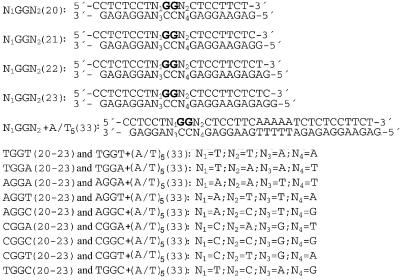Abstract
Antitumor cisplatin [cis-diamminedichloroplatinum(II)] forms on DNA predominantly intrastrand cross-links between neighboring purine residues. Several discoveries suggested that the toxicity of cisplatin originated from these lesions. The formation of 1,2-GG intrastrand cross-link of cisplatin leads to marked conformational alterations in DNA including a directional, rigid bend toward the major groove and local unwinding. These altered structures attract various cellular proteins. This phenomenon has been postulated to mediate antitumor properties of cisplatin. Importantly, the binding affinity of several proteins that specifically recognize 1,2-GG intrastrand cross-link to platinated DNA is modulated by the nature of the base pairs that immediately flank the platinated d(GpG) site. However, the influence of sequence context on DNA bending and unwinding due to the formation of the 1,2-GG intrastrand cross-link has not been extensively investigated. In the present study we have employed electrophoretic retardation (phasing) assay to analyze bending and unwinding induced by the single, site-specific 1,2-GG intrastrand cross-link immediately flanked by various bases formed by cisplatin in nine oligodeoxyribonucleotide duplexes. The results indicate that bending and unwinding of DNA as a consequence of the formation of the major adduct of cisplatin is, in the first approximation, independent of the base pairs flanking the platinated d(GpG) site.
INTRODUCTION
Sufficient evidence has accumulated to identify DNA as the relevant pharmacological target of antitumor cisplatin [cis-diamminedichloroplatinum(II)] (1,2). Cisplatin forms on DNA predominantly intrastrand cross-links (CLs) between neighboring purine residues, mainly between guanine bases (3,4). In these CLs the platinum atom binds to the N7 nitrogen atoms of the purine residues. Several discoveries (5) suggested that the toxicity of cisplatin originated from these intrastrand CLs although relative antitumor efficacy of these and other CLs formed on DNA by this drug still remains unknown (6,7). Hence, a number of studies turned to investigating the structure, biochemical consequences and cellular responses of 1,2-GG intrastrand CLs formed on DNA by cisplatin.
The formation of major 1,2-GG intrastrand CL by cisplatin leads to marked conformational alterations in DNA (8,9). These adducts induce a directional, rigid bend of the helix axis toward the major groove at the site of platinum binding and local unwinding. In addition, severe perturbation of hydrogen bonding within the 5′ coordinated GC base pair, widening and flattening of the minor groove opposite the cisplatin adduct, creation of a hydrophobic notch, global distortion extending over 4–5 bp and additional helical parameters characteristic of A-form DNA have been also reported.
Among the alterations of secondary and tertiary structure of DNA to which it may be subject, the role of bending and unwinding of DNA is increasingly recognized as of potential importance in regulating replication and transcription functions through specific DNA–protein interactions. Some structures altered by 1,2-GG intrastrand CLs, such as stable directional bending and unwinding, attract various damaged-DNA binding proteins (10–12). This binding of the cellular proteins has been postulated to mediate the antitumor properties of cisplatin (11–13). Interestingly, the binding affinity of several proteins that specifically recognize 1,2-GG intrastrand CL, such as, for instance, abundant nuclear HMGB1 protein and TATA-binding protein (TBP), to DNA containing this adduct is modulated by the nature of the base pairs that immediately flank the platinated d(GpG) site in this intrastrand CL (14–16). In addition, formation of cisplatin 1,2-intrastrand CLs on DNA is flanking-sequence independent (17). In spite of these observations and the fact that DNA bending and unwinding have been postulated to play an important role in the recognition of the major 1,2-GG intrastrand CLs of cisplatin by the proteins, and consequently in other downstream cellular events underlying the mechanism of antitumor effects of cisplatin, the influence of sequence context on these structural distortions has not been extensively investigated.
In the present study we have employed electrophoretic retardation (phasing) assay to analyze bending and unwinding induced by the single, site-specific 1,2 intrastrand CL of cisplatin formed in oligodeoxyribonucleotide duplexes between neighboring guanine residues immediately flanked by various bases. The results indicate that bending and unwinding of DNA double helix as a consequence of the formation of the 1,2-GG intrastrand CLs by cisplatin is, in the first approximation, independent of the base pairs flanking the platinated d(GpG) site.
MATERIALS AND METHODS
Chemicals
Cisplatin was purchased from Sigma. The diaqua species were generated from cisplatin (0.5 mM) by the addition of 1.9 mol equivalent of AgNO3 in 10 mM NaClO4 at 37°C for 24 h in the dark. The AgCl precipitate was removed by centrifugation (15 000 g). The synthetic oligodeoxyribonucleotides (Fig. 1) were synthesized and purified as described previously (18). T4 DNA ligase and T4 polynucleotide kinase were purchased from New England Biolabs (Beverly, MA). Acrylamide, bis(acrylamide), urea and NaCN were from Merck KgaA (Darmstadt, Germany). Dimethyl sulfate (DMS) was from Sigma. [γ-32P]ATP was from Amersham (Arlington Heights, IL).
Figure 1.
Sequences of the synthetic oligodeoxyribonucleotide duplexes used in the present study with their abbreviations. The top and bottom strands of each pair are designated top and bottom, respectively, in the text. The bold letters in the top strands of the duplexes indicate the location of the intrastrand CL after modification of the oligonucleotides by cisplatin in the way described in the Materials and Methods.
Platination of oligonucleotides
The single-stranded oligonucleotides (the top strands of the duplexes shown in Fig. 1) were reacted in stoichiometric amounts with diaqua derivative of cisplatin. The platinated oligonucleotides were repurified by ion-exchange fast protein liquid chromatography (FPLC). It was verified by platinum flameless atomic absorption spectrophotometry (FAAS) and by the measurements of the optical density that the modified oligonucleotides contained one platinum atom. It was also verified using DMS footprinting of platinum on DNA (19,20) that in the platinated top strands of N1GGN2(20–23) and N1GGN2 + (A/T)5(33) duplexes (Fig. 1) the N7 position of both neighboring guanine residues were not accessible for reaction with DMS. Briefly, platinated and non-modified top strands (5′ end-labeled with 32P) were reacted with DMS. DMS methylates the N7 position of guanine residues in DNA, producing alkali labile sites (21). However, if N7 is coordinated to platinum, it cannot be methylated. The oligonucleotides were then treated with hot piperidine and analyzed by denaturing 24% polyacryalmide gel electrophoresis. For the non-modified oligonucleotides, shortened fragments, due to the cleavage of the strand at one or two methylated guanines, were observed in the gel. However, no such bands were detected for the oligonucleotides modified by cisplatin. These results indicate that one cisplatin molecule was coordinated to neighboring guanine residues forming 1,2-GG intrastrand CL in the top strands of N1GGN2(20–23) and N1GGN2 + (A/T)5(33) duplexes. The platinated strands were allowed to anneal with unplatinated complementary strands (the bottom strands in Fig. 1) in 50 mM NaCl plus 10 mM Tris–HCl/EDTA buffer, pH 7.4. FPLC purification and FAAS measurements were carried out on a Pharmacia Biotech FPLC System with MonoQ HR 5/5 column and a Unicam 939 AA spectrometer equipped with a graphite furnace, respectively.
Ligation and electrophoresis of oligonucleotides
Unplatinated single strands (top strands in Fig. 1) or these strands containing a unique intrastrand CL were 5′ end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. Then these single-stranded oligonucleotides were annealed with their phosphorylated complementary strands. Unplatinated or intrastrand CL containing duplexes were allowed to react with T4 DNA ligase. The resulting samples along with ligated unplatinated duplexes were subsequently examined on 8% native polyacrylamide [mono:bis(acrylamide) ratio = 29:1] electrophoresis gels. Other details of these experiments were as described previously (22,23).
RESULTS
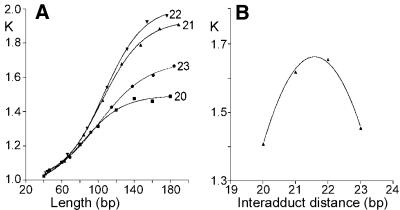
The oligodeoxyribonucleotide duplexes N1GGN2(20–23) (20–23 bp, Fig. 1) used in the bending and unwinding studies of the present work contained central base pair sequence d(GpG)/d(CpC) at which intrastrand CL was formed between guanine residues. The duplexes differed in base residues N1 and N2 (and N3 and N4 in the complementary strand), whereas the nucleotide sequences of these duplexes outside this central sequence N1GGN2 were identical for all duplexes of the same length tested in the present work. All sequences were also designed to leave a 1 nt overhang at their 5′ ends in double-stranded form. These overhangs facilitate polymerization of the monomeric oligonucleotide duplexes by T4 DNA ligase in only one orientation, and maintain a constant interadduct distance throughout the resulting multimer. Autoradiogram of electrophoresis gels revealing resolution of the ligation products of 20–23 bp duplexes AGGA unplatinated or containing a unique 1,2-GG intrastrand CL of cisplatin is shown in Figure 2 as an example. A significant retardation was observed for the multimers of all platinated duplexes. Decreased gel electrophoretic mobility may result from a decrease in the DNA end-to-end distance (24). Various platinum(II) complexes have been shown to form DNA adducts which decrease gel mobility of DNA fragments due to either stable curvature of the helix axis or increased isotropic flexibility (23,25–30). DNA multimers of identical length and number of stable bend units, but with differently phased bends, have different end-to-end distances. The DNA bends of a multimer must, therefore, be spaced evenly and phased with the DNA helical repeat in order to add constructively. Such constructively phased bends add in plane, yielding short end-to-end distances and the most retarded gel migration. In other words, gel electrophoresis of multimers of oligonucleotide duplexes that only differ in length and contain a stable curvature induced by the same platinum adduct should exhibit a phase effect, i.e. the maximum retardation should be observed for the multimers having the bends in phase with helix screw. In contrast, the normal electrophoretic mobility should be observed for the multimers having the bends separated by a half-integral number of DNA turns. The K factor is defined as the ratio of calculated to actual length. The calculated length is based on a multimer’s mobility, and is obtained from a calibration curve constructed from the mobilities of unplatinated multimers. The variation of the K factor versus sequence length obtained for multimers of the duplexes AGGA (20–23 bp long), non-modified and containing the unique 1,2-d(GG) intrastrand CL of cisplatin, is shown in Figure 3A. Maximum retardation was observed for the cross-linked duplex 22-bp long. This observation suggests that the natural 10.5-bp repeat of B-DNA and that of DNA perturbed by the cisplatin intrastrand CL are different as a consequence of DNA unwinding (31). Interestingly, the 21-bp curves had only a slightly smaller slope than 22-bp curves, whereas the 20- and 23-bp curves differed more pronouncedly (Fig. 3A). This asymmetry is also consistent with a significant DNA unwinding due to the formation of the intrastrand CL by cisplatin.
Figure 2.
Autoradiogram of the ligation products of double-stranded oligonucleotides AGGA(20–23) containing a unique 1,2-GG intrastrand CL formed by cisplatin between central guanine residues in the top strand. The ligation products were separated on an 8% polyacrylamide gel (lanes Pt). NoPt, unplatinated oligomers,.
Figure 3.
(A) Plots showing the relative mobility K versus sequence length curves for the oligomers AGGA(20–23) containing single 1,2-GG intrastrand CL of cisplatin, denoted as 20, 21, 22 and 23, respectively. (B) Plots showing the relative mobility K versus interadduct distance in base pairs for the duplexes AGGA(20–23) intrastrand cross-linked by cisplatin with a total length of 120 bp. The experimental points represent the average of three independent electrophoresis experiments. The curves represent the best fit of these experimental points to the equation K = ad2 + bd + c (31).
The exact helical repeat of the intrastrand cross-linked duplex and, from it, the unwinding angle were calculated by interpolation with the use of the K versus interadduct distance curve as described in the previous papers for the 1,2-GG intrastrand CL of cisplatin in the duplexes containing the central TGGT sequence (31). The maximum of these curves constructed for the duplexes intrastrand cross-linked by cisplatin with a total length of 120 bp (shown for the AGGA duplex in Fig. 3B) were determined to be 21.57 ± 0.01. Total sequence lengths other than 120 bp (110 and 130 bp) were examined and gave identical results. To convert the interadduct distance in base pairs corresponding to the curve maximum into a duplex unwinding angle in degrees, the value is compared with that of the helical repeat of B-DNA, which is 10.5 ± 0.05 bp (32,33). The difference between the helical repeat of B-DNA and the DNA containing 1,2-GG intrastrand CL of cisplatin, therefore, is [(21.57 ± 0.01) – 2(10.5 ± 0.05)] = 0.57 ± 0.06 bp. There are 360°/10.5 bp, so the DNA unwinding due to one intrastrand CL of cisplatin is 20 ± 2°.
The evaluation of the relationship between interadduct distance and phasing for self-ligated multimers composed of the identical number of monomeric duplexes (bend units) resulted in a bell-shaped pattern (Fig. 3B) characteristic for bending (23,25–30). The quantitation of the bend angle of the 1,2-GG intrastrand CL of cisplatin was performed in the way described previously (23,25–30) utilizing the empirical equation
K – 1 = (9.6 × 10–5 L2 – 0.47)(RC)2 1
where L represents the length of a particular oligomer with relative mobility K, and RC the curvature relative to a DNA bending induced at the tract of A residues (A tract) (25,34). Application of Equation 1 to the 120-bp multimers of the 22-bp oligomers of AGGA containing the single intrastrand CL of cisplatin leads to a mean curvature of 0.873, relative to the A tract. The average bend angle per two helix turns can be calculated by multiplying the relative curvature by the absolute value of the A tract bend (20°) (23,25,34,35). The results indicate that the bend induced by the 1,2-GG intrastrand CL of cisplatin flanked by adenine residues is ~35°. Application of Eqation 1 to the 110- and 130-bp multimers of the 22-bp oligomers yielded the same result. We assigned the bend direction by reference to an A tract, which is bent by ∼20° toward the minor groove (34) using the same procedure as described previously (28). The duplex AGGA + (A/T)5(33) (Fig. 1) was used which also contained, besides the single 1,2-GG intrastrand CL of cisplatin, the A tract located ‘in phase’ from the CL (the center of the platinated site and the center of the A tract were separated by 11 bp) (Fig. 1). In the cross-linked multimers, the CLs or the A tracts were separated by 33 bp, corresponding to about three helical turns after the incorporation of the estimated 20° of unwinding at the lesion (see above). The cross-linked multimers of AGGA + (A/T)5(33) were, in all cases, less retarded than their unplatinated counterparts (not shown). Hence, the effective bend of the helix axis at the center of the cisplatin intrastrand CL is in the opposite direction to that at the center of the A tract, i.e. the 1,2-GG intrastrand CL formed by cisplatin in the AGGA sequence bends DNA toward the major groove. Other details of the calculations of the unwinding and bending angles have been described previously (23,25–30).
Also produced in ligations of monomers investigated in this work were separate bands arising from small DNA circles that migrate close to the top of the gel (see the bands marked by an asterisk in Fig. 2, lane Pt for the 22mer as example). The occurrence of small DNA circles was even better evident if cisplatin was removed from the products of the ligation reaction by NaCN (not shown). The highest tendency to yield DNA circles was observed for the 22-bp intrastrand cross-linked multimers confirming a close match between the 22-bp sequence repeat and the helix screw (25,36). Interestingly, the ligation products of the intrastrand cross-linked 22-bp duplexes contained several types of DNA circles.
The bending and unwinding angles induced by the 1,2-GG intrastrand CL were also determined in the same way as for the duplex AGGA (see above) in another eight N1GGN2 duplexes where N1 and N2 were dA, dC and T (see Fig. 1 for their nucleotide sequences). The sequences containing flanking deoxyriboguanosines (where N1 and N2 are dG) were not investigated because of synthetic difficulties encountered when trying to prepare site-specifically modified oligonucleotides (15). The results indicate that bending and unwinding angles for all nine duplexes tested in the present work were in the ranges of (33–35) ± 2° toward the major groove and (15–20) ± 2°. Hence, there was no marked difference in DNA bending and unwinding induced by the 1,2-GG intrastrand CL if the bases immediately flanking the platinated d(GpG) site on both its sides varied.
DISCUSSION
The death of tumor cells treated with cisplatin is believed to be mediated by the recognition of its 1,2 intrastrand CLs by cellular proteins (13,37–39). Several models have been proposed to explain the role of platinated DNA-binding proteins in mediating antitumor effects of cisplatin. For instance, one possible pathway is through ‘repair shielding’. The protein that binds to the CL of cisplatin inhibits its repair by physically blocking the access to other (repair) proteins. In this way the cisplatin adducts persist in DNA, thereby potentiating their toxicity. Another mechanism is through the ‘hijacking’ of nuclear factors essential for cellular function. Cisplatin–DNA adducts hijack proteins away from their normal binding sites, thereby disrupting fundamental cellular processes. For instance the TBP, which is essential for transcription initiation in eukaryotes, recognizes and binds to the minor groove of a consensus sequence, TATAAA, known as the TATA box (40). In spite of the fact that this consensus sequence is intrinsically bent toward minor groove, the structure of the TATA box in the human TBP–TATA complex is strikingly similar to the structure of 1,2-GG intrastrand CL, i.e. it is bent toward major groove. Therefore, it is not surprising that the TBP binds selectively to and is sequestered by cisplatin-damaged DNA (41,42). Thus, while the structure of the TATA box in the TBP–TATA complex is an induced fit, TBP binds to the 1,2-GG intrastrand CL to a pre-formed, bent DNA in a lock-and-key fashion. In this way, cisplatin turns the GG sequence into a potential binding site for TBP.
Studies using HMG-domain and TBP proteins have demonstrated that that the sequence context surrounding 1,2-GG intrastrand CL of cisplatin can dramatically change the binding affinity for cisplatin-modified DNA (14–16). For example, affinity of the HMGB1 domain A for the duplex in which adenine residues immediately flank the platinated d(GpG) site was two orders of magnitude higher than for the duplex in which the platinated site was flanked by cytosine residues (14). It is also believed that important structural elements that contribute to the recognition of 1,2 intrastrand CLs by cellular proteins are bending and unwinding (11–13,31). Thus, a natural and so far unanswered question was whether the different affinity of the DNA-binding proteins to the major 1,2-GG intrastrand CLs flanked by different base pairs was due to different bending and unwinding. We have demonstrated in the present paper that DNA bending and unwinding due to the major adduct formed by cisplatin are flanking-base independent so that factors other than prebent and/or unwound target must be responsible for the effect of sequence context on the affinity of platinated DNA-binding proteins to 1,2-GG intrastrand CL of cisplatin.
Several such factors have been already proposed. It has been shown recently that the 1,2-GG intrastrand CL induces substantial thermal and thermodynamic destabilization of the host duplex (43–45). Both the extent and thermodynamic origin of this destabilization are modulated by the nature of the base pairs flanking the CL and this modulatory effect correlates with the impact of the flanking-base sequence on CL-induced energetic destabilization of the host duplex. Other factors that could also contribute to the specificities with which the platinated DNA-binding proteins bind to the 1,2-GG intrastrand CL of cisplatin are specific hydrogen bonding and/or van der Waals interactions between the proteins and the bases flanking the platinated d(GpG) site (11,44). Crystallographic studies will provide more insights into how the sequence context surrounding 1,2-GG intrastrand CL of cisplatin affects the binding affinity of platinated DNA-binding proteins for cisplatin-modified DNA.
Acknowledgments
ACKNOWLEDGEMENTS
This research was supported by the Internal Grant Agency of the Ministry of Health of the Czech Republic (grant no. NL6058-3/2000), Grant Agency of the Czech Republic (grant no. 305/02/1552) and Grant Agency of the Academy of Sciences of the Czech Republic (grant no. A5004101). J.K. is the international research scholar of the Howard Hughes Medical Institute.
REFERENCES
- 1.Johnson N.P., Butour,J.-L., Villani,G., Wimmer,F.L., Defais,M., Pierson,V. and Brabec,V. (1989) Metal antitumor compounds: the mechanism of action of platinum complexes. Prog. Clin. Biochem. Med., 10, 1–24. [Google Scholar]
- 2.Reedijk J. and Teuben,J.M. (1999) Platinum-sulfur interactions involved in antitumor drugs, rescue agents and biomolecules. In Lippert,B. (ed.), Cisplatin. Chemistry and Biochemistry of a Leading Anticancer Drug. Wiley-VCH, Weinheim, Germany, pp. 339–362.
- 3.Eastman A. (1987) The formation, isolation and characterization of DNA adducts produced by anticancer platinum complexes. Pharmacol. Ther., 34, 155–166. [DOI] [PubMed] [Google Scholar]
- 4.Fichtinger-Schepman A.M.J., Van der Veer,J.L., Den Hartog,J.H.J., Lohman,P.H.M. and Reedijk,J. (1985) Adducts of the antitumor drug cis-diamminedichloroplatinum(II) with DNA: formation, identification and quantitation. Biochemistry, 24, 707–713. [DOI] [PubMed] [Google Scholar]
- 5.van de Vaart P.J.M., Belderbos,J., de Jong,D., Sneeuw,K.C.A., Majoor,D., Bartelink,H. and Begg,A.C. (2000) DNA-adduct levels as a predictor of outcome for NSCLC patients receiving daily cisplatin and radiotherapy. Int. J. Cancer, 89, 160–166. [PubMed] [Google Scholar]
- 6.Leng M. and Brabec,V. (1994) DNA adducts of cisplatin, transplatin and platinum-intercalating drugs. In Hemminki,K., Dipple,A., Shuker,D.E.G., Kadlubar,F.F., Segerbäck,D. and Bartsch,H. (eds), DNA Adducts: Identification and Biological Significance. IARC Scientific Publications, No. 125, International Agency for Research on Cancer, Lyon, France, pp. 339–348. [PubMed]
- 7.Brabec V. (2000) Chemistry and structural biology of 1,2-interstrand adducts of cisplatin. In Kelland,L.R. and Farrell,N.P. (eds), Platinum-Based Drugs in Cancer Therapy. Humana Press Inc., Totowa, NJ, pp. 37–61.
- 8.Jamieson E.R. and Lippard,S.J. (1999) Structure, recognition and processing of cisplatin-DNA adducts. Chem. Rev., 99, 2467–2498. [DOI] [PubMed] [Google Scholar]
- 9.Gelasco A. and Lippard,S.J. (1999) Anticancer activity of cisplatin and related complexes. In Clarke,M.J. and Sadler,P.J. (eds), Metallopharmaceuticals I. DNA Interactions. Springer, Berlin, Germany, pp. 1–43.
- 10.Zlatanova J., Yaneva,J. and Leuba,S.H. (1998) Proteins that specifically recognize cisplatin-damaged DNA: a clue to anticancer activity of cisplatin. FASEB J., 12, 791–799. [DOI] [PubMed] [Google Scholar]
- 11.Ohndorf U.M., Rould,M.A., He,Q., Pabo,C.O. and Lippard,S.J. (1999) Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature, 399, 708–712. [DOI] [PubMed] [Google Scholar]
- 12.Zamble D.B. and Lippard,S.J. (1999) The response of cellular proteins to cisplatin-damaged DNA. In Lippert,B. (ed.), Cisplatin. Chemistry and Biochemistry of a Leading Anticancer Drug. Wiley-VCH, Weinheim, Germany, pp. 73–110.
- 13.Kartalou M. and Essigmann,J.M. (2001) Recognition of cisplatin adducts by cellular proteins. Mutat. Res., 478, 1–21. [DOI] [PubMed] [Google Scholar]
- 14.Dunham S.U. and Lippard,S.J. (1997) DNA sequence context and protein composition modulate HMG-domain protein recognition of ciplatin-modified DNA. Biochemistry, 36, 11428–11436. [DOI] [PubMed] [Google Scholar]
- 15.Cohen S.M., Mikata,Y., He,Q. and Lippard,S.J. (2000) HMG-domain protein recognition of cisplatin 1,2-intrastrand d(GpG) cross-links in purine-rich sequence contexts. Biochemistry, 39, 11771–11776. [DOI] [PubMed] [Google Scholar]
- 16.Wei M., Cohen,S.M., Silverman,A.P. and Lippard,S.J. (2001) Effects of spectator ligands on the specific recognition of intrastrand platinum-DNA cross-links by high mobility group box and TATA-binding proteins. J. Biol. Chem., 276, 38774–38780. [DOI] [PubMed] [Google Scholar]
- 17.Burstyn J.N., HeigerBernays,W.J., Cohen,S.M. and Lippard,S.J. (2000) Formation of cis-diamminedichloroplatinum(II) 1,2-intrastrand cross-links on DNA is flanking-sequence independent. Nucleic Acids Res., 28, 4237–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brabec V., Reedijk,J. and Leng,M. (1992) Sequence-dependent distortions induced in DNA by monofunctional platinum(II) binding. Biochemistry, 31, 12397–12402. [DOI] [PubMed] [Google Scholar]
- 19.Lemaire M.A., Schwartz,A., Rahmouni,A.R. and Leng,M. (1991) Interstrand cross-links are preferentially formed at the d(GC) sites in the reaction between cis-diamminedichloroplatinum(II) and DNA. Proc. Natl Acad. Sci. USA, 88, 1982–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brabec V. and Leng,M. (1993) DNA interstrand cross-links of trans-diamminedichloroplatinum(II) are preferentially formed between guanine and complementary cytosine residues. Proc. Natl Acad. Sci. USA, 90, 5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maxam A.M. and Gilbert,W. (1980) Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol., 65, 499–560. [DOI] [PubMed] [Google Scholar]
- 22.Koo H.S., Wu,H.M. and Crothers,D.M. (1986) DNA bending at adenine.thymine tracts. Nature, 320, 501–506. [DOI] [PubMed] [Google Scholar]
- 23.Bellon S.F. and Lippard,S.J. (1990) Bending studies of DNA site-specifically modified by cisplatin, trans-diamminedichloroplatinum(II) and cis-Pt(NH3)2(N3-cytosine)Cl+. Biophys. Chem., 35, 179–188. [DOI] [PubMed] [Google Scholar]
- 24.Koo H.S. and Crothers,D.M. (1988) Calibration of DNA curvature and a unified description of sequence-directed bending. Proc. Natl Acad. Sci. USA, 85, 1763–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice J.A., Crothers,D.M., Pinto,A.L. and Lippard,S.J. (1988) The major adduct of the antitumor drug cis-diamminedichloroplatinum(II) with DNA bends the duplex by 40° toward the major groove. Proc. Natl Acad. Sci. USA, 85, 4158–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leng M. (1990) DNA bending induced by covalently bound drugs. Gel electrophoresis and chemical probe studies. Biophys. Chem., 35, 155–163. [DOI] [PubMed] [Google Scholar]
- 27.Brabec V., Sip,M. and Leng,M. (1993) DNA conformational distortion produced by site-specific interstrand cross-link of trans-diamminedichloroplatinum(II). Biochemistry, 32, 11676–11681. [DOI] [PubMed] [Google Scholar]
- 28.Huang H.F., Zhu,L.M., Reid,B.R., Drobny,G.P. and Hopkins,P.B. (1995) Solution structure of a cisplatin-induced DNA interstrand cross-link. Science, 270, 1842–1845. [DOI] [PubMed] [Google Scholar]
- 29.Malinge J.M., Perez,C. and Leng,M. (1994) Base sequence-independent distorsions induced by interstrand cross-links in cis-diaminedichloroplatinum (II)-modified DNA. Nucleic Acids Res., 22, 3834–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasparkova J., Mellish,K.J., Qu,Y., Brabec,V. and Farrell,N. (1996) Site-specific d(GpG) intrastrand cross-links formed by dinuclear platinum complexes. Bending and NMR studies. Biochemistry, 35, 16705–16713. [DOI] [PubMed] [Google Scholar]
- 31.Bellon S.F., Coleman,J.H. and Lippard,S.J. (1991) DNA unwinding produced by site-specific intrastrand cross-links of the antitumor drug cis-diamminedichloroplatinum(II). Biochemistry, 30, 8026–8035. [DOI] [PubMed] [Google Scholar]
- 32.Wang J.C. (1979) Helical repeat of DNA in solution. Proc. Natl Acad. Sci. USA, 76, 200–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhodes D. and Klug,A. (1980) Helical periodicity of DNA determined by enzyme digestion. Nature, 286, 573–578. [DOI] [PubMed] [Google Scholar]
- 34.Zinkel S. and Crothers,D.M. (1987) DNA bend direction by phase sensitive detection. Nature, 328, 178–181. [DOI] [PubMed] [Google Scholar]
- 35.Koo H.S., Drak,J., Rice,J.A. and Crothers,D.M. (1990) Determination of the extent of DNA bending by an adenine thymine tract. Biochemistry, 29, 4227–4234. [DOI] [PubMed] [Google Scholar]
- 36.Ulanovsky L., Bodner,M., Trifonov,E.N. and Choder,M. (1986) Curved DNA: design, synthesis and circularization. Proc. Natl Acad. Sci. USA, 83, 862–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jordan P. and Carmo-Fonseca,M. (2000) Molecular mechanisms involved in cisplatin cytotoxicity. Cell. Mol. Life Sci., 57, 1229–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen S.M. and Lippard,S.J. (2001) Cisplatin: from DNA damage to cancer chemotherapy. Prog. Nucleic Acid Res. Mol. Biol., 67, 93–130. [DOI] [PubMed] [Google Scholar]
- 39.Brabec V. (2002) DNA modifications by antitumor platinum and ruthenium compounds: their recognition and repair. Prog. Nucleic Acid Res. Mol. Biol., 71, 1–68. [DOI] [PubMed] [Google Scholar]
- 40.Orphanides G., Lagrange,T. and Reinberg,D. (1996) The general transcription factors of RNA polymerase II. Genes Dev., 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- 41.Vichi P., Coin,F., Renaud,J.P., Vermeulen,W., Hoeijmakers,J.H.J., Moras,D. and Egly,J.M. (1997) Cisplatin- and UV-damaged DNA lure the basal transcription factor TFIID/TBP. EMBO J., 16, 7444–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coin F., Frit,P., Viollet,B., Salles,B. and Egly,J.M. (1998) TATA binding protein discriminates between different lesions on DNA, resulting in a transcription decrease. Mol. Cell. Biol., 18, 3907–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poklar N., Pilch,D.S., Lippard,S.J., Redding,E.A., Dunham,S.U. and Breslauer,K.J. (1996) Influence of cisplatin intrastrand crosslinking on the conformation, thermal stability and energetics of a 20-mer DNA duplex. Proc. Natl Acad. Sci. USA, 93, 7606–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pilch D.S., Dunham,S.U., Jamieson,E.R., Lippard,S.J. and Breslauer,K.J. (2000) DNA sequence context modulates the impact of a cisplatin 1,2-d(GpG) intrastrand cross-link on the conformational and thermodynamic properties of duplex DNA. J. Mol. Biol., 296, 803–812. [DOI] [PubMed] [Google Scholar]
- 45.Hofr C., Farrell,N. and Brabec,V. (2001) Thermodynamic properties of duplex DNA containing a site-specific d(GpG) intrastrand crosslink formed by an antitumor dinuclear platinum complex. Nucleic Acids Res., 29, 2034–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]