Abstract
Background
As cell-free nanotherapeutics, extracellular vesicles derived from mesenchymal stem cells (MSC-EVs) have shown potential therapeutic action against liver diseases. However, their effects on autoimmune hepatitis (AIH) are not yet well understood.
Methods and results
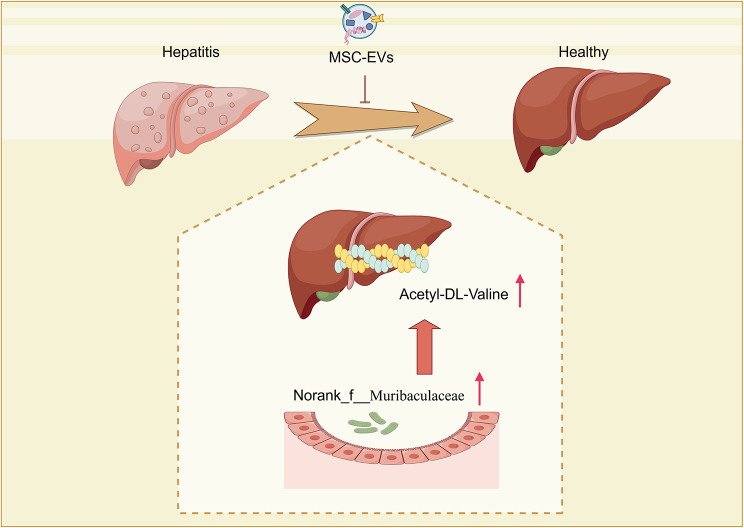
In this study, we utilized a well-established concanavalin A (Con A)-induced fulminant hepatitis mouse model to investigate the effects of MSC-EVs on AIH. We found that MSC-EVs provide significant protection against Con A-induced hepatitis in C57BL/6 male mice, with their effectiveness being critically dependent on the gut microbiota. MSC-EVs modulate the composition of the gut microbiota, particularly by increasing the abundance of norank_f__Muribaculaceae, and impact liver metabolic profiles, leading to significant amelioration of liver injury. The identification of Acetyl-DL-Valine as a protective metabolite underscores the therapeutic potential of targeting gut‒liver axis interactions in liver diseases.
Conclusion
Overall, our data demonstrate that MSC-EVs exhibit nanotherapeutic potential in Con A-induced hepatitis and provide new insights into the treatment of autoimmune hepatitis.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13287-024-04013-7.
Keywords: Extracellular vesicles, Mesenchymal stromal cells, Con A-induced hepatitis, Gut microbiota
Introduction
Autoimmune hepatitis (AIH), characterized by immune-mediated liver inflammation and cytokine production, is a progressive liver condition that can progress to cirrhosis, liver failure, or even tumors. An elevated aminotransferase level is associated with the condition, as well as hypergammaglobulinemia with high levels of IgG and the production of characteristic autoantibodies [1].The histopathology of AIH typically manifests as interface hepatitis, characterized by dense portal mononuclear cell infiltration, including lymphocytes, monocytes/macrophages, and plasma cells [2].
However, the underlying mechanisms of this disease remain largely unknown, and treatment options are limited. While corticosteroids and azathioprine are effective combinations for treating AIH, these therapies, like those for other autoimmune diseases, often come with significant drawbacks [3]. Prolonged steroid use can lead to significant side effects, including immunosuppression, osteoporosis, and sodium retention [4]. Furthermore, the discontinuation of steroid treatment often results in disease relapse in the majority of patients [5]. Therefore, exploring alternative therapeutic approaches offers a novel strategy that could reduce side effects and enhance treatment efficacy.
As mesenchymal stem cells (MSCs) develop specific secretion abilities, they are becoming an important cell therapy for the treatment of liver diseases [6, 7]. Previously, we showed that MSCs can effectively treat ischemia-type biliary lesions after liver transplantation (LT) [8]and after hepatitis B virus-related acute-to-chronic liver failure [9]. Furthermore, our experimental data revealed that MSCs protect the liver in a Concanavalin A (ConA)-induced hepatitis mouse model, a well-established model of human autoimmune hepatitis caused by T cells and inducible by interferon-gamma (IFN-gamma) [10, 11].
The extracellular vesicles (EVs) produced by MSCs include exosomes (with diameters ranging from 30 to 150 nm), microvesicles (with diameters between 150 and 500 nm), and apoptotic bodies (ranging from 800 to 500 nm in diameter) [12]. MSC-EVs can replicate the biological potential of MSCs, suggesting that they could replace cell therapy and enable cell-free therapy [13, 14]. Nevertheless, the therapeutic possibilities and fundamental mechanisms of MSC-EVs in Con A-induced hepatitis are still not well understood.
Recently, the concept of the gut‒liver axis, which describes the reciprocal interactions between the liver and gut, has attracted significant research interest. Changes in the gut microbiota, including reduced microbial diversity, loss of beneficial bacteria, and an increase in opportunistic pathogens, are commonly observed in patients with liver diseases [15]. Compared with that in healthy controls, intestinal dysbiosis has been detected in patients with AIH, with studies indicating reduced species richness and evenness in their fecal microbiome [16]. Changes in the gut microbiota are linked to the development of autoimmune hepatitis (AIH). A study by Qingqing Liu et al.. demonstrated that probiotics and prebiotics, alone or in combination, can alter the gut microbiota, restore immune balance, and preserve the integrity of the intestinal barrier, effectively mitigating liver injury induced by Con A [17].
Therefore, we hypothesized that treating Con A-induced hepatitis with MSC-EVs could normalize the microbiota composition and function, thereby reducing inflammation to normal levels. To test this hypothesis, we explored how MSC-EVs affect disease conditions and the composition of the gut microbiome and function in a ConA-induced hepatitis model via 16 S rRNA gene sequencing. This approach allowed us to evaluate the response of the gut microbiome to MSC-EVs treatment and its role in liver inflammation and disease development.
Materials and methods
Human UC-MSC isolation and identification
The method for isolating UC-MSCs received approval from the Research Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University and was executed under sterile conditions following previously established standardized protocols [18]. With the consent of the parents, we stored fresh umbilical cords from newborns at 4 °C after they had severed. The cords were washed twice with PBS to eliminate any residual blood. The cleaned cords were then cut into 10 mm³ pieces and incubated in a digestion solution containing type I collagenase and 0.1% hyaluronidase with 3 mM CaCl2 at 37 °C for 4 h. The digested tissues were transferred to DMEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (PAN-Biotech, Aidenbach, Germany) and incubated at 37 °C in a 5% CO2 humidified environment. After 3 days, the culture medium was replaced to remove nonadherent cells, and subsequently, the medium was replaced every 4 days. After 14 days, colonies of fibroblast-like cells appeared, which were subsequently trypsinized and moved to new flasks for continued expansion. For further analysis, the MSCs were treated at 37 °C with 0.05% trypsin-EDTA (GIBCO, Invitrogen, Inc., Carlsbad, CA, USA) and stored at 80 °C.
Phenotypic characterization of human umbilical cord-derived mesenchymal stem cells (hUC-MSCs) was conducted via flow cytometry. When the cells reached 70–80% confluence, they were harvested and rinsed twice with PBS supplemented with 1% bovine serum albumin (BSA; Gibco, USA) to avoid nonspecific antigen attachment. The cells were incubated for 30 min at 4 °C in the dark with monoclonal antibodies, including PE-conjugated anti-CD73, FITC-conjugated anti-CD90, PE-conjugated anti-CD105, FITC-conjugated anti-CD34, FITC-conjugated anti-CD45, and FITC-conjugated anti-HLA-DR. Flow cytometry was carried out via CytoFLEX flow cytometers (Beckman Coulter), and the data were analyzed with CytoExpert software (Beckman Coulter).
hUC-MSC-EV isolation, purification, and identification
After 70–80% confluence was reached, the medium was changed to 10% exosome-free FBS with 1.0 g/L glucose, and the cells were incubated for another 48 h. The cell culture supernatant was collected into a centrifuge tube and centrifuged at 300 × g for 10 min at 4 °C. The supernatant was then transferred and centrifuged at 3000 × g for 30 min at 4 °C. The supernatant was filtered through a 0.22 μm membrane filter. The resulting supernatant was transferred to ultracentrifuge tubes, balanced with PBS to the nearest milligram, and ultracentrifuged at 100,000 × g for 1 h and 10 min at 4 °C. The pellet was collected (leaving 1 ml at the bottom), resuspended in 3 ml of PBS, and transferred to a centrifuge tube. The volume was replenished with PBS, and the mixture was ultracentrifuged at 100,000 × g for 1 h at 4 °C for washing. The pellet was collected again (with the supernatant completely removed), resuspended in 500 µl of PBS from the exosome pellet obtained from 400 ml of MSC supernatant, and then transferred to an EP tube. The concentration of exosomal proteins was measured via a bicinchoninic acid (BCA) protein assay kit (KeyGEN BioTECH, Jiangsu, China).
hUC-MSC-EVs were visualized via transmission electron microscopy (TEM), with images captured at 120 kV via an FEI Tecnai T12 Spirit transmission electron microscope (FEI Company, Hillsboro, OR, USA). In summary, purified hUC-MSC-EVs were treated with 2.5% glutaraldehyde for 1 h, followed by ultracentrifugation and resuspension in 100 µl of PBS. A formvar carbon-coated grid was then coated with 10 µl of hUC-MSC-EVs, and TEM images were produced after negative staining with 3% aqueous phosphotungstic acid.
To monitor the size distribution of the hUC-MSC-EVs, nanoparticle tracking analysis (NTA) was performed via a Particle Metrix ZetaView® instrument (Particle Metrix GmbH, Inning, Germany). A microscope was used to observe hUC-MSC-EVs diluted 100-fold in PBS at a concentration of approximately 2 µg/µl. A video was recorded for documentation, and ZetaView was employed for data analysis.
Mice
C57BL/6 mice, obtained from GemPharmatech Co., Ltd. (Nanjing, China), were maintained in a specific pathogen-free environment. To minimize confounding effects, we implemented rigorous control measures.They were housed in a temperature-controlled setting and provided exclusively autoclaved water and sterilized food. A total of eight-week-old male animals weighing 22–26 g were used for the in vivo studies. A random allocation of C57BL/6 mice was performed for each group. The mice were randomly divided into three groups: the control group (n = 6), Con A + PBS group (n = 7), and Con A + MSC-EV group (n = 7). The random numbers for group allocation were generated via the Rand() function in Microsoft Excel. Additionally, the order of the experimental treatments and measurements was randomized to avoid any systematic bias. The mice were anesthetized with ketamine. Mice showing signs of liver failure that are incompatible with survival will be euthanized via cervical dislocation following deep anesthesia. The Sun Yat-sen University Institutional Animal Care and Use Committee reviewed and approved all the animal protocols. This work has been reported in line with the ARRIVE guidelines 2.0.
Pretreatment with a cocktail of broad-spectrum antibiotics (Abx)
The Con A + Abx and Con A + Abx + MSC-EV group mice (n = 4 per group) were given a cocktail of 200 mg/kg vancomycin, 200 mg/kg metronidazole, 200 mg/kg neomycin, or 200 mg/kg ampicillin by oral gavage daily for 3 days.
Con A-induced hepatitis and hUC-MSC-EV injection
A single dose of Con A (15 mg/kg) was administered through the tail vein. Thirty minutes after Con A injection, PBS or hUC-MSC-EVs (100 µg/100 µl, approximately 1 × 109 nanoparticles) suspended in 150 µl of PBS were intravenously (i.v.) injected through the tail vein. Further analysis of the blood and liver tissues was carried out 12 h later.
Acetyl-DL-Valine administration
The Acetyl-DL-Valine administration group received i.p. injections of 50 mg/kg Acetyl-DL-Valine dissolved in normal saline 30 min before tail vein injection of ConA to induce the hepatitis model.
Hematoxylin and eosin (HE)
An overnight fixation in 4% paraformaldehyde was followed by paraffin embedding of mouse liver tissues from the corresponding locations. Hematoxylin and eosin (H&E) staining was conducted on the Sect. (4 μm thick).
Measurement of serum alanine aminotransferase (ALT) and aspartate transaminase (AST)
Twelve hours after Con A injection, serum samples were collected. The levels of ALT and AST in the serum were measured via an automatic biochemical analyzer (Hitachi, Tokyo, Japan).
Collection and DNA extraction of fecal samples
The sterilized tubes containing mouse feces were stored at -80 °C until use. Genomic DNA from the microbial community was extracted via the E.Z.N.A.® Soil DNA Kit (Omega Bio-Tek, Norcross, GA, USA) following the manufacturer’s instructions. To quantify the DNA concentration and purity, a NanoDrop 2000 was used (Thermo Fisher Scientific, Wilmington, DE).
Sequencing and analysis of 16 S rRNA
The V3-V4 hypervariable region of the bacterial 16 S rRNA gene was amplified via the primer pairs 338 F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’) with an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, USA). The resulting amplicons were purified via the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences) and quantified via QuantiFluor-ST (Promega).
The purified amplicons were combined in equimolar amounts and sequenced via the Illumina MiSeq PE300 platform (Illumina, San Diego, USA) following standard protocols provided by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The raw reads have been deposited in the NCBI Sequence Read Archive (SRA) database under Accession Number PRJNA1161608. The multiplexed 16 S rRNA sequencing reads were processed via fastp version 0.19.6 [19] and merged via FLASH version 1.2.11 [20].
The high-quality sequences subsequently underwent denoising through the DADA2 [21] plugin integrated within the QIIME2 [22] (version 2020.2) pipeline, which utilized the suggested parameters. This process achieves single nucleotide resolution by analyzing error profiles present within the samples. The sequences that have been denoised by DADA2 are commonly referred to as amplicon sequence variants (ASVs). The analysis and classification of the 16 S rRNA raw reads were based on ASV. To reduce the impact of sequencing depth on alpha and beta diversity, each sample’s sequence count was rarefied to 20,000. This approach maintained an average Good’s coverage of 97.90%. Taxonomic classification of ASVs was carried out via the naive Bayes consensus taxonomy classifier in QIIME 2, with reference to the SILVA 16 S rRNA database (v138) [23].
Liquid Chromatography‒Mass Spectrometry (LC‒MS) analysis
Fifty milligrams of fecal samples were obtained from the same samples utilized for 16 S rRNA sequencing. Metabolite extraction was conducted using 400 µL of a methanol solution at a 4:1 (v/v) ratio. The resulting mixture was incubated at − 20 °C and subsequently homogenized with a high-throughput tissue grinder (Wonbio-96c, Shanghai Wanbo Biotechnology Co., Ltd.) at a frequency of 50 Hz for six minutes. This procedure was followed by 30 s of vortex mixing and ultrasonication at 40 kHz for 30 min at a temperature of 5 °C. The samples were then incubated at − 20 °C for an additional 30 min to facilitate protein precipitation. Following this, the samples were centrifuged at 13,000×g and 4 °C for 15 min, after which the supernatants were carefully transferred to vials designated for LC‒MS analysis.
To maintain quality control, the supernatants from each sample were combined. LC‒MS detection was executed with an AUHPLC-Q Exactive system. The chromatographic parameters were established as follows: an ACQUITY UPLC HSS T3 column (100 mm × 2.1 mm, 1.8 μm; Waters, Milford) was used. Mobile phase A was composed of 95% water and 5% acetonitrile (with 0.1% formic acid), while mobile phase B was composed of 47.5% acetonitrile, 47.5% isopropanol, and 5% water (with 0.1% formic acid). The flow rate was maintained at 0.40 mL/min, with an injection volume of 2 µL, and the column temperature was maintained at 40 °C. The raw data were processed, aligned, and identified via Progenesis QI metabonomics software.The raw data can be accessed publicly through the MetaboLights repository (http://www.ebi.ac.uk/metabolights) [24]under accession number MTBLS11105.
Statistical and bioinformatics analyses
The data analysis for this investigation was performed via IBM SPSS version 21.0, with the results expressed as the means ± standard deviations (SDs). For comparisons involving multiple groups, Tukey’s post hoc test was employed, whereas Student’s t test was utilized for the analysis of two groups. Graphical representations and additional analyses were produced via GraphPad Prism version 5.01.
Power analysis was conducted using G*Power (Version 3.1.9.7, Germany) for the t-test based on our previous data with this model [10, 11]. An alpha level of 0.05, a power of 80%, and an effect size of 0.8 (one-tailed) were employed. The power analysis indicated that a minimum of 6 mice per group is required.
The relative abundances of bacterial taxa at both the phylum and genus levels were compared across groups on the basis of ASV data, employing one-way nonparametric ANOVA (Kruskal‒Wallis H test), with a significance threshold established at P < 0.05. The alpha diversity of the gut microbiome was assessed via the ACE, Chao1, Shannon, Simpson, and Sobs indices. The resemblance between microbial communities across various samples was assessed through principal coordinate analysis (PCoA), utilizing both Bray‒Curtis and Jaccard metric dissimilarity, implemented via the Vegan v2.4.3 package. Statistical significance was determined via the nonparametric Wilcoxon rank-sum test and the Kruskal‒Wallis test. The features that contributed most to the differences between the control and treatment groups were identified. The LEfSe tool [25] (http://huttenhower.sph.harvard.edu/LEfSe) was used to determine the taxa of bacteria, ranging from phylum to genus, that exhibited significant abundance across the various groups, with a threshold set for the linear discriminant analysis (LDA) score greater than 3.5 and a p value of less than 0.05. The therapeutic outcomes (AST and ALT) and the relative abundances of genera were analyzed using Pearson’s rank correlation analysis to obtain the corresponding correlation coefficient (Corr) matrix and correlation P-value matrix. The statistical analysis was performed using Python 2.7 and R 3.3.1.
To evaluate global metabolic alterations among groups, partial least squares discriminant analysis (PLS-DA) was employed, with all metabolite variables subjected to Pareto scaling prior to analysis. The validity of the model was assessed through the calculation of R² and Q², which reflect the interpretability and predictability of the model, respectively, thereby reducing the potential for overfitting. The variable importance in projection (VIP) was calculated within the PLS-DA framework, with significant differences identified by VIP values exceeding 1 and P values falling below 0.05.
Differentially abundant metabolites between the two groups were mapped to their respective biochemical pathways through metabolic enrichment and pathway analysis via the KEGG database (http://www.genome.jp/kegg/). These metabolites are categorized according to their associated pathways or functions. The enrichment analysis typically investigates a collection of metabolites within a functional node to ascertain their presence. The focus of the analysis evolved from individual metabolites to a group-level annotation approach. The Python package scipy.stats (https://docs.scipy.org/doc/scipy/) was employed to identify significantly enriched pathways via Fisher’s exact test.
Pearson correlation coefficients were computed to explore the relationships between fecal metabolite concentrations and the relative abundances of genera, resulting in both a correlation coefficient matrix and a P value matrix. Only correlations with P < 0.05 were deemed significant. Throughout all the statistical evaluations, a P value threshold of < 0.05 was considered indicative of statistical significance.
Results
Phenotypic analysis of hUC-MSCs
The hUC-MSCs exhibited a fibroblast-like morphology and high expression levels of specific antigens (CD73, CD90, and CD105, ≥ 95% positive) (Fig. S1A, B, C) but minimal expression of hematopoietic lineage markers (CD11b, CD19, CD34, CD45, and HLA-DR, ≤ 2% positive) (Fig. S1D, E, F, G, H). The morphology of the MSCs was spindle shaped, which was consistent with that of the fibroblasts (Fig. S1I). Following the induction of osteogenesis and adipogenesis, Alizarin red staining revealed mineral accumulation and bone nodule formation (Fig. S1J). Oil Red O staining revealed that the hUC-MSCs contained numerous neutral lipid droplets in their cytoplasm (Fig. S1K).
Assessment of MSC-EV characteristics
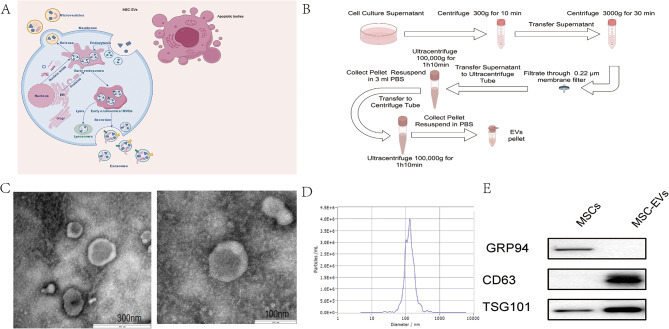
Ultracentrifugation was used to isolate hUC-MSC-EVs from hUC-MSC-conditioned medium (hUC-MSC-CM), which were then subjected to TEM, nanoparticle tracking analysis (NTA), and Western blotting. The obtained MSC-EVs exhibited a rounded shape enveloped by a double-layered membrane structure, as observed by TEM (Fig. 1A, B). Furthermore, NTA revealed that the average diameter of the hUC-MSC-EVs was 128 nm (Fig. 1C). Western blot analysis also confirmed the presence of MSC-EV markers such as TSG101 in the hUC-MSC-EVs, whereas the absence of glucose-regulated protein 94 (Grp94) verified the absence of contamination from endoplasmic reticulum (ER)-derived vesicles (Fig. 1D).
Fig. 1.
Characterization of MSC-EVs. (A) EVs produced by MSCs include exosomes (30–150 nm), microvesicles (150–500 nm), and apoptotic bodies (500–800 nm) in diameter. (B) EV preparation via ultracentrifugation. (C) The morphologies of two typical electric vehicles were captured via transmission electron microscopy (scale bars = 300 nm and 100 nm). (D) TEM was used to detect the size of the MSC-EVs. (E) Western blot analysis showing the expression levels of MSC-EV markers (GRP94, CD63, and TSG101) in both MSCs and MSC-EVs.Full-length blots/gels are presented in Supplementary Fig. S2
MSC-EVs significantly protected against Con A-induced hepatitis in mice via the gut microbiota
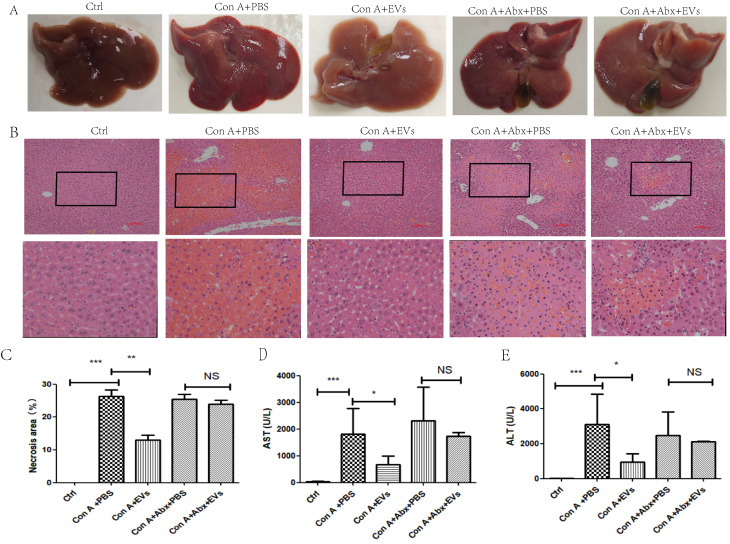
Gross liver appearance and histological analysis at 12 h demonstrated that MSC-EVs effectively ameliorated Con-A-induced hepatocyte necrosis, disseminated hemorrhage, and collagen deposition. Hepatocyte necrosis, disseminated hemorrhage, and collagen deposition caused by Con-A were effectively ameliorated by MSC-EVs on the basis of gross liver appearance and histological analysis at 12 h (Fig. 2A, B, C). At 12 h after Con A administration, the MSC-EV group presented significant decreases in serum AST and ALT activities, indicating the immediate and effective amelioration of Con A-induced liver hepatitis by MSC-EVs (Fig. 2D, E). These results confirmed that MSC-EVs effectively protected mice from Con A-induced hepatitis in vivo. However, Con A-induced hepatitis model mice were administered a cocktail of antibiotics (Abx) to clear the intestinal flora. Compared with Con-EVs alone, MSC-EVs did not effectively ameliorate Con A-induced hepatocyte necrosis, disseminated hemorrhage, or collagen deposition (Fig. 2A, B, C). Additionally, the Con A + Abx + MSC-EV group did not have significantly lower serum AST or ALT activities than the Con A + MSC-EV group did (Fig. 2D, E). These findings suggest that the addition of antibiotics may compromise the therapeutic efficacy of MSC-EVs in mitigating Con A-induced liver injury.
Fig. 2.
MSC-EVs significantly alleviated symptoms of Con A-induced hepatitis; however, when mice were administered a combination of antibiotics to clear the intestinal flora, the MSC-EVs failed to effectively mitigate Con A-induced liver injury. (A) Representative gross appearance of livers 12 h after Con A treatment as indicated. groups. (B) Representative hematoxylin and eosin (H&E) staining Photographs and (C) quantification of necrotic areas in liver tissues 12 h after Con A administration in the indicated groups. (D) Serum AST and ALT (E) levels were measured 12 h after Con A treatment in the indicated groups. The data in C-E are presented as the means ± standard deviations (means ± SDss) (n = 4–7 for each group), with statistical significance indicated by asterisks: *p < 0.05, **p < 0.01, ***p < 0.001
MSC-EVs modulate the gut microbiota and increase the abundance of norank_f__Muribaculaceae at the genus level
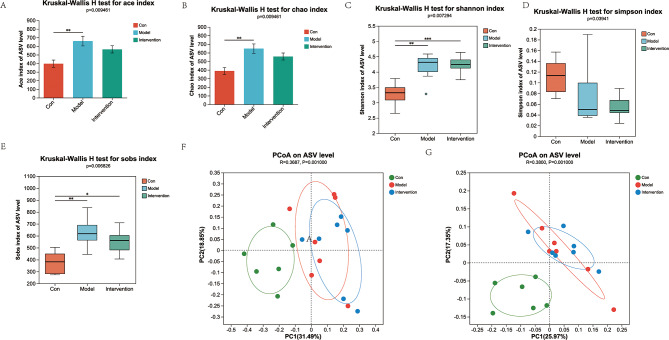
Considering the role of the gut microbiota in mice, we hypothesized that MSC-EVs could impact the microbial composition. To investigate this hypothesis, we conducted 16 S rRNA gene sequencing analysis on fecal samples collected from three groups of mice: control, Con A + PBS, and Con A + MSC-EVs (n = 6–7 individuals/group). The microbial diversity was measured and compared at the ASV level. Alpha diversity indices were measured via various methods (ACE, Chao1, Shannon, Simpson, and Sobs indices) (Fig. 3A-E). Significant differences in alpha diversity were observed among the control, Con A + PBS, and Con A + MSC-EV groups (P = 0.009461, P = 0.009461, P = 0.007294, P = 0.03941, and P = 0.006826 for each index, respectively). On the basis of the Bray‒Curtis and Jaccard metric distances, PCoA was used to analyze beta diversity, and the microbiota of these three groups were distinctively separated (R = 0.3687, P = 0.001000, and R = 0.3800, P = 0.001000, respectively) (Fig. 3F‒G).
Fig. 3.
Gut microbiota diversity among the three groups in terms of alpha and beta diversity
Alpha diversity based on the ACE (A), Chao1 (B), Shannon (C), Simpson (D), and Sobs (E) indices. Differences in the data were calculated via a two-sided Mann‒Whitney test. The horizontal bar within the box represents the median. The top and bottom of the box represent the 75th and 25th quartiles, respectively. (F) PCoA via Bray‒Curtis and (G) Jaccard metric distances of beta diversity. Differences in the data were calculated via the ANOSIM test. n = 6–7 individuals/group. Each point represents an individual mouse. All the exact P values are given. analysis. *P < 0.05, **P < 0.01, and ***P < 0.001. ANOSIM, analysis of similarities; PCoA, principal coordinate.
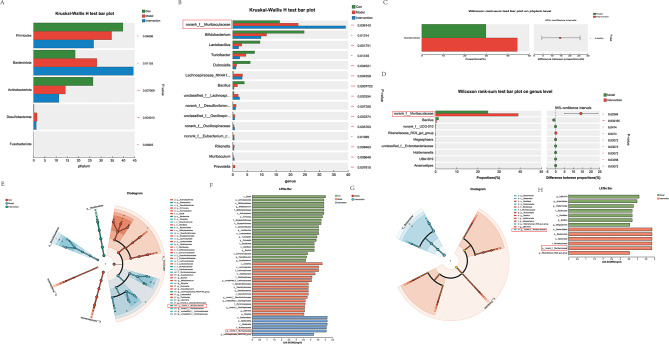
At the phylum level (Fig. 4A), significant differences were observed among the groups. Specifically, the relative abundance of Bacteroidia was greater in both the Con A + PBS and Con A + MSC-EV groups than in the control group. Analysis at the genus level (Fig. 4B) further revealed a notable increase in the relative abundance of norank_f__Muribaculaceae in both the Con A + PBS and Con A + MSC-EV groups compared with the control. Within the Con A-challenged groups, distinct differences were noted between the mice treated with PBS and those treated with MSC-EVs. At the phylum level (Fig. 4C), the Con A + MSC-EV group presented a greater relative abundance of Bacteroidia than did the Con A + PBS group. Similarly, at the genus level (Fig. 4D), the Con A + MSC-EV group presented increased levels of norank_f__Muribaculaceae compared with those in the Con A + PBS group.
Fig. 4.
MSC-EVs modulate the gut microbiota and appear to increase the abundance of the norank_f__Muribaculaceae genus. Fecal bacterial DNA from control mice, Con A + PBS-treated mice, and Con A + MSC-EV-treated mice (n = 6–7 individuals/group) was analyzed via 16 S rRNA gene sequencing. (A) The gut microbiota differed among the indicated groups at the phylum level. Compared with that in the control group, the relative abundance of Bacteroidia in the Con A + PBS and Con A + MSC-EV groups increased. (B) The gut microbiota differed among the indicated groups at the genus level. Compared with that in the control group, the relative abundance of norank_f__Muribaculaceae in the Con A + PBS and Con A + MSC-EV groups increased. (C) The gut microbiota differed among the indicated groups at the phylum level. The relative abundance of Bacteroidia was greater in the Con A + MSC-EV group than in the Con A + PBS group. (D) The gut microbiota differed among the indicated groups at the genus level. The relative abundance of norank_f__Muribaculaceae increased in the Con A + MSC-EV group compared with the Con A + PBS group. (E, F) Cladograms generated by LEfSe depict taxonomic associations between microbiome communities from control, Con A + PBS, and Con A + MSC-EV mice (LDA > 3.5). Red, blue, and green bars indicate taxa predominant in the control, Con A + PBS, and Con A + MSC-EV groups, respectively. (G, H) Cladograms generated by LEfSe depict taxonomic associations between microbiome communities from Con A + PBS and Con A + MSC-EV mice (LDA > 3.5). Red bars indicate taxa predominant in Con A + PBS-treated mice, and blue bars indicate taxa predominant in Con A + MSC-EV-treated mice
In addition, several genera with high discriminative power (linear discriminant analysis (LDA) score > 3.5) were identified on the basis of linear discriminant analysis effect size (LEfSe). In the control group, genera such as Bifidobacterium, Lactobacillus, Turicibacter, Dubosiella, Bacillus, Faecalibaculum, and Aquabacterium were notably predominant. Conversely, in the Con A + PBS group, genera such as unclassified_f__Lachnospiraceae, norank_f__Desulfovibrionaceae, unclassified_f__Oscillospiraceae, norank_f__Oscillospiraceae, UBA1819, and Bilophila showed pronounced predominance. The Con A + MSC-EV group exhibited a notable predominance of the genera norank_f__Muribaculaceae and _Lachnospiraceae_NK4A136_group (Fig. 4E, F).
Furthermore, to evaluate the microbiome response to MSC-EVs, the taxa that differentiated between the Con A + PBS group and the Con A + MSC-EV group were assessed (Fig. 4G, H). In the Con A + PBS group, the genera UBA1819, Anaerostipes, Holdemanella, Bacillus, and Megasphaera were predominant. Conversely, in the Con A + MSC-EV group, the genera norank_f__Muribaculaceae and Rikenellaceae_RC9_gut_group were notably predominant.
As shown in Fig. S3 Pearson’s correlation analysis indicated that changes in AST and ALT were negatively associated with changes in the abundance of the genus norank_f__Muribaculaceae (r = -0.69166, p = 0.00614 and r = -0.69716, p = 0.00558, respectively).
These findings underscore the modulatory effects of MSC-EVs on the gut microbiota composition, particularly in enhancing the abundance of norank_f__Muribaculaceae at the genus level in Con A-induced liver injury models.
Effects of MSC-EVs on liver metabolites
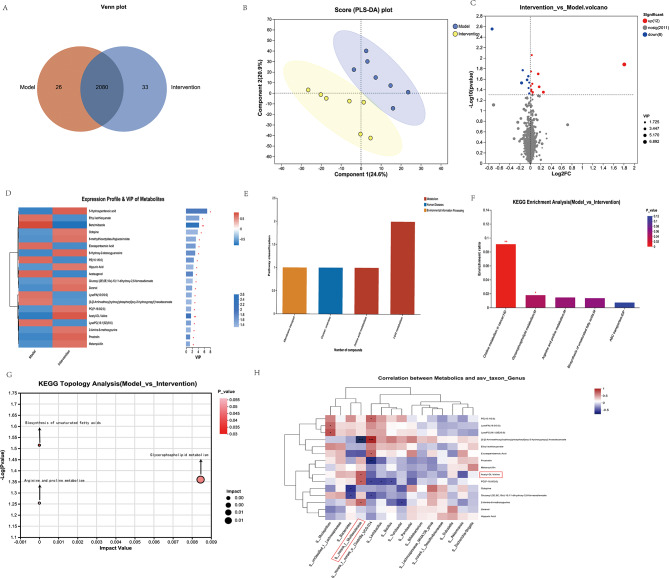
Liver metabolites were measured in all the samples via liquid chromatography‒tandem mass spectrometry (LC‒MS) to identify those associated with MSC-EV-induced alleviation in the Con A group. A total of 2106 metabolites were detected in the Con A + PBS group, and 2113 were detected in the Con A + MSC-EV group. The Venn diagram indicated that different groups presented distinct metabolite changes. Furthermore, the abundance of 59 metabolites significantly differed between the Con A + PBS group and the Con A + MSC-EV group (Fig. 5A). The difference in metabolite levels between Con A + PBS and Con A + MSC-EVs was then determined via partial least-squares discriminant analysis (PLS-DA). The score plot of the PLS-DA results clearly revealed separation between the two groups (Fig. 5B). Volcano plot showing differentially abundant metabolites between the Con A + PBS group and the Con A + MSC-EV group. Compared with the Con A + PBS group, the Con A + MSC-EV group presented an upregulation of 12 metabolites, a downregulation of 8 metabolites, and no significant changes in 2011 metabolites (Fig. 5C). The following top 20 metabolites were identified as potential discriminant markers between the Con A + PBS group and the Con A + MSC-EV group (Variable Importance in the Projection (VIP) > 1.0): 5-hydroxypentanoic acid (VIP = 6.8951), ethyl isothiocyanate (VIP = 4.2988), benzimidazole (VIP = 4.2021), octopine (VIP = 3.8176), 8-methylthiooctyldesulfoglucosinolate (VIP = 3.1891), eicosapentaenoic acid (VIP = 3.0786), 8-hydroxy-2-desoxyguanosine (VIP = 2.9641), PE (16:1/0:0) (VIP = 2.4922), hippuric acid (VIP = 2.3305), achenugenol (VIP = 2.2026), glucosyl (2E,6E,10x)-10,11-dihydroxy-2,6-farnesadienoate (VIP = 2.0944), zeranol (VIP = 1.9236), LysoPA (16:0/0:0) (VIP = 1.7724), [3-[2-aminoethoxy “Lipid metabolism” was ranked first in the “metabolism” category, followed by “amino acid metabolism” (Fig. 5E). Two metabolites, LysoPC(16:1(9Z)/0:0) and eicosapentaenoic acid, were assigned to “lipid metabolism,” whereas one metabolite, octopine, was assigned to “membrane transport.”
Fig. 5.
A comprehensive comparative metabolic analysis was performed to compare the liver metabolite profiles between the Con A + PBS and Con A + MSC-EV groups. (A) Composition of liver metabolites between the Con A + PBS and Con A + MSC-EV groups of mice is represented by a Venn diagram of metabolites. (B) PLS-DA score plot of liver metabolites between the Con A + PBS and Con A + MSC-EV groups of mice. (C) A volcano plot was generated to illustrate the distribution of all the metabolites identified via LC‒MS between the Con A + PBS and Con A + MSC-EV groups. (D) Heatmap of distinct metabolites between the Con A + PBS and Con A + MSC-EV groups. Differentiated groups are illustrated by the horizontal axis, metabolites by the vertical axis, and quantities of metabolites by colored blocks at different locations. A significant change was considered as VIP > 1 and p > 0.05 for the metabolites. (E) KEGG pathway classification: detected and annotated metabolites. The x-axis displays level-2 terms of the KEGG pathway, whereas the y-axis shows the number of identified metabolites. (F) Differentially abundant metabolites in the Con A + PBS group versus the Con A + MSC-EV group were analyzed for enriched metabolic pathways. On the X-axis, the significantly enriched pathways for the participating metabolites are represented. The y-axis represents the enrichment ratio, which represents the ratio between metabolites enriched in the pathway and all metabolites identified in the pathway. The colors represent the significance of enrichment; the deeper the color is, the more significant the enrichment. (G) Topological analysis of metabolic pathways between the Con A + PBS and Con A + MSC-EV groups. The x-axis denotes the relative significance of metabolites within the pathway and their corresponding impact values. The y-axis represents the enrichment significance of metabolites in the pathway, quantified as − log10 (P value). The bubble size indicates the impact value, whereas the bubble color signifies the P value. Larger bubbles indicate greater importance of the pathway. (H) Alterations in liver metabolite and fecal genus abundances. A heatmap shows the Pearson correlation coefficients between changes in fecal metabolite concentrations and variation in the relative abundances of the top 15 enriched bacterial genera. Each cell color intensity indicates the strength of the correlation between changes in liver metabolites and shifts in genus abundance. Statistical significance was adjusted for multiple comparisons via Benjamini‒Hochberg false discovery rate correction. *P < 0.05, **P < 0.01, and ***P < 0.001
Analysis of metabolite pathways revealed significant differences in the following pathways between the Con A + PBS group and the Con A + MSC-EV group: choline metabolism in cancer and glycerophospholipid metabolism (Fig. 5F). A topological analysis was conducted to assess the impact of these pathways, as indicated by the size of the bubbles (Fig. 5G). Glycerophospholipid metabolism, the biosynthesis of unsaturated fatty acids, and arginine and proline metabolism were the main pathways altered between the Con A + PBS group and the Con A + MSC-EV group. (Fig. 5G).
Correlations between liver metabolites and the fecal microbiota
Pearson correlation coefficients were calculated to explore the relationships between the relative abundances of genus-level constituents of the fecal microbiota and selected liver metabolites. In the fecal microbiome, liver metabolites and the abundance of genera were significantly correlated. As shown in Fig. 5H, Pearson’s correlation analysis revealed that the abundance of the genus g__norank_f__Muribaculaceae was negatively associated with the level of [3-[2-aminoethoxy(hydroxy)phosphoryl]oxy-2-hydroxypropyl] hexadecanoate (r = -0.7897, P = 0.00078). Additionally, it was positively associated with the levels of acetyl-DL-valine (r = 0.5812, P = 0.02927), PC (P-16:0/2:0) (r = 0.5758, P = 0.03119), and 2-amino-6-methoxypurine (r = 0.5532, P = 0.04019).
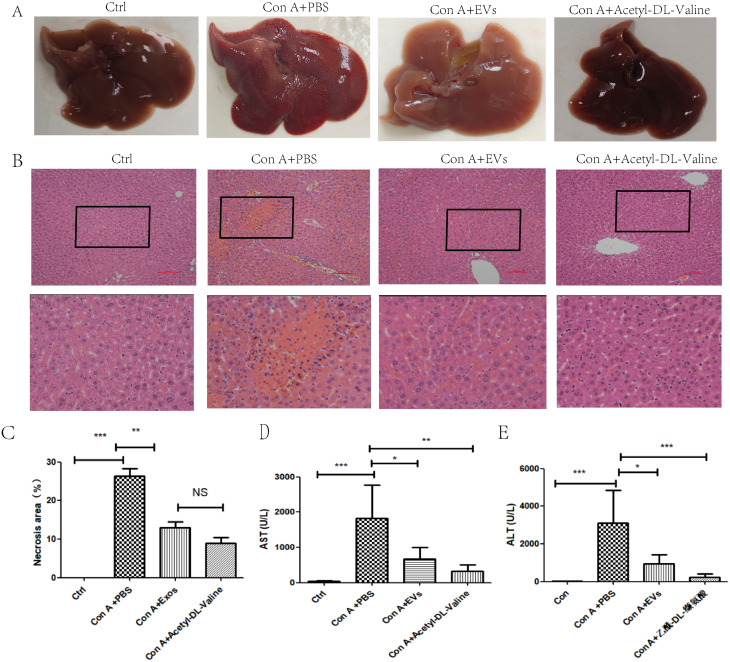
Acetyl-DL-Valine significantly protected against Con A-induced hepatitis in mice
Gross liver appearance and histological analysis at 12 h demonstrated that Acyl-DL-Valine effectively improved hepatocyte necrosis, disseminated hemorrhage, and collagen deposition in Con A-induced hepatitis mice (Fig. 6A, B, C). At 12 h post-Con A administration, the Con A + Acetyl-DL-Valine group presented significant decreases in serum AST and ALT activities, indicating the immediate and effective amelioration of Con A-induced hepatitis by Acetyl-DL-Valine (Fig. 6D, E). These results confirm that Acetyl-DL-Valine effectively protects mice from Con A-induced hepatitis in vivo.
Fig. 6.
Acetyl-DL-Valine significantly alleviated symptoms in Con A-induced hepatitis. (A) Representative gross images of livers from the indicated groups 12 h after Con A treatment are shown. (B) Representative hematoxylin and eosin (H&E) staining (C) The number of necrotic areas in the liver tissues 12 h after Con A administration was quantified in the indicated groups. Serum AST (D) and ALT (E) levels were assessed 12 h after Con A treatment in the specified groups. For each group in C-E, the means ± standard deviations are presented (n = 4–7 individuals for each group), with asterisks symbolizing statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
MSC-EV-mediated therapy is anticipated to offer an innovative, cell-free, noninvasive, less immunogenic, and nontoxic alternative strategy for liver treatment, while also providing important mechanistic insights into the reparative functions of liver cells [26].MSC-EVs have been extensively studied by researchers in the field of liver diseases. MSC-EVs have the capacity to shift the polarization balance towards type-2 regulatory macrophages [27], modulate the ratios of Th17 to Treg cells, diminish neutrophil infiltration [28], and mitigate oxidative stress [29, 30]. MSC-EVs help reduce mitochondrial damage and inflammation by stabilizing mitochondrial DNA [31]. MSC-EVs modulate CD154 expression of intrahepatic CD4 + T cells during liver IRI through the Ca2+-calcineurin-NFAT1 signaling pathway [32]. MSC-EVs alleviate sepsis-associated liver dysfunction by inhibiting macrophage glycolysis-mediated inflammatory response [33]. Additionally, MSC-EVs can downregulate hepatic stellate cell activation and reduce liver injury in mice infected with S. japonicum [34].In our research, we concentrate on the impacts of MSC-EVs on autoimmune hepatitis via the regulation of gut microbiota and metabolites.
In this study, we elucidated a novel role for MSC-EVs in ameliorating Con A-induced hepatitis in a gut microbiota-dependent manner. Our findings indicate that MSC-EVs directly modulate the gut microbiota, resulting in a greater abundance of norank_f__Muribaculaceae at the genus level and increased levels of Acetyl-DL-Valine in the liver.
Con A-induced autoimmune hepatitis (AIH) in murine experimental models effectively replicates human AIH [35]. In vitro research has demonstrated that Con A exerts direct toxicity on hepatocytes, causing their apoptosis. The pathological characteristics of AIH include elevated serum levels of ALT and AST, as well as extensive liver tissue necrosis [36].
Our previous research demonstrated significant therapeutic effects of UC-MSCs in a ConA-induced hepatitis mouse model. Given this finding, it is pertinent to explore whether MSC-EVs could similarly alleviate symptoms in such models. Additionally, research has conclusively demonstrated that the extracellular vesicles of macrophages inhibit the production of cytokines in macrophages after concanavalin A-induced hepatitis [37]. Ryo Tamura et al. demonstrated that exosomes derived from bone marrow mesenchymal stem cells (MSCs) can effectively suppress Con-A-induced liver injury in mice [38].Our research represents the first investigation demonstrating that UC-MSC-EVs effectively alleviate symptoms of Con A-induced hepatitis. Importantly, our findings indicate that this therapeutic effect is dependent on the gut microbiota. According to this novel discovery, the gut microbiota plays a crucial role in mediating the therapeutic efficacy of UC-MSC-EVs when treating hepatitis induced by ConA.
A critical finding of our research is the dependence of the therapeutic effects of MSC-EVs on the gut microbiota. When mice with Con A-induced hepatitis were treated with a cocktail of antibiotics to deplete the intestinal flora, the beneficial effects of MSC-EVs were notably compromised. The results of the histological and biochemical analyses revealed that the antibiotic-treated group did not exhibit significant improvements in hepatocyte necrosis, hemorrhage, or collagen deposition or reductions in the serum AST and ALT levels compared with those in the Con A + MSC-EV group. These findings suggest that the presence of the gut microbiota is essential for the therapeutic efficacy of MSC-EVs in liver injury. In a study conducted by Bo Yi et al.., the therapeutic effects of MSC-EVs in a liver trauma model were investigated, revealing that MSC-exos promote tissue repair in rats with liver trauma by modulating gut microbiota and serum metabolites [39].In a model of Con A-induced hepatitis, MSC-EVs have the potential to modulate the gut microbiota. Although this has not been previously reported, the gut microbiota is believed to play a role in the amelioration of concanavalin A-induced autoimmune hepatitis by koumine [40].
To investigate the function of the gut microbiome more deeply, 16 S rRNA gene sequencing was conducted on stool samples from the control group, the Con A + PBS group, and the Con A + MSC-EV group. The microbial composition of the groups differed significantly according to the alpha and beta diversity analyses. Notably, MSC-EV treatment led to an increase in the relative abundance of the genus norank_f__Muribaculaceae. The norank_f__Muribaculaceae is a prominent genus found in warm-blooded animals, including humans, mice, and guinea pigs [41]. These anaerobic bacteria flourish on host glycans, α-glucans, or plant glycans such as hemicellulose and pectin. They are involved in bacterial‒host interactions that maintain homeostasis and inhibit pathogenic bacterial growth [42]. Furthermore, polysaccharides from Agaricus bisporus improve ulcerative colitis in mice by increasing the level of norank_f__Muribaculaceae [43]. Alcoholic liver injury is alleviated by cytodin-3-O-glucoside, which increases the abundance of norank_f__Muribaculaceae [44]. Furthermore, fecal microbiota transplantation (FMT) significantly improves LPS/D-gal-induced liver metabolite disorders by increasing the abundance of norank_f__Muribaculaceae [45]. In our study, norank_f__Muribaculaceae, identified as a beneficial bacterium, exhibited heightened responsiveness under ConA treatment. This responsiveness was further enhanced by the influence of MSC-EVs, thereby facilitating the repair of liver injury.
Metabolomic analysis via LC‒MS identified 2113 metabolites in the Con A + MSC-EV group, with 59 metabolites showing significant differences in abundance compared with those in the Con A + PBS group. The notable metabolites included LysoPC(16:1(9Z)/0:0), eicosapentaenoic acid, and octopine, which are involved in metabolic pathways related to lipid metabolism and amino acid metabolism. KEGG pathway analysis revealed significant alterations in glycerophospholipid metabolism and the biosynthesis of unsaturated fatty acids, suggesting that MSC-EVs influence key metabolic pathways to exert their therapeutic effects. Pearson correlation analysis revealed significant associations between liver metabolites and the relative abundances of specific gut microbiota genera. For example, the genus norank_f__Muribaculaceae was positively correlated with metabolites such as acetyl-DL-valine, PC (P-16:0/2:0), and 2-amino-6-methoxypurine but negatively correlated with [3-[2-aminoethoxy(hydroxy)phosphoryl]oxy-2-hydroxypropyl] hexadecanoate. These correlations underscore the complex interplay between the gut microbiota and liver metabolites, suggesting that MSC-EVs may mediate their protective effects through the modulation of both microbial and metabolic pathways.
Acetyl-DL-valine has emerged as a significant metabolite with protective effects against Con A-induced hepatitis. Mice treated with Acetyl-DL-Valine showed substantial improvements in liver pathology and significant reductions in serum AST and ALT levels, confirming its immediate and effective protective role. These findings highlight Acetyl-DL-Valine as a potential therapeutic agent for liver injury, warranting further investigation into its mechanisms of action and therapeutic applications. Although the specific effects of Acetyl-DL-Valine on AIH have not been extensively studied, as an acetylated amino acid, it potentially exhibits biological activities akin to those of other acetylated amino acids. These compounds are involved in metabolic regulation, the antioxidant stress response, and anti-inflammatory reactions. For example, N-acetyl-L-cysteine (NAC) has been widely studied for its anti-inflammatory properties. It acts as a precursor to glutathione, a critical antioxidant that helps reduce oxidative stress and inflammation in various tissues. Studies have demonstrated that NAC can significantly reduce inflammation in conditions such as respiratory diseases, liver disorders, and sepsis [46]. Similarly, N-acetylglutamine enhances immune function and reduces inflammation by increasing glutamine availability, facilitating glutathione synthesis, which in turn helps maintain the cellular redox balance and alleviates inflammatory responses in immune cells and tissues under stress [47].
Numerous studies suggest that short-chain fatty acids (SCFAs), particularly butyrate, significantly influence inflammation modulation in the liver. For instance, research indicates that butyrate can diminish inflammation by inhibiting the activation of NFκB, a key player in inflammatory processes, while also facilitating the resolution of inflammatory responses [48, 49].In addition, certain metabolites produced by gut microbiota, such as lactate and acetate, play a significant role in influencing liver metabolism and inflammation. Lactate, which is generated during glycolysis in inflammatory conditions, acts as a signaling molecule that can modulate immune responses and inflammation [48]. On the other hand, acetate has been linked to enhanced metabolic health and a decrease in fat accumulation in the liver [50]. In our study, the intestinal metabolite Acetyl-DL-valine may be absorbed into the bloodstream and then enter the liver to exert anti-inflammatory effects.
The Con A-induced hepatitis model presents several limitations worth noting. Firstly, this model, characterized by T-lymphocyte-mediated hepatic injury, does not trigger B cell activation during the experimental period [51]. Consequently, we postulate that the development of autoantibodies is unlikely, indicating potential discrepancies in the underlying mechanisms of AIH. Secondly, while Con A induces transient inflammatory cell infiltration within the liver, the hepatic damage observed is subject to gradual repair over time [52]. Therefore, we euthanized the animals 12 h after model induction to analyze the blood and liver tissues. Lastly, it is critical to highlight that excessively high concentrations of Con A could lead to irreversible liver damage, resulting in rapid mortality among the mice [35]. Therefore, meticulous control of the Con A dosage is imperative for the successful induction of liver injury.
Besides the limitations of the model, there are several limitations to this study. In addition to the mechanisms previously discussed, hUC-MSC-MSC-EVs also contain protein, mRNA, and ncRNA components that might contribute to their immunoregulatory actions. Furthermore, it is worthwhile to further investigate how the intestinal norank_f__Muribaculaceae influences the levels of Acetyl-DL-Valine in the liver. Additionally, further research is needed to explore how Acetyl-DL-Valine affects CoA-induced hepatitis. Future research is essential to overcome these limitations and to determine whether MSC-EVs have an impact on patients with AIH, as well as to investigate whether there are alterations in the gut microbiota of these patients following MSC-EVs treatment.
Conclusions
In summary, our study provides the first evidence that MSC-EVs offer significant protection against Con A-induced hepatitis, with their efficacy being critically dependent on the gut microbiota. MSC-EVs modulate the composition of the gut microbiota, particularly by increasing the abundance of norank_f__Muribaculaceae, and impact liver metabolic profiles, leading to significant amelioration of liver injury. The identification of Acetyl-DL-Valine as a protective metabolite underscores the therapeutic potential of targeting gut‒liver axis interactions in liver diseases. Future research should delve deeper into the mechanisms underlying these interactions and explore the clinical potential of MSC-EVs and related metabolites in liver injury therapies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to express our gratitude to Figdraw (www.figdraw.com) for supplying the materials used in the graphical abstract. We also thank Springer Nature AJE Curie (https://china.aje.com/cn/curie) for language editing.
Abbreviations
- MSC
Mesenchymal stem cell
- hUC-MSCs
Human umbilical cord-derived mesenchymal stem cells
- EVs
Extracellular vesicles
- hUC
Human umbilical cord-derived
- Con
Concanavalin A
- AIH
Autoimmune hepatitis
- BCA
Bicinchoninic acid
- TEM
Transmission electron microscopy
- NTA
Nanoparticle tracking analysis
- Abx
Antibiotics
- HE
Hematoxylin and eosin
- ALT
Alanine aminotransferase
- AST
Aspartate transaminase
- Grp
Glucose-regulated protein
- ER
Endoplasmic reticulum
- ANOSIM
analysis of similarities
- PCoA
principal coordinate
- LEfSe
Linear discriminant analysis effect size
- VIP
Variable Importance in the Projection
- LC-MS
Liquid chromatography tandem mass spectrometry
- PLS-DA
Partial least-squares discriminant analysis
- LDA
Linear discriminant analysis
- ASV
Amplicon sequence variant
- SRA
Sequence Read Archive
- SCFAs
Short-chain fatty acids
Author contributions
W. C.,Q.Z. Y.X. conceived the study and supervised the project. F.Y.,W.C. B.N., X.L.,Y.H.,J.C.,Y.H.,H.Z. and C.Y. performed the experiments. L.Y. and J.L. collected the human umbilical cord samples. F.Y. and L.Y. prepared the figures. F.Y. and W.C. contributed to the experimental design and data analysis. F.Y. drafted the manuscript. All the authors discussed the results and revised the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (82460130, 82260110, 82425010, 82170674, and 82270652), the Natural Science Foundation of Guangdong Province (2024A1515013044), the China Postdoctoral Science Foundation (2019M653904XB), the Guangzhou Science and Technology Planning Project (202102010310, 2024A03J0272, and 202206010072), the Guangdong Xinjiang Aid Special Program (KSYJ2022002), the Kashgar Prefecture Science and Technology Planning Project (KS2023013), the National Key Research and Development Program of China (2022YFA1106700), and Xinjiang Autonomous Region Regional Collaborative Innovation Special Project (Science and Technology Assistance to Xinjiang Plan)2024E02062. Kashgar Region Cadre and Talent Aid Xinjiang Coordination Projects (KSRC-2022001, KSRC-2022002).
Data availability
The data will be available upon request. The raw data generated by 16 S rRNA gene sequencing have been uploaded to the NCBI BioProject database (Submission ID: SUB14729617, Accession number: PRJNA1161608). The raw LC‒MS data have been uploaded to the online MetaboLights repository (ID: MTBLS11105). The data will be released to the public when the paper is published.
Declarations
Ethics approval and consent to participate
All experiments were performed in accordance with the institutional guidelines of the Third Affiliated Hospital of Sun Yat-sen University. The use of animals in this study was approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University and was conducted in accordance with the ARRIVE guidelines 2.0. The approved study is titled “Mechanism of mesenchymal stem cells in relieving ConA-induced liver failure”, with approval granted on December 25, 2023 (Approval number: IACUC-F3-24-0102). The umbilical cords were obtained from healthy mothers at the Third Affiliated Hospital of Sun Yat-sen University with parental consent. The procedure for isolating UC-MSCs was approved by the Research Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University. The approved study is titled “A Multicenter Randomized Controlled Clinical Study on the Treatment of Hepatitis B-Related Acute-on-Chronic Liver Failure with Allogeneic Human Umbilical Cord Mesenchymal Stem Cells”, with approval granted on December 30, 2020 (Approval number: 2020-14). Informed consent was obtained from all the subjects and/or their legal guardian(s) under the ethical committee of the Third Affiliated Hospital of Sun Yat-sen University.
Consent for publication
All the authors confirm their consent for publication.
Competing interests
The authors declare that they have no competing financial interests or personal relationships that could have influenced the work reported in this paper.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fan Yang, Beibei Ni, and Xiaoqi Liang are co-first authors.
Contributor Information
Yan Xu, Email: xuyan55@mail.sysu.edu.cn.
Qi Zhang, Email: zhangq27@mail.sysu.edu.cn.
Wenjie Chen, Email: chenwj5@mail.sysu.edu.cn.
References
- 1.Mack CL, Adams D, Assis DN, Kerkar N, Manns MP, Mayo MJ, Vierling JM, Alsawas M, Murad MH, Czaja AJ. <ArticleTitle Language=“En”>Diagnosis and management of Autoimmune Hepatitis in adults and children: 2019 Practice Guidance and Guidelines from the American Association for the study of Liver diseases. Hepatology. 2020;72:671–722. [DOI] [PubMed] [Google Scholar]
- 2.Longhi MS, Ma Y, Mieli-Vergani G, Vergani D. Aetiopathogenesis of autoimmune hepatitis. J Autoimmun. 2010;34:7–14. [DOI] [PubMed] [Google Scholar]
- 3.Kornerup LS, Grønbæk L, Kjær MB, Grønbæk H. Chap. 18 - Current paradigm and treatment options for autoimmune hepatitis. In: Seto W-K, Eslam M, editors. Comprehensive Guide to Hepatitis Advances. Academic; 2023. pp. 429–41.
- 4.Chotiyarnwong P, McCloskey EV. Pathogenesis of glucocorticoid-induced osteoporosis and options for treatment. Nat Reviews Endocrinol. 2020;16:437–47. [DOI] [PubMed] [Google Scholar]
- 5.Harrison L, Gleeson D. Stopping immunosuppressive treatment in autoimmune hepatitis (AIH): Is it justified (and in whom and when)? Liver Int. 2019;39:610–20. [DOI] [PubMed] [Google Scholar]
- 6.Lee CW, Chen YF, Wu HH, Lee OK. Historical Perspectives and Advances in Mesenchymal Stem Cell Research for the Treatment of Liver Diseases. Gastroenterology. 2018;154:46–56. [DOI] [PubMed] [Google Scholar]
- 7.Luan Y, Kong X, Feng Y. Mesenchymal stem cells therapy for acute liver failure: Recent advances and future perspectives. Liver Res. 2021;5:53–61. [Google Scholar]
- 8.Zhang YC, Liu W, Fu BS, Wang GY, Li HB, Yi HM, Jiang N, Wang G, Zhang J, Yi SH, Li H, Zhang Q, Yang Y, Chen GH. Therapeutic potentials of umbilical cord-derived mesenchymal stromal cells for ischemic-type biliary lesions following liver transplantation. Cytotherapy. 2017;19:194–9. [DOI] [PubMed] [Google Scholar]
- 9.Lin BL, Chen JF, Qiu WH, Wang KW, Xie DY, Chen XY, Liu QL, Peng L, Li JG, Mei YY, Weng WZ, Peng YW, Cao HJ, Xie JQ, Xie SB, Xiang AP, Gao ZL. Allogeneic bone marrow-derived mesenchymal stromal cells for hepatitis B virus-related acute-on-chronic liver failure: A randomized controlled trial. Hepatology. 2017;66:209–19. [DOI] [PubMed] [Google Scholar]
- 10.Liu Q, Chen X, Liu C, Pan L, Kang X, Li Y, Du C, Dong S, Xiang AP, Xu Y, Zhang Q. Mesenchymal stem cells alleviate experimental immune-mediated liver injury via chitinase 3-like protein 1-mediated T cell suppression. Cell Death Dis. 2021;12:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan L, Liu C, Liu Q, Li Y, Du C, Kang X, Dong S, Zhou Z, Chen H, Liang X, Chu J, Xu Y, Zhang Q. Human Wharton’s jelly-derived mesenchymal stem cells alleviate concanavalin A-induced fulminant hepatitis by repressing NF-κB signaling and glycolysis. Stem Cell Res Ther. 2021;12:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lotfy A, AboQuella NM, Wang H. Mesenchymal stromal/stem cell (MSC)-derived exosomes in clinical trials. Stem Cell Res Ther. 2023;14:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikfarjam S, Rezaie J, Zolbanin NM, Jafari R. Mesenchymal stem cell derived-exosomes: a modern approach in translational medicine. J Transl Med. 2020;18:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janockova J, Slovinska L, Harvanova D, Spakova T, Rosocha J. New therapeutic approaches of mesenchymal stem cells-derived exosomes. J Biomed Sci. 2021;28:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider KM, Elfers C, Ghallab A, Schneider CV, Galvez EJC, Mohs A, Gui W, Candels LS, Wirtz TH, Zuehlke S, Spiteller M, Myllys M, Roulet A, Ouzerdine A, Lelouvier B, Kilic K, Liao L, Nier A, Latz E, Bergheim I, Thaiss CA, Hengstler JG, Strowig T, Trautwein C. Intestinal Dysbiosis Amplifies Acetaminophen-Induced Acute Liver Injury. Cell Mol Gastroenterol Hepatol. 2021;11:909–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei Y, Li Y, Yan L, Sun C, Miao Q, Wang Q, Xiao X, Lian M, Li B, Chen Y, Zhang J, Li Y, Huang B, Li Y, Cao Q, Fan Z, Chen X, Fang JY, Gershwin ME, Tang R, Ma X. Alterations of gut microbiome in autoimmune hepatitis. Gut. 2020;69:569–77. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q, Yang H, Kang X, Tian H, Kang Y, Li L, Yang X, Liu H, Ren P, Kuang X, Tong M, Fan W. A Synbiotic Ameliorates Con A-Induced Autoimmune Hepatitis in Mice through Modulation of Gut Microbiota and Immune Imbalance. Mol Nutr Food Res. 2023;67:e2200428. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Ni B, Liu Q, He F, Li L, Zhong X, Zheng X, Lu J, Chen X, Lin H, Xu R, He Y, Zhang Q, Zou X, Chen W. Human umbilical cord-derived mesenchymal stem cells ameliorate experimental colitis by normalizing the gut microbiota. Stem Cell Res Ther. 2022;13:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S, Zhou Y, Chen Y. Gu, fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, 2nd, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, C.H.D., Williamson AD, Willis ZZ, Xu Y, Zhang. Q. Zhu, R. Knight, J.G. Caporaso, Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat Biotechnol, 37 (2019) 852–857. [DOI] [PMC free article] [PubMed]
- 23.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yurekten O, Payne T, Tejera N, Amaladoss FX, Martin C, Williams M, O’Donovan C. MetaboLights: open data repository for metabolomics. Nucleic Acids Res. 2023;52:D640–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Psaraki A, Ntari L, Karakostas C, Korrou-Karava D, Roubelakis MG. Extracellular vesicles derived from mesenchymal stem/stromal cells: The regenerative impact in liver diseases. Hepatology. 2022;75:1590–603. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Zhang X, Zhang H, Song P, Pan W, Xu P, Wang G, Hu P, Wang Z, Huang K, Zhang X, Wang H, Zhang J. Mesenchymal Stem Cells Derived Extracellular Vesicles Alleviate Traumatic Hemorrhagic Shock Induced Hepatic Injury via IL-10/PTPN22-Mediated M2 Kupffer Cell Polarization. Front Immunol. 2021;12:811164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu T, Zhang J, Cai J, Xiao J, Sui X, Yuan X, Li R, Li Y, Yao J, Lv G, Chen X, Chen H, Zeng K, Liu Y, Chen W, Chen G, Yang Y, Zheng J, Zhang Y. Extracellular vesicles derived from mesenchymal stromal cells as nanotherapeutics for liver ischaemia-reperfusion injury by transferring mitochondria to modulate the formation of neutrophil extracellular traps. Biomaterials. 2022;284:121486. [DOI] [PubMed] [Google Scholar]
- 29.Sitbon A, Delmotte P-R, Pistorio V, Halter S, Gallet J, Gautheron J, Monsel A. Mesenchymal stromal cell-derived extracellular vesicles therapy openings new translational challenges in immunomodulating acute liver inflammation. J Translational Med. 2024;22:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao J, Zheng J, Cai J, Zeng K, Zhou C, Zhang J, Li S, Li H, Chen L, He L, Chen H, Fu H, Zhang Q, Chen G, Yang Y, Zhang Y. Extracellular vesicles derived from human umbilical cord mesenchymal stem cells alleviate rat hepatic ischemia-reperfusion injury by suppressing oxidative stress and neutrophil inflammatory response. Faseb j. 2019;33:1695–710. [DOI] [PubMed] [Google Scholar]
- 31.Zhao M, Liu S, Wang C, Wang Y, Wan M, Liu F, Gong M, Yuan Y, Chen Y, Cheng J, Lu Y, Liu J. Mesenchymal Stem Cell-Derived Extracellular Vesicles Attenuate Mitochondrial Damage and Inflammation by Stabilizing Mitochondrial DNA. ACS Nano. 2021;15:1519–38. [DOI] [PubMed] [Google Scholar]
- 32.Zheng J, Lu T, Zhou C, Cai J, Zhang X, Liang J, Sui X, Chen X, Chen L, Sun Y, Zhang J, Chen W, Zhang Y, Yao J, Chen G, Yang Y. Extracellular Vesicles Derived from Human Umbilical Cord Mesenchymal Stem Cells Protect Liver Ischemia/Reperfusion Injury by Reducing CD154 Expression on CD4 + T Cells via CCT2. Adv Sci (Weinh). 2020;7:1903746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pei L, Li R, Wang X, Xu D, Gong F, Chen W, Zheng X, Liu W, Zhao S, Wang Q, Mao E, Chen E, Chen Y, Yang Z. MSCs-derived extracellular vesicles alleviate sepsis-associated liver dysfunction by inhibiting macrophage glycolysis-mediated inflammatory response. Int Immunopharmacol. 2024;128:111575. [DOI] [PubMed] [Google Scholar]
- 34.Dong L, Pu Y, Chen X, Qi X, Zhang L, Xu L, Li W, Ma Y, Zhou S, Zhu J, Li Y, Wang X, Su C. hUCMSC-extracellular vesicles downregulated hepatic stellate cell activation and reduced liver injury in S. japonicum-infected mice. Stem Cell Res Ther. 2020;11:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao J, Sun W, Xu H. Pathogenesis of Concanavalin A induced autoimmune hepatitis in mice. Int Immunopharmacol. 2022;102:108411. [DOI] [PubMed] [Google Scholar]
- 36.Terziroli Beretta-Piccoli B, Mieli-Vergani G, Vergani D. Autoimmmune hepatitis. Cell Mol Immunol. 2022;19:158–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawata R, Oda S, Koya Y, Kajiyama H, Yokoi T. Macrophage-derived extracellular vesicles regulate concanavalin A-induced hepatitis by suppressing macrophage cytokine production. Toxicology. 2020;443:152544. [DOI] [PubMed] [Google Scholar]
- 38.Tamura R, Uemoto S, Tabata Y. Immunosuppressive effect of mesenchymal stem cell-derived exosomes on a concanavalin A-induced liver injury model. Inflamm Regen. 2016;36:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi B, Pan J, Yang Z, Zhu Z, Sun Y, Guo T, Zhao Z. Mesenchymal stem cell-derived exosomes promote tissue repair injury in rats with liver trauma by regulating gut microbiota and metabolism. Mol Cell Probes. 2024;75:101958. [DOI] [PubMed] [Google Scholar]
- 40.Que W, Lin H, Li X, Zhang B, Liu M, Hu X, Fu J, Cheng Y, Qiu H. Koumine ameliorates concanavalin A-induced autoimmune hepatitis in mice: involvement of the Nrf2, NF-κB pathways, and gut microbiota. Int Immunopharmacol. 2023;114:109573. [DOI] [PubMed] [Google Scholar]
- 41.Zou Y, Liang N, Zhang X, Han C, Nan X. Functional differentiation related to decomposing complex carbohydrates of intestinal microbes between two wild zokor species based on 16SrRNA sequences. BMC Vet Res. 2021;17:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shao J, Li Z, Gao Y, Zhao K, Lin M, Li Y, Wang S, Liu Y, Chen L. Construction of a Bacteria-Metabolites Co-Expression Network to Clarify the Anti-Ulcerative Colitis Effect of Flavonoids of Sophora flavescens Aiton by Regulating the Host-Microbe Interaction. Front Pharmacol. 2021;12:710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W, Wang L, Yuan Q, Hao W, Wang Y, Wu D, Chen X, Wang S. Agaricus bisporus polysaccharides ameliorate ulcerative colitis in mice by modulating gut microbiota and its metabolism. Food Funct. 2024;15:1191–207. [DOI] [PubMed] [Google Scholar]
- 44.Zhu L, Cao F, Hu Z, Zhou Y, Guo T, Yan S, Xie Q, Xia X, Yuan H, Li G, Luo F, Lin Q. Cyanidin-3-O-Glucoside Alleviates Alcoholic Liver Injury via Modulating Gut Microbiota and Metabolites in Mice, Nutrients, 16 (2024). [DOI] [PMC free article] [PubMed]
- 45.Yuan C, Fan J, Jiang L, Ye W, Chen Z, Wu W, Huang Q, Qian L. Integrated Analysis of Gut Microbiome and Liver Metabolome to Evaluate the Effects of Fecal Microbiota Transplantation on Lipopolysaccharide/D-galactosamine-Induced Acute Liver Injury in Mice. Volume 15. Nutrients; 2023. [DOI] [PMC free article] [PubMed]
- 46.Koo AN, Rim HP, Park DJ, Kim J-H, Jeong SY, Lee SC. Glutathione-mediated intracellular release of anti-inflammatory N-acetyl-L-cysteine from mesoporous silica nanoparticles. Macromol Res. 2013;21:809–14. [Google Scholar]
- 47.Newsholme P, Diniz VLS, Dodd GT, Cruzat V. Glutamine metabolism and optimal immune and CNS function. Proc Nutr Soc. 2023;82:22–31. [DOI] [PubMed] [Google Scholar]
- 48.Fang Y, Li Z, Yang L, Li W, Wang Y, Kong Z, Miao J, Chen Y, Bian Y, Zeng L. Emerging roles of lactate in acute and chronic inflammation. Cell Communication Signal. 2024;22:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomasova L, Grman M, Ondrias K, Ufnal M. The impact of gut microbiota metabolites on cellular bioenergetics and cardiometabolic health. Nutr Metabolism. 2021;18:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hernández MAG, Canfora EE, Jocken JWE, Blaak EE. The Short-Chain Fatty Acid Acetate in Body Weight Control and Insulin Sensitivity. Nutrients; 2019. p. 11. [DOI] [PMC free article] [PubMed]
- 51.Hardtke-Wolenski M, Noyan F, Jaeckel E. Requirements and challenges of a preclinical autoimmune hepatitis mouse model. Dig Dis. 2011;29:402–10. [DOI] [PubMed] [Google Scholar]
- 52.Erhardt A, Biburger M, Papadopoulos T, Tiegs G. IL-10, regulatory T cells, and Kupffer cells mediate tolerance in concanavalin A-induced liver injury in mice. Hepatology. 2007;45:475–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data will be available upon request. The raw data generated by 16 S rRNA gene sequencing have been uploaded to the NCBI BioProject database (Submission ID: SUB14729617, Accession number: PRJNA1161608). The raw LC‒MS data have been uploaded to the online MetaboLights repository (ID: MTBLS11105). The data will be released to the public when the paper is published.