Abstract
Abstract
Detecting alterations in plasmid structures is often performed using conventional molecular biology. However, these methods are laborious and time-consuming for studying the conditions inducing these mutations, which prevent real-time access to cell heterogeneity during bioproduction. In this work, we propose combining both flow cytometry and fluorescence-activated cell sorting, integrated with mechanistic modelling to study conditions that lead to plasmid recombination using a limonene-producing microbial system as a case study. A gene encoding GFP was introduced downstream of the key enzymes involved in limonene biosynthesis to enable real-time kinetics monitoring and the identification of cell heterogeneity according to microscopic and flow cytometric analyses. Three different plasmid configurations (one correct and two incorrect) were identified through cell sorting based on subpopulations expressing different levels of GFP at 10 and 50 µM IPTG. Higher limonene production (530 mg/L) and lower subpopulation proportion carrying the incorrect plasmid (12%) were observed for 10 µM IPTG compared to 50 µM IPTG (96 mg/L limonene and more than 70% of cell population carrying the incorrect plasmid, respectively) in 100 mL production culture. We also managed to derive exploratory hypotheses regarding the plasmid recombination region using the model and successfully validated them experimentally. Additionally, the results also showed that limonene production was proportional to GFP fluorescence intensity. This correlation could serve as an alternative to using biosensors for a high-throughput screening process. The developed method enables rapid identification of plasmid recombination at single-cell level and correlates the heterogeneity with bioproduction performance.
Key points
• Strategy to study plasmid recombination during bioproduction.
• Different plasmid structures can be identified and monitored by flow cytometry.
• Mathematical modelling suggests specific alterations in plasmid structures.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00253-024-13273-5.
Keywords: Cell heterogeneity, Limonene, Plasmid recombination, Mathematical modelling, Flow cytometry, Fluorescence-activated cell sorting
Introduction
Cell heterogeneity during bioproduction can emerge in an isogenic culture through the segregation into subpopulations with different characteristics triggered by intrinsic or extrinsic sources. Intrinsic heterogeneity comprehends cell cycle, age, metabolic reactions, and stochasticity of gene expression, while extrinsic heterogeneity originates from fluctuations in the environment (Heins and Weuster-Botz 2018). Heterogeneity in bioproduction can adversely impact productivity and yield due to the generation of less productive subpopulations. Therefore, methods to identify and isolate these subpopulations are essential to define strategies aiming at mitigating their growth.
Identification and monitoring of heterogeneity can be performed by employing fluorescent dyes for cell staining and the use of reporter strains in combination with flow cytometry (Davey and Kell 1996; Joux and Lebaron 2000; Alonso et al. 2012; Delvigne and Goffin 2014; Boy et al. 2020). Fluorescence signals from dyes or fluorescent proteins can be used to distinguish subpopulations based on fluorescence intensity. Heins et al. (2020) developed triple reporter strains by combining three different fluorescent proteins to provide information related to cell growth rate, oxygen limitation and general stress response of single cells. In addition, cells from different subpopulations can be isolated through cell sorting for further analysis (Davey and Kell 1996). For example, fluorescence-activated cell sorting (FACS) has been used to study cell viability, cell engineering and overproducing cells (Mattanovich and Borth 2006). Xiao et al. (2016) applied FACS to isolate different subpopulations with different abundances of free fatty acid based on the fluorescence intensity of cells stained with Nile Red. Based on this approach, they were able to identify more productive cells and designed a genetic circuit to control this population.
For heterologous protein synthesis, plasmid stability throughout the culture process is essential to maintain desirable productivity. Plasmid stability can be affected by plasmid size, chi sequences (5’-GCTGGTGG-3’), direct and inverted repeats and polyA sequences (Oliveira et al. 2009). Therefore, under specific conditions, the host may modify the structure of the plasmids, for example, by inserting or deleting DNA sequences (Oliveira et al. 2009); in certain cases, the plasmid may be lost (Corchero and Villaverde 1998). The analysis of structural changes from deletions in plasmids can be performed by conventional molecular biology, such as colony PCR (Bao et al. 2019), restriction enzyme digestion, and verification of the fragment lengths (Friehs 2004). However, these methods are commonly used to identify recombination events rather than studying the conditions that lead to these mutations since they can become laborious and time-consuming (Bao et al. 2019). These methods may require the analysis of numerous cell colonies to identify subpopulations carrying incorrect plasmids, which prevent rapid access to cell heterogeneity dynamics.
To overcome these limitations, we present a workflow for studying and characterizing cell subpopulations carrying different plasmid structures due to plasmid recombination events by integrating flow cytometry, FACS and mechanistic modelling (Fig. 1). In genetic circuits where plasmid recombination is expected, different levels of a reporter gene expressed downstream of the key enzymes could indicate subpopulations carrying deletions or mutations in the plasmid. This approach would enable rapid identification of plasmid recombination at cell level since it would reduce the need for laborious conventional genetic techniques. It can provide the proportion of different plasmid structures in real-time while analysing samples by flow cytometry. This method could potentially be extended to study the stability of genetic circuits that lead to gene expression inactivation in the genome or plasmid by adding a fluorescent protein at specific positions to track the region of interest.
Fig. 1.
Workflow of the method developed combines fluorescent protein, flow cytometry, FACS and mathematical modelling to study conditions that lead to plasmid recombination. The strategies commonly used involve colony PCR or plasmid purification for restriction enzyme digestion followed by gel electrophoresis and sequencing. The method proposed here was developed to reduce these laborious and time-consuming steps. 1. Identification of plasmid recombination by gel electrophoresis or sequencing. 2. Development of strategies to study the conditions that lead to plasmid recombination, such as position and number of fluorescent proteins, medium composition, etc. 3. Performing experiments and evaluation of fluorescent protein production using flow cytometry. 4. Development of a mechanistic model to correlate all the experimental parameters and test the correlation between plasmid recombination and production performance. 5. Confirmation and identification of subpopulations carrying the correct and incorrect plasmids. This information can be used to refine the strategies to study the conditions that lead to plasmid recombination
Specifically, a green fluorescent protein (GFP) can be added to the plasmid of interest to monitor recombination events, expression heterogeneity and dynamics of key enzymes. Flow cytometry can be used to examine the proportion of different subpopulations expressing different levels of GFP over time. Cell sorting followed by restriction enzyme analysis and sequencing can then be employed to confirm which subpopulation carries the correct plasmid. Mathematical modelling can be developed in parallel to capture the physiological phenomena of bioproduction and derive mechanistic insights into the potential correlation of GFP heterogeneity with production performance. To our knowledge, the use of reporter genes to identify and quantify subpopulations with correct and incorrect plasmids using flow cytometry/cell sorting and to correlate these findings with product formation has not been reported in the literature.
To evaluate the proposed method, we sought to investigate heterogeneity originating from plasmid recombination during the production of limonene, a cyclic monoterpene that can be used as a fragrance, flavour additive, antimicrobial, medicinal compound, fuel, and biomaterial (Jongedijk et al. 2016). Alonso-Gutierrez et al. (2013) constructed a plasmid system for limonene production by using heterologous enzymes to compose the mevalonate pathway. This system has been subsequently applied to produce pinene and linalool (Rinaldi et al. 2022; Bao et al. 2019). However, these works have reported problems with the stability of this plasmid due to repeated sequences, which may lead to recombination. Bao et al. (2019) and Rinaldi et al. (2022) identified regions in the plasmid with a high probability of recombination due to homologous sequences (identical promoters and terminators). They indicated that the metabolic burden caused by this plasmid can induce cells to use these homologous regions to delete genes, resulting in a low product formation. The approach developed in this work combines flow cytometry and FACS for real-time monitoring of plasmid recombination and based on the modelling insights, bioproduction was successfully correlated to cell heterogeneity level. Additionally, it can be readily generalised to other studies aimed at monitoring instability in genetic circuits arising from plasmid recombination or any mutation leading to gene inactivation and verifying the correlation with the performance of bioproduction.
Materials and methods
Strain and plasmids
E. coli MG1655 (F- lambda- ilvG- rfb-50 rph-1) was used as the host for limonene production harbouring the plasmid pJBEI-6409 (Alonso-Gutierrez et al. 2013). mGFPmut2 from pEB1-mGFPmut2 plasmid (Balleza et al. 2018) was added into pJBEI-6409 downstream of limonene synthase. Plasmids were purchased from Addgene. DNA fragments were amplified using Q5 high-fidelity DNA polymerase (NEB). 2X ClonExpress Mix (Vazyme) was employed for plasmid assembly and NEB 10-beta electrocompetent E. coli was used as the host for cloning. Oligonucleotide synthesis was performed by IDT (Singapore) and gene sequencing by 1st Base (Singapore). Snapgene software was used for the analysis of plasmid sequences. Purified plasmids were digested with NcoI, and the fragments were analysed by electrophoresis using 0.8% agarose gel. ATCGCTTTGTTCAGTGCTTC and GCCTGGTATCTTTATAGTCCT primers (5’-3’) were used for sequencing to verify plasmid recombination.
Cell cultivations and conditions
A minimum medium (MM) was adapted from Sunya et al. (2012) and consisted of: 0.75 g/L (NH4)2SO4, 8 g/L (NH4)2HPO4, 0.13 g/L NH4Cl, 8 g/L K2HPO4, 0.8 g/L Na2HPO4, 6 g/L citric acid, 1 g/L MgSO4.7H2O, 0.02 g/L MnSO4.H2O, 0.008 g/L CoCl2.6H2O, 0.004 g/L ZnSO4.7H2O, 0.004 g/L Na2MoO4.2H2O, 0.002 g/L CuCl2.2H2O, 0.001 g/L H3BO3, 0.04 g/L FeSO4.7H2O, 0.01 g/L thiamine-HCl, 0.03 g/L CaCl2.
To prepare the production medium, MM was supplemented with 10 g/L glycerol, 1 g/L glucose, 40 mg/L histidine and 1.92 g/L yeast synthetic drop-out medium supplements without histidine (Sigma-Aldrich) (AAN). This supplement is composed of all standard amino acids at 76 mg/L, except for leucine, which is at 380 mg/L. Adenine (18 mg/L), inositol (76 mg/L), uracil (76 mg/L) and p-aminobenzoic acid (8 mg/L) are also present in this supplement. After conducting preliminary tests using LB and various production media with different compositions to produce limonene, a production medium with the addition of AAN resulted in the highest and most stable production (data not shown). The concentration of antibiotic was 30 μg/mL chloramphenicol.
For limonene production, inocula were prepared by inoculating a single colony (freshly transformed) into 10 or 30 mL of LB liquid medium and culturing overnight at 37 °C with 220 rpm in a 50 mL tube or 250 mL shake flask, respectively. Next, 0.9 mL of 10 mL inoculum was added into 29.1 mL production medium in 250 mL shake flask and 3 mL of the 30 mL inoculum was added into 97 mL production medium in 500 mL shake flask. Additionally, a different inoculum preparation was also tested, a single colony was grown in 2 mL LB for 4 h, then 10% (v/v) was added into 3 mL MM supplemented with 10 g/L glycerol until OD reached a value of 4 (37 °C, 220 rpm, 50 mL tube). This inoculum (0.9 mL) was also tested in 29.1 mL production medium (250 mL shake flask). The cultures had an initial pH of 6.3 and were cultivated at 30 °C with 220 rpm shaking; biological duplicates of all cultures were performed. In addition, 20% (v/v) methyl decanoate was added to trap limonene. Induction with IPTG was performed during inoculation (0, 10, 20, 50, and 100 µM). The range of IPTG concentrations was selected according to the results described in Alonso-Gutierrez et al. (2013).
Microplate reader
Experiments were carried out in a Biotek H1M microplate reader operating the Gen5 software. The plate was cultured for 16 h at 30 °C, with orbital shaking. The following measurements were performed every 20 min: optical density at 600 nm, green fluorescence at excitation 485 nm, emission 525 nm and gain 75. The medium preparation and composition used for microplate were the same as described for shake flasks (Cell cultivations and conditions). Images were taken on a Leica DMi8 fluorescence microscope using a 100 × oil immersion objective.
Analytical procedures
Limonene was measured using GC-FID (Agilent 8890) based on the conditions described in Camargo et al. (2020). HP-5 column, split 10:1, He as the carrier gas, initial temperature of 40 °C, hold time of 3 min, 10 °C/min until 100 °C, then 60 °C/min until 220 °C. Samples were taken from the methyl decanoate layer, centrifuged for 1 min at 13,000 rpm to remove cell debris. The supernatant was diluted into ethyl acetate before injection.
Glucose, glycerol and acetate were measured using HPLC (Agilent LC1260 HPLC System), Aminex HPX-87H column, 0.6 mL/min, 50 °C, 5 mM H2SO4, UV 210 nm and RI detection. Samples were taken from cell cultivation, centrifuged for 1 min at 13,000 rpm. The supernatants were filtered with 0.22 μm pore size, then diluted accordingly before analysis. Initial concentrations of glycerol and glucose (time zero) were also estimated by HPLC and displayed in the results.
Cell growth in shake flasks was monitored by reading the optical density at 600 nm (Eppendorf BioPhotometer Plus). Samples were centrifuged for 1 min at 13,000 rpm, washed and diluted in 0.9% NaCl before analysis.
Flow cytometry and fluorescence-activated cell sorting (FACS)
GFP and propidium iodide (PI) fluorescence were monitored with BD Accuri C6 Flow Analyzer with blue laser (473 nm) using the channels FL1 (520/30 nm) and FL3 (670/14 nm). Samples were taken from cell cultivation, washed, and diluted to approximately 106 cell/mL in 0.9% NaCl. 10,000 events were recorded using the threshold of FSC-H 10,000 and SSC-H 1000. For PI staining, 10 µL from 1 mg/mL PI solution was added into 1 mL 0.9% NaCl containing approximately 106 cell/mL, 5 min at 37 °C (Maertens et al. 2020). Control for dead cells was prepared by incubating 1 mL of cells (106 cell/mL) at 100 °C for 15 min. FlowJo software was used for data analysis.
Cell sorting was performed using a Mo-Flo XDP cell sorter (Beckman Coulter) equipped with 100mW 488 nm laser (Coherent Scientific). Briefly, cells were gated using forward and side scatter light signals, cell clumps were excluded by forward scatter area versus height pulse measurement. GFP fluorescence was measured through a 530/40 nm bandpass filter and the GFP positive and negative cells were sorted.
Computational modelling
An in silico mathematical model was developed to describe the key cellular physiological phenomena such as cell growth, nutrient consumption, overflow metabolism and reassimilation, and limonene production, represented in a cascade of ordinary differential equations (ODEs). The limonene-producing pathway was represented by a simplified IPTG induction model that was directly linked to the limonene production level. The individual enzymes along the limonene-producing pathway were excluded for simplicity as there was no modification to the genetic circuit of the strain. To capture the heterogeneity of the key enzymes, a GFP reporter gene was expressed downstream to the geranyl pyrophosphate synthase (GPPS) and limonene synthase (LS) which were thought to be the key bottlenecks of the pathway, regulated under the same promoter system. The heterogeneity of the enzyme was described and modelled as a conversion of the proportions among the three different subpopulations (-GFP, +GFP, and ++ GFP). This model was used to capture the batch culture bioproduction of limonene at different IPTG induction levels and the intercorrelation between variables to provide insights into how heterogeneity affects the limonene production level.
The model was developed and implemented in Python 3. The ODEs were solved using the ode solver adopted from the SciPy package, and the parameter estimation was implemented using global optimizer differential evolution followed by Nelder-Mead local optimizer available from SciPy package. The mathematical model formulations and parameters are provided in Supplementary Table S2-S3.
Results
Identification of cell heterogeneity
Bao et al. (2019) and Rinaldi et al. (2022) identified regions in pJBEI-6409 plasmid with high probability of recombination (R1 and R2, Fig. 2a). To verify the possibility of monitoring cell heterogeneity under different conditions, a GFP was added into pJBEI-6409 downstream of limonene synthase (primers used for plasmid assembly are presented in the supplementary material, Table S1). The position of GFP brings the possibility of tracking changes in the plasmid due to recombination events. If recombination occurs in the R1 region, the expression of GFP may be modified since fewer genes would be expressed. In the case of recombination occurring in the R2 region, GFP will be lost and cells will not fluoresce.
Fig. 2.
Identification of cell heterogeneity using a fluorescent protein (a) Addition of GFP into pJBEI-6409 plasmid and regions with high probability of recombination due to identical sequences (R1 and R2). Acetoacetyl-CoA synthase (atoB), HMG-CoA (hydroxymethylglutaryl-CoA) synthase (HMGS), HMG-CoA reductase (HMGR), mevalonate kinase (MK), phosphomevalonate kinase (PMK), diphosphomevalonate decarboxylase (PMD), isopentenyl diphosphate isomerase (idi), geranyl pyrophosphate synthase (GPPS), limonene synthase (LS), GFP, origin of replication (Ori), chloramphenicol acetyltransferase (CmR) and lacI repressor (lacI) (Table S4). (b) Microplate results showing the effect of different concentrations of IPTG on GFP expression. (c) Microscope images from samples taken from microplate reader after 16 h for the conditions using 10, 50 and 100 µM IPTG (black bars correspond to 27 µm). (d) GFP proportions and intensities for two subpopulations (-GFP, + GFP) from flow cytometry for the conditions using 10, 50 and 100 µM IPTG at 16 h
Microplate experiments were performed to evaluate the GFP expression as a function of IPTG concentration. A range of IPTG concentrations (0, 10, 20, 50, and 100 µM) was used to evaluate the system behaviours under different induction burdens. Higher IPTG concentrations increase the expression of heterologous enzymes, which may cause cellular stress. Consequently, cells may redesign the plasmid to minimize metabolic burden. Since GFP is expressed downstream of limonene synthase under the control of trc promoter, different profiles of fluorescence are expected. Figure 2b shows the influence of four IPTG concentrations on the population’s average GFP fluorescence normalised to growth measured by a microplate reader. Lower IPTG concentrations triggered an exponential increase in GFP expression followed by a gradually decelerating expression rate. Intriguingly, increasing the IPTG concentration above 50 µM IPTG caused a negative impact on GFP/OD with a drop in expression after reaching the peak at 5 h.
Samples were taken at the end of cell cultivation (16 h) and were analysed using a fluorescence microscope and flow cytometer. Microscope images showed an increase in cell heterogeneity with increasing IPTG concentration (Fig. 2c). Under the induction of 50 and 100 µM IPTG, the presence of some brighter cells was observed. However, the number of cells not expressing GFP also increased and became more distinguishable than under 10 µM IPTG induction. Figure 2d shows the rising proportion of the subpopulation not expressing GFP with increasing IPTG concentrations. Therefore, the addition of GFP into the system at appropriate regions can be used to monitor cell heterogeneity under different concentrations of inducer. Cells that were not fluorescent could indicate that the plasmids for these cells could have been modified, possibly in the R2 region (Fig. 2a).
Confirmation of plasmid recombination using cell sorting
To verify whether the different subpopulations observed in the previous experiment carry mutations in the plasmid, an experiment using 10 and 50 µM IPTG was performed in shake flasks for 24 h in 30 mL medium. Cells from different subpopulations were isolated by cell sorting and the plasmids of cells from each subpopulation were analysed. Subpopulations expressing different levels of GFP were evaluated: -GFP (cells not expressing GFP), + GFP (basal expression level) and + + GFP (GFP expression under induction). The gates to identify these subpopulations were based on the control condition (without inducer, basal GFP expression). The control condition corresponded to cells that were weakly expressing GFP in the absence of the inducer. Induced cells were expected to express more GFP and have a higher fluorescence intensity. Based on this, the + GFP gate corresponded to cells from the control condition, while cells with higher fluorescence intensity were considered induced and represented by the + + GFP gate. Finally, cells with lower fluorescence intensity than the control were considered not expressing GFP (-GFP gate). Figure 3a shows the GFP proportions from flow cytometry for both conditions. For the condition using 10 µM IPTG, cells were isolated from the densest region of -GFP and between + GFP and + + GFP gates, for 50 µM IPTG, cells from the densest region of -GFP and + + GFP were isolated (Fig. 3a, indicated by yellow circles). After isolation, cells were grown in LB overnight and each subpopulation was then analysed by flow cytometry (Fig. 3b). Cells isolated from the -GFP gate consistently remained in the same region. Most of the cells from the region between + GFP and + + GFP (10 µM IPTG condition) and cells from + + GFP (50 µM IPTG condition) were fluorescent, approximately 87% and 93%, respectively.
Fig. 3.
Verification of plasmid recombination by cell sorting (a) GFP intensity for the three subpopulations (-GFP, + GFP, + + GFP) from flow cytometry for the conditions using 10 and 50 µM IPTG, 10.6 cells were sorted from each of the subpopulations indicated by yellow circles. (b) Cell distributions from LB cultures inoculated with cells sorted from each subpopulation were analysed and compared for the two different IPTG concentrations. (c) The plasmids from the cells grown in LB cultures were purified and digested with NcoI. (d) Plasmids from + + GFP and -GFP gates were sent for sequencing and the arrows show the sequenced regions (R1 and R2 are the regions in the plasmid that recombined, Fig. 2a)
The plasmids from the cells grown in LB cultures were purified and digested with NcoI to verify plasmid sizes (Fig. 3c). It can be observed that the plasmids were correct for cells originated from the region between + GFP and + + GFP gates under 10 µM IPTG. However, for cells originated from -GFP subpopulations, plasmid digestion indicated the same band size for both conditions but smaller than the correct plasmid. Finally, the plasmids of cells originated from + + GFP gate for the condition using 50 µM IPTG had a different size, smaller than the correct plasmid, but bigger than the plasmid from -GFP subpopulation. These results indicate that each subpopulation showed a variation in their plasmids, and this could lead to different performances in limonene production. Since the cells used for this analysis originated from the densest regions according to flow cytometry analysis, this indicates that these plasmid structures were present in most of the cells.
Simulations of gel electrophoresis using NcoI as a cut site (Snapgene software) were performed on plasmid structures resulting from plasmid recombination in the position R1 followed by R2 (Fig. 2a). This was performed to verify the resulting size of the bands and compare them with the experimental data (Fig. S1). It was observed that the bands had the same size as those in the experiment (Fig. 3c). This suggests that the recombination occurred in R1 for the plasmids originated from + + GFP gate (50 µM condition), which resulted in the loss of four intermediate enzymes (MK, PMK, PMD, and idi) and R2 for the plasmids originated from -GFP gate (10 and 50 µM conditions) which leads to the loss of six enzymes along the downstream of the limonene-producing pathway and GFP gene. Plasmids from + + GFP gate (50 µM condition) and -GFP gate (10 and 50 µM conditions) were sent for sequencing and the recombinations in regions R1 and R2, respectively, were confirmed (Fig. 3d).
Monitoring cell heterogeneity during limonene production using flow cytometry
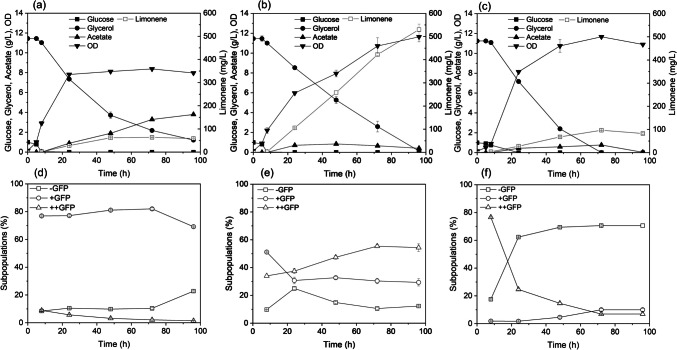
Batch cultures for limonene bioproduction were performed in shake flasks to examine the cell heterogeneity at the single-cell level using flow cytometry. This is important to determine if the system with GFP could correlate the expression dynamics of enzymes under different IPTG inductions with limonene production and to monitor the variation of subpopulations over time using flow cytometry. Figure 4a-c presents the influence of IPTG concentrations on limonene production and compares it with the control (no IPTG) (Fig. 4a) in 100 mL medium. 10 µM IPTG induction achieves better performance than 50 µM for limonene production, reaching a maximum of 530 mg/L of limonene after 96 h (Fig. 4b). On the other hand, 50 µM IPTG induction triggers much slower productivity and stops production after 72 h with a maximum of 96 mg/L (Fig. 4c). The addition of IPTG to 50 µM was observed to impose a delay in cell growth based on the OD values and glucose measurement. For the condition using 10 µM IPTG, the production of limonene seems to be correlated with the consumption of glycerol, both demonstrated linear behaviour throughout the process. Preliminary tests using concentrations of IPTG higher than 10 µM in shake flasks provided lower levels of limonene, therefore, 10 µM IPTG was the best inducer concentration under the conditions tested (data not shown).
Fig. 4.
The impact of different IPTG concentrations on the performance of limonene production and cell heterogeneity. Profiles of glucose, glycerol, acetate, OD, and limonene for the control condition (no IPTG) (a) and conditions with 10 µM IPTG (b), and 50 µM IPTG (c) in 100 mL medium culture for 96 h. Flow cytometry data showing different expressions of GFP (-GFP, + GFP and + + GFP) for control (no IPTG) (d), 10 µM IPTG (e) and 50 µM IPTG (f)
Figure 4d-f shows the dynamic changes of subpopulations expressing different levels of GFP (-GFP, + GFP and + + GFP). For the control, GFP expression profile remained the same during cell cultivation, with 70 to 80% of cells in + GFP gate, potentially due to the slight leakiness of the promoter. For the conditions with 10 µM IPTG induction, the cells were slowly moving from + GFP to + + GFP gate (achieving more than 50%) with 12% of cells not expressing GFP at 96 h. At 8 h, most of the cells were highly fluorescent for the conditions with 50 µM IPTG (+ + GFP, 76%). However, this subpopulation subsequently decreased continuously to ~ 6% at 96 h with a simultaneous increase in -GFP subpopulation to about 70%. According to PI staining results, most of the cells remained intact during the whole process (supplementary material, Fig. S2).
The condition using 10 µM IPTG provided a stable fluorescent subpopulation with higher intensity than the control. When exposed to 50 µM IPTG, a high proportion of the population not expressing GFP was observed after 8 h. These results corroborate with the data presented in Fig. 4b-c where a high and stable GFP expression can be correlated with higher limonene production.
The conditions using 10 µM IPTG and control (no IPTG) were previously performed with pJBEI-6409 without GFP insertion. These replicates are presented in the supplementary material (Fig. S3) and the results were similar to the plasmid containing GFP (Fig. 4a-b). This implies that the addition of GFP had not considerably influenced the kinetics for limonene production, therefore these replicates (using plasmid with and without GFP) support the reproducibility of the experimental conditions applied.
The impact of culture conditions on cell heterogeneity and limonene production
Figure 5a-b presents the results for limonene production when supplied with different culture conditions in 30 mL medium. Two types of inocula were tested, the first using LB and the second using MM supplemented with 10 g/L of glycerol (MM 10 g). These inocula were used to inoculate shake flasks containing the production medium (PM) (Fig. 5, denoted with LB PM and MM 10 g PM, respectively). In addition, a LB inoculum was also used to inoculate shake flasks containing a medium identical to PM in composition, excluding glucose (LB PM (without glucose)). Production culture inoculated with LB (LB PM) provided higher production than MM 10 g PM. For LB PM (without glucose), we observed that the lack of glucose negatively affected the limonene production.
Fig. 5.
The performance of limonene production at different culture conditions. (a) Limonene production and (b) yield after 48 h: LB inoculum and production medium without IPTG induction (Control (LB PM)), LB inoculum with 10 µM IPTG induction in the production medium (LB PM), LB inoculum inoculated in the production medium without glucose and with 10 µM IPTG induction (LB PM (without glucose)), minimum medium inoculum supplemented with 10 g/L glycerol and 10 µM IPTG induction in the production medium (MM 10 g PM), in 30 mL medium culture. (c) Flow cytometry data showing different expressions of GFP at 8, 24 and 48 h. Three subpopulations expressing different levels of GFP were observed, -GFP (low or no expression), + GFP (basal expression level) and + + GFP (expression under IPTG induction)
An earlier study has shown that catabolite repression caused by glucose can improve the overall yield of recombinant protein regulated by the lac promoter (Donovan et al. 1996). The presence of glucose could reduce lac promoter activity by decreasing the level of cyclic adenosine monophosphate (cAMP) and consequently the formation of the complex responsible for increasing the promoter affinity for RNA polymerase (cAMP-catabolite activator protein) (Donovan et al. 1996). Therefore, in our study, glucose could potentially enhance plasmid stability by decreasing cell burden caused by overexpression.
Flow cytometry data presented in Fig. 5c shows how the proportions of GFP expression are modified over time in different medium compositions. For LB PM (without glucose), we observed a higher proportion of cells not expressing GFP (-GFP subpopulation), achieving nearly 50% of the whole cell population at 48 h (third column). For LB PM, the percentage of the population not expressing GFP was only 15% which was approximately 3 times lower at 48 h (second column). MM 10 g PM (fourth column) provided a higher percentage of -GFP subpopulation (40%) in comparison with the condition using LB as the inoculum (LM PM). The conditions that provided a higher percentage of cells expressing GFP (+ + GFP) were expected to produce more limonene, which corroborated with the limonene production level demonstrated in Fig. 5a-b, indicating that GFP expression can be correlated to limonene production.
Mechanistic modelling for limonene bioproduction
In parallel, to better gain insights into the mechanism underlying limonene production, in silico modelling has been deployed to capture the physiological phenomena including the biomass formation, nutrients consumption, acetate formation due to overflow metabolism and reassimilation, and limonene production from acetyl-CoA (represented by acetate measurement) under different IPTG inductions. The limonene-producing pathway was omitted and instead represented solely by an IPTG induction model, as the same gene circuit was utilized throughout the entire study. First, a preliminary model based on ODEs was constructed to describe the batch culture conditions using cells with plasmid without GFP under the control condition and with 10 µM IPTG induction (refer to Fig. S3-S4 for more details). While attempting to capture the experimental profiles, the model hypothesized that there could be an acetate inhibitory effect imposed on cell growth during the control condition and the alleviation of this inhibitory effect when induced by 10 µM IPTG, which mitigates acetate accumulation by converting acetyl-CoA into other intermediates/products along the limonene production pathway. In limonene bioproduction, acetyl-CoA can be converted into acetoacetyl-CoA and/or acetate. Acetoacetyl-CoA is the first metabolite in the limonene production pathway (Fig. S5), and it was assumed to be correlated with acetate concentration, as a lower acetate concentration suggests that acetyl-CoA is being converted into acetoacetyl-CoA instead of acetate.
The same model framework was then employed to recapitulate the physiological phenomena of limonene bioproduction of independent experimental data (Fig. 4) when induced by different IPTG concentrations (control at 0 µM, 10 µM, and 50 µM) and with GFP gene expressed downstream of limonene synthase (Fig. 6). The expression profiles were found to be similar to the earlier results with only minor variations in expression levels which implies that the inclusion of GFP gene had a negligible effect on the plasmid system. In our attempt to describe the conversion process from acetate level to limonene production levels under the control condition and when induced by 10 µM, and 50 µM IPTG, the same model hypotheses regarding the growth inhibitory effect imposed by high acetate accumulation, and the alleviation of growth inhibition under IPTG induction due to the conversion of acetyl-CoA (represented here by acetate concentration) into the limonene-producing pathway remain relevant. These hypotheses were validated by the observations of a much higher biomass and lower acetate when increasing IPTG concentration from 10 µM to 50 µM, which was counterintuitive to the common perception that higher IPTG could impose a higher metabolic burden, and potentially lead to lower cellular growth. Meanwhile, we realized that the decrease in acetate accumulation when exposed to 50 µM IPTG did not result in an equivalent higher limonene production level as proposed by the model simulation result. The inclusion of the conversion factor from acetate to limonene accurately represented the high production level at 10 µM but failed to capture the low limonene production at 50 µM, despite providing a good representation of all acetate levels. We thus hypothesized that there could be potential mutations or malfunctions that occurred to the intermediate enzymes or loss of some intermediate enzymes after the acetoacetyl-CoA synthase gene along the limonene-producing pathway.
Fig. 6.
In silico mechanistic model simulations (a) to capture the phenomena at different IPTG inductions (0 µM (ctrl), 10 µM, and 50 µM) and correlation between the proportion of GFP subpopulation to limonene bioproduction performance. Dashed lines represent simulations at 0 µM (ctrl), and dotted lines and solid lines represent simulations at 10 µM and 50 µM respectively. (b) A plot showing the correlation between the + + GFP proportion and the limonene production. Linear fit 1 and Linear fit 2 represent the linear fitting of the experimental data before and after excluding the outlier (denoted by the arrow) with Pearson correlation coefficients of 0.47 and 0.78, respectively. The solid line indicates the fitting using a Hill equation after excluding the outlier with R2 (coefficient of determination) of 0.95. (c) A schematic diagram illustrating the different key cellular phenomena captured by the developed mechanistic model. The details of the corresponding ODEs and parameters are provided in Supplementary Table S2-S3
In this study, the GFP reporter inserted downstream of the key enzyme was used to estimate enzyme dynamics. From the flow cytometry analysis, we detected cells with different fluorescence intensities. Based on this, three different subpopulations were proposed (-GFP, + GFP, + + GFP) under control conditions without induction and when exposed to 10 or 50 µM IPTG. More specifically, the -GFP subpopulation represents the low or non-expressing cells, + GFP represents the subpopulation that expressed GFP during the control condition without IPTG induction, mainly due to basal leakiness, and + + GFP corresponds to the subpopulation of high-expressing cells when subjected to 10 or 50 µM IPTG. When analyzing the proportions of the three GFP subpopulations retrieved from the flow cytometry (Fig. 6a), it is intriguing to realize that there is a pronounced initial spike in the proportion of + + GFP upon induction with 50 µM of IPTG, followed by a sharp decline, accompanied by a simultaneous increase in the -GFP proportion from 24 h onward. Concurrently, we observed a gradual increase in + + GFP proportion when induced with lower IPTG concentration (10 µM). Since the GFP expression could indirectly represent the dynamics of the last two enzymes (GPPS and LS) along the limonene-producing pathway, it thus corroborates our earlier model hypothesis that there could be a potential loss of some intermediate or downstream enzymes at 50 µM IPTG, leading to low acetate accumulation and yet achieving low limonene production. To further verify the model hypothesis, we then proceeded to perform cell sorting and the hypothesis was validated experimentally with the detection of smaller plasmid sizes with missing intermediate enzymes using cells originating from + + GFP gate at 50 µM IPTG induction (Fig. 3c-d).
To close the gap between acetate and limonene, building on the earlier observations and insights, we modelled the conversion kinetics of the three GFP subpopulations under the control condition and in response to the different IPTG concentrations, and proposed a potential association between the + + GFP proportion dynamics and the measured limonene production. Upon integrating the influence of + + GFP dynamics into the limonene equation, we successfully bridged the missing link between the acetate accumulation level and the limonene production level and profoundly improved the model fit. This suggests that + + GFP dynamics could be correlated to the limonene production and be used to explain the observed low acetate yet low limonene level at 50 µM IPTG induction. To verify this relationship, we also computed the Pearson correlation coefficient (r) using the combined experimental data of + + GFP and limonene at the three conditions, obtaining a correlation value of 0.47 (ranging from -1 to 1), indicating a moderate yet positive association between the two variables. This relatively low r value is attributed to an outlier at ~ 80 + + GFP (the arrow in Fig. 6b) obtained from the peak at 8 h under 50 µM IPTG induction before the sudden drop in proportion due to plasmid recombination. This initial spike followed by a sharp decline in + + GFP proportion, reflecting enzyme dynamics, did not lead to a corresponding increase in limonene production, could be explained by a delay in product formation after enzyme generation. After excluding this outlier, we obtained a higher r value of 0.78 with the linear fit and a R2 (coefficient of determination) value of 0.95 upon fitting using a Hill equation, consistent with the model’s equation (Fig. 6b). In a nutshell, we have demonstrated that the developed in silico mechanistic model was not only able to capture the experimental observations well but also provide valuable exploratory hypotheses to be tested experimentally and explanatory insights into the observed phenomena based on the underlying mechanisms and interactions. The representative schematic diagram of the developed mechanistic model is illustrated in Fig. 6c. The corresponding model formulations and parameters are provided in Supplementary Table S2-S3.
Discussion
Evaluation of subpopulations carrying mutations in genetic circuits is routinely performed by the analysis of several colonies (Bao et al. 2019; Chlebek et al. 2023). For example, in Bao et al. (2019), plasmid recombination was evaluated by harvesting cell cultures at particular time points, followed by dilution and plating, performing colony PCR of 100 colonies. Rinaldi et al. (2022) assessed plasmid recombination by collecting samples from cell cultivation followed by plasmid purification and restriction enzyme digestion. These procedures require prolonged time for the formation of colonies (commonly overnight) and preparation of PCR reactions, or plasmid purification and digestion, and this is repeated for each time point from each condition.
In this work, we propose that the addition of a GFP reporter gene in key positions of a genetic circuit in combination with flow cytometry can enable a reliable and rapid strategy to study the stability of these systems. The use of conventional molecular biology techniques such as restriction enzyme digestion followed by gel electrophoresis are implemented to further confirm the plasmid recombination event and to identify the subpopulation carrying the correct plasmid (Fig. 3). Nevertheless, the number of analyses can be considerably reduced after correlating the fluorescence intensity with plasmid structures using a modelling approach. It should be noted that if cells with different plasmid structures provide similar fluorescence intensity, identification of different subpopulations may not be possible. However, the position and number of fluorescent proteins (use of multiple reporter genes) could be adjusted to provide better resolution.
Figure 3b shows that cells isolated from the densest region between + GFP and + + GFP generated 13% of cells not expressing GFP after LB overnight culture (10 µM IPTG condition). According to gel results after restriction enzyme digestion, only the correct size bands were observed. This indicates that by performing gel analysis using plasmids extracted from the cell population, it may not be possible to identify plasmid recombination events due to the low concentration of some of the modified plasmids. However, 13% of a whole population with the incorrect plasmid could considerably impact product formation.
GFP reporter genes in combination with flow cytometry and cell sorting have been used to identify and monitor cell heterogeneity based on the frequency of plasmid loss in Saccharomyces cerevisiae (Hegemann et al. 1999), E. coli (Bahl et al. 2004) or DNA uptake in mammalian cells (Batard et al. 2001). This combination has also been used to monitor plasmid expression level heterogeneity in Cupriavidus necator (Boy et al. 2020, 2022a, b). Noda et al. (2011) presented a method to detect mutations at specific gene locus in cultured human cells using flow cytometry and cell sorting. Their system was designed to evaluate mutations in mammalian cells after exposure to ionizing radiation and it took a week to determine the mutation frequency. The procedure presented here differs from those abovementioned methods because it was designed to rapidly identify and monitor changes in the plasmid structure, enabling real-time evaluation of cell heterogeneity for bioproduction.
GFP has also been utilised as a reporter to study promoter activity by flow cytometry (Cao and Kuipers 2018; Boy et al. 2020), thus, the expression of GFP, in this work, can also provide indirect information related to the expression of limonene synthase since both are under the control of the same promoter. Notably, limonene synthase is responsible for the last step in the synthesis of limonene which was deemed to be one of the key bottlenecks along the pathway (Cheng et al. 2019). Thus, the intensity and dynamics of GFP expression could be correlated to limonene synthase, providing insights into the limonene production level. According to Figs. 4 and 5 and modelling results (Fig. 6), limonene production was identified to be correlated with the proportion of the subpopulation named + + GFP. More importantly, the correlation of bioproducts with key enzymes from the bioproduction pathway could serve as an alternative to the use of biosensors for the indication of productive cells by flow cytometry or during the high-throughput screening process. Although biosensors can serve as invaluable tools for monitoring and optimizing bioprocesses, the primary obstacles associated with their development, including specificity, sensitivity, and dynamic range, often necessitate an extended timeframe for their readiness and practical deployment (Hicks et al. 2020). It is also important to note that the addition of GFP was not the reason for plasmid recombination using the plasmid pJBEI-6409, the instability of this plasmid has already been discussed (Bao et al. 2019; Rinaldi et al. 2022).
The model-driven approach rooted in mechanistic modelling has been extensively used in gaining exploratory insights into optimizing bioproduction and narrowing the design space (Yeoh et al. 2021; Yeoh and Poh 2023; Benito-Vaquerizo et al. 2022; Du et al. 2022). Utilizing the developed mechanistic model, we attempted to describe the experimental process based on fundamental concepts and to verify if we could reproduce the experimental profiles using those assumptions or underlying hypotheses through model fitting. In this study, leveraging the developed mechanistic model, we unravelled a missing link between acetate accumulation and limonene production levels during high IPTG induction (50 µM), which was proposed to be due to possible missing intermediate enzymes along the limonene-producing pathway. This hypothesis was subsequently validated experimentally via cell sorting, revealing a plasmid recombination event. Furthermore, the model successfully bridged the link between acetate and limonene production levels after integrating the influence of + + GFP dynamics into limonene production, effectively recovering the experimental profiles. This suggests a potential correlation between + + GFP subpopulation dynamics and limonene production. Meanwhile, we posited that there could be an acetate inhibition on the cell growth due to the high accumulation of acetate during the batch control condition and the relief or recovery of acetate inhibition on growth when induced by IPTG concentrations. In bioproduction, overflow metabolism commonly occurs when encountering an excess of carbon source, leading to the production of metabolic byproducts, particularly acetate in E. coli. The high accumulation of acetate (exceeding 5 g/L) or prolonged exposure can impose growth inhibition on the cells, thereby limiting the cell density and impacting the production yield (Chong et al. 2013; Koh et al. 1992). In this study, the acetate concentration reached the highest level at ~ 4 g/L which approximates the inhibitory concentration found in the literature. We also observed that the growth inhibition of acetate could be reversible, relieved under IPTG induction that directs the acetate accumulation toward the limonene-producing pathway (Fig. S4), confirmed by a follow-up independent study using higher IPTG concentration (Fig. 6).
In this study, we have developed a method utilizing flow cytometry, cell sorting and mechanistic modelling to study cell heterogeneity arising from plasmid recombination. This approach aims to gain a deeper understanding of the conditions that lead to the mutations and how the resulting heterogeneity impacts the bioproduction yield. To evaluate the effectiveness of this approach, a limonene-producing microbial system was selected as a case study. A GFP reporter gene was inserted downstream to the key bottleneck enzymes along the pathway to track cell heterogeneity using different inocula, with and without supplementing glucose in the medium composition and under different inducer concentrations. Mechanistic modelling was employed in parallel to capture the different physiological phenomena and propose exploratory hypotheses and explanatory insights into the underlying interdependencies. Built on the modelling insights, we were able to verify the potential region of plasmid recombination and validate experimentally, and more importantly, correlate the proportion of GFP subpopulation from flow cytometry with the performance of limonene bioproduction. Utilizing this method offers a more effective manner to study the conditions responsible for plasmid recombination compared to relying solely on gel analysis. The advantage lies in reducing the need for performing gel electrophoresis and developing product-sensitive biosensors. In short, we have successfully demonstrated the feasibility of our developed approach, by incorporating a GFP reporter close to the key enzymes and analysing the dynamics of their proportion using flow cytometry and cell sorting, complemented by insights from in silico mechanistic modelling, to study cell heterogeneity originated from plasmid recombination. This methodology can be generalised to other bioproduction studies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This research is supported by the National Research Foundation, Prime Minister’s Office, Singapore under its Campus for Research Excellence and Technological Enterprise (CREATE) programme. We thank the Life Sciences Institute Flow Cytometry lab for assistance with cell sorting.
Author contributions
LG, JWY, GSH, SA, SG, HL, CLP, NG, JLF. conceptualised and designed the research for this study. LG conducted experiments. JWY developed the mechanistic model. LG, JWY, GSH, SA, SG, HL, CLP, NG, JLF analysed data. LG, JWY wrote the manuscript. All authors read and approved the manuscript.
Funding
This work is funded by the EcoCTs and CNegSAF projects with the support of the National Research Foundation, Prime Minister’s Office, Singapore under its Campus for Research Excellence and Technological Enterprise (CREATE) programme.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Declarations
This article does not contain any studies with animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lucas Gelain and Jing Wui Yeoh contributed equally to this work.
Contributor Information
Nathalie Gorret, Email: ngorret@insa-toulouse.fr.
Jee Loon Foo, Email: jeeloon.foo@nus.edu.sg.
References
- Alonso S, Rendueles M, Díaz M (2012) Physiological heterogeneity of Pseudomonastaetrolens during lactobionic acid production. Appl Microbiol Biotechnol 96(6):1465–1477. 10.1007/s00253-012-4254-2 [DOI] [PubMed] [Google Scholar]
- Alonso-Gutierrez J, Chan R, Batth TS, Adams PD, Keasling JD, Petzold CJ, Lee TS (2013) Metabolic engineering of Escherichiacoli for limonene and perillyl alcohol production. Metab Eng 19:33–41. 10.1016/j.ymben.2013.05.004 [DOI] [PubMed] [Google Scholar]
- Bahl MI, Sørensen SJ, Hestbjerg Hansen L (2004) Quantification of plasmid loss in Escherichiacoli cells by use of flow cytometry. FEMS Microbiol Lett 232(1):45–49. 10.1016/S0378-1097(04)00015-1 [DOI] [PubMed] [Google Scholar]
- Balleza E, Kim JM, Cluzel P (2018) Systematic characterization of maturation time of fluorescent proteins in living cells. Nat Methods 15(1):47–51. 10.1038/nmeth.4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao SH, Zhang DY, Meng E (2019) Improving biosynthetic production of pinene through plasmid recombination elimination and pathway optimization. Plasmid 105:102431. 10.1016/j.plasmid.2019.102431 [DOI] [PubMed] [Google Scholar]
- Batard P, Jordan M, Wurm F (2001) Transfer of high copy number plasmid into mammalian cells by calcium phosphate transfection. Gene 270(1–2):61–68. 10.1016/s0378-1119(01)00467-x [DOI] [PubMed] [Google Scholar]
- Benito-Vaquerizo S, Nouse N, Schaap PJ, Hugenholtz J, Brul S, López-Contreras AM, Martins Dos Santos VAP, Suarez-Diez M (2022) Model-driven approach for the production of butyrate from CO2/H2 by a novel co-culture of C. autoethanogenum and C. beijerinckii. Front Microbiol n13:1064013. 10.3389/fmicb.2022.1064013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy C, Lesage J, Alfenore S, Gorret N, Guillouet SE (2020) Plasmid expression level heterogeneity monitoring via heterologous eGFP production at the single-cell level in Cupriavidusnecator. Appl Microbiol Biotechnol 104(13):5899–5914. 10.1007/s00253-020-10616-w [DOI] [PubMed] [Google Scholar]
- Boy C, Lesage J, Alfenore S, Guillouet SE, Gorret N (2022) Co-expression of an isopropanol synthetic operon and eGFP to monitor the robustness of Cupriavidusnecator during isopropanol production. Enzyme Microb Technol 161:110114. 10.1016/j.enzmictec.2022.110114 [DOI] [PubMed] [Google Scholar]
- Boy C, Lesage J, Alfenore S, Guillouet SE, Gorret N (2022) Study of plasmid-based expression level heterogeneity under plasmid-curing like conditions in Cupriavidusnecator. J Biotechnol 10(345):17–29. 10.1016/j.jbiotec.2021.12.015 [DOI] [PubMed] [Google Scholar]
- Camargo FP, Sarti A, Alécio AC, Sabatini CA, Adorno MAT, Duarte ICS, Silva MBAV (2020) Limonene quantification by gas chromatography with mass spectrometry (GC-MS) and its effects on hydrogen and volatile fatty acids production in anaerobic reactors. Química Nova 43(7):844–850. 10.21577/0100-4042.20170557 [Google Scholar]
- Cao H, Kuipers OP (2018) Influence of global gene regulatory networks on single cell heterogeneity of green fluorescent protein production in Bacillussubtilis. Microb Cell Fact 17:134. 10.1186/s12934-018-0985-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng BQ, Wei LJ, Lv YB, Hua Q (2019) Elevating limonene production in oleaginous yeast Yarrowialipolytica via genetic engineering of limonene biosynthesis pathway and optimization of medium composition. Biotechnol Bioproc E 24:500–506. 10.1007/s12257-018-0497-9 [Google Scholar]
- Chlebek JL, Leonard SP, Kang-Yun C, Yung MC, Ricci DP, Jiao Y, Park DM (2023) Prolonging genetic circuit stability through adaptive evolution of overlapping genes. Nucleic Acids Res 51(13):7094–7108. 10.1093/nar/gkad484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong H, Yeow J, Wang I, Song H, Jiang R (2013) Improving acetate tolerance of Escherichiacoli by rewiring its global regulator cAMP receptor protein (CRP). PLoS One 8(10):e77422. 10.1371/journal.pone.0077422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corchero JL, Villaverde A (1998) Plasmid maintenance in Escherichiacoli recombinant cultures is dramatically, steadily, and specifically influenced by features of the encoded proteins. Biotechnol Bioeng 58(6):625–632 [PubMed] [Google Scholar]
- Davey HM, Kell DB (1996) Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol Rev 60(4):641–696. 10.1128/mr.60.4.641-696.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvigne F, Goffin P (2014) Microbial heterogeneity affects bioprocess robustness: dynamic single-cell analysis contributes to understanding of microbial populations. Biotechnol J 9(1):61–72. 10.1002/biot.201300119 [DOI] [PubMed] [Google Scholar]
- Donovan RS, Robinson CW, Glick BR (1996) Review: optimizing inducer and culture conditions for expression of foreign proteins under the control of the lac promoter. J Ind Microbiol 16(3):145–154. 10.1007/BF01569997 [DOI] [PubMed] [Google Scholar]
- Du YH, Wang MY, Yang LH, Tong LL, Guo DS, Ji XJ (2022) Optimization and scale-up of fermentation processes driven by models. Bioengineering (Basel) 9(9):473. 10.3390/bioengineering9090473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friehs K (2004) Plasmid copy number and plasmid stability. Adv Biochem Eng Biotechnol 86:47–82. 10.1007/b12440 [DOI] [PubMed] [Google Scholar]
- Hegemann JH, Klein S, Heck S, Güldener U, Niedenthal RK, Fleig U (1999) A fast method to diagnose chromosome and plasmid loss in Saccharomycescerevisiae strains. Yeast 15(10B):1009–1019. 10.1002/(SICI)1097-0061(199907)15:10B%3c1009::AID-YEA396%3e3.0.CO;2-I [DOI] [PubMed] [Google Scholar]
- Heins AL, Reyelt J, Schmidt M, Kranz H, Weuster-Botz D (2020) Development and characterization of Escherichia coli triple reporter strains for investigation of population heterogeneity in bioprocesses. Microb Cell Fact 19(1):14. 10.1002/elsc.202100162 [DOI] [PMC free article] [PubMed]
- Heins AL, Weuster-Botz D (2018) Population heterogeneity in microbial bioprocesses: origin, analysis, mechanisms, and future perspectives. Bioprocess Biosyst Eng 41(7):889–916. 10.1007/s00449-018-1922-3 [DOI] [PubMed] [Google Scholar]
- Hicks M, Bachmann TT, Wang B (2020) Synthetic biology enables programmable cell-based biosensors. ChemPhysChem 21(2):132–144. 10.1002/cphc.201900739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongedijk E, Cankar K, Buchhaupt M, Schrader J, Bouwmeester H, Beekwilder J (2016) Biotechnological production of limonene in microorganisms. Appl Microbiol Biotechnol 100(7):2927–2938. 10.1007/s00253-016-7337-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joux F, Lebaron P (2000) Use of fluorescent probes to assess physiological functions of bacteria at single-cell level. Microbes Infect 2(12):1523–1535. 10.1016/s1286-4579(00)01307-1 [DOI] [PubMed] [Google Scholar]
- Koh BT, Nakashimada U, Pfeiffer M, Yap MG (1992) Comparison of acetate inhibition on growth of host and recombinant E. coli K12 strains. Biotech Lett 14:1115–1118. 10.1007/BF01027012 [Google Scholar]
- Maertens H, Demeyere K, De Reu K, Dewulf J, Vanhauteghem D, Van Coillie E, Meyer E (2020) Effect of subinhibitory exposure to quaternary ammonium compounds on the ciprofloxacin susceptibility of Escherichia coli strains in animal husbandry. BMC Microbiol 20:155. 10.1186/s12866-020-01818-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattanovich D, Borth N (2006) Applications of cell sorting in biotechnology. Microb Cell Fact 5:12. 10.1186/1475-2859-5-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda A, Hirai Y, Kodama Y, Kretzschmar WW, Hamasaki K, Kusunoki Y, Mitani H, Cullings HM, Nakamura N (2011) Easy detection of GFP-positive mutants following forward mutations at specific gene locus in cultured human cells. Mutat Res 721(1):101–107. 10.1016/j.mrgentox.2010.12.010 [DOI] [PubMed] [Google Scholar]
- Oliveira PH, Prather KJ, Prazeres DM, Monteiro GA (2009) Structural instability of plasmid biopharmaceuticals: challenges and implications. Trends Biotechnol 27(9):503–511. 10.1016/j.tibtech.2009.06.004 [DOI] [PubMed] [Google Scholar]
- Rinaldi MA, Tait S, Toogood HS, Scrutton NS (2022) Bioproduction of linalool from paper mill waste. Front Bioeng Biotechnol 10:892896. 10.3389/fbioe.2022.892896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunya S, Delvigne F, Uribelarrea JL, Molina-Jouve C, Gorret N (2012) Comparison of the transient responses of Escherichiacoli to a glucose pulse of various intensities. Appl Microbiol Biotechnol 95(4):1021–1034. 10.1007/s00253-012-3938-y [DOI] [PubMed] [Google Scholar]
- Xiao Y, Bowen C, Liu D, Zhang F (2016) Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nat Chem Biol 12:339–344. 10.1038/nchembio.2046 [DOI] [PubMed] [Google Scholar]
- Yeoh JW, Poh CL (2023) Designing a model-driven approach towards rational experimental design in bioprocess optimization. Methods Mol Biol 2553:173–187. 10.1007/978-1-0716-2617-7_9 [DOI] [PubMed] [Google Scholar]
- Yeoh JW, Jayaraman SSO, Tan SG-D, Jayaraman P, Holowko MB, Zhang J, Kang C-W, Leo HL, Poh CL (2021) A model-driven approach towards rational microbial bioprocess optimization. Biotechnol Bioeng 2:305–318. 10.1002/bit.27571 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.