.M. and Yang,V.W. (1998) Identification of the DNA sequence that interacts with the gut-enriched Krüppel-like factor. Nucleic Acids Res., 26, 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W., Shields,J.M., Sogawa,K., Fujii-Kuriyama,Y. and Yang,V.W. (1998) The gut-enriched Krüppel-like factor suppresses the activity of the CYP1A1 promoter in an Sp1-dependent fashion. J. Biol. Chem., 273, 17917–17925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sogawa K., Imataka,H., Yamasaki,Y., Kusume,H., Abe,H. and Fujii-Kuriyama,Y. (1993) cDNA cloning and transcriptional properties of a novel GC box-binding protein, BTEB2. Nucleic Acids Res., 21, 1527–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W., Geiman,D.E., Shields,J.M., Dang,D.T., Mahatan,C., Kaestner,K.H., Biggs,J.R., Kraft,A.S. and Yang,V.W. (2000) The gut-enriched Krüppel-like factor (Krüppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J. Biol. Chem., 275, 18391–18398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yet S.F., Folta,S.C., Jain,M.K., Hsieh,C.M., Maemura,K., Layne,M.D., Zhang,D., Marria,P.B., Yoshizumi,M., Chin,M.T., Perrella,M.A. and Lee,M.E. (1998) Human EZF, a Krüppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J. Biol. Chem., 273, 1026–1031. [DOI] [PubMed] [Google Scholar]
- 25.Shi H., Zhang,Z., Wang,X., Liu,S. and Teng,C.T. (1999) Isolation and characterization of a gene encoding human Krüppel-like factor 5 (IKLF): binding to the CAAT/GT box of the mouse lactoferrin gene promoter. Nucleic Acids Res., 27, 4807–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duprey P., Chowdhury,K., Dressler,G.R., Balling,R., Simon,D., Guenet,J.L. and Gruss,P. (1988) A mouse gene homologous to the Drosophila gene caudal is expressed in epithelial cells from the embryonic intestine. Genes Dev., 2, 1647–1654. [DOI] [PubMed] [Google Scholar]
- 27.James R. and Kazenwadel,J. (1991) Homeobox gene expression in the intestinal epithelium of adult mice. J. Biol. Chem., 266, 3246–3251. [PubMed] [Google Scholar]
- 28.James R., Erler,T. and Kazenwadel,J. (1994) Structure of the murine homeobox gene cdx-2. Expression in embryonic and adult intestinal epithelium. J. Biol. Chem., 269, 15229–15237. [PubMed] [Google Scholar]
- 29.Freund J.N., Domon-Dell,C., Kedinger,M. and Duluc,I. (1998) The Cdx-1 and Cdx-2 homeobox genes in the intestine. Biochem. Cell Biol., 76, 957–969. [DOI] [PubMed] [Google Scholar]
- 30.Silberg D.G., Furth,E.E., Taylor,J.K., Schuck,T., Chiou,T. and Traber,P.G. (1997) CDX1 protein expression in normal, metaplastic and neoplastic human alimentary tract epithelium. Gastroenterology, 113, 478–486. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian V., Meyer,B. and Evans,G.S. (1998) The murine Cdx1 gene product localises to the proliferative compartment in the developing and regenerating intestinal epithelium. Differentiation, 64, 11–18. [DOI] [PubMed] [Google Scholar]
- 32.Silberg D.G., Swain,G.P., Suh,E.R. and Traber,P.G. (2000) Cdx1 and cdx2 expression during intestinal development. Gastroenterology, 119, 961–971. [DOI] [PubMed] [Google Scholar]
- 33.Maulbecker C.C. and Gruss,P. (1993) The oncogenic potential of deregulated homeobox genes. Cell Growth Differ., 4, 431–441. [PubMed] [Google Scholar]
- 34.Soubeyran P., Haglund,K., Garcia,S., Barth,B.U., Iovanna,J. and Dikic,I. (2001) Homeobox gene Cdx1 regulates Ras, Rho and PI3 kinase pathways leading to transformation and tumorigenesis of intestinal epithelial cells. Oncogene, 20, 4180–4187. [DOI] [PubMed] [Google Scholar]
- 35.Suh E., Chen,L., Taylor,J. and Traber,P.G. (1994) A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol. Cell. Biol., 14, 7340–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polakis P. (2000) Wnt signaling and cancer. Genes Dev., 14, 1837–1851. [PubMed] [Google Scholar]
- 37.Lickert H., Domon,C., Huls,G., Wehrle,C., Duluc,I., Clevers,H., Meyer,B.I., Freund,J.N. and Kemler,R. (2000) Wnt/β-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development, 127, 3805–3813. [DOI] [PubMed] [Google Scholar]
- 38.Ziemer L.T., Pennica,D. and Levine,A.J. (2001) Identification of a mouse homolog of the human BTEB2 transcription factor as a β-catenin-independent Wnt-1-responsive gene. Mol. Cell. Biol., 21, 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schug J. and Overton,G.C. (1997) TESS: Transcription Element Search Software on the WWW, Technical Report CBIL-TR-1997-1001-v0.0. Computational Biology and Informatics Laboratory, School of Medicine, University of Pennsylvania. http://www.cbil.upenn.edu/tess/index.html.