Abstract
In this paper, we demonstrate the use of synthetic polyamide probes to fluorescently label heterochromatic regions on human chromosomes for discrimination in cytogenetic preparations and by flow cytometry. Polyamides bind to the minor groove of DNA in a sequence-specific manner. Unlike conventional sequence-specific DNA or RNA probes, polyamides can recognize their target sequence without the need to subject chromosomes to harsh denaturing conditions. For this study, we designed and synthesized a polyamide to target the TTCCA-motif repeated in the heterochromatic regions of chromosome 9, Y and 1. We demonstrate that the fluorescently labeled polyamide binds to its target sequence in both conventional cytogenetic preparations of metaphase chromosomes and suspended chromosomes without denaturation. Chromosomes 9 and Y can be discriminated and purified by flow sorting on the basis of polyamide binding and Hoechst 33258 staining. We generate chromosome 9- and Y-specific ‘paints’ from the sorted fractions. We demonstrate the utility of this technology by characterizing the sequence of an olfactory receptor gene that is duplicated on multiple chromosomes. By separating chromosome 9 from chromosomes 10–12 on the basis of polyamide fluorescence, we determine and differentiate the haplotypes of the highly similar copies of this gene on chromosomes 9 and 11.
INTRODUCTION
The technology to analyze and sort human chromosomes by flow cytometry has played an important role in genomics, human genetics and comparative cytogenetics since its advent in the 1980s (1,2). The loss or gain of DNA caused by gross chromosomal rearrangements associated with genetic disorders can be assessed by flow cytometry and sorting (3–7). DNA sequence libraries constructed from flow-sorted chromosomes laid much of the groundwork for the assembly of the human genome sequence (8–11), and flow sorting continues to play a critical role in analyses of sequences that are duplicated on multiple chromosomes (12,13). Flow sorting is also the key technology used to generate chromosome-specific collections of DNA sequences, which can be used as a probe in fluorescence in situ hybridization (FISH) to fluorescently ‘paint’ specific chromosomes (14,15). These paints facilitate the detection of chromosome abnormalities (16,17) and reconstruction of chromosome rearrangements that have occurred during the evolution of primates (18,19).
Despite extensive searches, the choice of stains for chromosome discrimination by flow cytometry is very limited. Currently, the most discriminatory and robust staining regime employs two DNA-specific dyes, Hoechst 33258 (HO) and chromomycin A3 (CA), whose intensities are measured after separate excitation in a two-laser flow cytometer (20). Because HO and CA bind preferentially to A–T and G–C base pairs, respectively, chromosomes are distinguished by differences in overall DNA content and base pair ratio. Most human chromosomes are remarkably well resolved by this stain combination. However, the 9–12 group and various chromosomes of other species are poorly resolved due to similarity in their size and base pair composition (6,21–23).
A strategy for using DNA probes to label specific chromosomes for sorting would be highly desirable. Many short, highly repetitive sequences have been identified that when hybridized to conventional cytogenetic preparations using FISH produce bright, chromosome-specific signals (24,25). Flow cytometry and FISH have been combined to quantify specific sequences in interphase nuclei (26), most notably to resolve nuclei carrying zero, one or two copies of the Y chromosome (27,28) or with other chromosome abnormalities (29). However, attempts to conduct FISH on chromosomes isolated in suspension have not been particularly successful, despite the efforts of several laboratories (30–34). Chromosomes in suspension fall apart and clump together when subjected to the harsh denaturing conditions required for FISH.
In this study, we explore the use of polyamides to discriminate chromosomes by flow cytometry. Polyamides are synthetic ligands that bind in the minor groove of DNA in a sequence-specific manner (35–39). Unlike DNA-, RNA- or PNA-probes, polyamides can recognize and bind to specific sequences without requiring denaturation of the target (39–42). In theory, a repertoire of polyamides could be developed to recognize chromosome-specific repetitive sequences in any species.
To demonstrate this approach, we have designed and tested a polyamide targeted to a sequence motif that is highly repeated in the heterochromatic regions of chromosomes 9, Y, 1 and a few other chromosomes (43). We show here that this fluorescently labeled polyamide binds to its target sequence in both conventional cytogenetic preparations of metaphase chromosomes and suspended chromosomes prepared for flow cytometry. The labeling procedure is extremely simple, because no denaturation step is required. We demonstrate the utility of this approach to analyze the sequences of an olfactory receptor gene that has recently been duplicated onto multiple chromosomes.
MATERIALS AND METHODS
Fluorescence in situ hybridization
Methanol- and acetic acid-fixed mitotic chromosome spreads were prepared from the cell line CGM1 by standard cytogenetic techniques and dropped onto glass slides as described elsewhere (44). The metaphase preparations were denatured in 70% formamide/2× SSC at 70°C for 2 min and immediately dehydrated in a series of 70, 85, 90 and 100% ice-cold ethanol for 3 min each and then air-dried. For hybridizations of a fluorescein-labeled oligonucleotide directed against the TTCCA heterochromatic repeat (5′-CATTCCATTCCATTCCATTC-3′; synthesized by Gibco-BRL), we used a method of FISH modified for the use of a short, synthetic oligomer (45). Approximately 2 ng/µl oligomer was hybridized along with unlabeled herring sperm competitor DNA (∼600 ng/µl) overnight at 37°C in 10% formamide, 2× SSC and 10% dextran sulfate in 10-µl total volumes. The slides were subsequently washed at 42°C three times in 10% formamide/2× SSC at 37°C for 10 min each, followed by three times in 2× SSC for 10 min each. For hybridizations using biotinylated degenerate oligo-primed (DOP)-PCR-amplified flow-sorted chromosomes (see below), the probe was prepared as described (44) and hybridized overnight at 37°C in 50% formamide/2× SSC/50% dextran sulfate. Washes were performed as described above in 50% formamide/2× SSC followed by 2× SSC. Detection with avidin–fluorescein was performed by standard techniques, as described elsewhere (44). Chromosomes were mounted in an antifade solution [1 mg/ml p-phenylenediamine dihydrochloride, 0.1× D-PBS (pH 7.6) and 90% glycerol] containing 250 ng/µl DAPI. Images were acquired as described previously (46).
Polyamide synthesis
Abbreviations. The following abbreviations are used to describe the hairpin polyamides (47): N-methylpyrrole (Py), N-methylimidazole (Im), β-alanine (β), γ-aminobutyric acid (γ) and 3,3′-diamino-N-methyldipropylamine (DA). Additional abbreviations used are: AEEA, 2[(2-aminoethoxy)ethoxy] acetic acid; and C6-F, 6-(fluorescein-5-carboxamido)-hexanoic acid.
Synthesis. The automated syntheses of Im2Py2-γ-Py4-β-DA-AEEA-C6-F (S-PA-F, for short polyamide) and Py4-γ-Im2Py2-β-Im2Py2-β-DA-AEEA-C6-F (L-PA-F, for long polyamide) were performed on an Advanced ChemTech 348 MPS system. Programs were written according to the instrument manufacturer’s instructions for protecting group (Boc) removal, addition of each coupling unit (monomer, dimer or trimer) to the growing chain and capping.
Materials. Boc-β-alanine-(4-carbonylaminomethyl)-benzyl-ester-co-poly(styrene-divinyl-benzene) resin (Boc-β-Ala-PAM) (0.22 mmol/g) was purchased from Peptides International. 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) and Boc-β-alanine were purchased from Advanced ChemTech. N,N-diisopropylethylamine (DIEA) and N,N′-dicyclohexylcarbodiimide (DCC) were purchased from Acros. 1-Methyl-2-pyrrolidinone (NMP), acetic anhydride, dichloromethane (DCM), trifluoroacetic acid (TFA), acetic acid and methanol were purchased from Fisher. Tetrahydrofuran (THF), DA, N,N-dimethylformamide (DMF), triethylamine, N-hydroxysuccinimide (NHS) and 1-methyl-2-pyrrole were purchased from Aldrich. 2[(2-Boc-aminoethoxy)ethoxy] acetic acid (Boc-AEEA-OH) was purchased from Applied Biosystems. 6-(Fluorescein-5-carboxamido)hexanoic acid succinimidyl ester (C6-F-NHS) was purchased from Molecular Probes. Boc-γ-aminobutyric acid was purchased from Nova Biochem. HPLC buffer [0.05 M triethylammonium acetate (TEAA)] was prepared from acetic acid and triethylamine and adjusted to pH 4.5 or 8.3. The syntheses of Boc-PyOH, Boc-ImOH, ImOH, Im2OH, Boc-γ-Im2OH and Boc-β-Im2OH were carried out according to literature procedures (47).
Preparation of Im2Py2-γ-Py4-β-DA-AEEA-C6-F (S-PA-F). Boc-β-Ala-PAM resin (1.0 g, 0.22 mmol/g) was treated with 80% (v/v) TFA in DCM for 30 min to remove the Boc protecting group. The solution was drained and the resin washed sequentially with DCM and NMP. Boc-PyOH monomer (1.0 ml, 0.5 M in NMP) was dissolved in NMP (2.0 ml) and activated with HBTU (1.0 ml, 0.5 M in NMP) and DIEA (1.0 ml, 1 M in NMP) for 10 min. The activated monomer was added to the resin, and the mixture was agitated for 2 h. The solution was drained and the activation/coupling process repeated twice more (‘triple coupling’). Following the third coupling, the resin was washed sequentially with NMP and DCM. Unreacted amines were capped with acetic anhydride (362 µl) and DIEA (625 µl) in DCM (5.5 ml) for 30 min, followed by washing with DCM. Addition of subsequent coupling units was accomplished by repeating the deprotection/activation and coupling/capping cycle. Py, β and γ units were added as monomers; however, due to reduced coupling efficiency of Im to Im, the final coupling was performed using Im2OH dimer.
Cleavage of the polyamide from the resin was achieved by treating the resin with a solution of DA (1.5 ml) dissolved in THF (10 ml) in a 25-ml round-bottomed flask fitted with a condenser and heated under nitrogen to 60°C for 16 h. The mixture was filtered, and the resin washed with methanol. The combined filtrates were evaporated in vacuo, and the resulting oil was dissolved in water (2 ml) and methanol (2 ml). The solution was cooled in an ice bath and carefully acidified to pH 4.5 with acetic acid. The crude product was purified in a single aliquot by reversed phase HPLC [Waters PrepLC2000; µBondapak C18 PrepPak cartridge (25 × 100 mm); flow rate 10 ml/min] using gradient elution (0.05 M TEAA pH 4.5 and methanol; gradient 30–70% methanol over 60 min). Fractions (20 ml) were collected and evaporated in vacuo to yield 82.2 mg (29%) of Im2Py2-γ-Py4-β-DA. Analytical HPLC of the product showed a single UV-absorbing peak. The product was also analyzed by mass spectrometry using an HP1100MSD operating in positive ion mode and using electrospray injection: calculated (M+H+) 1266, observed 1266.
The purified Im2Py2-γ-Py4-β-DA was treated with Boc-AEEA-NHS (prepared by treatment of Boc-AEEA-OH with NHS and DCC) in DMF for 2 h, followed by evaporation of the solvent in vacuo and treatment of the crude product with 80% (v/v) TFA in DCM for 30 min to remove the Boc protecting group. This provided a hydrophilic nine-atom spacer for subsequent coupling with fluorescent dye. The solvent was evaporated in vacuo, and the crude product was treated with C6-F-NHS (20 mg) in DMF (1 ml) containing DIEA (73 µl) for 1 h. The solvent was again evaporated in vacuo. The crude Im2Py2-γ-Py4-β-DA-AEAA-C6-F was dissolved in water (2 ml) and methanol (2 ml) and purified in a single aliquot by reversed phase HPLC as above, but using gradient elution (0.05 M TEAA pH 8.3 and methanol; gradient 30–70% methanol over 60 min). Fractions (20 ml) were collected and evaporated in vacuo to yield the purified product. Analytical HPLC showed a single UV-absorbing peak. The product was also analyzed by mass spectrometry as above: calculated (M+H+) 1883, observed 1883.
Py4-γ-Im2Py2-β-Im2Py2-β-DA-AEEA-C6-F (L-PA-F). The approach employed above was used in the synthesis of Py4-γ-Im2Py2-β-Im2Py2-β-DA-AEEA-C6-F. The trimers Boc-γ-Im2OH and Boc-β-Im2OH were used at appropriate points in the sequence assembly. The final purified product was analyzed by mass spectrometry as above: calculated (M+H+) 2444, observed 2444.
Polyamide labeling of conventional cytogenetic preparations
Stock concentrations of S-PA-F and L-PA-F were made by reconstituting lyophilized polyamide in equal volumes of DMF and 0.1 M NaHCO3 (pH 9). Polyamide was freshly diluted in Dulbecco’s phosphate-buffered saline (PBS) to 1 µM each day prior to use and was applied directly to slides carrying methanol/acetic acid-fixed chromosomes. After covering with a 22 × 22 mm2 coverslip sealed around the edges with rubber cement, slides were placed in a humidified environment and put in a 37°C incubator overnight. Following incubation, coverslips were removed, and slides were briefly rinsed in a phosphate–NP-40 buffer [100 mM phosphate buffer (pH 8.0) and 0.05% (v/v) NP-40] at room temperature. Slides were then mounted in antifade solution lacking DAPI counterstain.
Preparation and flow sorting of chromosomes
Chromosome suspensions were prepared from the lymphoblast cell line CGM1 using a modification (7) of the polyamine-based method originally described by Sillar and Young (48) and Lalande et al. (49). Briefly, cells cultured in RPMI medium containing 20% fetal bovine serum were arrested at mitosis by incubation in 0.1 µg/ml colcemid for ∼16 h. After centrifugation, the cells were resuspended in 40 mM KCl for 10 min and then centrifuged again. The pellet was resuspended in cold buffer containing 80 mM KCl, 20 mM NaCl, 15 mM Tris–HCl pH 7.2, 2 mM EDTA, 0.5 mM EGTA, 7 mM β-mercaptoethanol, 0.2 mM spermine, 0.5 mM spermidine and 0.12% digitonin, and incubated on ice for 10 min. The suspension was vortexed vigorously for 2 min and then stored for up to 90 days at 4°C before use.
Prior to flow analysis, the chromosomes were stained for ≥2 h with a combination of either 40 µg/ml CA and 2 µg/ml HO or 1 µM S-PA-F and 2 µg/ml HO. Sodium citrate and sodium sulfite were added to chromosomes stained with CA and HO 15–30 min before flow analysis at final concentrations of 10 and 25 mM, respectively, to improve chromosome resolution (50).
Flow cytometry
Chromosomes were analyzed on an Influx flow sorter (Cytopeia, Inc., Seattle, WA) (51,52). One laser was tuned to emit ultraviolet light (351–364 nm, 250 mW) to excite HO, and HO fluorescence was measured after passing through a 425-nm long-pass filter and a 458-nm rejection-band filter. A second laser was tuned to 458 nm (250 mW) to excite CA, and CA fluorescence was measured after passing through a 500-nm long-pass filter and a 458-nm rejection-band filter. Alternatively, fluorescein fluorescence was measured following excitation at 488 nm (250 mW) after passing through a 530/40 band-pass filter. The fluorescence pulses from the individual chromosomes were integrated by a data acquisition system, and the measurements of fluorescence intensity of ∼200 000 chromosomes were collected in listmode at a rate of ∼1000 chromosomes/s (53).
To prepare chromosome paints or for sequence analysis, 2000–2500 chromosomes of each type were flow-sorted in 0.2-ml PCR tubes containing 10 µl of sterile water. In the case of chromosomes 9–12, which sort as a group with HO and CA, 6000 chromosomes were collected.
Preparation of chromosome paints (DOP-PCR amplification)
Flow-sorted chromosomes were frozen, thawed and then subjected to DOP-PCR to randomly amplify DNA sequences in the sorted chromosomes according to the method of Telenius et al. (54) and Rabbitts et al. (22). Reagents were added to the 10 µl of sorted chromosomes to achieve final concentrations of 2 µM 6MW primer (5′-CCGACTCGAGNNNNNNATGTGG-3′) (where N = any base), 5 µl per sample of 1× DOP-PCR buffer (0.05 M KCl, 10 mM Tris–HCl, 0.1% Triton X-100), 160 µM MgCl2, 200 µM dNTPs in a 50-µl total volume. The mixture was then heated for 5 min at 95°C to denature the chromosomes, after which 0.3 U/µl (f.c.) Taq DNA polymerase was added to each tube. Polymerase addition was followed by five cycles of 94°C (1 min), 30°C (1.5 min), a 3-min ramp to 72°C, 3 min at 72°C, followed by 30 cycles of 94, 62 and 72°C for 1, 1 and 3 min, respectively, concluding with a final extension cycle of 72°C for 10 min.
The chromosomes were labeled with biotin in a second DOP-PCR using 5 µl of the above primary DOP-PCR product in a 50-µl reaction containing 200 µM dGTP, dCTP, dTTP, 100 µM dATP, 100 µM biotin-14-dATP (Invitrogen Life Tech. catalog no. 19524016) and 1× DOP-PCR buffer, 2 mM MgCl2, 2 µM primer and 0.5 U/µl Taq DNA polymerase. An initial 3-min denaturation period at 94°C was performed, followed by 30 cycles of 94°C for 30 s, 62°C for 45 s and 72°C for 2 min. A final extension cycle of 72°C was performed for 5 min.
Sequence analysis
For PCR amplification and sequence analysis, 2500 chromosomes were flow-sorted into a sterile PCR tube containing 10 µl of sterile water. For Figure 5, a 25-µl PCR using primers F16147 (CAAGAAGTCAGAATCAGAAGG) and R17372 (TATTTTCACTCCCTCATCTCA) was carried out in the same tube into which chromosomes were sorted. An aliquot of 0.5 µl of product from the first reaction was used as template in a second reaction with primers F16192 (GATCTTTCTCAATAGTGGTCT) and R17307 (AATGTAGTACCTCAAATCCTT). For other analyses of the OR-A gene (not shown), primers F4708 (ATTGAGGCAATGTATGTGGAAG) and OLF-AR (ACACTGAGAAGCCGAGATAACTGAA) were used in the first reaction, and OLA10 (CCAACTTCACTATATTTTGTG) and OLA4 (TCTGACTTCCTTCTCCTTCTC) were used for the second reaction. PCRs were performed using 0.7 U of Expand High-Fidelity Polymerase, 1× Expand HF buffer No. 2 with MgCl2 (Boehringer Mannheim), 0.4 µM each primer and 200 µM each dNTP. The first reaction consisted of a 2-min denaturation step at 94°C followed by 40 cycles of 94°C for 30 s, 52°C (55°C for F4708 and OLF-AR) for 30 s and 72°C for 90 s; a final 7-min extension step was carried out at 72°C. The second PCR consisted of 35 cycles with an annealing temperature of 60°C. PCR products were purified using a Sephacryl-300 column and sequenced using ABI Big-Dye Terminator chemistry. Sequencing reactions were performed using a 2-min denaturation at 96°C followed by 25 cycles of 96°C for 10 s, 50°C for 5 s and 72°C for 4 min. Sequencing primers were F16192, R17307, F16666 (ACATCTCCTTTTCAGAGTGGA) and R16913 (TTTCAATTTCTTCTCTTCTGT) or OLA10 and OLA4. PCR and sequencing reactions were performed in a PTC-100 thermocycler.
RESULTS
Design of polyamides to recognize a pentamer repeated in heterochromatic regions of human chromosomes
We designed polyamides to recognize the pentameric sequence 5′-TTCCA-3′, which is tandemly repeated within the heterochromatic regions of several chromosomes (43,55–57). To characterize the specificity of a probe for this repeat, we first hybridized a 20-nt fluorescein-labeled (TTCCA)4 oligomer to denatured human chromosomes using a slight modification of conventional FISH methodology. The (TTCCA)4 oligomer produced an intense fluorescent signal on the heterochromatic region of chromosome 9, as well as chromosomes Y, 1, 16 and the acrocentric chromosomes (Fig. 1A).
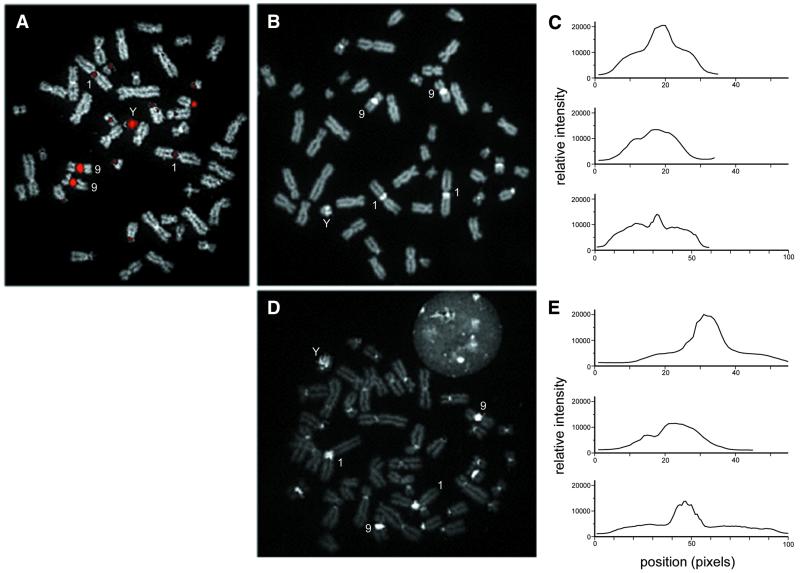
Figure 1.
Comparison of the binding of DNA oligomer and polyamides designed to recognize TTCCA repeats in metaphase chromosomes from a normal male. (A) Conventional FISH of the 20-nt (TTCCA)4 oligomer to denatured chromosome preparations. Hybridization sites are labeled with avidin–fluorescein (pseudocolored red), and chromosomes are counterstained with DAPI (gray). (B and D) Binding of the polyamides S-PA-F and L-PA-F to non-denatured chromosomes, respectively. The polyamides are responsible for all visible signals (intensity displayed as gray scale). (C and E) Relative intensity profile plots for chromosomes 9, Y and 1 from (B) and (D), respectively. The plotted intensities represent the sum of signal intensities in a seven-pixel-wide swath running from the p-terminus to q-terminus for each chromosome.
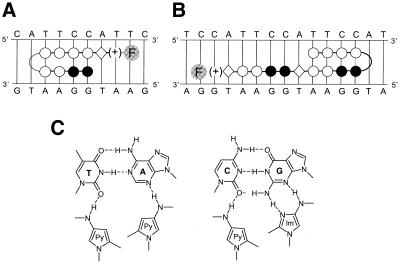
Figure 2A and B illustrates the two polyamides that we synthesized to target this TTCCA-repeat. Our designs were based on the work of Dervan and colleagues (36,38,58–62). The building blocks for polyamides are the monomeric aromatic amino acids Im and Py. Two oligomers composed of Im and Py are connected together with γ, which creates a turn in the polyamide. The molecule folds into a ‘hairpin’ structure, such that two polyamide oligomers bind in a side-by-side, anti-parallel manner in the minor groove. A pairing of Im opposite Py (Im/Py) targets a G–C base pair, while a Py/Py pairing targets both T–A and A–T base pairs. In addition to the Im and Py rings, a C-terminal β and DA are incorporated into the molecule. These residues enhance DNA-binding affinity and specificity (59,63). In addition, they display a strong preference for flanking A–T or T–A base pairs over G–C or C–G base pairs (58,62). Finally, a fluorescein derivative (C6-F) was attached via an AEEA linker on the end of the polyamide as a fluorescent tag. Hence, the short polyamide Im2Py2-γ-Py4-β-DA-AEEA-C6-F (S-PA-F) should target the sequence 5′HWWWCCW-3′, and the long polyamide Py4-γ-Im2Py2-β-Im2Py2-β-DA-AEEA-C6-F (L-PA-F) should target the sequence 5′-WWWCCWWWCCW-3′, where W indicates either A or T.
Figure 2.
Molecular recognition of the 5′-TTCCA-3′ target sequence by the polyamides S-PA-F and L-PA-F. (A) Binding model for the complex formed between S-PA-F and 5′-ATTCCA-3′. (B) Binding model for the complex formed between L-PA-F and 5′-ATTCCATTCCA-3′. (A and B) Filled circles, Im; open circles, Py; diamond, β; (+), DA. Curvature of the polyamide chain to form a hairpin is achieved with a γ linker, and fluorescence is conferred by a fluorescein derivative (AEEA-C6-F) (represented by the F in a gray circle). (C) Hydrogen bonding patterns of nucleotide base pairs with polyamide subunits. Dashed lines represent hydrogen bonds.
TTCCA-targeted polyamides bind to heterochromatic regions in conventional cytogenetic preparations
Figure 1B shows the binding of S-PA-F to a cytogenetic preparation. The polyamide probe produces a fluorescent pattern very similar to that of the (TTCCA)4 oligomer. However, in the case of the polyamide, non-denatured chromosomes were simply incubated in a solution containing the polyamide probe overnight and then rinsed before mounting. The polyamide probe results in an intense signal on the targeted regions of chromosome 9, Y, 1, 16 and the acrocentric chromosomes. The fluorescence signals along the length of each chromosome in Figure 1B suggest that S-PA-F is able to access binding sites throughout the chromosomes in addition to the repeated arrays detected by (TTCCA)4. Relative differences among the intensities of chromosomes 9, Y and 1 are graphically illustrated in Figure 1C. Both the size and the intensity of the labeled region on chromosome 9 exceed that of chromosome 1. The intensity per unit length of the labeled regions on chromosome Y and 1 are similar, but a greater proportion of chromosome Y’s length is labeled by the polyamide. In cytogenetic preparations, the longer polyamide L-PA-F yields a better signal:noise ratio, as binding outside of the heterochromatic regions is significantly reduced (Fig. 1D and E).
Flow cytometric analysis of polyamide-labeled chromosomes
Figure 3B shows a conventional flow karyotype of chromosomes isolated from a normal male cell line (CGM1) that have been stained with the DNA-specific fluorochromes HO and CA and analyzed by flow cytometry. The resulting plot shows distinct populations representing the various human chromosomes. Chromosomes 9–12 are not resolved.
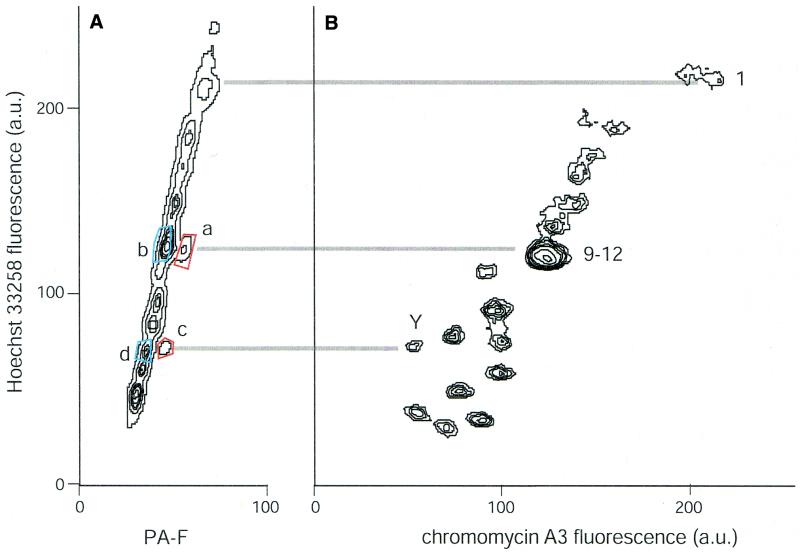
Figure 3.
Bivariate flow analyses of chromosomes treated with HO and the polyamide S-PA-F (A) or CA (B). Numbers represent the chromosome(s) responsible for the peaks in the flow karyotypes, with lines between the two plots drawn to indicate the positions of chromosomes 1, 9 and Y on the HO scales. Blue- and red-boxed areas in (A) denote sorting windows used to sort chromosomes analyzed further in Figures 4 and 5. Each plot represents measurements of ∼200 000 chromosomes.
Figure 3A shows the fluorescence distribution of the same chromosomes, stained instead with S-PA-F and HO. The HO-intensity distribution is similar in Figure 3A and B, indicating that the polyamide does not deleteriously affect the chromosomes or relative HO staining. Distinct populations of chromosomes displaying enhanced fluorescent signal on the fluorescein axis are evident. HO correlation between the two flow karyotypes suggests that the fluorescein-bright populations in Figure 3A represent chromosomes 1, 9 and Y. In this setting, S-PA-F gave better signal:noise ratio than L-PA-F (not shown).
Cytogenetic analysis of flow-sorted chromosomes using DOP-PCR
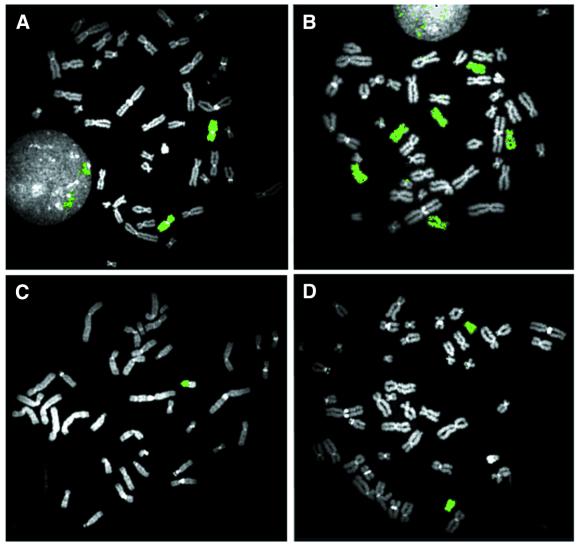
We verified the identity of chromosomes sorted on the basis of S-PA-F and HO staining by using DOP-PCR to prepare chromosome ‘paints’ from sorted material. Chromosomes were sorted from the boxed regions indicated in Figure 3A. Sorted chromosomes (2000 per reaction) were universally amplified and biotinylated using DOP-PCR and then used as conventional FISH probes. Figure 4 shows the sites to which the sorted material hybridized on a normal male metaphase spread. In Figure 4A, the chromosomes isolated using sort window a, with HO intensity corresponding to chromosomes 9–12 and bright S-PA-F staining, paint chromosome 9 specifically. Paints made of material purified using sort window b (HO intensity of 9–12, but S-PA-F-dim) stain chromosomes 10, 11 and 12, but not chromosome 9 (Fig. 4B). In Figure 4C, the polyamide-bright chromosomes in sort window c corresponding to chromosome Y indeed label chromosome Y specifically. The polyamide-dim chromosomes with a similar HO intensity (sort window d) correspond to chromosome 18 (Fig. 4D).
Figure 4.
Chromosome paints generated from chromosomes sorted on the basis of the intensity of bound S-PA-F polyamide and HO. The chromosomes used to produce the paints in (A)–(D) correspond to the sort windows a–d in Figure 3A. In each case, 2000 chromosomes were sorted, amplified and biotinylated by DOP-PCR, hybridized to metaphase chromosomes and detected with fluorescein–avidin. The fluorescein-bright chromosomes in sort window a yield a chromosome 9-specific paint (A); the fluorescein-dim chromosomes with similar HO fluorescence intensities (sort window b) paint chromosomes 10, 11 and 12 (B). The fluorescein-bright chromosomes in sort window c yield a chromosome Y-specific paint (the long arm is unlabeled due to the suppression of the hybridization of highly repetitive sequences in the paint probe) (C); the fluorescein-dim chromosomes with a similar HO fluorescence intensity (sort window d) paint chromosome 18 (D). All metaphases were counterstained with DAPI.
Use of polyamide staining and flow cytometry to resolve paralogous sequences
We used polyamide staining and flow cytometry to separate chromosomes carrying slightly different copies of a duplicated block of DNA known to contain sequences similar to olfactory receptor genes (12). This sequence is present on many chromosomes in subtelomeric locations (12). In individual CGM1, it is present on nine chromosomes, including a copy on one homolog of chromosome 9 and a copy on each homolog of chromosome 11 (12). In order to evaluate the sequence variation among these copies and determine whether any of the paralogous genes encode functional proteins, it is necessary to separate chromosomes by flow cytometry. The copies on chromosomes 9 and 11 could not be resolved for sequence analysis using the standard HO and CA staining strategy. For example, when the mixture of chromosomes 9–12 was used as a template for PCR amplification and sequencing, both a G and T are observed at a site within this block (at site 17015 within GenBank accession no. L78442). It is impossible to assign the variants to chromosome 9 or 11. Therefore, we separated these chromosomes from each other based on the intensity of bound S-PA-F. Figure 5, middle and bottom, shows the sequence generated from the sorted fractions representing chromosomes 9 and 10–12, respectively. The copy with the T at position 17015 derives from chromosome 9, and the copies with the G at this position derive from chromosome 11 in this individual. Other samples of chromosomes 9 and 10–12 were flow sorted, PCR amplified and sequenced to examine variants within the gene itself (not shown). We found three nucleotide differences within the OR-A genes on chromosomes 9 and 11 and could resolve the two haplotypes using the flow-sorted material (9 versus 11: T versus C at nt 4896; G versus A at nt 5113; C versus T at nt 5730).
Figure 5.
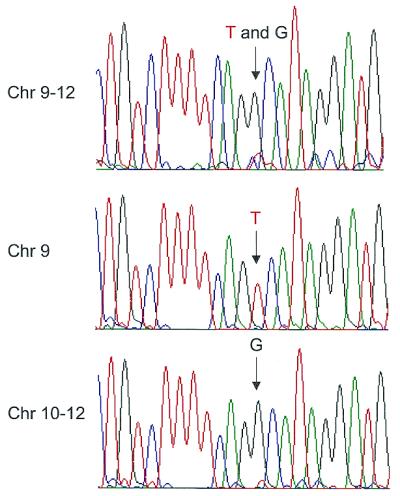
Sequence analysis of a duplicated block encompassing the OR-A olfactory receptor gene derived from chromosomes 9 and 11 of the CGM1 cell line. Previous FISH analyses had demonstrated the presence of a copy of this block on one homolog of chromosome 9 and both homologs of chromosome 11 in this individual in addition to six other sites (both homologs of 3, 15 and 19). Sorted chromosomes were subjected to PCR to amplify a region of OR-A corresponding to bases 16 147–17 307 in GenBank accession no. L78442. The PCR product was used as template for the sequencing reaction after removal of PCR primers and nucleotides. (Top) The sequence generated when chromosomes 9–12 were sorted together on the basis of HO and CA staining. The arrow indicates the site of a single-nucleotide variation among the copies of OR-A found on these chromosomes. Two peaks, corresponding to a thymine (T) (red) and guanine (G) (black), are observed at nt 17 015. (Middle) Sequence of the block on chromosome 9, obtained by PCR amplification of chromosomes sorted on the basis of S-PA-F polyamide and HO fluorescence intensities (sort window a; Fig. 3A). A single peak corresponding to thymine is observed at nt 17 015. (Bottom) Sequence of the block on chromosome 11, obtained by PCR amplification of chromosomes 10–12 sorted on the basis of polyamide and HO fluorescence (sort window b; Fig. 3A). The arrow denotes a single peak, corresponding to a guanine at this position. Other nucleotides represented are cytosine (C) (blue) and adenine (A) (green).
DISCUSSION
In this paper, we demonstrate the utility of sequence-specific, minor groove-binding polyamides in two novel molecular cytogenetics applications.
First, we demonstrate that sequence-specific polyamides can produce bright sequence-specific labels in conventional cytogenetic preparations of human chromosomes. The procedure is considerably simpler than conventional FISH, as it is not necessary to denature chromosomes. The ease with which polyamides can be used to mark human chromosomes could supplant conventional FISH probes for cytogenetic analyses of chromosome aneuploidy in the pre-natal clinical setting and in cancer research. It should be straightforward to develop polyamides that recognize chromosome-specific α- and/or β-satellite sequences (25,62,64). Given that each polyamide carries only a single fluorochrome, this approach will be most suitable for labeling sequences repeated thousands of times within a concentrated region of the chromosome of interest, such as is the case with satellite repeats.
Secondly, we demonstrate the first application of sequence-specific labeling of human chromosomes for discrimination by flow cytometry. This approach is particularly useful for discriminating chromosomes that cannot be resolved using the conventional HO and CA staining regime. We note that chromosome 9 can be discriminated from chromosomes 10–12 by the addition of netropsin to HO- and CA-stained chromosomes (43,65). Netropsin interferes with the binding of HO to the extensive heterochromatic region on chromosome 9, such that the overall HO fluorescence of chromosome 9 is reduced significantly relative to these other similarly sized chromosomes. Polyamide labeling offers an alternative way to resolve chromosome 9, and we expect that polyamides designed to label other human chromosomes and specific chromosomes of other species will improve the ability to discriminate and purify chromosomes by flow cytometry for a variety of molecular analyses.
In our studies, this capability has been especially useful for resolving copies of sequences that have recently duplicated onto multiple chromosomes (13). Some of the most interesting regions of the human genome are similarly complicated by large, recent duplications (46,66–69), and the ability to conduct large-scale sequence analyses of duplicated segments derived from different chromosomes from multiple individuals will greatly facilitate efforts to understand the evolutionary dynamics and functions of these regions.
Also, it should be possible to develop procedures to label specific chromosomes in whole-cell nuclei for flow cytometric quantification of chromosome copy number. Recent developments by Dervan and coworkers (41,70,71) and Laemmli and coworkers (42) suggest that it may even be possible to label live cells with fluorescently tagged polyamides. This capability would allow functional assays of aneuploid subpopulations to be conducted either during or after flow analysis and sorting.
A variety of modifications can be made to polyamides to improve their target specificity. Longer polyamides, while difficult to synthesize, could potentially achieve better sequence specificity than the short polyamides used here. We are intrigued by the different behavior of the two polyamides in the two applications demonstrated here. In cytogenetic preparations, L-PA-F gave better signal:noise than S-PA-F, whereas the shorter polyamide resulted in more optimal labeling of chromosomes prepared for flow analysis. We suspect that different accessibility of target sequences in the two preparations is responsible for the different behaviors of the two polyamides. These observations illustrate that experimenting with variations on polyamide structure is worthwhile. Laemmli and coworkers showed that subtle variations in the structure of polyamides targeted for telomeric repeat had significant effects on binding efficiency (42). Furthermore, the polyamides used in our study could not discriminate A–T from T–A base pairs. The hydroxypyrrole (Hp) polyamide subunit is capable of distinguishing A–T from T–A base pairs. Although polyamides containing the Hp moiety are still difficult to synthesize routinely (38,62), we anticipate significant improvements in sequence specificity and expanded application of polyamides with their incorporation.
FINANCIAL DISCLOSURE
Authors G.J.v.d.E. and B.J.T. have significant financial interests in Cytopeia, Inc., source of the Influx cell sorter used in this study.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Robert Kaiser, Karin Hughes, Ed Ramos, Jimmy Eng and Steve Gygi for discussions and advice. This work was supported in part by a grant from Prolinx, Inc.
REFERENCES
- 1.Carrano A.V., Gray,J.W., Langlois,R.G., Burkhart-Schultz,K.J. and Van Dilla,M.A. (1979) Measurement and purification of human chromosomes by flow cytometry and sorting. Proc. Natl Acad. Sci. USA, 76, 1382–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langlois R.G., Yu,L.C., Gray,J.W. and Carrano,A.V. (1982) Quantitative karyotyping of human chromosomes by dual beam flow cytometry. Proc. Natl Acad. Sci. USA, 79, 7876–7880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris P., Cooke,A., Boyd,E., Young,B.D. and Ferguson-Smith,M.A. (1987) The potential of family flow karyotyping for the detection of chromosome abnormalities. Hum. Genet., 76, 129–133. [DOI] [PubMed] [Google Scholar]
- 4.Gray J.W., Trask,B., van den Engh,G., Silva,A., Lozes,C., Grell,S., Schonberg,S., Yu,L.C. and Golbus,M.S. (1988) Application of flow karyotyping in prenatal detection of chromosome aberrations. Am. J. Hum. Genet., 42, 49–59. [PMC free article] [PubMed] [Google Scholar]
- 5.Trask B., van den Engh,G., Nussbaum,R., Schwartz,C. and Gray,J. (1990) Quantification of the DNA content of structurally abnormal X chromosomes and X chromosome aneuploidy using high resolution bivariate flow karyotyping. Cytometry, 11, 184–195. [DOI] [PubMed] [Google Scholar]
- 6.Carter N.P., Ferguson-Smith,M.E., Affara,N.A., Briggs,H. and Ferguson-Smith,M.A. (1990) Study of X chromosome abnormality in XX males using bivariate flow karyotype analysis and flow sorted dot blots. Cytometry, 11, 202–207. [DOI] [PubMed] [Google Scholar]
- 7.Mefford H., van den Engh,G., Friedman,C. and Trask,B.J. (1997) Analysis of the variation in chromosome size among diverse human populations by bivariate flow karyotyping. Hum. Genet., 100, 138–144. [DOI] [PubMed] [Google Scholar]
- 8.Davies K.E., Young,B.D., Elles,R.G., Hill,M.E. and Williamson,R. (1981) Cloning of a representative genomic library of the human X chromosome after sorting by flow cytometry. Nature, 293, 374–376. [DOI] [PubMed] [Google Scholar]
- 9.Van Dilla M.A., Deaven,L.L., Albright,K.L., Allen,N.A., Aubuchon,M.R., Bartholdi,M.F., Brown,N.C., Campbell,E.W., Carrano,A.V., Clark,L.M. et al. (1986) Human chromosome-specific DNA libraries: construction and availability. BioTechnology, 4, 537–552. [Google Scholar]
- 10.Montgomery K.T., LeBlanc,J.M., Tsai,P., McNinch,J.S., Ward,D.C., de Jong,P.J., Kucherlapati,R. and Krauter,K.S. (1993) Characterization of two chromosome 12 cosmid libraries and development of STSs from cosmids mapped by FISH. Genomics, 17, 682–693. [DOI] [PubMed] [Google Scholar]
- 11.Kim U.J., Shizuya,H., Sainz,J., Garnes,J., Pulst,S.M., de Jong,P. and Simon,M.I. (1995) Construction and utility of a human chromosome 22-specific Fosmid library. Genet. Anal., 12, 81–84. [DOI] [PubMed] [Google Scholar]
- 12.Trask B.J., Friedman,C., Martin-Gallardo,A., Rowen,L., Akinbami,C., Blankenship,J., Collins,C., Giorgi,D., Iadonato,S., Johnson,F. et al. (1998) Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum. Mol. Genet., 7, 13–26. [DOI] [PubMed] [Google Scholar]
- 13.Mefford H.C., Linardopoulou,E., Coil,D., van den Engh,G. and Trask,B.J. (2001) Comparative sequencing of a multicopy subtelomeric region containing olfactory receptor genes reveals multiple interactions between non-homologous chromosomes. Hum. Mol. Genet., 10, 2363–2372. [DOI] [PubMed] [Google Scholar]
- 14.Fuscoe J.C., Collins,C.C., Pinkel,D. and Gray,J.W. (1989) An efficient method for selecting unique-sequence clones from DNA libraries and its application to fluorescent staining of human chromosome 21 using in situ hybridization. Genomics, 5, 100–109. [DOI] [PubMed] [Google Scholar]
- 15.Telenius H., Pelmear,A.H., Tunnacliffe,A., Carter,N.P., Behmel,A., Ferguson-Smith,M.A., Nordenskjold,M., Pfragner,R. and Ponder,B.A. (1992) Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosom. Cancer, 4, 257–263. [DOI] [PubMed] [Google Scholar]
- 16.Pinkel D., Landegent,J., Collins,C., Fuscoe,J., Segraves,R., Lucas,J. and Gray,J. (1988) Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc. Natl Acad. Sci. USA, 85, 9138–9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schrock E., du Manoir,S., Veldman,T., Schoell,B., Wienberg,J., Ferguson-Smith,M.A., Ning,Y., Ledbetter,D.H., Bar-Am,I., Soenksen,D. et al. (1996) Multicolor spectral karyotyping of human chromosomes. Science, 273, 494–497. [DOI] [PubMed] [Google Scholar]
- 18.Wienberg J., Jauch,A., Stanyon,R. and Cremer,T. (1990) Molecular cytotaxonomy of primates by chromosomal in situ suppression hybridization. Genomics, 8, 347–350. [DOI] [PubMed] [Google Scholar]
- 19.Jauch A., Wienberg,J., Stanyon,R., Arnold,N., Tofanelli,S., Ishida,T. and Cremer,T. (1992) Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc. Natl Acad. Sci. USA, 89, 8611–8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray J.W., Langlois,R.G., Carrano,A.V., Burkhart-Schultz,K. and Van Dilla,M.A. (1979) High resolution chromosome analysis: one and two parameter flow cytometry. Chromosoma, 73, 9–27. [Google Scholar]
- 21.Trask B., van den Engh,G., Mayall,B. and Gray,J.W. (1989) Chromosome heteromorphism quantified by high-resolution bivariate flow karyotyping. Am. J. Hum. Genet., 45, 739–752. [PMC free article] [PubMed] [Google Scholar]
- 22.Rabbitts P., Impey,H., Heppell-Parton,A., Langford,C., Tease,C., Lowe,N., Bailey,D., Ferguson-Smith,M. and Carter,N. (1995) Chromosome specific paints from a high resolution flow karyotype of the mouse. Nature Genet., 9, 369–375. [DOI] [PubMed] [Google Scholar]
- 23.Langford C.F., Fischer,P.E., Binns,M.M., Holmes,N.G. and Carter,N.P. (1996) Chromosome-specific paints from a high-resolution flow karyotype of the dog. Chromosome Res., 4, 115–123. [DOI] [PubMed] [Google Scholar]
- 24.Willard H.F. (1985) Chromosome-specific organization of human alpha satellite DNA. Am. J. Hum. Genet., 37, 524–532. [PMC free article] [PubMed] [Google Scholar]
- 25.Pinkel D., Gray,J.W., Trask,B., van den Engh,G., Fuscoe,J. and van Dekken,H. (1986) Cytogenetic analysis by in situ hybridization with fluorescently labeled nucleic acid probes. Cold Spring Harb. Symp. Quant. Biol., 51, 151–157. [DOI] [PubMed] [Google Scholar]
- 26.Trask B., van den Engh,G., Landegent,J., in de Wal,N.J. and van der Ploeg,M. (1985) Detection of DNA sequences in nuclei in suspension by in situ hybridization and dual beam flow cytometry. Science, 230, 1401–1403. [DOI] [PubMed] [Google Scholar]
- 27.Trask B., van den Engh,G., Pinkel,D., Mullikin,J., Waldman,F., van Dekken,H. and Gray,J. (1988) Fluorescence in situ hybridization to interphase cell nuclei in suspension allows flow cytometric analysis of chromosome content and microscopic analysis of nuclear organization. Hum. Genet., 78, 251–259. [DOI] [PubMed] [Google Scholar]
- 28.Arkesteijn G.J., Erpelinck,S.L., Martens,A.C. and Hagenbeek,A. (1995) Chromosome specific DNA hybridization in suspension for flow cytometric detection of chimerism in bone marrow transplantation and leukemia. Cytometry, 19, 353–360. [DOI] [PubMed] [Google Scholar]
- 29.Kwak T., Nishizaki,T., Ito,H., Kimura,Y., Murakami,T. and Sasaki,K. (1994) Flow-cytometric quantification in human gliomas of alpha satellite DNA sequences specific for chromosome 7 using fluorescence in situ hybridization. Cytometry, 17, 26–32. [DOI] [PubMed] [Google Scholar]
- 30.Trask B. (1985) Studies of chromosomes and nuclei using flow cytometry. Thesis, University of Leiden, Leiden, The Netherlands.
- 31.Dudin G., Cremer,T., Schardin,M., Hausmann,M., Bier,F. and Cremer,C. (1987) A method for nucleic acid hybridization to isolated chromosomes in suspension. Hum. Genet., 76, 290–292. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen B.T., Lazzari,K., Abebe,J., Mac,I., Lin,J.B., Chang,A., Wydner,K.L., Lawrence,J.B., Cram,L.S., Weier,H.U. et al. (1995) In situ hybridization to chromosomes stabilized in gel microdrops. Cytometry, 21, 111–119. [DOI] [PubMed] [Google Scholar]
- 33.Hausmann M., Dudin,G., Aten,J.A., Heilig,R., Diaz,E. and Cremer,C. (1991) Slit scan flow cytometry of isolated chromosomes following fluorescence hybridization: an approach of online screening for specific chromosomes and chromosome translocations. Z Naturforsch [C], 46, 433–441. [DOI] [PubMed] [Google Scholar]
- 34.Macas J., Dolezel,J., Gualberti,G., Pich,U., Schubert,I. and Lucretti,S. (1995) Primer-induced labeling of pea and field bean chromosomes in situ and in suspension. BioTechniques, 19, 402–404, 407–408. [PubMed] [Google Scholar]
- 35.Trauger J.W., Baird,E.E. and Dervan,P.B. (1996) Extended hairpin polyamide motif for sequence-specific recognition in the minor groove of DNA. Chem. Biol., 3, 369–377. [DOI] [PubMed] [Google Scholar]
- 36.Trauger J.W., Baird,E.E. and Dervan,P.B. (1996) Recognition of DNA by designed ligands at subnanomolar concentrations. Nature, 382, 559–561. [DOI] [PubMed] [Google Scholar]
- 37.White S., Baird,E.E. and Dervan,P.B. (1996) Effects of the A.T/T.A degeneracy of pyrrole–imidazole polyamide recognition in the minor groove of DNA. Biochemistry, 35, 12532–12537. [DOI] [PubMed] [Google Scholar]
- 38.Kielkopf C.L., White,S., Szewczyk,J.W., Turner,J.M., Baird,E.E., Dervan,P.B. and Rees,D.C. (1998) A structural basis for recognition of A.T and T.A base pairs in the minor groove of B-DNA. Science, 282, 111–115. [DOI] [PubMed] [Google Scholar]
- 39.White S., Szewczyk,J.W., Turner,J.M., Baird,E.E. and Dervan,P.B. (1998) Recognition of the four Watson–Crick base pairs in the DNA minor groove by synthetic ligands. Nature, 391, 468–471. [DOI] [PubMed] [Google Scholar]
- 40.McBryant S.J., Baird,E.E., Trauger,J.W., Dervan,P.B. and Gottesfeld,J.M. (1999) Minor groove DNA–protein contacts upstream of a tRNA gene detected with a synthetic DNA binding ligand. J. Mol. Biol., 286, 973–981. [DOI] [PubMed] [Google Scholar]
- 41.Mapp A.K., Ansari,A.Z., Ptashne,M. and Dervan,P.B. (2000) Activation of gene expression by small molecule transcription factors. Proc. Natl Acad. Sci. USA, 97, 3930–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maeshima K., Janssen,S. and Laemmli,U.K. (2001) Specific targeting of insect and vertebrate telomeres with pyrrole and imidazole polyamides. EMBO J., 20, 3218–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moyzis R.K., Albright,K.L., Bartholdi,M.F., Cram,L.S., Deaven,L.L., Hildebrand,C.E., Joste,N.E., Longmire,J.L., Meyne,J. and Schwarzacher-Robinson,T. (1987) Human chromosome-specific repetitive DNA sequences: novel markers for genetic analysis. Chromosoma, 95, 375–386. [DOI] [PubMed] [Google Scholar]
- 44.Trask B. (1999) Fluorescence in situ hybridization. In Birren,B.G., Hieter,P., Slapholz,S., Myers,R.M., Riethman,H., Roskams,J. (eds), Genome Analysis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. 4, pp. 303–413.
- 45.O’Keefe C.L., Warburton,P.E. and Matera,A.G. (1996) Oligonucleotide probes for alpha satellite DNA variants can distinguish homologous chromosomes by FISH. Hum. Mol. Genet., 5, 1793–1799. [DOI] [PubMed] [Google Scholar]
- 46.Trask B.J., Massa,H., Brand-Arpon,V., Chan,K., Friedman,C., Nguyen,O.T., Eichler,E., van den Engh,G., Rouquier,S., Shizuya,H. et al. (1998) Large multi-chromosomal duplications encompass many members of the olfactory receptor gene family in the human genome. Hum. Mol. Genet., 7, 2007–2020. [DOI] [PubMed] [Google Scholar]
- 47.Baird E.E. and Dervan,P.B. (1996) Solid phase synthesis of polyamides containing imidazole and pyrrole amino acids. J. Am. Chem. Soc., 118, 6141–6146. [DOI] [PubMed] [Google Scholar]
- 48.Sillar R. and Young,B.D. (1981) A new method for the preparation of metaphase chromosomes for flow analysis. J. Histochem. Cytochem., 29, 74–78. [DOI] [PubMed] [Google Scholar]
- 49.Lalande M., Kunkel,L.M., Flint,A. and Latt,S.A. (1984) Development and use of metaphase chromosome flow-sorting methodology to obtain recombinant phage libraries enriched for parts of the human X chromosome. Cytometry, 5, 101–107. [DOI] [PubMed] [Google Scholar]
- 50.van den Engh G., Trask,B., Lansdorp,P. and Gray,J. (1988) Improved resolution of flow cytometric measurements of Hoechst- and chromomycin-A3-stained human chromosomes after addition of citrate and sulfite. Cytometry, 9, 266–270. [DOI] [PubMed] [Google Scholar]
- 51.van den Engh G. and Farmer,C. (1992) Photo-bleaching and photon saturation in flow cytometry. Cytometry, 13, 669–677. [DOI] [PubMed] [Google Scholar]
- 52.Asbury C.L., Esposito,R., Farmer,C. and van den Engh,G. (1996) Fluorescence spectra of DNA dyes measured in a flow cytometer. Cytometry, 24, 234–242. [DOI] [PubMed] [Google Scholar]
- 53.van den Engh G. and Stokdijk,W. (1989) Parallel processing data acquisition system for multilaser flow cytometry and cell sorting. Cytometry, 10, 282–293. [DOI] [PubMed] [Google Scholar]
- 54.Telenius H., Carter,N.P., Bebb,C.E., Nordenskjold,M., Ponder,B.A. and Tunnacliffe,A. (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics, 13, 718–725. [DOI] [PubMed] [Google Scholar]
- 55.Gosden J.R., Mitchell,A.R., Buckland,R.A., Clayton,R.P. and Evans,H.J. (1975) The location of four human satellite DNAs on human chromosomes. Exp. Cell Res., 92, 148–158. [DOI] [PubMed] [Google Scholar]
- 56.Frommer M., Prosser,J., Tkachuk,D., Reisner,A.H. and Vincent,P.C. (1982) Simple repeated sequences in human satellite DNA. Nucleic Acids Res., 10, 547–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwarzacher-Robinson T., Cram,L.S., Meyne,J. and Moyzis,R.K. (1988) Characterization of human heterochromatin by in situ hybridization with satellite DNA clones. Cytogenet. Cell Genet., 47, 192–196. [DOI] [PubMed] [Google Scholar]
- 58.Swalley S.E., Baird,E.E. and Dervan,P.B. (1997) A pyrrole-imidazole polyamide motif for recognition of eleven base pair sequences in the minor groove of DNA. Chem. Eur. J., 3, 1600–1607. [Google Scholar]
- 59.Swalley S.E., Baird,E.E. and Dervan,P.B. (1997) Discrimination of 5′-GGGG-3′ and 5′-GGCC-3′ sequences in the minor groove of DNA by eight-ring hairpin. J. Am. Chem. Soc., 119, 6953–6961. [Google Scholar]
- 60.Kielkopf C.L., Baird,E.E., Dervan,P.B. and Rees,D.C. (1998) Structural basis for G.C recognition in the DNA minor groove. Nature Struct. Biol., 5, 104–109. [DOI] [PubMed] [Google Scholar]
- 61.Hermann D.M., Turner,J.M., Baird,E.E. and Dervan,P.B. (1999) Cycle polyamide motif for recognition of the minor groove of DNA. J. Am. Chem. Soc., 121, 1121–1129. [Google Scholar]
- 62.Urbach A.R. and Dervan,P.B. (2001) Toward rules for 1:1 polyamide:DNA recognition. Proc. Natl Acad. Sci. USA, 98, 4343–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parks M.E. and Dervan,P.B. (1996) Simultaneous binding of a polyamide dimer and an oligonucleotide in the minor and major grooves of DNA. Bioorg. Med. Chem., 4, 1045–1050. [DOI] [PubMed] [Google Scholar]
- 64.Chen C., Wu,B., Wei,T., Egholm,M. and Strauss,W.M. (2000) Unique chromosome identification and sequence-specific structural analysis with short PNA oligomers. Mamm. Genome, 11, 384–391. [DOI] [PubMed] [Google Scholar]
- 65.Meyne J., Bartholdi,M.F., Travis,G. and Cram,L.S. (1984) Counterstaining human chromosomes for flow karyology. Cytometry, 5, 580–583. [DOI] [PubMed] [Google Scholar]
- 66.Eichler E.E. (2001) Segmental duplications: what’s missing, misassigned and misassembled—and should we care? Genome Res., 11, 653–656. [DOI] [PubMed] [Google Scholar]
- 67.Bailey J.A., Yavor,A.M., Massa,H.F., Trask,B.J. and Eichler,E.E. (2001) Segmental duplications: organization and impact within the current human genome project assembly. Genome Res., 11, 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eichler E.E. (2001) Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet., 17, 661–669. [DOI] [PubMed] [Google Scholar]
- 69.Bailey J.A., Yavor,A.M., Viggiano,L., Misceo,D., Horvath,J.E., Archidiacono,N., Schwartz,S., Rocchi,M. and Eichler,E.E. (2002) Human-specific duplication and mosaic transcripts: the recent paralogous structure of chromosome 22. Am. J. Hum. Genet., 70, 83–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dickinson L.A., Gulizia,R.J., Trauger,J.W., Baird,E.E., Mosier,D.E., Gottesfeld,J.M. and Dervan,P.B. (1998) Inhibition of RNA polymerase II transcription in human cells by synthetic DNA-binding ligands. Proc. Natl Acad. Sci. USA, 95, 12890–12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gottesfeld J.M., Neely,L., Trauger,J.W., Baird,E.E. and Dervan,P.B. (1997) Regulation of gene expression by small molecules. Nature, 387, 202–205. [DOI] [PubMed] [Google Scholar]