Abstract
The specific formylation of initiator methionyl-tRNA (Met-tRNA) by methionyl-tRNA formyltransferase (MTF) is important for the initiation of protein synthesis in Escherichia coli. The determinants for formylation are located in the acceptor stem and in the dihydrouridine (D) stem of the initiator tRNA (tRNAfMet). Here, we have used ethylation interference analysis to study the interactions between the Met-tRNAfMet and MTF in solution. We have identified three clusters of phosphates in the tRNA that, when ethylated, interfere with binding of MTF. Interference due to ethylation of phosphates in the acceptor stem and in the D stem is most likely due to the close proximity of the protein as seen in the crystal structure of the MTF.fMet-tRNAfMet complex. The third cluster of phosphates, whose ethylation interferes with binding of MTF, is dispersed along the anticodon stem, which is distal to the sites of tRNA protein contacts. Interestingly, these latter positions correspond to sites of increased cleavages by RNase V1 in RNA footprinting experiments. Together, these results suggest that in addition to the protein, which binds to the substrate tRNA in an induced fit mechanism, the tRNA also undergoes induced structural changes during its binding to MTF.
INTRODUCTION
Initiator tRNAs are used exclusively for the initiation step of protein synthesis and not for the elongation step (1). In eubacteria, mitochondria and chloroplasts, the initiator tRNAs are used as formylmethionine-tRNA (fMet-tRNA) whereas in the cytoplasm of eukaryotes and in archaea they are used as methionyl-tRNA (Met-tRNA). In Escherichia coli, the formylation of initiator Met-tRNA by Met-tRNA formyltransferase (MTF) is very important for initiation (2,3). The formyl group provides a positive determinant for the initiation factor IF2 (4), which allows IF2 to select the initiator fMet-tRNA from other tRNAs and a second negative determinant, which blocks the binding of the elongation factor EF-Tu to the initiator tRNA (5,6). The formylation reaction is highly specific. MTF formylates only the initiator Met-tRNA and no other aminoacyl-tRNA including the elongator Met-tRNA (7,8). The primary determinants for formylation are clustered in the acceptor stem of E.coli initiator tRNA (Fig. 1) (9–11). A secondary determinant is the A11:U24 base pair in the dihydrouridine (D) stem (9,12).
Figure 1.
Cloverleaf structure of E.coli initiator Met-tRNAN. Nucleotides playing a major role in formylation are shaded dark gray, whereas those playing a minor role are shaded light gray. RNase cleavages that are decreased in the presence of MTF are indicated in red, whereas those that are increased are indicated in green. Arrows, RNase T2 cleavage; arrowheads, RNase V1 cleavage. Figure is reproduced from Ramesh et al. (20).
The crystal structure of E.coli MTF shows the organization of the enzyme in two domains connected by a linker region (13). The N-terminal domain consists of a Rossmann fold and is strikingly homologous to E.coli glycinamide ribonucleotide formyltransferase (GARF), another enzyme that, like MTF, utilizes N10-formyltetrahydrofolate as a formyl group donor in formylation reactions. Four highly conserved amino acid residues are part of the N-terminal domain and are thought to be involved in catalysis. The C-terminal domain of MTF (amino acids E210–V314), which is structurally homologous to the anticodon-binding domain of two aminoacyl-tRNA synthetases from E.coli (14) and yeast (15), is not present in GARF. A second distinctive feature of MTF in all eubacteria is the presence of a 16 amino acid insertion loop (T34–S49 in E.coli) within the N-terminus of the enzyme. Previously, we have used a combination of biochemical techniques to investigate the role of each of the separate domains of MTF in recognition of the initiator Met-tRNA. Cross-linking (16), mutational analysis and binding studies (17) indicate that the C-terminal domain is mostly involved in general non-specific interactions with tRNA, whereas the 16 amino acid insertion loop is important for specific recognition of the initiator tRNA substrate (18,19). Furthermore, by using a combined protein/RNA-footprinting approach, we found that a segment of the 16 amino acid insertion loop of MTF changes conformation upon substrate binding in an induced fit mechanism (20). This induced structural change was shown to depend on the formation of a ‘functional’ complex between MTF and initiator Met-tRNA. RNA footprinting experiments showed that MTF specifically protects the acceptor stem and the 3′-end region of the initiator Met-tRNA against double and single strand-specific nucleases. However, because of the inherent limitations of enzymatic probes, which do not cover the entire tRNA molecule, we could not obtain any information on the D stem, which contains a secondary determinant for MTF recognition (Fig. 1).
In this paper, we have used ethylation interference analysis to probe the effect of ethylation of backbone phosphates of the initiator Met-tRNA substrate on its binding to MTF. The analysis involves ethylation of the initiator Met-tRNA, followed by (i) binding of the Met-tRNA to MTF, (ii) separation of bound and unbound forms of the Met-tRNA and (iii) comparison of the ethylation sites in the bound and unbound forms. Because of the relatively unstable ester linkage in Met-tRNA, for the ethylation interference analysis we used an analog of the initiator Met-tRNA [3′-amino 3′-deoxyA (Met-tRNAN)] in which the amino acid methionine is attached to the tRNA by a stable amide linkage. The specific advantage of ethylation interference analysis is that the ethylating reagent ethylnitrosourea (ENU) reacts at comparable rates with all the solvent-accessible phosphates of nucleic acids and, therefore, allows scanning of the entire molecule. Our results have identified mainly three clusters of phosphates that, when ethylated, interfere with binding of MTF. They are located in the D stem, the acceptor stem and along the anticodon stem. The results of this study in combination with those from RNA footprinting obtained earlier (20) allow us to draw conclusions about the importance of tRNA structure changes in binding to the enzyme. Comparison of the effects of ethylation on binding of MTF to Met-tRNAN, which is a substrate of MTF and with which it forms a productive complex, and uncharged tRNAN, which is not a substrate of MTF but with which it forms a non-productive complex, also supports the idea that MTF binds first to the D stem of the tRNA through the C-terminal domain in a mostly non-specific manner (13,17).
MATERIALS AND METHODS
Strains, plasmids and purified enzymes
Cloning, expression and purification of MTF and methionyl-tRNA synthetase (MetRS) as C-terminal 6× His fusion proteins have been described earlier (19). The purified enzymes were stored in 20 mM imidazole–HCl pH 7.6, 150 mM NaCl, 10 mM 2-mercaptoethanol and 50% glycerol. Escherichia coli UT481 containing the plasmid pEC7 and overexpressing tRNA-nucleotidyl transferase was provided by M. P. Deutscher (University of Miami School of Medicine). The enzyme was purified as described (21).
Preparation of 5′-[32P]-labeled methionyl-tRNA containing 3′-amino 3′-deoxyA (Met-tRNAN) at the 3′ end
The cloned initiator tRNA2fMet was purified from E.coli B by electrophoresis of total tRNA on a 15% native polyacrylamide gel (22). The purity and integrity of the tRNA, assessed by aminoacylation assay, was >95%. Incorporation of Met-tRNAN into the A76 position of the tRNA by exchange reaction using tRNA-nucleotidyl transferase has been reported before (20). An aliquot of 1 µg of this purified initiator tRNA analog (tRNAN) was dephosphorylated with calf intestinal alkaline phosphatase and 5′-end labeled with [γ-32P]ATP and T4 polynucleotide kinase (23) in a 12 µl reaction. After heat inactivation of the kinase (20 min at 65°C), the reaction mixture was chilled on ice for 5 min. For the aminoacylation, reaction conditions were adjusted to 150 mM NH4Cl, 10 mM MgCl2, 10 µg/ml BSA, 1 mM EDTA, 20 mM imidazole–HCl pH 7.6, 2 mM ATP and 100 µM l-methionine. MetRS (250 ng) was added, and the reaction mixture was incubated for 20 min at 37°C. The 5′-[32P]-Met-tRNAN was purified by electrophoresis on a 12% polyacrylamide–8 M urea gel, located by autoradiography and eluted into a buffer containing 0.3 M sodium acetate pH 5.5 and 1 mM EDTA for 5 h at 20°C. The tRNA was recovered by ethanol precipitation and dissolved in distilled water to ∼30 000 c.p.m./µl.
Ethylation interference assay
Ethylation of backbone phosphates was essentially done as described (24), the protocol for studying the interference of ethylation on binding of the tRNA to MTF was modified from Schlegl et al. (25). Six picomoles of 5′-[32P]-labeled Met-tRNAN (∼1.4 × 107 c.p.m.) and 7 µl of a saturated solution of ENU in ethanol (∼750 mM) were incubated under either (i) native conditions in a buffer consisting of 5 mM MgCl2, 150 mM KCl and 150 mM sodium cacodylate pH 8.0 at 37°C for 3 h or (ii) denaturing conditions in a buffer consisting of 1 mM EDTA and 150 mM sodium cacodylate pH 8.0 at 90°C for 2 min (total reaction volume 35 µl). tRNA was precipitated with 3 vol of ethanol in the presence of 2 µg of glycogen as carrier. The pellet was washed twice in 80% ethanol, resuspended in distilled water (20 µl) and precipitated again.
For the isolation of a complex between MTF and the tRNA, the ethylated 5′-[32P]-labeled Met-tRNAN (0.045 µM) was renatured in binding buffer (20 mM imidazole–HCl pH 7.6, 150 mM KCl, 5 mM MgCl2, 5% glycerol and 1 mM 2-mercaptoethanol) at 55°C for 10 min and slowly cooled to 20°C. Varying amounts of MTF (between 0.3 and 1.0 µM) were added and the mixtures (10 µl) were left at 20°C for 10 min. After the addition of 1 µl of a 1% bromophenol blue solution, the mixtures were applied to a native 6% polyacrylamide gel (20 × 20 × 0.1 cm slab gel). Electrophoresis was carried out in 0.5× TBE buffer at 150 V for 30 min at room temperature. Bound and unbound tRNAs were located by autoradiography and the tRNA bands from the bandshifts that gave a ratio of ~1:1 (bound versus unbound tRNA) were excised. tRNAs were eluted into a buffer containing 0.3 M sodium acetate pH 5.5, 1 mM EDTA and 0.1% SDS to denature the protein and to facilitate extraction of the tRNA from the bound fraction and recovered by ethanol precipitation.
The tRNA samples were resuspended in 25 µl of 0.1 M Tris–HCl pH 9.0 and incubated for 5 min at 50°C to facilitate cleavage at sites of ethylation. The tRNA fragments were precipitated in 0.3 M sodium acetate pH 5.5 and 3 vol of ethanol, washed twice in 80% ethanol and resuspended in 8 M urea/0.03% (w/v) bromophenol blue and xylene cyanol. Electrophoresis was performed on a 15% polyacrylamide–7 M urea gel (40 × 33 × 0.04 cm). Products of partial cleavage with alkali and RNase T1 served as markers to identify the fragments derived from ethylated tRNA (26). The gel was fixed, dried and subjected to autoradiography. Radioactivity was quantified by PhosphorImaging (Molecular Dynamics) and is given as the percentage of tRNA cleaved at one specific site as compared with the total amount of tRNA loaded per lane.
Quantitation
As a quantitative measure of the extent of ethylation interference on binding of Met-tRNAN to MTF, we use the term ‘interference factor’, which is the ratio of ethylation-dependent cleavage at a given phosphate in the unbound (free) form to that in the bound (complexed) form. If at position x the ratio of unbound to bound tRNA is 1:1, then the factor is 1. Thus, an interference factor of 1 means that the presence of an ethyl group at this phosphate had no influence on this particular tRNA for binding to MTF. A factor >4 implies a strong interference of the ethyl group, since most of the tRNA with the ethyl group at the given position was not bound by MTF. The cut-off number has been set at 1.5, which means that if the ratio of cleavages in unbound to bound fraction is 3:2, it is not considered significant.
RESULTS
Tertiary interactions in Met-tRNA2fMet determined by ethylation of the tRNA under native and denaturing conditions
The reagent typically used for alkylation of the phosphate backbone of nucleic acids is ENU. It reacts at nearly equal rates with all the solution-accessible phosphate groups of the nucleic acid backbone, forming phosphotriester groups at the non-esterified oxygens of the otherwise phosphodiester backbone. Two stereoisomeric triesters can be generated with the ethyl groups positioned in either the major or the minor groove (27). The phosphotriester products are labile when heated under alkaline conditions, so backbone scission can be induced at sites of ethylation. Because of the presence of an ethyl group on the tRNA fragment, the products of cleavage can be distinguished from random alkaline cleavage due to a slight mobility shift on a sequencing gel run under denaturing conditions.
In view of the chemical instability of the ester linkage between methionine and tRNA in Met-tRNA2fMet, an initiator tRNA analog, tRNAN, in which the 3′-terminal A residue is replaced by Met-tRNAN, was used (28). Upon aminoacylation of tRNAN by MetRS, the methionine, which is initially attached to the 2′-OH group through an ester linkage, migrates to the 3′ position and forms a stable amide linkage (29).
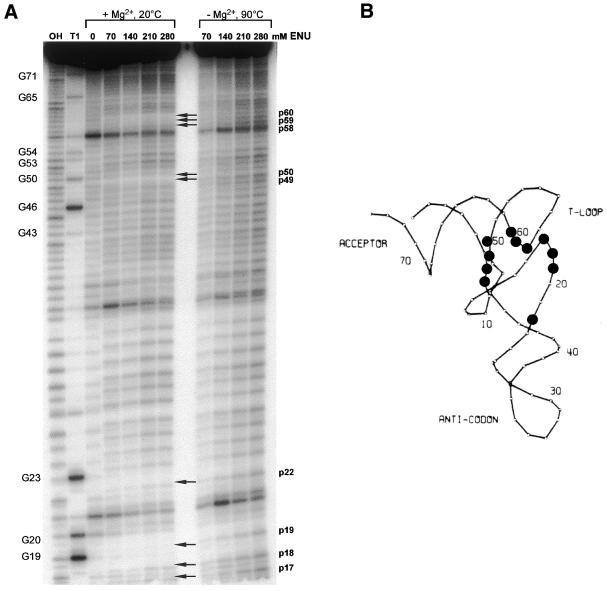
5′-[32P]-labeled Met-tRNAN was ethylated either under conditions that preserve the native structure of the tRNA (Mg2+, low temperature) or under denaturing conditions (without Mg2+, high temperature), such that all potential sites were modified at a low level (on average less than one phosphate alkylation at some random site per tRNA molecule) with approximately equal frequency. Figure 2A shows the sites of ethylation with increasing concentration of ENU, both under native and denaturing conditions. Comparison of the fragmentation pattern shows that several of the phosphodiester bonds (Fig. 2A, arrows) are protected from the ethylation-dependent cleavage in the natively folded tRNA compared with the denatured tRNA. Figure 2B shows a wireframe representation of the three-dimensional structure of the tRNA indicating these unreactive phosphates in natively folded tRNA: phosphates p17–p19, p22 in the D loop, p58–p60 in the T-loop, p49, p50 near the variable loop and p7, p8 between acceptor stem and D stem. They are centered in those areas of the tRNA where extensive tertiary interactions are present. In fact, ethylation of the phosphate backbone by ENU has been used extensively to investigate the tertiary structure of tRNAs in solution. Among others, uncharged tRNA1fMet has been analyzed by backbone ethylation (30). The results obtained with this related species were very similar to the results reported here, using Met-tRNAN. This indicates that neither the nucleotide at position 46, which is the only difference between tRNA1fMet and tRNA2fMet, nor the presence of the adenosine analog at A76 and the amino acid affect the general tertiary conformation of the tRNA.
Figure 2.
(A) Autoradiogram of a 15% denaturing polyacrylamide gel of phosphate alkylation experiments with ENU on 5′-[32P]-labeled Met-tRNAN using either conditions that preserve the tertiary structure of the tRNA (with Mg2+ ions and 20°C) or conditions where the tRNA is denatured (no Mg2+ ions and 90°C). Lane OH, alkaline ladder; lane T1, sequencing ladder generated by cleavage with RNase T1. Increasing amounts of ENU (70, 140, 210 and 280 mM) were used for ethylation. Lane 0, incubation control without the addition of ENU. (B) Phosphates that are unreactive under native conditions [indicated by arrows in (A)] are marked by spheres in the three-dimensional wireframe model of the tRNA.
The appearance of the typical ethylation pattern under native conditions was routinely used in the following experiments to ensure that the tRNA was not misfolded due to any of the preparative procedures, i.e. radioactive labeling, 3′-end modification with Met-tRNAN or purification on denaturing polyacrylamide gel.
Effect of ethylation on the formation of MTF.Met-tRNAN complex
We have used ethylation interference analysis to study the interaction between the MTF enzyme and an analog of its natural substrate, Met-tRNA. This analysis involves mild ethylation of end-labeled Met-tRNAN followed by binding to MTF. Unbound and bound tRNA populations are separated on a non-denaturing gel, isolated and cleaved at ethylated phosphates. The fragments are run on a denaturing polyacrylamide gel and the ratio of cleavage at each phosphodiester bond between the unbound and the bound form is determined. tRNA molecules with ethylations at sites that interfere with binding to MTF will be excluded from the bound RNA pool and will be enriched in the unbound pool. If the conditions used for binding are such that an approximately equal amount of unbound and bound tRNA is yielded, a significant increase in the ratio of cleavage at a particular phosphodiester bond between unbound and bound tRNAs from the value of 1 means an interference on binding of the tRNA to the protein due to ethylation.
5′-[32P]-labeled Met-tRNAN was ethylated under both native and denaturing conditions. The ethylated Met-tRNAN was then incubated with MTF to allow complex formation. In order to achieve the desired 1:1 ratio between bound and unbound tRNA, small-scale titrations with varying amounts of MTF (0.1–5 µM) were performed. Eventually, a range between 0.3 and 1 µM MTF was selected and used for large-scale binding reactions. Concentrations of 0.5–1 µM MTF yielded between 35 and 65% bound material as detected by gel shift analysis (data not shown). Met-tRNAN in the bound and unbound fraction was eluted and analyzed after cleavage at the ethylated phosphates and the fragmentation patterns of bound and unbound tRNA pools were compared.
Figure 3 shows a typical interference pattern obtained with Met-tRNAN ethylated under native conditions. In order to visualize the region of the 3′ end of the acceptor stem with better resolution, an autoradiograph of a longer run of the sequencing gel is displayed. In the bound fraction (Fig. 3, lane C, complex), there is a reduction in intensities of the bands corresponding to cleavages at phosphates p69–p71 in the acceptor stem, p53 in the T stem, p42–p44 at the 3′ end of the anticodon stem, p23–p30 in the D stem/loop area and the 5′ end of the anticodon stem (Fig. 3, arrowheads), and p11, p12 in the D stem (data not shown). Correspondingly, cleavage at these sites is increased in the unbound fraction (Fig. 3, lane F, free), compared with ethylated Met-tRNAN that has not been subjected to binding to MTF (lane U, unselected).
Figure 3.
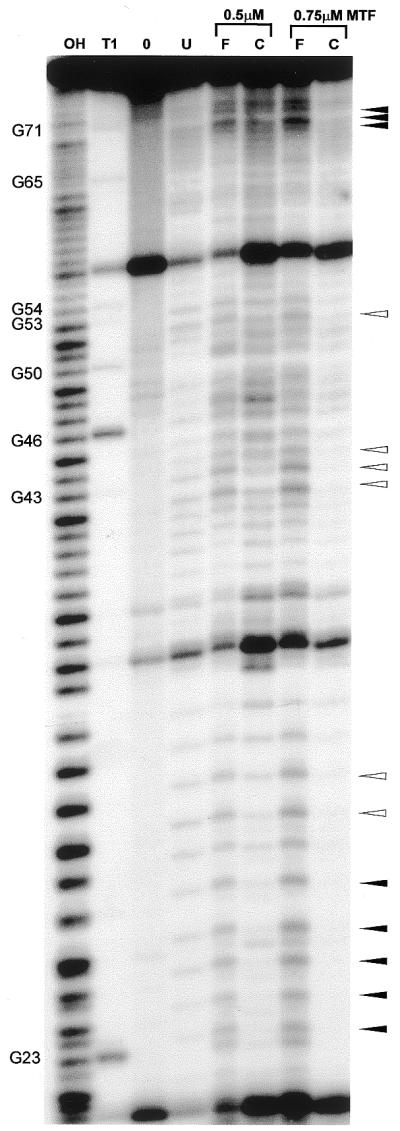
Ethylation interference analysis on MTF.Met-tRNAN complex formation. The 5′-[32P]-labeled tRNA was ethylated under native conditions prior to incubation with 0.5 and 0.75 µM MTF. Following native gel electrophoresis to separate unbound and bound tRNA, an alkaline incubation step was used to cleave the tRNA at the ethylated phosphates. An autoradiogram of a 15% denaturing polyacrylamide gel is shown. Lane F, free tRNA (unbound); lane C, tRNA complexed to MTF (bound). Lane U, ethylated 5′-[32P]-labeled Met-tRNAN before selection by binding to MTF; lane OH, alkaline ladder; lane T1, sequencing ladder generated by cleavage with RNase T1; lane 0, incubation control without the addition of ENU.
Similar results were obtained with the Met-tRNAN ethylated under denaturing conditions (data not shown). Exceptions were those positions where the initial comparisons between ethylation under the two folding states had indicated tertiary interactions. For instance, p7 showed very strong interference under denaturing conditions, but the reactivity of this phosphate is greatly reduced under native conditions. It seems likely that in this case interference is mostly due to problems in folding of the ethylated Met-tRNAN before binding to MTF. A few positions could not be interpreted due to the intrinsic fragility of the tRNA molecule in the loop regions, mostly involving alternating pyrimidine–purine sequences. An appropriate control with untreated tRNA was run in parallel. Each experiment was repeated at least three times.
In order to assess the role of the amino acid attached to the tRNAN in the formation of a complex between MTF and Met-tRNAN, we also used uncharged tRNAN in the ethylation interference experiments (data not shown). The uncharged tRNAN is not a substrate for MTF, however, it binds to MTF with similar affinity and forms a non-productive complex. Positions where interference is seen in both charged and uncharged tRNAN are indicated in Figure 3 by filled arrowheads whereas those that show interference only with charged tRNAN are indicated by empty arrowheads.
Quantitative analysis of ethylation interference
Figure 4A shows a quantitative analysis of the interference results (described above) using the method outlined in the Materials and Methods. By far the strongest interference is seen in the D stem region, with five phosphates characterized by discrimination factors of between 4 and 6 or higher. p29 and p30 as well as p43 on both sides of the anticodon stem belong to an intermediate category of interference (2.5–3.5). As indicated in the Materials and Methods, the cut-off number for interference was set at 1.5. This leaves p11, p12, p22, p28, p42, p44, p53, p69, p70 and p71 as positions showing weak but significant interference in binding due to ethylation.
Figure 4.
Quantitation of the results obtained from the ethylation interference experiments using (A) Met-tRNAN or (B) uncharged tRNAN. The definition of interference is given in the Materials and Methods. The plot represents the average of three independent experiments. Due to spontaneous scission of the tRNA at certain sites, data on some of the phosphate positions are missing: these are p13–p15, p19–p21, p33 and p55–p57. In addition, the extreme ends of the tRNA (p1–p7 and p72–p76) could not be resolved on the gels. Positions where interference is seen in both charged and uncharged tRNA are marked by filled arrowheads; those that essentially show interference with charged tRNA are marked with empty arrowheads on top of the graphs.
Figure 4B shows a quantitative analysis of interference results for tRNAN which had been ethylated under native conditions and selected by MTF binding as described above. In general, results obtained with uncharged tRNAN are similar to those obtained with Met-tRNAN: strong to medium interference in the D stem, p10–p12 and from p23 to p27, and medium interference in the acceptor stem at p70 and p71. Most interestingly, binding of uncharged tRNAN showed little or no interference due to ethylation in the anticodon stem (p29, p30, p43, p44) and at p53. These latter positions are marked with arrows on top of each column in Figure 4B.
DISCUSSION
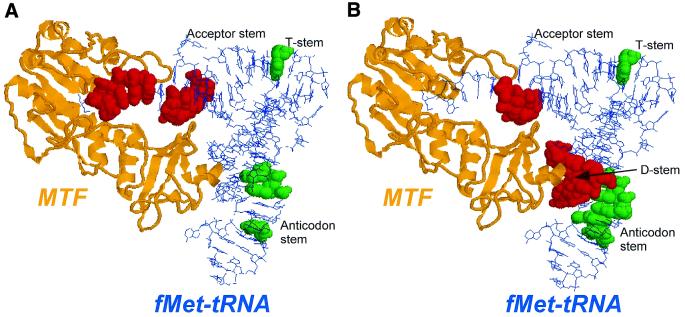
There are several possible interpretations of how ethylation of the backbone phosphates of a tRNA interfere with its binding to proteins: (i) direct steric hindrance due to the bulky modification or reduction in binding affinity due to loss of a critical electrostatic contact; (ii) a long range structural effect, i.e. a change in the structure of the tRNA as a consequence of ethylation; and (iii) restriction of the tRNA conformation such that a conformational change of the tRNA that is necessary for binding cannot occur. The availability of a substantial amount of biochemical data based on the use of mutant MTFs, mutant tRNAs and results of RNase footprinting experiments (18–20) and the co-crystal structure (31) of the MTF.fMet-tRNA complex (Fig. 5) allow us to interpret the results of ethylation interference analyses reported here. Figure 5B illustrates the results of ethylation interference studies in the framework of the co-crystal structure of MTF complexed with fMet-tRNA. Two of the three clusters of phosphates in the Met-tRNAN, ethylation of which interferes with binding to MTF, coincide with regions of tRNA known to carry the determinants for formylation in the acceptor stem and the D stem (colored red). The third cluster of phosphates, ethylation of which interferes with binding, but less strongly, is located in the anticodon stem (colored green). In addition, ethylation of p53 in the T stem also interferes with binding but weakly (colored green).
Figure 5.
X-ray structure of E.coli MTF.fMet-tRNA complex (31) drawn using RASMOL version 2.6 with coordinates obtained from the Protein Data Bank (PDB code 2FMT). MTF is colored gold, the tRNA is in blue wireframe, except for those positions which (A) show decreased (red spacefill) or increased (green spacefill) RNase V1 and T2 cleavage. (B) Same color code for MTF and tRNA, phosphates which show ethylation interference are highlighted in either red spacefill (matching the decreased RNase cleavage) or green spacefill (matching increased RNase cleavage).
In the crystal structure of the MTF.fMet-tRNA complex, MTF binds to the inner side of the L-shape of the tRNA. The very strong interference (Fig. 4A) due to ethylation of phosphates in the D stem is consistent with the co-crystal structure of the complex showing that MTF approaches the D stem very closely and makes a number of contacts with the backbone phosphates and the O2′-oxygens of the ribose (Fig. 5B). The D stem contacts are exclusively with the C-terminal domain of MTF. This domain consists of a positive electrostatic potential on the surface pointing towards the catalytic center of MTF and binds tRNA on its own, although non-specifically. This observation along with other biochemical evidence has led to the suggestion that the C-terminal domain of MTF is involved in the initial binding of the tRNA, through mostly non-specific interactions, and in the orientation of the 3′ end of Met-tRNA towards the catalytic center of MTF. In a productive complex, this initial binding is followed by alignment of the acceptor stem of the tRNA, which contains the most critical determinants for formylation, with amino acid residues in MTF involving an induced fit mechanism. The very strong interference due to ethylation of phosphates in the D stem of Met-tRNAN and the finding that uncharged tRNAN, which is not a substrate for MTF but which forms a non-productive complex, also shows the strong interference are consistent with the idea that MTF binds first to the D stem of the tRNA in a mostly non-specific manner. Interestingly, a recent study on bovine mitochondrial MTF recognition of the tRNAMet indicated that the 11:24 base pair in the D stem and the amino acid linked to the tRNAMet are the major determinants for formylation (32). This would suggest that the initial binding step(s) is conserved among all MTF.tRNA complexes including those that are evolutionarily distant.
The co-crystal structure of the MTF.fMet-tRNAfMet complex shows no contact between MTF and the anticodon stem–loop region of the tRNA, yet there is significant interference due to ethylation of phosphates in the anticodon stem with binding to MTF (Figs 4A and 5B). Interestingly, this coincides with sites of increased RNase V1 cleavage of the tRNA in the anticodon stem upon binding to MTF (Fig. 5A). Together, these results suggest that binding of MTF to the D and the acceptor stem induces a conformational change of the tRNA in the anticodon stem. The most likely explanation is that this conformational change contributes towards the correct binding of the tRNA to MTF and is hindered by ethylation of the phosphates in the anticodon stem. The nature and extent of the conformational change of the tRNA upon binding to MTF is not known because the crystal structure of the free tRNAfMet is available only to a resolution of ∼3.5 Å. Interestingly, there is essentially no interference due to ethylation of phosphates in the anticodon stem for the binding of uncharged tRNAN (Fig. 4B). Thus, this conformational change occurs in a functional MTF.Met-tRNAN complex but not in a non-functional Met-tRNAN complex. In other words, correct binding of the amino acid and the acceptor stem region of the tRNA to MTF is necessary to induce a conformational change in the anticodon stem. It is possible that the T stem also undergoes a subtle conformational change as indicated by the weak interference on binding due to ethylation of p53 (Figs 4A and 5B). It is worth noting that p53 is also a site of increased RNase VI cleavage in a functional MTF.Met-tRNAN complex (Fig. 5A).
Biochemical studies based on footprinting of MTF and tRNA and structural analysis showed previously that MTF undergoes an induced fit upon binding to the substrate Met-tRNAN. The current work suggests that the tRNA also undergoes a conformational change, possibly subtle, in the process. Thus, there is a mutually induced fit in the formation of a specific complex between the Met-tRNAN and MTF. Mutually induced fit is well established in RNA–protein interactions (33) and is also seen in the case of tRNAs and their interactions with various proteins (34,35).
The conformational change in the anticodon stem of Met-tRNAN upon binding to MTF is distal to the site of contact between fMet-tRNA and MTF. This is similar to E.coli EF-Tu.Phe-tRNAPhe.GTP or E.coli IF2.fMet-tRNA interactions. In the crystal structure of the EF-Tu.Phe-tRNAPhe.GTP ternary complex (5), the sites of protein–tRNA interaction are restricted to the acceptor stem and the T stem–loop region of the tRNA. Interestingly, this leads to enhanced cleavage of the anticodon stem by RNase V1 and a change in the anticodon loop conformation as detected by immobilization of a spin label placed at position 37 following the anticodon sequence. Similarly, for the IF2.fMet-tRNA interaction, footprinting experiments have localized the regions of interaction to the acceptor stem and part of the T stem (30). Here too, binding of IF2 leads to enhanced cleavages of the anticodon stem by RNase V1. A recent study on a tRNA-like structure at the end of bromo mosaic virus RNA also suggests that the tRNA-like structure undergoes a conformational change when it is bound to TyrRS (36), at a site distal to that of TyrRS binding. It would thus appear that the induction of conformational changes distal to the site of binding is a common theme in tRNA–protein interactions.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the reviewers and the reviewing editor for constructive and helpful comments and suggestions. We thank Caroline Köhrer for comments and suggestions on this manuscript and Annmarie McInnis for patience and care in the preparation of this manuscript. This work was supported by grant R37 GM17151 from the National Institutes of Health.
REFERENCES
- 1.Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- 2.Varshney U., Lee,C.P., Seong,B.L. and RajBhandary,U.L. (1991) Mutants of initiator tRNA that function both as initiators and elongators. J. Biol. Chem., 266, 18018–18024. [PubMed] [Google Scholar]
- 3.Guillon J.M., Mechulam,Y., Schmitter,J.M., Blanquet,S. and Fayat,G. (1992) Disruption of the gene for Met-tRNAfMet formyltransferase severely impairs growth of Escherichia coli. J. Bacteriol., 174, 4294–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sundari R.M., Stringer,E.A., Schulman,L.H. and Maitra,U. (1976) Interaction of bacterial initiation factor 2 with initiator tRNA. J. Biol. Chem., 251, 3338–3345. [PubMed] [Google Scholar]
- 5.Nissen P., Kjeldgaard,M., Thirup,S., Polekhina,G., Reshetnikova,L., Clark,B.F. and Nyborg,J. (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu and a GTP analog. Science, 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- 6.Seong B.L. and RajBhandary,U.L. (1987) Mutants of Escherichia coli formylmethionine tRNA: a single base change enables initiator tRNA to act as an elongator in vitro. Proc. Natl Acad. Sci. USA, 84, 8859–8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcker K. and Sanger,F. (1964) N-Formyl-methionyl-S-RNA. J. Mol. Biol., 8, 835–840. [DOI] [PubMed] [Google Scholar]
- 8.Dickerman H.W., Steers,E., Redfield,B.G. and Weissbach,H. (1967) Methionyl soluble ribonucleic acid transformylase. I. Purification and partial characterization. J. Biol. Chem., 242, 1522–1525. [PubMed] [Google Scholar]
- 9.Lee C.P., Seong,B.L. and RajBhandary,U.L. (1991) Structural and sequence elements important for recognition of Escherichia coli formylmethionine tRNA by methionyl-tRNA transformylase are clustered in the acceptor stem. J. Biol. Chem., 266, 18012–18017. [PubMed] [Google Scholar]
- 10.Guillon J.M., Meinnel,T., Mechulam,Y., Lazennec,C., Blanquet,S. and Fayat,G. (1992) Nucleotides of tRNA governing the specificity of Escherichia coli methionyl-tRNAfMet formyltransferase. J. Mol. Biol., 224, 359–367. [DOI] [PubMed] [Google Scholar]
- 11.Lee C.P., Dyson,M.R., Mandal,N., Varshney,U., Bahramian,B. and RajBhandary,U.L. (1992) Striking effects of coupling mutations in the acceptor stem on recognition of tRNAs by Escherichia coli Met-tRNA synthetase and Met-tRNA transformylase. Proc. Natl Acad. Sci. USA, 89, 9262–9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramesh V., Varshney,U. and RajBhandary,U.L. (1997) Intragenic suppression in tRNA: evidence for crosstalk between the D and the T stems. RNA, 3, 1220–1232. [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitt E., Blanquet,S. and Mechulam,Y. (1996) Structure of crystalline Escherichia coli methionyl-tRNAfMet formyltransferase: comparison with glycinamide ribonucleotide formyltransferase. EMBO J., 15, 4749–4758. [PMC free article] [PubMed] [Google Scholar]
- 14.Commans S., Plateau,P., Blanquet,S. and Dardel,F. (1995) Solution structure of the anticodon-binding domain of Escherichia coli lysyl-tRNA synthetase and studies of its interaction with tRNALys. J. Mol. Biol., 253, 100–113. [DOI] [PubMed] [Google Scholar]
- 15.Ruff M., Krishnaswamy,S., Boeglin,M., Poterszman,A., Mitschler,A., Podjarny,A., Rees,B., Thierry,J.C. and Moras,D. (1991) Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp. Science, 252, 1682–1689. [DOI] [PubMed] [Google Scholar]
- 16.Gite S. and RajBhandary,U.L. (1997) Lysine 207 as the site of cross-linking between the 3′-end of Escherichia coli initiator tRNA and methionyl-tRNA formyltransferase. J. Biol. Chem., 272, 5305–5312. [DOI] [PubMed] [Google Scholar]
- 17.Gite S., Li,Y., Ramesh,V. and RajBhandary,U.L. (2000) Escherichia coli methionyl-tRNA formyltransferase: role of amino acids conserved in the linker region and in the C-terminal domain on the specific recognition of the initiator tRNA. Biochemistry, 39, 2218–2226. [DOI] [PubMed] [Google Scholar]
- 18.Ramesh V., Gite,S. and RajBhandary,U.L. (1998) Functional interaction of an arginine conserved in the sixteen amino acid insertion module of Escherichia coli methionyl-tRNA formyltransferase with determinants for formylation in the initiator tRNA. Biochemistry, 37, 15925–15932. [DOI] [PubMed] [Google Scholar]
- 19.Ramesh V., Gite,S., Li,Y. and RajBhandary,U.L. (1997) Suppressor mutations in Escherichia coli methionyl-tRNA formyltransferase: role of a 16-amino acid insertion module in initiator tRNA recognition. Proc. Natl Acad. Sci. USA, 94, 13524–13529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramesh V., Mayer,C., Dyson,M.R., Gite,S. and RajBhandary,U.L. (1999) Induced fit of a peptide loop of methionyl-tRNA formyltransferase triggered by the initiator tRNA substrate. Proc. Natl Acad. Sci. USA, 96, 875–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cudny H. and Deutscher,M.P. (1986) High-level overexpression, rapid purification and properties of Escherichia coli tRNA nucleotidyltransferase. J. Biol. Chem., 261, 6450–6453. [PubMed] [Google Scholar]
- 22.Mandal N. and RajBhandary,U.L. (1992) Escherichia coli B lacks one of the two initiator tRNA species present in E. coli K-12. J. Bacteriol., 174, 7827–7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silberklang M., Gillum,A.M. and RajBhandary,U.L. (1977) The use of nuclease P1 in sequence analysis of end group labeled RNA. Nucleic Acids Res., 4, 4091–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vlassov V.V., Giege,R. and Ebel,J.P. (1981) Tertiary structure of tRNAs in solution monitored by phosphodiester modification with ethylnitrosourea. Eur. J. Biochem., 119, 51–59. [DOI] [PubMed] [Google Scholar]
- 25.Schlegl J., Gegout,V., Schlager,B., Hentze,M.W., Westhof,E., Ehresmann,C., Ehresmann,B. and Romby,P. (1997) Probing the structure of the regulatory region of human transferrin receptor messenger RNA and its interaction with iron regulatory protein-1. RNA, 3, 1159–1172. [PMC free article] [PubMed] [Google Scholar]
- 26.Donis-Keller H., Maxam,A.M. and Gilbert,W. (1977) Mapping adenines, guanines and pyrimidines in RNA. Nucleic Acids Res., 4, 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papavassiliou A.G. (1995) Useful considerations for the interpretation of ethylation interference results. Methods Mol. Cell. Biol., 5, 245–246. [Google Scholar]
- 28.Fraser T.H. and Rich,A. (1973) Synthesis and aminoacylation of 3′-amino-3′-deoxy transfer RNA and its activity in ribosomal protein synthesis. Proc. Natl Acad. Sci. USA, 70, 2671–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sprinzl M. (1986) Isomers of aminoacyl- and peptidyl-tRNA in the peptidyl transferase reaction. In Hardesty,B. and Kramer,G. (eds), Structure, Function and Genetics of Ribosomes. Springer, Berlin, pp. 509–522.
- 30.Wakao H., Romby,P., Westhof,E., Laalami,S., Grunberg-Manago,M., Ebel,J.P., Ehresmann,C. and Ehresmann,B. (1989) The solution structure of the Escherichia coli initiator tRNA and its interactions with initiation factor 2 and the ribosomal 30 S subunit. J. Biol. Chem., 264, 20363–20371. [PubMed] [Google Scholar]
- 31.Schmitt E., Panvert,M., Blanquet,S. and Mechulam,Y. (1998) Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet. EMBO J., 17, 6819–6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeuchi N., Vial,L., Panvert,M., Schmitt,E., Watanabe,K., Mechulam,Y. and Blanquet,S. (2001) Recognition of tRNAs by methionyl-tRNA transformylase from mammalian mitochondria. J. Biol. Chem., 276, 20064–20068. [DOI] [PubMed] [Google Scholar]
- 33.Williamson J.R. (2000) Induced fit in RNA-protein recognition. Nature Struct. Biol., 7, 834–837. [DOI] [PubMed] [Google Scholar]
- 34.Ibba M. and Söll,D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem., 69, 617–650. [DOI] [PubMed] [Google Scholar]
- 35.Delagoutte B., Moras,D. and Cavarelli,J. (2000) tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J., 19, 5599–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fechter P., Giege,R. and Rudinger-Thirion,J. (2001) Specific tyrosylation of the bulky tRNA-like structure of brome mosaic virus RNA relies solely on identity nucleotides present in its amino acid-accepting domain. J. Mol. Biol., 309, 387–399. [DOI] [PubMed] [Google Scholar]