Abstract
Background
In the soy sauce fermentation industry, Aspergillus oryzae (A. oryzae) plays an essential role and is frequently subjected to high salinity levels, which pose a significant osmotic stress. This environmental challenge necessitates the activation of stress response mechanisms within the fungus. The Zn(II)2Cys6 family of transcription factors, known for their zinc binuclear cluster-containing proteins, are key regulators in fungi, modulating various cellular functions such as stress adaptation and metabolic pathways.
Results
Overexpression of AozC decreased growth rates in the presence of salt, while its knockdown enhanced growth, the number of spores, and biomass, particularly under conditions of 15% salt concentration, doubling these metrics compared to the wild type. Conversely, the knockdown of AozC via RNA interference significantly enhanced spore density and dry biomass, particularly under 15% salt stress, where these parameters were markedly improved over the wild type strain. Moreover, the overexpression of AozC led to a downregulation of the FAD2 gene, a pivotal enzyme in the biosynthesis of unsaturated fatty acids (UFAs), which are essential for preserving cell membrane fluidity and integrity under saline conditions. Transcriptome profiling further exposed the influence of AozC on the regulation of UFA biosynthesis and the modulation of critical stress response pathways. Notably, the regulatory role of AozC in the mitogen-activated protein kinase (MAPK) signaling and ABC transporters pathways was highlighted, underscoring its significance in cellular osmotic balance and endoplasmic reticulum homeostasis. These findings collectively indicate that AozC functions as a negative regulator of salt tolerance in A. oryzae.
Conclusion
This research suggest that AozC acts as a negative regulator in salt tolerance and modulates fatty acid biosynthesis in response to osmotic stress. These results provide insights into the regulatory mechanisms of stress adaptation in A. oryzae.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12934-024-02639-z.
Keywords: Aspergillus oryzae, Differentially expressed genes, Salt treatment, Fatty acid, Transcriptome, Transcription factor, Unsaturated fatty acids
Background
Aspergillus oryzae (A. oryzae), a filamentous fungus with a long history of safe use in the fermentation of traditional Asian foods such as miso [1], soy sauce, and sake, has emerged as a model organism for studying fungal physiology and genetic manipulation [2]. Its powerful metabolic capacity and ease of genetic manipulation make it an ideal host for various industries, including pharmaceuticals, food, cosmetics [3], production of enzymes and recombinant proteins [4], bio-based materials industries [5], and cell factory construction [6]. For example, the production of red rice fermented by A. oryzae exemplifies the potential of this fungus in generating novel skincare compounds with multifunctional properties, expanding its application beyond traditional fermentation into the cosmetics and healthcare industries [7]. However, environmental challenges such as salt stress, high ethanol concentrations and high temperatures during industrial fermentation threaten the cellular health and fermentation efficiency of these microorganisms [8]. For A. oryzae, in particular, the high salt concentrations and low pH values encountered in the soy sauce mash environment result in significant osmotic stress [9], which poses a significant challenge for the fungus to maintain cellular integrity and enzyme activity, ensuring the success of the fermentation process.
Organisms ranging from bacteria to fungi have evolved unique strategies to withstand saline conditions, adjusting cellular mechanisms to maintain membrane integrity and function. For example, halophilic fungi produce bioactive compounds to adapt to high salt environments [10], and cyanobacteria such as Synechococcus elongatus PCC 7942 regulate enzymatic processes to manage salt stress [11]. Yeast cells, including Zygosaccharomyces rouxii, enhance ergosterol content and alter fatty acid profiles to preserve membrane stability under such conditions [12]. These adaptations highlight the importance of maintaining membrane fluidity and stability, particularly through the action of unsaturated fatty acids (UFAs), which are vital for the stress tolerance of microorganisms like A. oryzae [13]. The presence of oleic and linoleic acids as major UFAs in the study emphasizes their importance in the antimicrobial activity against Streptococcus mutans biofilms [14]. Zygosaccharomyces rouxii, known for its osmo- and thermotolerance, modulates its ergosterol content and increases the unsaturated fatty acid content to maintain membrane integrity under stress [15]. The role of UFAs in preserving membrane fluidity and integrity is well-established [16], making them a critical target for genetic engineering to enhance the stress tolerance of industrially relevant fungi like A. oryzae [17]. By manipulating genes involved in UFA metabolism, we can potentially develop strains with improved resilience to osmotic stress, thereby enhancing the efficiency and quality of fermentation processes, as observed in the soy sauce industry [18]. The genetic flexibility afforded by advances in genetic engineering, coupled with insights from transcriptomic and metabolomic studies, presents a promising avenue for tailoring A. oryzae to better cope with the challenges of saline environments.
Zinc binuclear cluster-containing proteins, typified by the Zn(II)2Cys6 domain architecture, are specific to fungi and are implicated in a spectrum of biological processes [19]. These proteins are engaged in a multitude of cellular functions, including but not limited to, the adaptation to environmental stress, cellular development, and metabolic regulation, making them essential components of fungal biology and evolution [20]. The Zn(II)2Cys6 family of transcription factors has garnered significant interest due to their dynamic involvement in stress response pathways [21]. In Tolypocladium guangdongense, Zhang et al. identified 54 Zn(II)2Cys6 genes with altered expression in response to light, including key regulators of developmental processes and metabolic pathways [20]. Under environmental stress, these factors have been shown to regulate the transcriptional responses and basal resistance to stress in fungal species. For example, under azole stress, Zn(II)2Cys6 transcription factors, such as ADS-1 in Neurospora crassa, is pivotal in orchestrating fungal responses to azole stress by regulating gene involved in drug efflux and ergosterol metabolism [22]. AozC is a member of the Zn(II)2Cys6 family of transcription factors, known for their crucial in the regulation of various cellular processes in fungi, including stress responses and secondary metabolism. Our interest in AozC was prompted by preliminary data indicating its differential expression patterns under salt stress conditions, suggesting a potential role in the adaptation of A. oryzae to high salinity environments [23]. Building on our prior research, which established a link between salt stress and the upregulation of genes involved in arginine and oleic acid synthesis, we hypothesize that these metabolic pathways may be integral to the salt tolerance of organism. The current study expands upon these insights by investigating the regulatory influence of AozC on these pathways and its broader implications for fungal adaptation. However, the precise mechanisms of these adaptations, especially regarding UFA homeostasis and their impact on membrane integrity, remain to be fully elucidated. In our current investigation, we have delved into the impact of the Zn(II)2Cys6 transcription factor AozC on the salt tolerance of A. oryzae by introducing the gene into A. oryzae. To further comprehend the molecular mechanisms by which AozC influences A. oryzae, we conducted transcriptomic sequencing on both overexpression and RNA interference (RNAi) of AozC strains of the transcription factor. Our findings have not only shed new light on the genetic and biochemical strategies that A. oryzae employs to counteract osmotic pressure, but also provided strategic insights for the development of fermentation strains with enhanced salt tolerance.
Materials and methods
Fungal strains and growth conditions
The A. oryzae strain 3.042 (CICC 40092) was obtained from the China Center of Industrial Culture Collection for genetic manipulation. The strain was cultivated on a dextrin–peptone–yeast extract (DPY) agar medium containing (per 1,000 ml of deionized water): 2% glucose, 1% peptone, 0.5% yeast extract, 0.5% KH2PO4, 0.05% MgSO4, 0.1% histidine, and 1.8% agar powder [24]. The A. oryzae strain was pre-cultured on DPY agar plates at 30 °C for 3 days, and conidia were harvested for further use, with concentration determined using a hemocytometer according to standard procedures. Biomass was measured by filtering a known volume of the culture, washing the spores, and then drying them to a constant weight at a specified temperature to determine the dry weight. Escherichia coli DH5α was employed for bacterial transformations and plasmid amplification. The Saccharomyces cerevisiae (S. cerevisiae) strain Y00000 (BY4741) was grown in Yeast Peptone Dextrose (YPD) medium at 28 °C for heterologous expression studies.
Expression analysis of AozC gene in A. oryzae under varying salt concentrations
To delineate the expression profile of the AozC gene across various developmental stages and in response to salt stress, quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) analysis was conducted. The DPY medium was supplemented with NaCl to achieve final concentrations of 0%, 5%, 10%, and 15% (w/v), and the incubation was conducted at 30 °C. Total RNA was extracted from A. oryzae strain 3.042 at three developmental time points (24, 48, and 72 h) under conditions of varying salt stress. The RNA was isolated utilizing the PrimeScript™ RT Reagent Kit from Takara (Dalian, China). To eliminate genomic DNA contamination, DNase I (Sigma, Aldrich, USA) was employed at 37 °C. The primers for AozC were designed based on the sequences of fatty acid desaturases from A. oryzae, with reference to GenBank Accession Numbers EIT74184.1. The qRT-PCR was performed on the CFX96 Real-Time PCR Detection System (Bio-Rad, CA, USA) using the Bio-Rad CFX Connect Optics Module software, with 18 S RNA as an endogenous control [25]. The list of all primers is provided in Additional file 1: Table S1. Cloning and sequence analysis of AozC gene from A. oryzae.
Cloning and sequence analysis of AozC gene from A. Oryzae
To clone the AozC gene from A. oryzae strain 3042, we first converted total RNA into complementary DNA (cDNA) using the M-MLV Reverse Transcriptase kit (TIANGEN, Beijing, China) as per the manufacturer’s protocol. Subsequently, gene-specific primers for AozC, listed in Supplementary Table S1, were utilized to amplify the gene of interest via PCR. The resulting PCR products were then cloned into the pMD19-T Vector (Takara, Dalian, China) for further analysis.
The conserved domain within the AozC gene was identified using the NCBI Conserved Domain Database (CDD) and visualized with Domain Graph 2.0 (DOG) [26]. Multiple sequence alignment of the AozC domains was conducted using the CLUSTALW algorithm within the MEGA 5.0 software suite [27]. For this, protein sequences from various Aspergillus species, including Aspergillus parasiticus (KJK61240), Aspergillus arachidicola (PIG88819), Aspergillus indologenus (PYI34919), Aspergillus fijiensis (RAK71911), Aspergillus terreus (XP_001208859), A. oryzae (XP_001826164), Aspergillus flavus (XP_002377833), Aspergillus nomius (XP_015402290), Aspergillus bombycis (XP_022388403), Aspergillus brunneoviolaceus (XP_025437062), Aspergillus uvarum (XP_025485649), Aspergillus aculeatinus (XP_025508167), and Aspergillus japonicus (XP_025522981), were retrieved from NCBI. Phylogenetic analysis and statistical maximum likelihood (ML) bootstrap tests were performed using the MEGA software package [27]. The bootstrap analysis, consisting of 1000 replicates, was applied to assess the robustness of the inferred phylogenies.
Functional characterization and growth assessment of AozC in S. Cerevisiae under salt stress
For the preliminary functional validation of the AozC gene from A. oryzae, we employed a molecular cloning approach to integrate the gene into the yeast expression vector. The AozC coding sequence was PCR-amplified using primers designed to introduce PML I and AFL II restriction sites, enabling seamless ligation into the pYES2 vector, which was digested with corresponding enzymes. The recombinant plasmid, designated pYES2-AozC, was sequenced to confirm the correct integration and orientation of gene. Utilizing the lithium acetate method, the plasmid was transformed into S. cerevisiae Y00000, after which transformants were selected on synthetic defined (SD) agar lacking leucine. Colony PCR and plasmid DNA extraction followed by restriction enzyme digestion verified the successful integration of AozC in the transformed yeast strains. To validate the expression of the AozC gene in the transformed yeast strains, we performed qRT-PCR. The cDNA was synthesized from total RNA extracted from the yeast cultures points post-induction. The relative expression levels were determined using the 2−ΔΔCt method, with the 18 S as an internal control to normalize the data. To assess the impact of AozC expression on yeast growth under salt stress, optical density (OD) values of the transformed S. cerevisiae were determined at 600 nm. The OD measurements were taken at 12-, 24-, and 36-hours post-inoculation, with the yeast broth diluted 1:3 with deionized water prior to assessment. A UV spectrophotometer was used for OD determination, with deionized water as the blank and the diluted yeast solution as the sample. The reported OD values were adjusted based on the dilution factor, and each time point was measured in triplicate to ensure accuracy and reproducibility.
Functional characterization of AozC in A. Oryzae
To investigate the role of the AozC gene in A. oryzae under salt stress, we employed a genetic approach involving both overexpression and RNAi strategies. The pEX1-AozC vector was constructed for the targeted manipulation of AozC expression levels. For the transformation, Agrobacterium tumefaciens, harboring the pEX1-AozC vector, was prepared using a heat shock assay. A. oryzae spores were inoculated and cultured in liquid DPY medium. Subsequently, the germinated spores were co-cultured with Agrobacterium in induction medium (IM) containing acetosyringone (AS) to facilitate the transfer of the recombinant vector into the fungal genome. Transformed strains, alongside the wild type (WT) strain 3.042, were assessed for their growth, biomass accumulation, and FAD2 gene expression in response to varying sodium chloride concentrations (0, 5%, 10%, and 15%). Specifically, spore suspensions of the overexpression and RNAi of AozC strains, as well as the WT, were inoculated onto DPY agar plates containing different NaCl concentrations and incubated at 30 °C for 72 h. Growth was monitored, and spore counts were taken after 72 h to evaluate the impact of salt stress on spore production. Additionally, the expression of the FAD2 gene, implicated in fatty acid biosynthesis, was quantified in these strains to determine the regulatory influence of AozC on stress-responsive pathways.
Determination of intracellular fatty acid compositions and levels
The intracellular fatty acid compositions and levels were determined following a refined approach to extract total lipids from both Aspergillus oryzae mycelia and Saccharomyces cerevisiae strains. For A. oryzae, the procedure involved the collection, washing, and freeze-drying of mycelia, followed by powdering and weighing for lipid extraction, as previously described [23]. S. cerevisiae cells were harvested from liquid culture by centrifugation, washed with distilled water, and processed in a similar manner to A. oryzae for lipid extraction. The extracted lipids from both organisms were subjected to a transmethylation reaction using a 2% H2SO4–MeOH solution in chloroform at 70 °C for 2 h. This step is essential for converting fatty acids into their methyl esters (FAMEs), which are compatible with the gas chromatography-mass spectrometry (GC-MS) system (Shimadzu, Kyoto, Japan). The FAMEs were separated and quantified using a Shimadzu QP2010 GC-MS system equipped with a Supelco SP-2340 fused silica capillary column. The GC-MS system was optimized for the resolution and detection of FAMEs, with the column providing high efficiency in separating complex lipid mixtures. Identification of FAMEs was performed by matching their mass spectra with those in a comprehensive spectral database, ensuring accurate compound identification. Retention times were compared with external standards to confirm the identity of individual fatty acid peaks. The relative abundance of each fatty acid component was determined by integrating the peak area of the most intense ion for each peak, as measured by the GC-MS system [28].
Transcriptome analysis
RNA was extracted from A. oryzae strains with either overexpression or RNAi of AozC strains. The RNA was isolated using a Fungal Total RNA Extraction Kit (Omega Bio-Tek, Norcross, United States). RNA concentrations were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States) to ensure adequate input for downstream applications. RNA integrity was assessed with a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, United States), confirming the integrity of the samples for further analysis. To ensure the reliability and reproducibility of our results, equal quantities of RNA from each of three individual cultures were pooled for cDNA library construction. The mRNA was enriched from the pooled total RNA using oligo(dT) magnetic beads (New England Biolabs, Ipswich, MA, United States) and fragmented at 94 °C for 5 min using a thermocycler. The fragmented mRNA was reverse transcribed into first-strand cDNA using random hexamer primers (Thermo Fisher Scientific) and subsequently synthesized into second-strand cDNA using DNA polymerase I and RNase H (Thermo Fisher Scientific). The cDNA fragments were purified with the QIAquick PCR Purification Kit (QIAGEN, Hilden, Germany), end-repaired, and ligated to Illumina sequencing adapters to create the cDNA library. The library was then sequenced on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, United States) [25]. Post-sequencing, raw reads containing adapters or low-quality bases were filtered using established criteria to obtain clean reads. Further cleaning was performed using the Bowtie2 software (Johns Hopkins University, Baltimore, MD, United States) to remove reads that mapped to the ribosome RNA (rRNA) database. The resulting clean reads were employed for assembly and transcriptome analysis [29]. These clean reads were aligned to the A. oryzae 3.042 reference genome using TopHat2, allowing up to two mismatches in the seed region. Gene expression levels were quantified with RSEM and normalized to FPKM (Fragments Per Kilobase of transcript per Million mapped reads) [30]. The edgeR package on the R package (version 3.4.2) was utilized to identify differentially expressed genes (DEGs) with a fold change ≥ 2 and an false discovery rate (FDR) < 0.05 [23]. To validate the RNA-seq results, a subset of DEGs was selected for confirmation via qPCR analysis. 18 S RNA was used as the internal control gene. The qPCR was performed using a SYBR® Green qPCR Mix (TaKaRa, China), and the relative expression levels of the selected genes normalized to 18 S RNA were calculated using 2-ΔΔCt method. Following validation, the identified DEGs were subjected to hierarchical clustering. Subsequently, a subsequent KEGG pathway enrichment analysis was performed to determine the metabolic pathways most significantly impacted by the modulation of AozC expression in A. oryzae [31]. Pathways with a Q-value ≤ 0.05, resulting from hypothesis testing that included p-value calculation and FDR correction, were considered significantly enriched. The top five enriched metabolic pathways were determined based on the enrichment factor.
Data analysis
This study was conducted with three independent experimental replications, and the results presented are the average values ± standard error (SE) derived from these replicates. To assess the data collected at equivalent time intervals, a one-way nested analysis of variance (ANOVA) was employed. Subsequent mean comparisons were executed using the Least Significant Difference (LSD) test, which is an appropriate post-hoc analysis for nested ANOVA designs. The statistical significance of differences between means was determined using a one-tailed Student’s t-test. All computations were performed using SAS 9.20 software (SAS Institute Inc., Cary, NC, USA), with the significance threshold set at p < 0.05.
Results
Sequence analysis and expression patterns of AozC
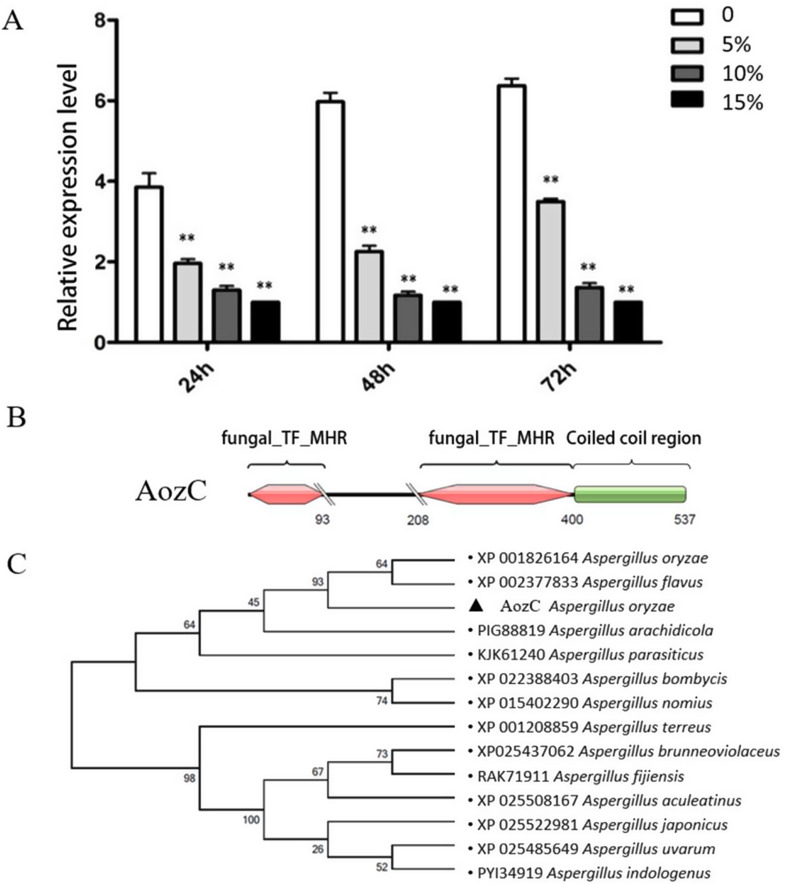
qRT-PCR was performed to analyze the expression profile of the AozC gene in A. oryzae during different growth phases and under various salt concentrations. Samples were collected at 24, 48, and 72 h, representing the adaptive, logarithmic, and stationary phases, respectively [32]. The mRNA levels were assessed under salt concentrations of 0%, 5%, 10%, and 15%. As depicted in Fig. 1A, AozC exhibited a significant upregulation from the early to the later growth phases and displayed a notable downregulation under increased salinity, suggesting a stage-specific and stress-responsive regulatory function.
Fig. 1.
Analysis of AozC gene expression and phylogenetic characterization. (A) The relative expression levels of the AozC gene across various salt concentrations (0%, 5%, 10%, and 15%) at different time points (24 h, 48 h, and 72 h) post-treatment. Each bar represents the mean expression level ± standard error (SE) derived from three independent biological replicates. The error bars indicate the variability among the replicates, and statistical significance was assessed using a one-way ANOVA with a Tukey’s post-hoc test for multiple comparisons (*p < 0.05, **p < 0.01, ***p < 0.001). (B) The conserved functional domain(s) within the AozC protein, highlighting the key structural features that define its role as a transcription factor in A. oryzae. The Zn cluster region is located at the N-terminus, which is consistent with the N-terminal position of most Zn(II)2Cys6 transcription factors. (C) The maximum likelihood method implemented in MEGA 5.0 software was utilized to construct a phylogenetic tree, comparing the AozC protein sequence with homologous sequences from other Aspergillus species. The consensus tree, depicted here, is supported by the results of 1000 bootstrap replications, indicating the reliability of the phylogenetic relationships presented
The open reading frame (ORF) of AozC, which encodes a protein comprising 178 amino acids (aa), was successfully cloned utilizing a set of specifically designed primers. Subsequent domain analysis revealed that the AozC protein features two identical conserved domains, designated as fungal-Transcription Factors (TF)-MHR. The first of these domains is situated within the region spanning from residue 1 to residue 297, as depicted in Fig. 1B. To further elucidate the evolutionary connections between the AozC protein from various Aspergillus species, a phylogenetic tree was constructed using a maximum likelihood approach based on aligned amino acid sequences. The phylogenetic analysis, illustrated in Fig. 1C, suggests a close evolutionary relationship between AozC and the protein XP_002377833 from A. flavus.
Overexpression of AozC diminishes salt tolerance and fatty acid profiles in S. Cerevisiae
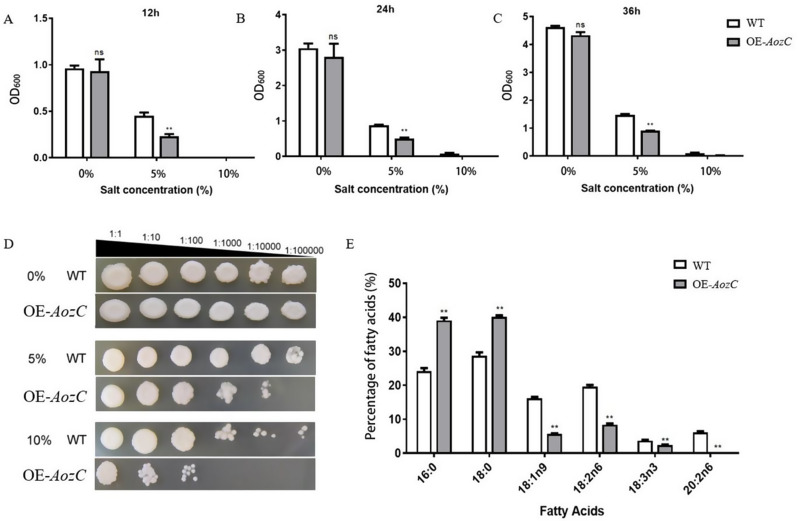
The impact of AozC overexpression on salt tolerance in S. cerevisiae was assessed by monitoring the growth kinetics of yeast strains under salt stress conditions. Optical density (OD) measurements at 12-, 24-, and 36-hours post-treatment revealed that strains overexpression AozC displayed significantly reduced growth rates compared to the wild-type when exposed to salt concentrations, as evidenced by diminished OD values (Fig. 2A-C). This indicates that the presence of AozC negatively affects the ability of yeast to tolerate saline conditions. Furthermore, a serial dilution assay confirmed the growth inhibition in transgenic strains, with a more pronounced suppression at 10% NaCl compared to the WT (Fig. 2D).
Fig. 2.
Phenotypic and molecular analysis of overexpression of AozC in S. cerevisiae under salt stress. (A-C) The optical density (OD) values of overexpression of AozC (pYES2-AozC) in S. cerevisiae strains were measured after exposure to liquid media with varying salt concentrations at 12 h (A), 24 h (B), and 36 h (C) of growth. (D) The phenotype of WT and overexpression of AozC in S. cerevisiae strains on solid media with different salt concentrations after 36 h of growth. The numerical annotations succeeding the strain identifiers, such as ‘1:1,’ ‘1:10,’ and ‘1:100’, indicate the dilution ratios of the S. cerevisiae cultures. (E) The content of fatty acids in WT and overexpression of AozC in S. cerevisiae strains was determined
To delve into the physiological effects of AozC overexpression, we analyzed the fatty acid composition of the yeast strains using gas chromatography. The results revealed a noteworthy reduction in the levels of unsaturated fatty acids (UFAs), including C18:1n9 (oleic acid), C18:2n6 (linoleic acid), C18:3n3 (alpha-linolenic acid), and C20:2n6 (eicosadienoic acid), within strains that overexpressed the AozC transcription factor. Concurrently, there was a corresponding elevation in the concentrations of saturated fatty acids, such as C16:0 (palmitic acids) and C18:0 (stearic acids) (Fig. 2E). This alteration in fatty acid profiles suggests a potential mechanism by which AozC overexpression impairs salt tolerance in yeast.
Overexpression of AozC diminishes salt tolerance and fatty acid profiles in A. oryzae.
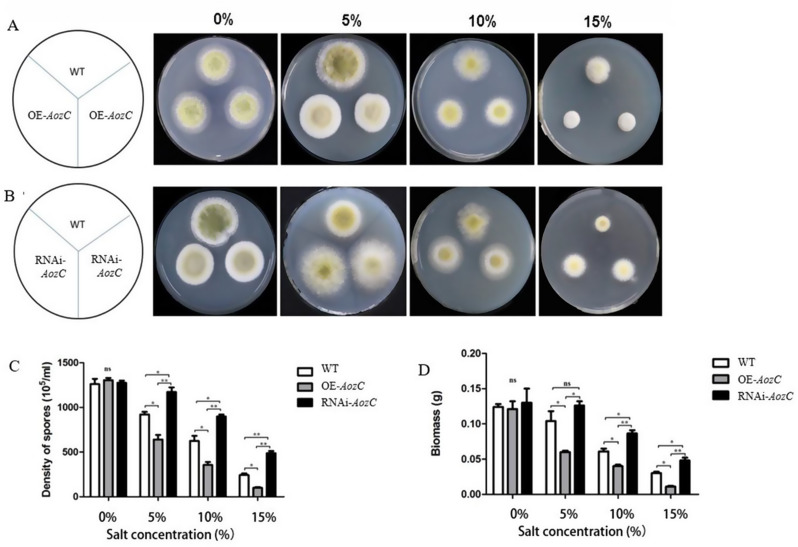
To verify the overexpression and RNAi of AozC, qRT-PCR analyses was performed. The results showed significant differences in the mRNA levels of AozC between the WT, overexpression and RNAi strains. Specifically, the overexpression strains exhibited markedly increased mRNA levels of AozC, while the RNAi strains displayed reduced mRNA levels, confirming the downregulation of the gene. Under salt stress, WT, overexpression and RNAi of AozC in A. oryzae exhibited some degree of growth inhibition, yet the extent of this inhibition varied significantly among them. Notably, as the salt concentration increased, the overexpression of AozC led to a more pronounced growth suppression compared to the WT, particularly as the salt concentration increased to 15% (Fig. 3).
Fig. 3.
Impact of salt stress on overexpressing and RNAi of AozC in A. oryzae strains. (A) The phenotype of WT and AozC transgenic strains under normal and salt treatment at 72 h post-inoculation (hpi). (B) The phenotype of WT and A. oryzae strains with RNAi targeting the AozC gene under normal and salt treatments at 72 hpi. (C) The spore density for WT, overexpressing and RNAi of AozC in A. oryzae strains were assessed in response to salt treatments. (D) The dry biomass for WT, overexpressing and RNAi of AozC in A. oryzae strains under salt stress conditions. The bars represent the average (± standard error, SE) of three biological repeats, indicating the reproducibility of the results. Asterisks indicate the presence of statistically significant differences between the treatment and the control samples based on Student’s t tests (ns No significant difference; *p < 0.05; **p < 0.01; ***p < 0.001)
In contrast, the RNAi of AozC strains, with reduced AozC expression, showed less growth inhibition under salt stress than the WT, indicating a potential adaptive advantage. Strikingly, as the salt concentration escalated to 15%, the RNAi of AozC strains not only maintained higher spore density and dry biomass but also exhibited approximately twice the spore count and biomass compared to the WT. This enhancement in growth parameters was also reflected in the mycelial diameter, which was notably larger in the RNAi of AozC strains under high salt conditions. The RNAi of AozC strains indicates that the downregulation of AozC may activate alternative pathways or mechanisms that improve the ability of strains to withstand salt stress. These contrasting phenotypes between the overexpression and RNAi of AozC strains underscore the complex regulatory role of AozC in the adaptation to salt stress (Fig. 3).
Reduction in unsaturated fatty acid content and FAD2 expression
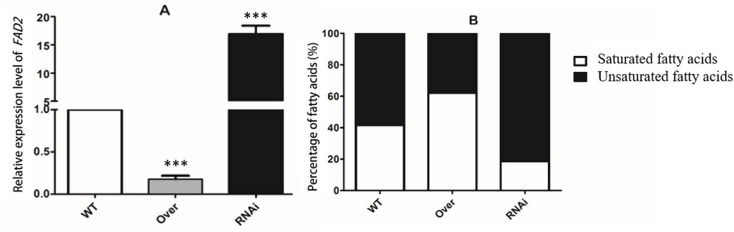
To further validate the RNA-seq findings, we conducted qRT-PCR analysis on the FAD2 gene. The qRT-PCR showing a consistent downregulation of FAD2 in the overexpression strains and an upregulation in the RNAi strains. An investigation of the fatty acid composition in the WT, overexpression and RNAi of AozC strains under salt stress conditions revealed significant findings. Specifically, the overexpression of AozC was found to be associated with a notable reduction in the content of UFA, consistent with the observed downregulation of the FAD2 gene, a key enzyme in UFA biosynthesis. In contrast, the RNAi of AozC resulted in upregulation of FAD2 expression and a corresponding increase in the intracellular content of UFAs. This finding suggests that AozC may play a critical role in the regulation of membrane fluidity and function under stress conditions by controlling the biosynthesis of UFAs. The negative regulatory relationship between AozC and FAD2 has implications for understanding the control of fatty acid composition in response to environmental stressors. The statistical analysis, represented by the error bars in the Fig. 4, confirms the reproducibility and significance of the observed differences.
Fig. 4.
Analysis of FAD2 expression and fatty acid content in WT, overexpression and RNAi of AozC in A. oryzae strains under normal conditions. (A) The relative expression levels of the FAD2 gene in WT, overexpression and RNAi of AozC in A. oryzae strains under normal growth conditions. (B) The intracellular content of saturated or unsaturated fatty acids, as a percentage of the total fatty acid content in WT, overexpression, and RNAi of AozC in A. oryzae under normal growth conditions. Asterisks indicate the presence of statistically significant differences between the treatment and the control samples based on Student’s t tests (ns No significant difference; *p < 0.05; **p < 0.01; ***p < 0.001)
Transcriptome overview
To further elucidate the molecular mechanisms underlying the function of AozC, we conducted a transcriptome analysis on the overexpression and RNAi of AozC in A. oryzae strains. The sequencing data from this analysis demonstrate a high mapping efficiency across all samples. In the WT, a total of 29,986,270 reads were generated, with 27,140,887 (90.51%) successfully mapped to the reference genome. Of these mapped reads, 27,058,859 (90.24%) were uniquely mapped, while a small fraction, 82,028 (0.27%), mapped to multiple locations (Table 1). For the overexpression of AozC strain, the total read count was higher at 41,514,850. These reads showed a mapping efficiency of 95.71%, with 39,733,790 reads aligned to the genome. The unique mapping rate was slightly higher than in the WT at 95.21%, corresponding to 39,525,625 reads. The number of multiple mappings was also higher in the overexpression strain, with 208,165 reads (0.50%) aligning to multiple sites. The RNAi of AozC strains exhibited the highest mapping efficiency among the three, with 96.38% of the total 40,872,100 reads mapped. Uniquely mapped reads were 39,247,380 (96.02%), and the multiple mappings were the lowest among the strains at 145,552 (0.36%).
Table 1.
Summary of sequencing data from overexpression and RNAi of AozC in A. oryzae strains
| Samples | Total Reads | Mapped Reads | Uniq Mapped Reads Multiple | Map Reads |
|---|---|---|---|---|
| WT | 29,986,270 |
27,140,887 (90.51%) |
27,058,859 (90.24%) | 82,028 (0.27%) |
| Over | 41,514,850 |
39,733,790 (95.71%) |
39,525,625 (95.21%) | 208,165 (0.50%) |
| RNAi | 40,872,100 |
39,392,932 (96.38%) |
39,247,380 (96.02%) | 145,552 (0.36%) |
RNAi: RNA interference
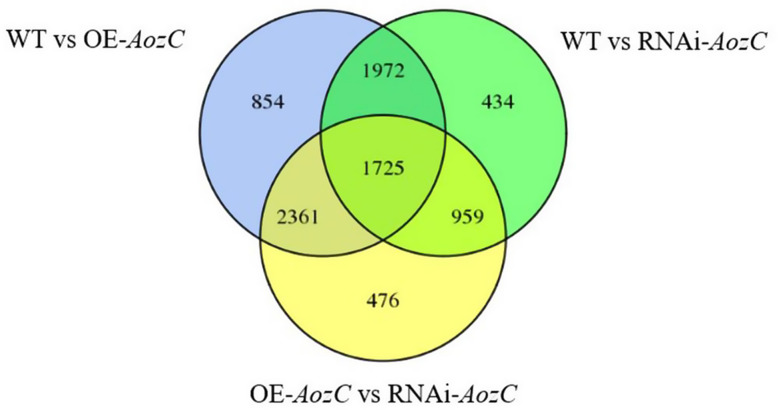
The transcriptome analysis of the WT, overexpression and RNAi of AozC in A. oryzae under normal conditions has yielded a distinct set of DEGs. The comparison between these strains was conducted to understand the impact of AozC gene expression modulation on the global transcriptional profile of the organism. In the comparison between the WT and the overexpression of AozC strains, a total of 6,912 DEGs were identified, with 3,101 genes upregulated and 3,811 genes downregulated (Table 2). This notable alteration in gene expression indicates that the overexpression of AozC has a profound impact on the cellular transcriptome, potentially altering the functional state of cell and response to environmental stimuli. When contrasting the WT with the RNAi of AozC strains, 5,090 DEGs were observed, comprising 2,553 upregulated and 2,537 downregulated genes. This indicates that the RNAi of AozC strains also results in a considerable change in gene expression, suggesting a critical role for this gene in regulating the transcriptional state of the cell. The direct comparison between the overexpression of AozC and the RNAi of AozC strains revealed 5,521 DEGs, with 3,222 genes upregulated and 2,299 genes downregulated. This comparison highlights the contrasting transcriptional responses elicited by the overexpression and RNAi of AozC. Notably, 1725 DEGs were consistently observed across all three comparisons, indicating that these genes may be central to the regulatory network influenced by AozC (Fig. 5).
Table 2.
Overview of differentially expressed genes among WT, overexpression and RNAi of AozC strains in A. Oryzae
| DEG Set | DEG Number | Mapped Reads | up-regulated | down-regulated |
|---|---|---|---|---|
| WT vs. Over | 6,912 |
27,140,887 (90.51%) |
3,101 | 3,811 |
| WT vs. RNAi | 5,090 |
39,733,790 (95.71%) |
2,553 | 2,537 |
| Over vs. RNAi | 5,521 |
39,392,932 (96.38%) |
3,222 | 2,299 |
Fig. 5.
Venn diagram displaying the distribution of differentially expressed genes among WT, overexpression and RNAi of AozC strains in A. oryzae
KEGG pathway enrichment analysis of differentially expressed genes
To delve deeper into the functions of the DEGs, we mapped them onto the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and conducted an enrichment analysis [31]. Among the DEGs between the WT and the overexpression of AozC strains, the pathways with the highest number of DEGs and the smallest q-values were identified as sphingolipid metabolism (ko00600), biosynthesis of UFAs (ko01040), mitogen-activated protein kinase (MAPK) signaling pathway (ko04011), steroid biosynthesis (ko00100), and ATP-binding cassette (ABC) transporters (ko02010) (Table 3).
Table 3.
The top 5 pathways with the highest rich factor
| Pathway | DEGs | All genes | Qvalue | Pathway ID |
|---|---|---|---|---|
| WT-vs-Over | ||||
| Sphingolipid metabolism | 16 | 26 | 0.00012 | ko00600 |
| Biosynthesis of unsaturated fatty acids | 10 | 20 | 0.000186 | ko01040 |
| MAPK signaling pathway | 25 | 65 | 0.005467 | ko04011 |
| Steroid biosynthesis | 14 | 41 | 0.008188 | ko00100 |
| ABC transporters | 5 | 12 | 0.008188 | ko02010 |
| WT-vs-RNAi | ||||
| Glutathione metabolism | 21 | 31 | 0.001257 | ko00480 |
| Biosynthesis of unsaturated fatty acids | 11 | 20 | 0.001257 | ko01040 |
| MAPK signaling pathway | 22 | 65 | 0.002514 | ko04011 |
| ABC transporters | 5 | 12 | 0.002746 | ko02010 |
| Pyruvate metabolism | 12 | 35 | 0.005013 | ko00620 |
| Over-vs-RNAi | ||||
| Biosynthesis of unsaturated fatty acids | 13 | 20 | 0.003051 | ko01040 |
| MAPK signaling pathway | 35 | 65 | 0.003734 | ko04011 |
| ABC transporters | 7 | 12 | 0.004234 | ko02010 |
| Pyruvate metabolism | 16 | 35 | 0.013524 | ko00620 |
| Glycine, serine and threonine metabolism | 24 | 53 | 0.017426 | ko00260 |
ABC: ATP-binding cassette; MAPK: Mitogen-activated protein kinase
When comparing the WT to the RNAi of AozC strains, the pathways with the lowest q-values were glutathione (GSH) metabolism (ko00480), biosynthesis of UFAs (ko01040), MAPK signaling pathway (ko04011), ABC transporters (ko02010), and pyruvate metabolism (ko00620). In the comparison between the overexpression and RNAi of AozC strains, besides the pathways mentioned above, the DEGs also participated in pyruvate metabolism (ko00620) and glycine, serine, and threonine metabolism (ko00260), with the smallest q-values observed. Notably, the top five pathways are all related to the biosynthesis of UFAs, further suggesting that the transcription factor may influence salt tolerance by regulating the content of UFAs.
Discussion
The molecular orchestration of cellular responses to environmental stress, particularly osmotic challenges like high salinity, is a complex process mediated by intricate genetic and biochemical mechanisms. In A. oryzae, a microorganism of paramount industrial value, the transcriptional modulation of stress response genes emerges as a pivotal determinant of its adaptability and productivity in fermentation processes. Our study delves into the role of the Zn(II)2Cys6 transcription factor, AozC, identified in this research as a key modulator in the salt tolerance of A. oryzae.
Through a comprehensive analysis, we have uncovered that AozC negatively influences the salt tolerance of A. oryzae by exerting regulatory control over biosynthesis pathways of UFAs. UFAs are crucial for the structural and functional integrity of biological membranes and also act as key regulators in various cellular processes [33]. They modulate the expression of inflammatory genes, including IL-1β. The strategic regulation of IL-1β and similar inflammatory mediators holds potential for therapeutic intervention and underscores the complexity of immune response regulation, which plays a role in conditions like type 2 diabetes and atherosclerosis [34, 35]. UFAs play a critical role in environmental adaptation, a phenomenon observed in deep-sea bacteria [36], and are known to bolster stress resistance in various organisms, including Komagataeibacter hansenii [37]. Research by Zhang et al. shows that UFA synthesis genes are upregulated under stress, aiding cells in coping with adverse conditions [38]. The overexpression of AozC resulted in a significant reduction of UFAs, essential for preserving membrane fluidity and functionality in saline conditions, as supported by our data and previous studies [23]. In contrast, the expression of RNAi of AozC in A. oryzae strains was associated with an increase in the production of UFAs (Fig. 4). Our results once again confirm the interplay between salt stress and the biosynthesis of unsaturated fatty acids, highlighting the necessity for a delicate balance in UFA production to ensure cellular homeostasis in response to osmotic challenges.
The identification and functional characterization of AozC have led us to classify it within a subgroup of transcription factors that are evolutionarily conserved across fungal species. However, the specific regulatory mechanisms and target genes under the purview of AozC in A. oryzae have not been previously described. The comparative analysis of gene expression patterns between WT and overexpression and RNAi of AozC strains revealed significant differences, particularly in the expression of the FAD2 gene, a well-known determinant in the biosynthesis of UFAs. As demonstrated in the oleaginous yeast Rhodotorula glutinis, overexpression of FAD2 enhances the production of lipids rich in linoleic acid, an example that parallels the potential regulatory impact of AozC on UFA levels in A. oryzae [39]. The downregulation of FAD2 in strains overexpression of AozC resulted in reduced UFA levels, which is hypothesized to impair membrane fluidity and function under saline conditions. Conversely, the upregulation of FAD2 in RNAi of AozC strains was associated with an increase in UFA production, suggesting a positive impact on salt tolerance. Our findings indicate that AozC is not only involved in the direct regulation of genes implicated in fatty acid biosynthesis but also interacts with a broader network of stress-responsive genes, thus orchestrating a coordinated cellular response to osmotic stress. While our study primarily focused on the FAD2 gene, the bioinformatics analysis of the transcriptomic data suggests that AozC may have a broader impact on the regulation of stress-response genes in A. oryzae. Further investigation is needed to fully elucidate the extent and specific identity of these stress-response regulators affected by AozC. The molecular mechanism of how AozC directly acts on the fatty acid synthesis pathway, as well as its specific interaction network in cells, remains to be further studied and verified.
In light of the KEGG pathway enrichment analysis, the modulation of the AozC transcription factor in A. oryzae has significant implications for several biological pathways. Specifically, the enrichment of DEGs in the overexpression of AozC strains within pathways such as sphingolipid metabolism, biosynthesis of UFAs, MAPK signaling, steroid biosynthesis, and ABC transporters is indicative of their critical roles in stress adaptation and survival (Table 3). The MAPK signaling pathway integral to the oxidative stress response in A. oryzae, is fundamental for transducing extracellular stress signals to the nucleus, thereby regulating stress responses and gene expression [40]. Our analysis indicates that AozC may play a role in modulating the cellular stress response by influencing these signal transduction mechanisms. Sphingolipids, enriched through the overexpression of AozC, are vital for lipid raft formation membrane microdomains critical for cellular signaling and stress adaptation [41]. The enrichment of the ABC transporter pathway is implicating AozC in the regulation of detoxification processes and resistance, as these transporters are involved in the efflux of a wide range of substances, including those that may accumulate under stress conditions, thus conferring resistance [42].
Moreover, the findings on the oxidative stress response in A. oryzae underscore the importance of UFAs in maintaining membrane integrity and fluidity. The modulation of UFAs in response to oxidative stress is a common strategy among fungi to counteract the harmful effects of reactive oxygen species [43]. The significant inhibitory impact of linoleic acid and gamma-linolenic acid on Candida krusei biofilms highlights the crucial role of PUFAs in modulating fungal responses to antifungal agents [44], which is reminiscent of the regulatory influence of AozC on UFA biosynthesis in A. oryzae. UFAs, influenced by the AozC transcription factor, can alter membrane fluidity and potentially influence lipid raft function, highlighting a role in maintaining cellular integrity under stress conditions [45]. The regulatory role of AozC in UFA biosynthesis may be central to its function as a key regulator of salt tolerance in A. oryzae. Zhu et al. have identified the Zn(II)2Cys6 transcription factor Bbotf1 in Beauveria bassiana as a key regulator linking oxidative stress responses to fatty acid assimilation. The study reveals essential function of Bbotf1 in activating fatty acid assimilation and maintaining lipid and iron homeostasis, thereby highlighting the broader role of Zn(II)2Cys6 factors in lipid metabolism and fungal adaptation to environmental stress [46]. Kakade et al. have identified a fungus specific Zn(II)2Cys6 transcription factor, ZCF32 as a crucial negative regulator in the biofilm development of Candida albicans, where it controls the expression of genes encoding adhesins, chitinases, and GPI-anchored proteins that constitute the biofilm matrix [47]. The emerging evidence from current research points to a significant and broad impact of Zn(II)2Cys6 transcription factors on fungal physiology, particularly in stress responses and metabolic regulation. Despite this, there is a limited direct association with specific pathways such as MAPK signaling documented in the existing literature, suggesting a need for further exploration in these areas.
Upon comparison of the WT to the RNAi of AozC strain, the GSH metabolism pathway was notably highlighted. This observation underscores the interconnected roles of GSH and UFAs in cellular processes that are pivotal to antioxidant defense, maintenance of membrane integrity, cellular signaling, and immune response [48]. The continued significance of the MAPK signaling and ABC transporter pathways in the overexpression of AozC versus RNAi of AozC comparison underscores the multifaceted influence of AozC on the cellular stress response network. The consistent association of the top five pathways with the biosynthesis of UFAs across all comparisons suggests the pivotal role of AozC in modulating salt tolerance by controlling the production of these critical membrane components. Future research should focus on the functional validation of the identified DEGs and their role in the adaptive response to salt stress. Additionally, the exploration of the molecular mechanisms by which AozC exerts its regulatory effects will be essential for fully understanding its biological significance and potential applications in strain engineering.
Conclusions
This study offers preliminary insights into the role of the AozC in salt tolerance of A. oryzae. Elevated AozC levels through overexpression were associated with a reduced growth rate as salt concentrations increased. In contrast, RNAi of AozC led to significant improvements in spore density and dry biomass at 15% salt concentration. Furthermore, overexpression of AozC caused a decrease in FAD2 gene expression, resulting in lowered unsaturated fatty acid production, which is crucial for maintaining cell membrane fluidity and integrity under saline conditions. These results suggest that the AozC transcription factor acts as a negative regulator in salt tolerance. Transcriptome analysis revealed DEGs involved in fatty acid biosynthesis and key stress response pathways. By revealing the role of AozC in regulating UFAs biosynthesis and stress response pathways, our study suggests potential strategies for improving its salt tolerance through genetic engineering. These findings could serve as a model for improving the salt tolerance of other industrially relevant fungi, thereby broadening their application in biotechnology.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
W.Y.: Conceptualization, software, writing—original draft preparation, writing— review and editing, validation. and methodology. Z.Z.: validation. Y.Z.: investigation. Y.T.: formal analysis and visualization. B.H.: data curation, supervision, project administration, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (32260017), Jiangxi Provincial Natural Science Foundation (20242BAB25334) and Youth Talent Support Program of Jiangxi Science & Technology Normal University (2022QNBJRC005).
Data availability
The sequencing data in this paper are available at NCBI/SRA database (https://www.ncbi.nlm.nih.gov/sra) under Bioproject Accession PRJNA1135475.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yayi Tu, Email: tuyayi@126.com.
Bin He, Email: hebin.li@foxmail.com.
References
- 1.Lv G, Xu Y, Tu Y, Cheng X, Zeng B, Huang J, He B. Effects of Nitrogen and Phosphorus limitation on fatty acid contents in aspergillus oryzae. Front Microbiol. 2021;12:739569–82. 10.3389/fmicb.2021.739569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daba GM, Mostafa FA, Elkhateeb WA. The ancient koji mold (aspergillus oryzae) as a modern biotechnological tool. Bioresources Bioprocess. 2021;8:52–62. 10.1186/s40643-021-00408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu T, Zhang C, Qin Z, Fan L, Jiang L, Zhao L. A novel GH Family 20 β-N-acetylhexosaminidase with both chitosanase and chitinase activity from aspergillus oryzae. Front Mol Biosci. 2021;8:684086–95. 10.3389/fmolb.2021.684086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang H, Song C, Liu C, Wang P. Synthetic Biology Tools for Engineering Aspergillus oryzae. J Fungi (Basel). 2024;10:34–46. 10.3390/jof10010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Li J, Shin HD, Du G, Chen J, Liu L. Metabolic engineering of aspergillus oryzae for efficient production of l-malate directly from corn starch. J Biotechnol. 2017;262:40–6. 10.1016/j.jbiotec.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 6.Sun Z, Wu Y, Long S, Feng S, Jia X, Hu Y, Ma M, Liu J, Zeng B. Aspergillus oryzae as a cell factory: research and applications in Industrial Production. J Fungi. 2024;10:248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M, Sun Y, Zhu L, Li L, Zhao Y. Study on the Skincare effects of Red Rice fermented by aspergillus oryzae in Vitro. Molecules. 2024;29:2066–73. 10.3390/molecules29092066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tegelaar M, Bleichrodt RJ, Nitsche B, Ram AFJ, Wösten HAB. Subpopulations of hyphae secrete proteins or resist heat stress in aspergillus oryzae colonies. Environ Microbiol. 2020;22:447–55. 10.1111/1462-2920.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao G, Liu C, Li S, Wang X, Yao Y. Exploring the flavor formation mechanism under osmotic conditions during soy sauce fermentation in aspergillus oryzae by proteomic analysis. Food Funct. 2020;11:640–8. 10.1039/C9FO02314C. [DOI] [PubMed] [Google Scholar]
- 10.Agrawal S, Chavan P, Dufossé L. Hidden treasure: Halophilic Fungi as a repository of bioactive lead compounds. J Fungi. 2024;10:290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang Y, Zhang M, Wang M, Zhang W, Qiao C, et al. Freshwater Cyanobacterium Synechococcus elongatus PCC 7942 Adapts to an environment with salt stress via Ion-Induced Enzymatic Balance of Compatible Solutes. Appl Environ Microbiol. 2020;86:e02904–02919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dihazi H, Kessler R, Eschrich K. High osmolarity glycerol (HOG) pathway-induced phosphorylation and activation of 6-phosphofructo-2-kinase are essential for glycerol accumulation and yeast cell proliferation under hyperosmotic stress. J Biol Chem. 2004;279:23961–8. 10.1074/jbc.m312974200. [DOI] [PubMed] [Google Scholar]
- 13.Wang D, Zhang M, Huang J, Zhou R, Jin Y, Wu C. Zygosaccharomyces rouxii combats salt stress by maintaining cell membrane structure and functionality. J Microbiol Biotechnol. 2020;30:62–70. 10.4014/jmb.1904.04006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdel-Aziz MM, Emam TM, Raafat MM. Hindering of cariogenic Streptococcus mutans biofilm by fatty acid array derived from an endophytic Arthrographis kalrae strain. Biomolecules. 2020;10:811–22. 10.3390/biom10050811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu X, Wang Y, Wang X, Wang W. Exogenous regulators enhance the yield and stress resistance of Chlamydospores of the Biocontrol Agent Trichoderma Harzianum T4. J Fungi (Basel). 2022;8:1017–26. 10.3390/jof8101017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mostofian B, Zhuang T, Cheng X, Nickels JD. Branched-chain fatty acid content modulates structure, fluidity, and phase in Model Microbial cell membranes. J Phys Chem B. 2019;123:5814–21. 10.1021/acs.jpcb.9b04326. [DOI] [PubMed] [Google Scholar]
- 17.Bajerski F, Wagner D, Mangelsdorf K. Cell membrane fatty acid composition of Chryseobacterium Frigidisoli PB4T, isolated from Antarctic Glacier Forefield Soils, in response to changing temperature and pH conditions. Front Microbiol. 2017;8:677–84. 10.3389/fmicb.2017.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah AM, Yang W, Mohamed H, Zhang Y, Song Y. Microbes: a hidden treasure of Polyunsaturated fatty acids. Front Nutr. 2022;9. 10.3389/fnut.2022.827837. [DOI] [PMC free article] [PubMed]
- 19.Wang W, Zhang K, Lin C, Zhao S, Guan J, Zhou W, Ru X, Cong H, Yang Q. Influence of Cmr1 in the regulation of antioxidant function melanin biosynthesis in Aureobasidium pullulans. Foods. 2023;12:2135–45. 10.3390/foods12112135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C, Huang H, Deng W, Li T. Genome-wide analysis of the zn(II)2Cys6 zinc cluster-encoding Gene Family in Tolypocladium guangdongense and its light-Induced expression. Genes. 2019;10:179–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bansal S, Mallikarjuna MG, Balamurugan A, Nayaka SC, Prakash G. Composition and Codon Usage Pattern Results in divergence of the Zinc Binuclear Cluster (Zn(II)2Cys6) sequences among Ascomycetes Plant Pathogenic Fungi. J Fungi. 2022;8:1134–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin Y, Zhang H, Zhang Y, Hu C, Sun X, Liu W, Li S. Fungal zn(II)2Cys6Transcription factor ADS-1 regulates Drug Efflux and Ergosterol Metabolism under antifungal azole stress. Antimicrob Agents Chemother. 2021;65:1316–20. 10.1128/aac.01316-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He B, Ma L, Hu Z, Li H, Ai M, Long C, Zeng B. Deep sequencing analysis of transcriptomes in aspergillus oryzae in response to salinity stress. Appl Microbiol Biotechnol. 2017;10:1007–17. 10.1007/s00253-017-8603-z. [DOI] [PubMed] [Google Scholar]
- 24.Imanaka H, Tanaka S, Feng B, Imamura K, Nakanishi K. Cultivation characteristics and gene expression profiles of aspergillus oryzae by membrane-surface liquid culture, shaking-flask culture, and agar-plate culture. J Biosci Bioeng. 2010;109:267–73. 10.1016/j.jbiosc.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 25.He B, Tu Y, Hu Z, Ma L, Dai J, Cheng X, Li H, Liu L, Zeng B. Genome-wide identification and expression profile analysis of the HOG gene family in aspergillus oryzae. World J Microbiol Biotechnol. 2018;34:35–48. 10.1007/s11274-018-2419-6. [DOI] [PubMed] [Google Scholar]
- 26.Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009;19:271–3. 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
- 27.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–8. 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, Tong Y, Shankar K, Baumgardner JN, Kang J, Badeaux J, Badger TM, Ronis MJJ. Lipid fatty acid Profile analyses in liver and serum in rats with nonalcoholic steatohepatitis using Improved Gas Chromatography – Mass Spectrometry Methodology. J Agric Food Chem. 2010;59:747–54. 10.1021/jf1038426. [DOI] [PubMed] [Google Scholar]
- 29.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–U354. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love MI, Huber W, Anders S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed]
- 31.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–4. 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He B, Hu Z, Ma L, Li H, Ai M, Han J, Zeng B. Transcriptome analysis of different growth stages of aspergillus oryzae reveals dynamic changes of distinct classes of genes during growth. BMC Microbiol. 2018;18:12–22. 10.1186/s12866-018-1158-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herndon JL, Peters RE, Hofer RN, Simmons TB, Symes SJ, Giles DK. Exogenous polyunsaturated fatty acids (PUFAs) promote changes in growth, phospholipid composition, membrane permeability and virulence phenotypes in Escherichia coli. BMC Microbiol. 2020;20:305–12. 10.1186/s12866-020-01988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian H, Yu H, Lin Y, Li Y, Xu W, Chen Y, Liu G, Xie L. Association between FADS Gene expression and polyunsaturated fatty acids in breast milk. Nutrients. 2022;14:457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monfort-Pires M, Crisma AR, Bordin S, Ferreira SRG. Greater expression of postprandial inflammatory genes in humans after intervention with saturated when compared to unsaturated fatty acids. Eur J Nutr. 2018;57:2887–95. 10.1007/s00394-017-1559-z. [DOI] [PubMed] [Google Scholar]
- 36.Allen EE, Bartlett DH. Structure and regulation of the omega-3 polyunsaturated fatty acid synthase genes from the deep-sea bacterium Photobacterium profundum strain SS9. Microbiology. 2002;148:1903–13. 10.1099/00221287-148-6-1903. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Yan P, Lei Q, Li B, Sun Y, Li S, Lei H, Xie N. Metabolic adaptability shifts of cell membrane fatty acids of Komagataeibacter Hansenii HDM1-3 improve acid stress resistance and survival in acidic environments. J Ind Microbiol Biotechnol. 2019;46:1491–503. 10.1007/s10295-019-02225-y. [DOI] [PubMed] [Google Scholar]
- 38.Zhang M, Yu Q, Liang C, Liu Z, Zhang B, Li M. Graphene oxide induces plasma membrane damage, reactive oxygen species accumulation and fatty acid profiles change in Pichia pastoris. Ecotoxicol Environ Saf. 2016;132:372–8. 10.1016/j.ecoenv.2016.06.031. [DOI] [PubMed] [Google Scholar]
- 39.Wu C-C, Ohashi T, Kajiura H, Sato Y, Misaki R, Honda K, Limtong S, Fujiyama K. Functional characterization and overexpression of ∆12-desaturase in the oleaginous yeast Rhodotorula toruloides for production of linoleic acid-rich lipids. J Biosci Bioeng. 2021;131:631–9. 10.1016/j.jbiosc.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Luo Z, Huang W, Wang G, Sun H, Chen X, Luo P, Liu J, Hu C, Li H, Shu H. Identification and characterization of p38MAPK in response to acute cold stress in the gill of Pacific white shrimp (Litopenaeus vannamei). Aquaculture Rep. 2020;17:100365–72. 10.1016/j.aqrep.2020.100365. [Google Scholar]
- 41.Bieberich E. Sphingolipids and lipid rafts: novel concepts and methods of analysis. Chem Phys Lipids. 2018;216:114–31. 10.1016/j.chemphyslip.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris A, Wagner M, Du D, Raschka S, Nentwig L-M, Gohlke H, Smits SHJ, Luisi BF, Schmitt L. Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5. Nat Commun. 2021;12:5254–66. 10.1038/s41467-021-25574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao H, Tu Y, Wang Y, Jiang C, Ma L, Hu Z, Wang J, Zeng B, He B. Oxidative stress response of Aspergillus Oryzae Induced by Hydrogen Peroxide and Menadione Sodium Bisulfite. Microorganisms. 2019;7:225–36. 10.3390/microorganisms7080225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamiu AT, Albertyn J, Sebolai O, Gcilitshana O, Pohl CH. Inhibitory effect of polyunsaturated fatty acids alone or in combination with fluconazole on Candida krusei biofilms in vitro and in Caenorhabditis elegans. Med Mycol. 2021;59:1225–37. 10.1093/mmy/myab055. [DOI] [PubMed] [Google Scholar]
- 45.Díaz M, Pereda de Pablo D, Valdés-Baizabal C, Santos G, Marin R. Molecular and biophysical features of hippocampal lipid rafts aging are modified by dietary n-3 long-chain polyunsaturated fatty acids. Aging Cell. 2023;22:e13867–13877. 10.1111/acel.13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu C, Sun J, Tian F, Tian X, Liu Q, Pan Y, Zhang Y, Luo Z. The Bbotf1 zn(II)2Cys6 transcription factor contributes to antioxidant response, fatty acid assimilation, peroxisome proliferation and infection cycles in insect pathogenic fungus Beauveria Bassiana. J Invertebr Pathol. 2024;204:108083–94. 10.1016/j.jip.2024.108083. [DOI] [PubMed] [Google Scholar]
- 47.Kakade P, Sadhale P, Sanyal K, Nagaraja V. ZCF32, a fungus specific zn(II)2 Cys6 transcription factor, is a repressor of the biofilm development in the human pathogen Candida albicans. Sci Rep. 2016;6:31124–36. 10.1038/srep31124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Usman K, Souchelnytskyi S, Al-Ghouti MA, Zouari N, Abu-Dieyeh MH. Proteomic analysis of T. Qataranse exposed to lead (pb) stress reveal new proteins with potential roles in pb tolerance and detoxification mechanism. Front Plant Sci. 2022;13:1009756–71. 10.3389/fpls.2022.1009756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data in this paper are available at NCBI/SRA database (https://www.ncbi.nlm.nih.gov/sra) under Bioproject Accession PRJNA1135475.