Abstract
Overlapping of genes, especially in an anti-parallel fashion, is quite rare in eukaryotic genomes. We have found a rare instance of exon overlapping involving CHRNE and MINK gene loci on chromosome 17 in humans. CHRNE codes for the ɛ subunit of the nicotinic acetylcholine receptor (AChRɛ) whereas MINK encodes a serine/threonine kinase belonging to the GCK family. To elucidate the evolutionary trail of this gene overlapping event, we examined the genomes of a number of primates and found that mutations in the polyadenylation signal of the CHRNE gene in early hominoids led to the overlap. Upon extending this analysis to genomes of other orders of placental mammals, we observed that the overlapping occurred at least three times independently during the course of mammalian evolution. Because CHRNE and MINK are differentially expressed, the potentially hazardous mutations responsible for the exon overlap seem to have escaped evolutionary pressures by differential temporo-spatial expression of the two genes.
INTRODUCTION
Gene loci rarely overlap in eukaryotic genomes with only a few exceptions to this rule. One is a gene-within-a-gene, in which a transcriptional unit is encoded within an extremely long intron. For example, two genes on chromosome 22 have been described to occur within other genes (1). Alternative frame usage of exons is another mechanism by which genes may overlap. In a recent report, it was found that a single exon gave rise to two different protein products, the XL domain of XLαs protein and ALEX, which actually interact with each other (2).
Several examples of naturally occurring antisense RNAs have been reported and they may serve to regulate expression of the protein-coding transcripts (3). However, the overlap in these cases is basically between a protein-coding gene and a non-protein-coding one, and not between genes that code for functional proteins.
Anti-parallel overlapping of exons of functional protein-coding genes is possible, although very few cases have been reported in mammalian genomes. For example, a gene locus on human chromosome 17 with overlapping exons on opposite strands gives rise to two different gene products, ear-1 and ear-7, which belong to the c-erbA family of proto-oncogenes (4). Similarly, the murine genes for the EP1 prostanoid receptor and PNK protein kinase overlap in a tail-to-tail fashion for 280 bp in the mouse genome (5).
In this report, we describe an evolutionarily conserved overlapping of exons involving two functionally distinct genes, MINK and CHRNE, in several mammalian genomes. MINK is a serine/threonine kinase that functions as an upstream activator of the JNK protein kinase pathway (6,7). CHRNE codes for the ɛ subunit of the nicotinic acetylcholine receptor (AChRɛ), which is expressed in the post-synaptic membrane at mature neuro-muscular junctions (8–10). In this report, based on an extensive examination of several mammalian genomes, we present the first evidence for the evolution of the occurrence of exon overlapping. We show data which suggest that sequential mutations in the polyadenylation signal of the AChRɛ gene resulted in the overlapping of its exons with that of the MINK gene.
MATERIALS AND METHODS
Genomic sources
The sources for genomic DNA (along with the accession nos for DDBJ/EMBL/GenBank) are as follows: mouse (Mus musclus, AB070514), brain of C57black6 strain; human (Homo sapiens, AB070507), embryonic kidney 293 cells; dog (Canis familiaris, AB070516), testis kindly provided by Tokyo Lovely Animal Hospital; ring-tailed lemur (Lemur catta, AB070513) and red-faced macaque (Macaca arctoides, AB070511), peripheral blood; white-handed gibbon (Hylobates lar, AB070510), orangutan (Pongo pygmaeus, AB070509) and bonobo (Pan paniscus, AB070508), Epstein–Barr virus-transfected lymphocytes as previously described (11). Skeletal muscle from rabbit (Oryctolagus cuniculus, AB070515), pig (Sus scrofa, AB070517), minke whale (Balaenoptera acutorostrata, AB070518), horse (Equus caballus, AB070519) and cow (Bos taurus, AB070520) were purchased from local markets. For all animals, DNA was extracted using the phenol/chloroform extraction method (12).
Polymerase chain reaction (PCR)
DNA fragments containing the MINK/CHRNE junctions were amplified by PCR from the genomic DNA sources indicated above. Degenerate primers were designed based on sequence homology between mouse and human genomes [positions of the primers with respect to the human genome BAC clone hRPK.17_H_5 (AC005973) are indicated in parentheses]. For forward primers, the following were generated: F1, 5′-CTGTGTGGATGCYGTGAACTT-3′ (nt 30 475–30 495); F2, 5′-GTGGCYGAGAGCACRAGAGA-3′ (nt 30 497–30 516); F3, 5′-GCGYATGGGGAAKGCCCTKGACAA-3′ (nt 30 552–30 685); F4, 5′-TCATCTTCCTYGGGGSYTACTTCAA-3′ (nt 30 733–300 757); F5, 5′-TGGTGGRAGCTGGTTGAATTGTCTTTAT-3′ (nt 31 475–31 501); and F6, 5′-CTTGCTGCTCACACTATATACAGATGC-3′ (nt 31 566–31 592). For reverse primers, the following were generated: R1, 5′-GTGTTTTTTGCCTCWGTCCGCT-3′ (nt 32 332–32 311); R2, 5′-TCTGGRGGMAGCAGCCAAGTTTACTT-3′ (nt 32 311–32 286); R3, 5′-TGACTCTGAACCGTAACTGCATCATGA-3′ (nt 32 283–32 257); R4, 5′-CCCTTMCCAGGAATTGAGTGG-3′ (nt 32 005–31 984); R5, 5′-TCCCCCCTTGAATGTACCAGA-3′ (nt 31 860–31 840); R6, 5′-ATATAGTGTGAGCAGCAAGTARCCCT-3′ (nt 31 584–301 559); and R7, 5′-TAAAGACAATTCAACCAGCTYCCACC-3′ (nt 31 501–31 476), where K stands for G and T, M for A and C, R for A and G, Y for C and T, and W for A and T. With several different combinations of the forward and reverse primers, DNA fragment PCR was performed using Takara ExTaq DNA polymerase (Takara, Otsu, Japan) according to the published method (13). The longest fragments were obtained by F1–R2 primer combination since R1 primer that should amplify the 5′ end of the MINK gene failed to amplify the right product. Thus these fragments contain the complete last exon of the CHRNE gene and the last exon of the MINK gene except for 21 bp at the 5′ end. The PCR fragments were cloned into pBlueScript SK– vector and several independent clones were sequenced.
RESULTS AND DISCUSSION
MINK and CHRNE gene loci overlap in the human genome
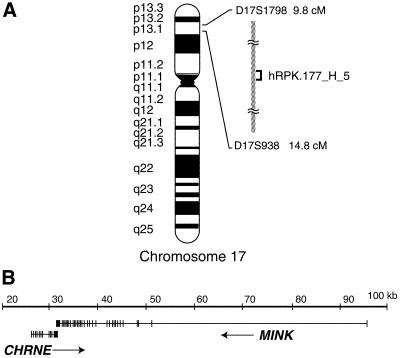
We have recently cloned and characterised a novel mammalian serine/threonine kinase, MINK (6,7). Subsequently, we mapped the human MINK gene in silico using sequence derived from a BAC clone (hRPK.177_H_5, GenBank accession no. AC005973) that contained the whole genomic region encoding MINK. Combining the information from genetic markers within and near this BAC clone, we mapped it to a region between 9.8 and 14.8 cM, which corresponds to Ch17p13.1 (Fig. 1A). The MINK gene spans >60 kb and consists of 32 exons (Fig. 1B). All the exon/intron boundaries matched the consensus sequence for splice sites (14). This BAC clone also contained the CHRNE locus encoding the AChRɛ subunit (8–10). To examine the location of CHRNE and MINK in detail, we compared the sequences of the genomic regions, CHRNE and MINK mRNAs, and all the available EST clones derived from the two genes. We found that the two genes overlapped for 280 bp in their terminal exons in a tail-to-tail manner (Fig. 1B). The last exon of CHRNE codes for the C-terminus of AChRɛ as well as its 3′ untranslated region (UTR). The overlapping occurs within the 3′ UTRs of CHRNE and MINK genes (Fig. 2A). These overlapping regions contain a conserved polyadenylation signal, AAUAAA, for MINK, and a second-preferred signal, AUUAAA, for CHRNE (at the potential polyadenylation site 3; Fig. 2A) (15,16). Since all of the EST clones that are derived from CHRNE contain the AUUAAA sequence, this is likely to be used as a polyadenylation signal.
Figure 1.
Gene structures of MINK and CHRNE in humans. (A) A schematic showing the location of the BAC clone, hRPK.177_H_5. It is located at Ch17 p13.1, between two markers, D17S1798 at 9.8 cM and D17S938 at 14.8 cM. (B) Gene organisation of MINK and CHRNE loci in the BAC clone shown in (A). Vertical bars represent exons. Arrows indicate the direction of transcription. CHRNE spans ∼6 kb and consists of 11 exons. MINK spans >60 kb and consists of 32 exons. Their last exons exhibit an anti-parallel overlap for 280 bp.
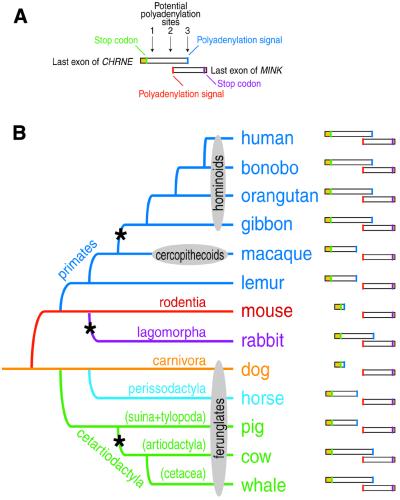
Figure 2.
Structure of MINK/CHRNE junctions in various mammalian species and their evolutionary relationship. (A) Structure of the MINK/CHRNE junction in the human genome. Last exons of human MINK and CHRNE genes are shown as rectangles. Coding regions are yellow and 3′ UTRs are white. The stop codon and the functional polyadenylation signal are shown as green and blue vertical stripes, respectively (CHRNE), or as purple and red vertical stripes, respectively (MINK). The three sites that can potentially give rise to polyadenylation signals are indicated by arrows. (B) A schematic showing the MINK/CHRNE junction in various placental mammals. The structure of the MINK/CHRNE junction is represented as in (A). An evolutionary tree based on molecular analyses is shown on the left (19–21,27). Each branch is indicated with the name of the order. Other classifications used in the text are shown with ovals. Asterisks represent occurrence of a gene overlapping event within a particular branch of the evolutionary tree.
MINK and CHRNE gene loci do not overlap in the mouse genome
We then examined the location of CHRNE and MINK loci in the mouse genome. The mouse genomic sequence for CHRNE is present in the public databases including the last exon of the mouse MINK gene (6,7). We found that CHRNE and MINK loci are located in close proximity but do not overlap (Fig. 2B). We verified this result by comparing genomic DNA sequences of the CHRNE/MINK junction and cDNA sequences of CHNRE and MINK transcripts (accession nos X55718 and AB035697, respectively) (6,17). While sequences of the 3′ UTR of MINK are conserved between human and mouse, those of CHRNE vary greatly (Table 1). The human CHRNE gene has a 3′ UTR of 970 bp that overlaps with the opposite strand of MINK, whereas its mouse counterpart contains a short 3′ UTR of only 80 bp due to use of an upstream alternative polyadenylation signal (corresponding to the potential polyadenylation site 1) (Fig. 2). MINK, in contrast, has an equally long 3′ UTR in both species.
Table 1. Degree of nucleotide sequence conservation in various CHRNE/MINK junctional regions.
| Sequence identity to human (%) | ||||
|---|---|---|---|---|
| CHRNE CDSa | CHRNE 3′ UTR | MINK 3′ UTR | MINK CDSa | |
| Bonobo | 99 | 97 | 98 | 98 |
| Orangutan | 97 | 90 | 96 | 97 |
| Gibbon | 97 | 93 | 97 | 98 |
| Macaque | 98 | 91 | 97 | 98 |
| Lemur | 92 | 73 | 93 | 98 |
| Mouse | 82 | 49 | 82 | 95 |
| Rabbit | 88 | 38 | 85 | 97 |
| Dog | 81 | 47 | 80 | 95 |
| Horse | 88 | 70 | 91 | 95 |
| Pig | 86 | 63 | 86 | 97 |
| Cow | 88 | 63 | 90 | 97 |
| Whale | 87 | 66 | 91 | 97 |
Sequence identity to the corresponding regions of the human CHRNE/MINK junction is indicated in each case. Since the length of the 3′ UTR of the CHRNE gene varies greatly among species, the region between the stop codon of the CHRNE gene and the potential polyadenylation site 1 was used for comparison.
aCDS, coding sequence.
Overlapping of MINK and CHRNE gene loci during the course of primate evolution
To investigate whether the overlapping of these two genes at their 3′ ends has any evolutionary basis, we examined the structure of the MINK/CHRNE junction in several primate genomes. We extracted genomic DNA from several primate species and obtained the DNA fragments containing the MINK/CHRNE junctions by PCR. Usually, the polyadenylation occurs at the CA site that is located just downstream of the polyadenylation signal. In addition, GT-rich sequence located downstream of the polyadenylation signal confirms the terminal position (15,16).
In lemur (prosimian) and macaque (cercopithecoid) genomes, the genes do not overlap but are barely separated by several base pairs because of the use of another alternative upstream polyadenylation signal (corresponding to the potential polyadenylation site 2) as shown in Figure 2. All hominoid species have an overlapping-type MINK/CHRNE junction (Fig. 2B). Thus the genes are likely to have overlapped after the cercopithecoid/hominoid split due to changes in the polyadenylation signal and subsequent utilisation of the polyadenylation signal located in the terminal exon of the MINK gene on the opposite strand (the potential polyadenylation site 3) (Fig. 2).
Occurrence of MINK and CHRNE genes overlapping in other mammalian genomes
We extended this analysis further to other orders of placental mammals. The most interesting aspect was found among the ferunglates (Fig. 2B). Historically, pigs belong to the order artiodactyla, together with cows, and are considered to be more distantly related to the order cetacea, which includes whales. Recent molecular evolutionary analysis suggests that pigs are rather independent and should form a new order consisting of suina and tylopoda, and that whales are more related to cows (18). Other groups have also found the same branching pattern and suggested the order cetartiodactyla to combine cetacea and artiodactyla (19–21). Our analysis also supports this newer hypothesis: MINK and CHRNE do not overlap in the pig genome, while they do so in whale and cow genomes in a similar fashion (Fig. 2B). We noticed that gene overlapping also occurs in the rabbit genome (Fig. 2B). Thus a gene overlapping event involving MINK and CHRNE occurred at least three times independently during the course of mammalian evolution (Fig. 2B).
Detailed analysis on the formation of the overlap
To examine further how three MINK/CHRNE overlapping events occurred in the course of mammalian evolution, we tracked changes in the polyadenylation signal sequences in the 3′ UTR of the CHRNE gene. There are three sites that can potentially give rise to a polyadenylation signal for the CHRNE gene: site 1, located 60–120 bp downstream of the stop codon of the coding sequence of the CHRNE gene; site 2, located 40 bp upstream of the polyadenylation signal of the MINK gene; and site 3, located 230 bp downstream of the polyadenylation signal of the MINK gene on the opposite strand of the terminal exon of MINK (Fig. 2A). The nucleotide sequences in site 3 that can function as a polyadenylation signal are completely conserved among all the species examined, while those in sites 1 and 2 are moderately conserved. Site 1 contains polyadenylation signal sequences in the mouse and dog genomes, as does site 2 in lemur, macaque, horse and pig genomes. Thus the MINK and CHRNE gene loci do not overlap in the genomes of these animals. In the other animals, there was no polyadenylation signal in the region corresponding to sites 1 and 2, and it was observed that site 3 is utilised in these cases. Thus hominoids, whale, cow and rabbit have the overlapping MINK/CHRNE loci.
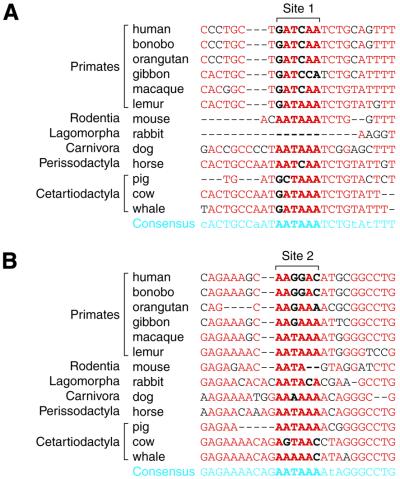
There are two major possibilities with respect to the changes in the polyadenylation signals. The two genes might have originally been separated but the polyadenylation signals in sites 1 and 2 disrupted, resulting in gene overlapping. Alternatively, the two genes might have originally been in an overlapping state but generation of a polyadenylation signal in either site 1 or 2 resulted in gene separation. To find out which case is more likely, we examined the nucleotide sequences at these two sites. We aligned nucleotide sequences around sites 1 and 2 in the genomes of all the animals examined. So that each of six orders examined could contribute equally in the estimation of the consensus nucleotide sequences, the contribution of each species in primates and cetartiodactyla was set to one-sixth and one-third, respectively. The nucleotide that appeared most frequently in each position was selected to generate the consensus sequences (Fig. 3). In both sites, the nucleotide sequence AATAAA was extracted as the consensus sequence. This means that the ancestral sequences in sites 1 and 2 most likely comprise polyadenylation signal sequences. Albeit we cannot exclude the possibility of polyadenylation signal generation in the course of mammalian evolution, it is more probable that the ancestral polyadenylation signal sequences in site 1 and/or 2 have collapsed and resulted in gene overlapping in some mammalian species.
Figure 3.
Comparison of nucleotide sequences around the potential polyadenylation sites for the CHRNE gene. (A) Nucleotide sequences around the potential polyadenylation site 1 for the CHRNE gene are aligned. Shown in blue are the consensus sequences where upper case represents >50% identities and lower case represents <49% identities. Sequences in site 1 are in bold type. Nucleotide sequences that match to the consensus sequences are shown in red while those that do not are in black. (B) Nucleotide sequences around the potential polyadenylation site 2 for the CHRNE gene are aligned in the same manner as in (A).
Provided that the two sites contained the ancestral polyadenylation signal sequences, the three independent gene overlapping events pointed out above are likely to have occurred in distinct ways, i.e. the ancestral polyadenylation signal sequences that led to gene overlapping got disrupted in different ways. First, in the rabbit genome, the entire site 1 was deleted and the fifth position in site 2 mutated to C. Secondly, in hominoid genomes, the first and fourth positions in site 1 changed to G and C, respectively, and the third position in site 2 mutated to G. Finally, in cow and whale genomes, the first position in site 1 changed to G and the sixth position in site 2 mutated to C.
The driving force for the overlap may be attributed to a faster evolutionary pace of the 3′ UTR of CHRNE (Table 1), which results in disruption of its upstream polyadenylation signal sequences. In contrast, the 3′ UTR of the MINK gene is extremely conserved. Its degree of conservation is as high as that of the coding sequence of the CHRNE gene (Table 1). This high degree of conservation in the 3′ UTR of the MINK gene lets a potential polyadenylation signal on the opposite strand be also conserved. Therefore, even when the upstream polyadenylation signal of the CHRNE gene is disrupted an alternative signal is provided by the conserved opposite strand of the MINK gene located downstream.
Theoretically, this exon overlapping may result in the formation of a heteroduplex RNA species from the transcripts of these two genes, which could lead to their degradation (22–25). In extreme cases, formation of antiparallel heteroduplex RNA may completely block the expression of both genes, as in the case of double strand RNA interference (26). However, such situations are avoided due to the fact that the MINK transcript is expressed mainly in post-natal developing brain and only at low levels in skeletal muscle (6), whereas the CHRNE transcript is solely expressed in mature skeletal muscle (8–10). In this context, the MINK/CHRNE exon overlap provides a unique example in which a potentially hazardous exon overlapping event has managed to escape evolutionary pressures by a temporo-spatial differential expression.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed at: Division of Food Engineering, National Food Research Institute, 2-1-12, Kannondai, Tsukuba, Ibaraki 305-8642, Japan. Tel: +81 298 38 8030; Fax: +81 298 38 8122; Email: dan@nfri.affrc.go.jp AB070507–AB070511, AB070513–AB070520
REFERENCES
- 1.Dunham I., Shimizu,N., Roe,B.A., Chissoe,S., Hunt,A.R., Collins,J.E., Bruskiewich,R., Beare,D.M., Clamp,M., Smink,L.J. et al. (1999) The DNA sequence of human chromosome 22. Nature, 402, 489–495. [DOI] [PubMed] [Google Scholar]
- 2.Klemke M., Kehlenbach,R.H. and Huttner,W.B. (2001) Two overlapping reading frames in a single exon encode interacting proteins—a novel way of gene usage. EMBO J., 20, 3849–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vanhee-Brossollet C. and Vaquero,C. (1998) Do natural antisense transcripts make sense in eukaryotes? Gene, 211, 1–9. [DOI] [PubMed] [Google Scholar]
- 4.Miyajima N., Horiuchi,R., Shibuya,Y., Fukushige,S., Matsubara,K., Toyoshima,K. and Yamamoto,T. (1989) Two erbA homologs encoding proteins with different T3 binding capacities are transcribed from opposite DNA strands of the same genetic locus. Cell, 57, 31–39. [DOI] [PubMed] [Google Scholar]
- 5.Batshake B. and Sundelin,J. (1996) The mouse genes for the EP1 prostanoid receptor and the PKN protein kinase overlap. Biochem. Biophys. Res. Commun., 227, 70–76. [DOI] [PubMed] [Google Scholar]
- 6.Dan I., Watanabe,N.M., Kobayashi,T., Fukagaya,Y., Kajikawa,E., Kimura,W.K., Nakashima,T.M., Matsumoto,K., Ninomiya-Tsuji,J. and Kusumi,A. (2000) Molecular cloning of MINK, a novel member of mammalian GCK family kinases, which is up-regulated during postnatal mouse cerebral development. FEBS Lett., 469, 19–23. [DOI] [PubMed] [Google Scholar]
- 7.Dan I., Watanabe,M.N. and Kusumi,A. (2001) The Ste20 group kinases as regulators of MAPK kinase cascades. Trends Cell Biol., 11, 220–230. [DOI] [PubMed] [Google Scholar]
- 8.Martinou J.C., Falls,D.L., Fischbach,G.D. and Merlie,J.P. (1991) Acetylcholine receptor-inducing activity stimulates expression of the epsilon-subunit gene of the muscle acetylcholine receptor. Proc. Natl Acad. Sci. USA, 88, 7669–7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beeson D., Brydson,M., Betty,M., Jeremiah,S., Povey,S., Vincent,A. and Newsom-Davis,J. (1993) Primary structure of the human muscle acetylcholine receptor. cDNA cloning of the gamma and epsilon subunits. Eur. J. Biochem., 215, 229–238. [DOI] [PubMed] [Google Scholar]
- 10.Lobos E.A. (1993) Five subunit genes of the human muscle nicotinic acetylcholine receptor are mapped to two linkage groups on chromosomes 2 and 17. Genomics, 17, 642–650. [DOI] [PubMed] [Google Scholar]
- 11.Ishida T. and Yamamoto,K. (1987) Survey of nonhuman primates for antibodies reactive with Epstein–Barr virus (EBV) antigens and susceptibility of their lymphocytes for immortalization with EBV. J. Med. Primatol., 16, 359–371. [PubMed] [Google Scholar]
- 12.Sambrook J., Russell,D.W. and Sambrook,J. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 13.Dieffenbach C.W. and Dveksler,G. (1995) PCR Primer: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Padgett R.A., Grabowski,P.J., Konarska,M.M., Seiler,S. and Sharp,P.A. (1986) Splicing of messenger RNA precursors. Annu. Rev. Biochem., 55, 1119–1150. [DOI] [PubMed] [Google Scholar]
- 15.Guo Z. and Sherman,F. (1996) 3′-end forming signals of yeast mRNA. Trends Biochem. Sci., 21, 477–481. [DOI] [PubMed] [Google Scholar]
- 16.Manley J.L. and Takagaki,Y. (1996) The end of the message—another link between yeast and mammals. Science, 274, 1481–1482. [DOI] [PubMed] [Google Scholar]
- 17.Arnason U., Gullberg,A., Gretarsdottir,S., Ursing,B. and Janke,A. (2000) The mitochondrial genome of the sperm whale and a new molecular reference for estimating eutherian divergence dates. J. Mol. Evol., 50, 569–578. [DOI] [PubMed] [Google Scholar]
- 18.Gardner P.D. (1990) Nucleotide sequence of the epsilon-subunit of the mouse muscle nicotinic acetylcholine receptor. Nucleic Acids Res., 18, 6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madsen O., Scally,M., Douady,C.J., Kao,D.J., DeBry,R.W., Adkins,R., Amrine,H.M., Stanhope,M.J., de Jong,W.W. and Springer,M.S. (2001) Parallel adaptive radiations in two major clades of placental mammals. Nature, 409, 610–614. [DOI] [PubMed] [Google Scholar]
- 20.Murphy W.J., Eizirik,E., Johnson,W.E., Zhang,Y.P., Ryder,O.A. and O’Brien,S.J. (2001) Molecular phylogenetics and the origins of placental mammals. Nature, 409, 614–618. [DOI] [PubMed] [Google Scholar]
- 21.Murphy W.J., Eizirik,E., O’Brien,S.J., Madsen,O., Scally,M., Douady,C.J., Teeling,E., Ryder,O.A., Stanhope,M.J., de Jong,W.W. and Springer,M.S. (2001) Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science, 294, 2348–2351. [DOI] [PubMed] [Google Scholar]
- 22.Hildebrandt M. and Nellen,W. (1992) Differential antisense transcription from the Dictyostelium EB4 gene locus: implications on antisense-mediated regulation of mRNA stability. Cell, 69, 197–204. [DOI] [PubMed] [Google Scholar]
- 23.Volk R., Koster,M., Poting,A., Hartmann,L. and Knochel,W. (1989) An antisense transcript from the Xenopus laevis bFGF gene coding for an evolutionarily conserved 24 kd protein. EMBO J., 8, 2983–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner R.W., Smith,J.E., Cooperman,B.S. and Nishikura,K. (1989) A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl Acad. Sci. USA, 86, 2647–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimelman D. and Kirschner,M.W. (1989) An antisense mRNA directs the covalent modification of the transcript encoding fibroblast growth factor in Xenopus oocytes. Cell, 59, 687–696. [DOI] [PubMed] [Google Scholar]
- 26.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 27.Arnason U., Gullberg,A. and Janke,A. (1998) Molecular timing of primate divergences as estimated by two non-primate calibration points. J. Mol. Evol., 47, 718–727. [DOI] [PubMed] [Google Scholar]