Abstract
Background
Vascular endothelial cell-derived exosomes are thought to mediate disease progression by regulating macrophage polarization. However, its mechanism in diabetes mellitus (DM)-related atherosclerosis (AS) progress is unclear.
Methods
High-glucose (HG) and oxLDL were used to induce human cardiac microvascular endothelial cells (HCMECs) to mimic DM-related AS model. The conditioned medium (CM) from HG+oxLDL-induced HCMECs was incubated with THP1-M0 monocytes treated with LPS or oxLDL. The mRNA levels of macrophage M1/M2 polarization markers, NEDD4L, IκBα and PPARγ were determined by qRT-PCR. Flow cytometry was used to analyze macrophage marker. Dil-labeled oxLDL and oil red O staining were performed to assess oxLDL uptake by THP1-M0 cells. The levels of inflammatory factors were examined using ELISA. Transmission electron microscope was used for observing foam cell formation and exosome morphology. The protein levels of p-Smad1/Smad1, p-Smad2/Smad2, p-IκBα/IκBα, p-P65/P65, anti-lipid metabolism-related markers, and NEDD4L were tested by western blot. The interaction between NEDD4L and IκBα or PPARγ was assessed by Co-IP assay.
Results
The CM of HG+oxLDL-induced HCMECs could promote macrophage M1 polarization, oxLDL uptake and foam cell formation, and exosome inhibiter GW4869 eliminated these effects. NEDD4L was overexpressed in exosomes from HG+oxLDL-induced HCMECs, which could be taken up by THP1-M0 cells. Exosomal NEDD4L knockdown inhibited macrophage M1 polarization, oxLDL uptake and foam cell formation by reducing the protein levels of p-Smad1/Smad1, p-Smad2/Smad2, p-IκBα/IκBα and p-P65/P65. NEDD4L could reduce IκBα and PPARγ expression through ubiquitination.
Conclusion
HG+oxLDL-induced HCMECs-derived exosomal NEDD4L could enhance the ubiquitination of IκBα and PPARγ to facilitate macrophage M1 polarization and oxLDL uptake, thus accelerating DM-related AS.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10565-024-09973-3.
Keywords: Atherosclerosis, Diabetes mellitus, Exosome, NEDD4L, Macrophages
Introduction
Atherosclerosis (AS) is a major complication of diabetes mellitus (DM), among which DM combined with coronary artery disease (CAD), myocardial infarction and cerebrovascular disease are the main causes of death (Ye et al. 2022; Malahfji and Mahmarian 2018; Heather et al. 2022; Sattar et al. 2023). The increase of blood glucose, blood pressure, and cholesterol lipids in DM patients can promote the damage of vascular endothelial cells, accelerate the formation of local atherosclerotic plaques, and then aggravate AS progress (Yang et al. 2024; Ding et al. 2024). Epidemiological statistics show that the risk of AS and other cardiovascular complications in patients with DM is several times higher than that in normal people (Mamudu et al. 2018; Liu et al. 2023). Therefore, elucidation of the underlying molecular mechanisms affecting DM-related AS is important to mitigate disease progression.
Exosomes are extracellular vesicles that regulate a variety of biological processes, which mediate intercellular communication by delivering RNA or protein to surrounding cells (Krylova and Feng 2023; Isaac et al. 2021). Exosomes have excellent stability and the detection of plasma exosomes can contribute to the early diagnosis of diseases and provide new treatment methods (Cai et al. 2022; Nila et al. 2022). Exosomes play a vital function in the process of DM and AS (Wang et al. 2021; Yang and Fan 2021). In AS, exosomes can serve as carriers for tight junctions between macrophages, smooth muscle cells, and endothelial cells (Raman and Khanal 2021). Hypoxic HUVECs-derived exosomes could promote the switch from M1 to M2 macrophages, thus accelerating wound healing in diabetics (Cheng et al. 2024). However, whether endothelial cells mediate macrophage activity through exosomes to affect DM-associated AS progression remain unclear.
NEDD4L is an E3 ubiquitin ligase, which participates in the regulation of cell biological functions by mediating the fate of substrate proteins (Lodes et al. 2022; Chen et al. 2023). NEDD4L promoted diabetic retinopathy progression by accelerating inflammation in retinal vascular endothelial cells (Cui and Ma 2024). In diabetic nephropathy, NEDD4L enhanced high-glucose (HG)-induced podocyte inflammatory injury (Zhang et al. 2024). Besides, overexpression of NEDD4L accelerated angiogenesis, migration and proliferation in HUVECs, so NEDD4L might facilitate the development of AS and other cardiovascular disease (Liu et al. 2024a). Song et al. reported that NEDD4L knockdown inhibited oxLDL uptake and M1/M2 ratio in oxLDL-induced macrophages (Song et al. 2021). Therefore, NEDD4L may paly vital role in the progression of DM-related disease and AS. This study found that NEDD4L was upregulated in the exosomes from HG+oxLDL-induced human cardiac microvascular endothelial cells (HCMECs), but whether exosomal NEDD4L derived from HG+oxLDL-induced HCMECs regulates macrophage activity to mediate DM-related AS is unclear. Our study revealed the molecular mechanism of exosomal NEDD4L derived from HG+oxLDL-induced HCMECs in macrophage polarization and oxLDL uptake to provide potential molecular targets for treating DM-associated AS.
Materials and methods
Cell culture, treatment and transfection
HCMECs (Procell, Wuhan, China) were cultured at 37°C with 5% CO2 in specific medium (Procell). To mimic DM-related AS model, HCMECs were treated with 25 mmol/L HG and 50 μg/mL oxLDL (Yeasen, Shanghai, China) for 24 h, with 25 mmol/L mannitol and 50 μg/mL native LDL (nLDL) as control. To explore whether exosome mediated macrophage polarization, HG+oxLDL-induced HCMECs were additionally treated with exosome inhibitor GW4869 (20 μM). For transfection, HCMECs were transfected with NEDD4L shRNA (sh-NEDD4L) or sh-NC using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
Human monocytes (THP-1, Procell) were grown in RPMI-1640 and treated with 100 ng/mL PMA (Sangon, Shanghai, China) to induce THP1-M0 macrophages. For detecting the effect of NEDD4L knockdown on the expression of IκBα and PPARγ, THP1-M0 cells were transfected with NEDD4L siRNA (si-NEDD4L) or si-NC using Lipofectamine 3000.
Co-culture system
The culture medium (containing FBS) of HCMECs under different treatment conditions were collected and mixed with RPMI-1640 to prepare conditioned medium (CM) for the incubation of THP1-M0 cells 24 h. In addition, the exosomes of HCMECs under different treatment conditions was collected and then incubated with THP1-M0 cells for 24 h. The medium for THP1-M0 cell culture was additionally supplemented with 1 μg/mL LPS or 50 μg/mL oxLDL/Dil-oxLDL for macrophage activation or foam cell formation, respectively.
qRT-PCR
Total RNA was isolated from THP1-M0 cells using TRIzol Reagent (Invitrogen), followed by reverse-transcribed into cDNA using PrimeScript RT Master Mix (Takara, Dalian, China). SYBR Green (Takara) was used for qRT-PCR with specific primers (Table 1). Relative expression was examined by 2−ΔΔCt methods.
Table 1.
Primer sequences used for qRT-PCR
| Name | Primers for PCR (5’−3’) | |
|---|---|---|
| NEDD4L | Forward | AACTTGCTGGTATTCCCCCG |
| Reverse | AGTGTTCAGCTGGACTAGCG | |
|
CD206 (MRC1) |
Forward | GCCTCGTTGTTTTGCGTCTT |
| Reverse | GAGAACAGCACCCGGAATGA | |
| CD80 | Forward | GGGAAATGTCGCCTCTCTGAA |
| Reverse | CCTGGGTCTCCAAAGGTTGT | |
|
iNOS (NOS2) |
Forward | ACAGGAGGGGTTAAAGCTGC |
| Reverse | GAGGCTCCGATCAATCCAGG | |
| IL-6 | Forward | CTTCGGTCCAGTTGCCTTCT |
| Reverse | TGGAATCTTCTCCTGGGGGT | |
| NFKBIA | Forward | AAGTGATCCGCCAGGTGAAG |
| Reverse | CTGCTCACAGGCAAGGTGTA | |
| PPARG | Forward | GGGATCAGCTCCGTGGATCT |
| Reverse | TGCACTTTGGTACTCTTGAAGTT | |
| Arg1 | Forward | TTCACACCAGCTACTGGCAC |
| Reverse | CCCAGGGATGGGTTCACTTC | |
| IL-10 | Forward | TTGCCTGGTCCTCCTGACTG |
| Reverse | TCACTCTGCTGAAGGCATCTC | |
| GAPDH | Forward | CAAATTCCATGGCACCGTCA |
| Reverse | GACTCCACGACGTACTCAGC |
Flow cytometry
After co-culturing, LPS/oxLDL-induced THP1-M0 cells were collected and then incubated with anti-CD80 (ab234229, Abcam, Cambridge, CA, USA) or anti-CD206 (ab270682, Abcam) for 1 h. Finally, the positive cells of CD80 and CD206 were analyzed by a flow cytometer with Cell Quest software.
ELISA
The levels of IL-6, TNF-α, IL-1β and IL-10 in the medium of LPS/oxLDL-induced THP1-M0 cells were measured using corresponding ELISA Kit (Beyotime, Shanghai, China) according to kit instructions.
Dil-oxLDL uptake staining
After incubated with Dil-oxLDL for 24 h, the uptake of Dil-oxLDL in co-cultured THP1-M0 cells was observed under a fluorescence microscope. Cell nuclei were stained with DAPI. Fluorescence intensity was analyzed using ImageJ software.
Oil red O staining
After culturing, oxLDL-induced THP1-M0 cells were fixed with 4% paraformaldehyde and stained with oil red O solution (Beyotime) for 10 min. Oil red O intensity was observed under a microscope.
Transmission electron microscope (TEM) analysis
Co-cultured oxLDL-induced THP1-M0 cells or exosomes were fixed with glutaraldehyde solution and dehydrated with a graded series of ethanol. Cells or exosomes were embedded in propylene oxide and solidified for 48 h, and the sections were analyzed under a TEM.
Extraction of exosomes and identification
Cell medium was centrifuged at 800 g for 10 min, 12,000 g for 30 min, and 100,000 g for 2 h. Finally, the exosome sediments from cell culture medium were resuspended with PBS. Western blot (WB) was used for detecting exosome marker protein expression, and NTA analysis was performed for assessing the diameter and concentration of exosomes.
WB
After electrophoresed, extracted proteins were transferred to PVDF membrane. Then, membrane was incubated with anti-NEDD4L (1:10000, 67276-1-Ig, Proteintech, Rosemont, IL, USA), anti-Smad1 (1:1000, #6944, CST, Danvers, MA, USA), anti-p-Smad1 (1:1000, #5753, CST), anti-Smad2 (1:1000, #5339, CST), anti-p-Smad2 (1:1000, #3108, CST), anti-IκBα (1:500, ab76429, Abcam), anti-p-IκBα (1:1000, ab133462, Abcam), anti-P65 (1:10000, ab32536, Abcam), anti-p-P65 (1:10000, ab86299, Abcam), anti-PPARγ (1:1000, ab178860, Abcam), anti-ABCG1 (1:1000, 13578-1-AP, Proteintech), anti-ABCA1 (1:1000, 26564-1-AP, Proteintech), anti-LDLR (1:1000, ab52818, Abcam), anti-GAPDH (1:200000, 60004-1-Ig, Proteintech), anti-Calnexin (1:20000, 10427-2-AP, Proteintech), anti-CD63 (1:1000, 25682-1-AP, Proteintech), anti-CD81 (1:2000, 27855-1-AP, Proteintech), anti-TSG101 (1:8000, 28283-1-AP, Proteintech), and secondary antibody. Finally, protein bands were detected by ECL reagent (Beyotime).
Exosome uptake assay
The isolated exosomes for HCMECs treated with Mannitol+nLDL or HG+oxLDL (ExoMannitol+nLDL or ExoHG+oxLDL) were incubated with 10 μM Dil solution. Then, Dil-labeled ExoMannitol+nLDL or ExoHG+oxLDL were co-cultured with THP1-M0 cells, and cell nuclei were stained with DAPI. The uptake of Dil-Exo by THP1-M0 cells was visualized under a fluorescence microscope.
Cycloheximide (CHX) treatment
THP1-M0 cells transfected with si-NC/si-NEDD4L were incubated with CHX solution (50 mg/mL). At each time point (0, 2, 4, and 8 h), cells were collected for WB analysis to detect IκBα and PPARγ protein expression.
Co-IP assay
The lysates of THP1-M0 cells were incubated with anti-IgG, anti-NEDD4L, anti-IκBα and anti-PPARγ with rotation. After incubated with Protein A/G agarose beads, immunoprecipitated proteins were detected by WB.
Ubiquitination assay
THP1-M0 cells were co-transfected with si-NC/si-NEDD4L and Myc-Ub expression vectors, followed by incubated with Myc-tag magnetic beads. WB was used for measuring the ubiquitination level of IκBα and PPARγ in immunoprecipitated proteins.
Statistical analysis
Data were expressed as mean ± SD and analyzed by GraphPad Prism 9.0 software. Statistical differences were assessed by Student’s t-test or ANOVA. P<0.05 was denoted statistically significant.
Results
Exosomes from HG+oxLDL-induced HCMECs promoted macrophage M1 polarization
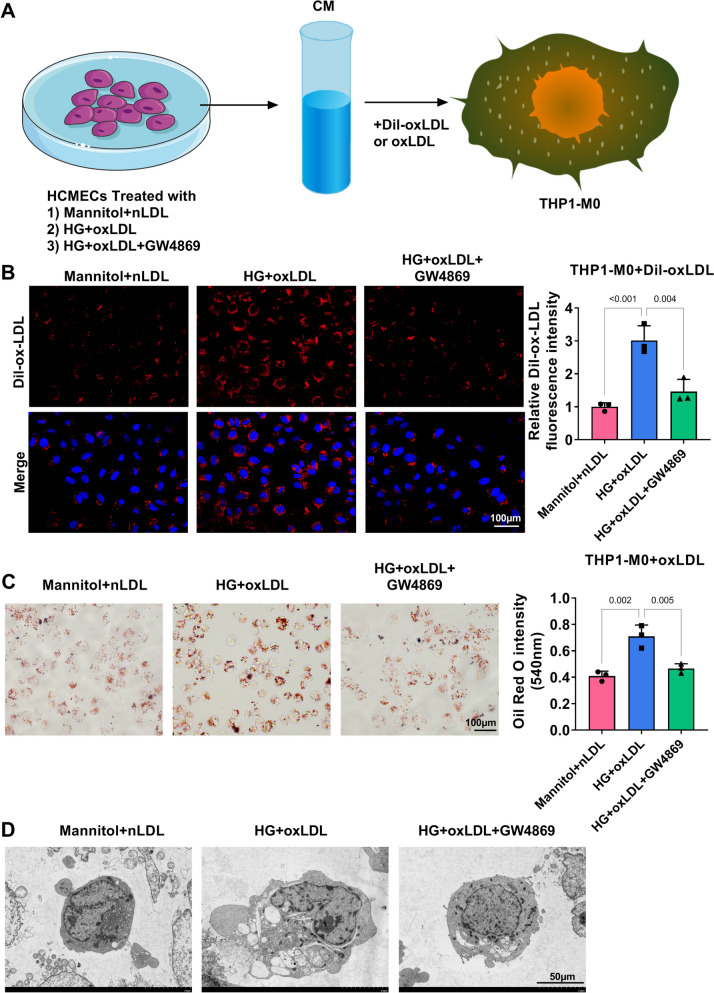
LPS treatment enhanced CD80, iNOS, and IL-6 mRNA levels, while reduced IL-10 mRNA level in THP-1-M0 cells (Supplementary Fig. 1A-B). LPS increased CD80 positive cells, IL-6, TNF-α and IL-1β levels, while decreased CD206 positive cells and IL-10 level in THP-1-M0 cells (Supplementary Fig. 1C-H). These data suggested that LPS could induce macrophage M1 polarization. To determine the role of exosomes from HG+oxLDL-treated HCMECs on macrophage polarization, CM from HCMECs treated with HG+oxLDL as well as GW4869 were collected and incubated with LPS-induced THP1-M0 cells (Fig. 1A). Through detecting the mRNA levels of M1 markers (CD80, iNOS and IL-6) and M2 markers (CD206, Arg-1 and IL-10), we found that HG+oxLDL treatment enhanced CD80, iNOS and IL-6 levels, while reduced CD206, Arg-1 and IL-10 levels in LPS-induced THP1-M0 cells. However, these effect could be abolished by exosome inhibitor GW4869 (Fig. 1B-C). Similarly, HG+oxLDL treatment promoted CD80 positive cells and decreased CD206 positive cells in LPS-induced THP1-M0 cells, while GW4869 treatment also eliminated these effect (Fig. 1D-E). Furthermore, HG+oxLDL increased IL-6, TNF-α, IL-1β and decreased IL-10 levels, while GW4869 reversed this effect (Fig. 1F-I). These data suggested that exosome derived from HG+oxLDL-induced HCMECs might accelerate macrophage M1 polarization.
Fig. 1.
Exosomes from HG+oxLDL-induced HCMECs mediated macrophage M1 polarization. A Flow chart of cell treatment and co-incubation. B-C qRT-PCR for detecting CD80, iNOS, IL-6, CD206, Arg-1 and IL-10 mRNA levels in LPS-induced THP1-M0 cells. D-E Flow cytometry for analyzing CD80 and CD206 positive cells in LPS-induced THP1-M0 cells. F-I ELISA was used to test the levels of IL-6, TNF-α, IL-1β and IL-10 in LPS-induced THP1-M0 cells. All experiments were performed in triplicate and P<0.05 was denoted statistically significant
Exosomes from HG+oxLDL-induced HCMECs promoted oxLDL uptake and foam cell formation
To determine whether exosomes from HG+oxLDL-treated HCMECs regulated oxLDL uptake and foam cell formation, CM from treated HCMECs were incubated with THP1-M0 cells induced with oxLDL or Dil-oxLDL (Fig. 2A). Through analyzing, we observed that the CM of HG+oxLDL-induced HCMECs promoted oxLDL uptake and enhanced lipid accumulation in oxLDL-induced THP1-M0 cells, while these effects were eliminated by GW4869 (Fig. 2B-C). Under TEM, the CM of HG+oxLDL-induced HCMECs increased lipid droplets and foam cell formation in oxLDL-induced THP1-M0 cells, and GW4869 treatment could abolish this effect (Fig. 2D). Thus, exosome derived from HG+oxLDL-induced HCMECs could promote foam cell formation.
Fig. 2.
Exosomes from HG+oxLDL-induced HCMECs mediated oxLDL uptake and foam cell formation. A Flow chart of cell co-incubation. B Fluorescence microscope was used to observe Dil-labeled oxLDL in THP1-M0 cells. C Oil red O staining to evaluate lipid deposition in oxLDL-induced THP1-M0 cells. D TEM for observing foam cell formation. All experiments were performed in triplicate and P<0.05 was denoted statistically significant
NEDD4L was upregulated in exosomes from HG+oxLDL-induced HCMECs
To carry out further analysis, exosomes were isolated from the CM of HCMECs treated with Mannitol+nLDL or HG+oxLDL, and their micromorphology was observed under TEM (Fig. 3A). Besides, the levels of CD63, CD81 and TSG101 were analyzed by WB to confirm the successful extraction of exosomes (Fig. 3B). NTA was used for analyzing the diameter and concentration of exosomes (Fig. 3C). The detection of NEDD4L mRNA level confirmed that NEDD4L expression was higher in HG+oxLDL-induced HCMECs (Fig. 3D), and this trend was also detected in exosomes (Fig. 3E). After Dil-labeled exosome was incubated with THP1-M0 cells, we confirmed the exosome uptake by THP1-M0 cells through observing fluorescent signal under TEM (Fig. 3F). After incubation, exosomes derived from HG+oxLDL-induced HCMECs could increase NEDD4L mRNA and protein levels in THP1-M0 cells (Fig. 3G-H).
Fig. 3.
NEDD4L expression in exosomes from HG+oxLDL-induced HCMECs. A TEM for observing the microstructure of exosomes from HCMECs treated with Mannitol+nLDL or HG+oxLDL. B WB for testing TSG101, CD81, CD63 and Calnexin protein expression in exosomes from HCMECs treated with Mannitol+nLDL or HG+oxLDL. C NTA for detecting the particle sizes and concentrations of exosomes from HCMECs treated with Mannitol+nLDL or HG+oxLDL. D NEDD4L mRNA level was tested by qRT-PCR in HCMECs treated with Mannitol+nLDL or HG+oxLDL. E NEDD4L mRNA level in exosomes from HCMECs treated with Mannitol+nLDL or HG+oxLDL was measured by qRT-PCR. F Fluorescence microscope was used to observe Dil-labeled Exo in THP1-M0 cells after co-culturing with ExoMannitol+nLDL or ExoHG+oxLDL. G-H qRT-PCR and WB for testing NEDD4L mRNA and protein levels in THP1-M0 cells after co-culturing with ExoMannitol+nLDL or ExoHG+oxLDL. All experiments were performed in triplicate and P<0.05 was denoted statistically significant
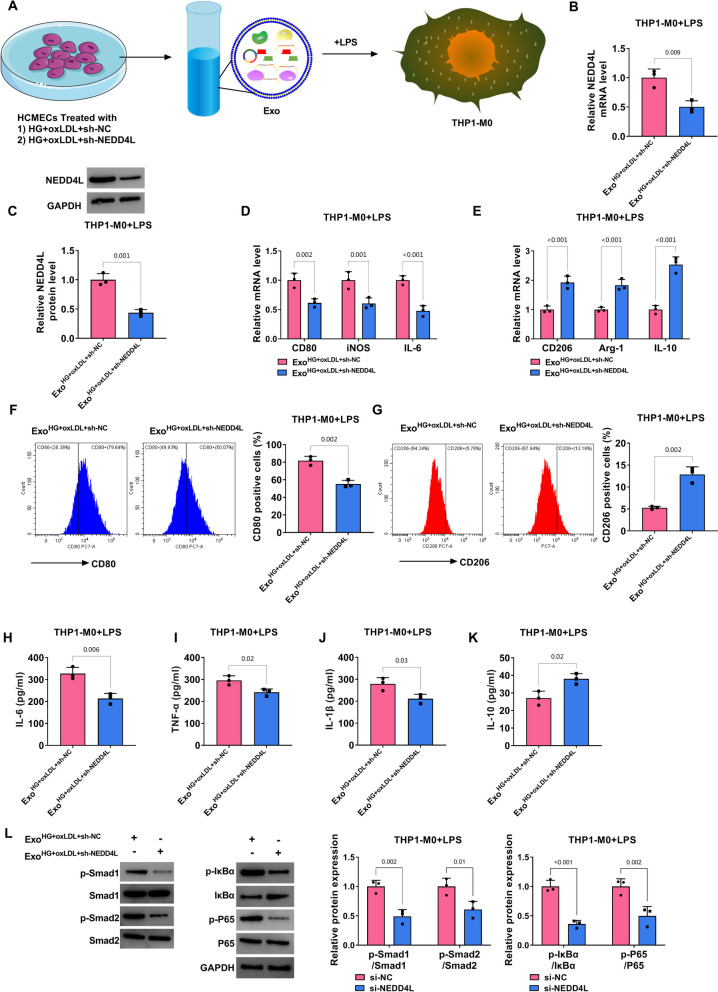
Exosomal NEDD4L knockdown inhibited macrophage M1 polarization mediated by HG+oxLDL-induced HCMECs
To confirm the role of exosomal NEDD4L derived from HG+oxLDL-induced HCMECs on macrophage M1 polarization, ExoHG+oxLDL+sh-NC/sh-NEDD4L was incubated with THP-1/M0 cells treated with LPS (Fig. 4A). Compared to ExoHG+oxLDL+sh-NC treatment, NEDD4L levels were decreased in LPS-induced THP-1/M0 cells treated with ExoHG+oxLDL+sh-NEDD4L (Fig. 4B-C). Further analysis revealed that ExoHG+oxLDL+sh-NEDD4L reduced CD80, iNOS and IL-6 levels, while enhanced CD206, Arg-1 and IL-10 levels in LPS-induced THP1-M0 cells (Fig. 4D-E). Besides, ExoHG+oxLDL+sh-NEDD4L decreased CD80 positive cells, IL-6, TNF-α and IL-1β levels, while increased CD206 positive cells and IL-10 level in LPS-induced THP1-M0 cells (Fig. 4F-K). In addition, ExoHG+oxLDL+sh-NEDD4L could inhibit the protein levels of p-Smad1/Smad1, p-Smad2/Smad2, p-IκBα/IκBα and p-P65/P65 in LPS-induced THP1-M0 cells (Fig. 4L).
Fig. 4.
Effect of exosomal NEDD4L knockdown from HG+oxLDL-induced HCMECs on macrophage M1 polarization. A Flow chart of cell treatment and co-incubation. B-C qRT-PCR and WB for detecting NEDD4L mRNA and protein levels in LPS-induced THP1-M0 cells. D-E qRT-PCR for testing CD80, iNOS, IL-6, CD206, Arg-1 and IL-10 mRNA levels in LPS-induced THP1-M0 cells. F-G CD80 and CD206 positive cells were analyzed by flow cytometry in LPS-induced THP1-M0 cells. H-K The levels of IL-6, TNF-α, IL-1β and IL-10 in LPS-induced THP1-M0 cells was analyzed by ELISA. L Protein levels of p-Smad1/Smad1, p-Smad2/Smad2, p-IκBα/IκBα and p-P65/P65 in LPS-induced THP1-M0 cells were examined by WB. All experiments were performed in triplicate and P<0.05 was denoted statistically significant
Exosomal NEDD4L knockdown from HG+oxLDL-induced HCMECs reduced oxLDL uptake and foam cell formation
Then, we assessed exosomal NEDD4L knockdown derived from HG+oxLDL-induced HCMECs on foam cell formation by incubating with ExoHG+oxLDL+sh-NC/sh-NEDD4L and THP-1/M0 cells treated with Dil-oxLDL or oxLDL (Fig. 5A). The measuring of NEDD4L mRNA and protein levels suggested that NEDD4L expression indeed downregulated in oxLDL-induced THP1-M0 cells co-cultured with ExoHG+oxLDL+sh-NEDD4L (Fig. 5B-C). ExoHG+oxLDL+sh-NEDD4L decreased oxLDL uptake, lipid accumulation and foam cell formation in oxLDL-induced THP1-M0 cells (Fig. 5D-F). Additionally, we detected the levels of anti-lipid metabolism-related proteins using WB, and confirmed that ExoHG+oxLDL+sh-NEDD4L enhanced PPARγ, ABCG1, ABCA1 and LDLR protein levels in oxLDL-induced THP1-M0 cells (Fig. 5G).
Fig. 5.
Effect of exosomal NEDD4L knockdown from HG+oxLDL-induced HCMECs on oxLDL uptake and foam cell formation. A Flow chart of cell co-incubation. B-C NEDD4L mRNA and protein levels in oxLDL-induced THP1-M0 cells were examined by qRT-PCR and WB. D Dil-labeled oxLDL in THP1-M0 cells was observed using fluorescence microscope. E Lipid deposition in oxLDL-induced THP1-M0 cells was assessed by Oil red O staining. F Foam cell formation was observed under TEM. G Protein levels of PPARγ, ABCG1, ABCA1 and LDLR in oxLDL-induced THP1-M0 cells were tested by WB. All experiments were performed in triplicate and P<0.05 was denoted statistically significant
NEDD4L mediated the ubiquitination of IκBα and PPARγ
After decreasing NEDD4L mRNA and protein expression using the transfection of si-NEDD4L in THP1-M0 cells, IκBα and PPARγ protein levels were upregulated by NEDD4L knockdown, while the mRNA level was unaffected (Fig. 6A-B). CHX treatment assay results revealed that silencing of NEDD4L enhanced the protein stability of IκBα and PPARγ (Fig. 6C-D). Further Co-IP assay was used to evaluate the interaction between NEDD4L and IκBα or PPARγ. As shown in Fig. 6E-F, NEDD4L could interact with IκBα and PPARγ, respectively. Furthermore, NEDD4L knockdown inhibited the ubiquitination level of IκBα and PPARγ (Fig. 6G-H). The above results confirmed that NEDD4L might promote IκBα and PPARγ ubiquitination to suppress their expression. Thus, we believed that exosomal NEDD4L derived from HG+oxLDL-induced HCMECs promoted macrophage M1 polarization, oxLDL uptake and foam cell formation by ubiquitinating IκBα/PPARγ and phosphorylating Smad1/Smad2 (Fig. 7).
Fig. 6.
Effect of NEDD4L on the ubiquitination of IκBα and PPARγ. A-B The mRNA and protein levels of NEDD4L, IκBα and PPARγ were detected by qRT-PCR and WB in THP1-M0 cells transfected with si-NC/si-NEDD4L. C-D WB was used for detected IκBα and PPARγ protein levels in THP1-M0 cells transfected with si-NC/si-NEDD4L and treated with CHX for indicated time points. E-F Co-IP for assessing the interaction between NEDD4L and IκBα or PPARγ. G-H Detection of ubiquitination levels of IκBα and PPARγ in THP1-M0 cells transfected with si-NC/si-NEDD4L. All experiments were performed in triplicate and P<0.05 was denoted statistically significant
Fig. 7.
The mechanism diagram of this study. Exosomal NEDD4L derived from HG+oxLDL-induced HCMECs could ubiquitinate IκBα/PPARγ and phosphorylate Smad1/Smad2, thus promoting macrophage M1 polarization, oxLDL uptake and foam cell formation
Discussion
Mannitol, a commonly used osmotic regulator, has a molecular structure similar to that of glucose but is not metabolized by cells. Cells may face changes in osmotic pressure in HG environment, and the use of mannitol as a control can model the changes in osmotic pressure caused by HG while excluding the effects of glucose metabolism on cells. In addition, mannitol does not participate in intracellular glucose metabolic pathways and therefore does not interfere with processes such as glycolysis, gluconeogenesis, or tricarboxylic acid cycle in cells. This makes mannitol an ideal control to assess the cell-specific effects of HG environment, especially in studies where metabolic interference needs to be excluded. nLDL is the product of oxidative modification of LDL, which has undergone significant changes in its structure and function. The use of nLDL as a control for oxLDL allows for the simulation of the oxidative modification of LDL in vivo and the assessment of the effect of the test substances under oxidative stress conditions. By comparing the effects of the test substances in the presence of nLDL and oxLDL, the potential mechanism of action in the environment of LDL with different degrees of oxidation can be revealed.
As a major complication of diabetes, AS is still an important cause of death in patients with DM. Macrophage polarization and pro-inflammatory activity are associated with the development of DM-related AS (Huang et al. 2023; Zhang et al. 2023). Studies have shown that insulin resistance in DM patients may accelerate vulnerable plaque formation by promoting macrophage inflammation and foam cell formation, leading to the occurrence of AS (Ward et al. 2018; Park et al. 2018). Therefore, exploring the analytical mechanisms affecting macrophage function may provide new ideas for treating DM-related AS. Vascular endothelial cell injury is considered to be one of the main causes of AS development (Shen et al. 2023). According to the report, endothelial cells-derived exosomes may regulate the AS progress (Bai et al. 2020). However, whether endothelial cells-derived exosomes regulate macrophage activity to mediate DM-related AS remains to be studied. In this, we cultured THP1-M0 cells with the CM from HG+oxLDL-induced HCMECs treated with or without exosome inhibitor GW4869. Our data indicated that co-cultured with the CM from HG+oxLDL-induced HCMECs could promote macrophage M1 polarization, oxLDL uptake and foam cell formation, while these effects were abolished by the treatment of GW4869. This information clearly suggested that exosomes derived from HG+oxLDL-induced HCMECs might promote macrophage M1 polarization, oxLDL uptake and foam cell formation, thus accelerating AS progression.
It had been reported that vascular smooth muscle cell-derived exosomal miR-221/222 from diabetic sources promoted inflammation and M1 polarization to accelerate AS plaque development (Yu et al. 2023). Therefore, exploring the effective RNA and protein in exosomes derived from HG+oxLDL-induced HCMECs is necessary. NEDD4L (also known as NEDD4-2), a member of the NEDD4 family of ubiquitin ligases, acts on the epithelial Na(+) channel to help maintain sodium balance and cellular homeostasis (Goel et al. 2015). Recently, NEDD4L roles in various human diseases have been demonstrated, such as diabetic kidney disease (Han et al. 2024) and skin cancer (Liu et al. 2024b). Given the positive role of NEDD4L in DM-related complications and AS (Cui and Ma 2024; Zhang et al. 2024; Liu et al. 2024a; Song et al. 2021), NEDD4L was chosen for this study. Here, NEDD4L was detected to be highly expressed in HG+oxLDL-induced HCMECs and their derived exosomes, which could be uptaken by THP1-M0 cells. Through further analysis, we determined that exosomal NEDD4L knockdown derived from HG+oxLDL-induced HCMECs could inhibit macrophage M1 polarization, oxLDL uptake and foam cell formation, revealing that endothelial cells-derived exosomes probably mediated macrophage activity via transferring NEDD4L, thus regulating DM-related AS process.
Previous study indicated that NEDD4L knockdown inhibited the ubiquitination of PPARγ, reduced the phosphorylation of Smad1/2, and promoted anti-lipid metabolism-related proteins (ABCA1, ABCG1 and LDLR) to repress oxLDL uptake and M1/M2 ratio in macrophages, thereby alleviating AS process (Song et al. 2021). Besides, NEDD4L had been confirmed to enhance the ubiquitination of IκBα to accelerate diabetic retinopathy progression (Cui and Ma 2024). In this, we detected that exosomal NEDD4L knockdown derived from HG+oxLDL-induced HCMECs decreased the phosphorylation of Smad1/2 and IκBα/P65, while increased PPARγ, ABCA1, ABCG1 and LDLR protein levels. Consistent with previous studies (Song et al. 2021; Cui and Ma 2024), we pointed out that NEDD4L inhibited IκBα and PPARγ expression through ubiquitination.
Of course, there are some limitations to this study. Although we have revealed the value of NEDD4L as a potential diagnostic marker for DM-associated AS, the feasibility of its clinical translation remains a great challenge. At present, due to resource limitations, we have not yet involved animal experiments. Further animal experiments are necessary, which may provide a rationale for the clinical application of NEDD4L. Targeted inhibition of NEDD4L may be a potential strategy for treating DM-associated AS, which also needs to be verified by further studies. In the future, we will further explore the regulatory role of NEDD4L in other potential downstream molecules or pathways in macrophages to further reveal the comprehensive mechanism of NEDD4L affecting inflammatory response and lipid metabolism. In addition, comparison with traditional biomarkers is an important part of assessing the diagnostic potential of NEDD4L. However, in the current study, our main goal was to initially validate the feasibility of exosome NEDD4L as a novel biomarker and lay the foundation for its subsequent studies. We have considered the comparison of NEDD4L with other traditional biomarkers in future plans to more fully evaluate its diagnostic value. Also, we are already in the process of collecting more detailed clinical information and plan to stratify patients according to disease severity in future studies to explore the relationship between NEDD4L expression levels and disease progression.
In conclusion, the increased expression of NEDD4L in HG+oxLDL-induced HCMECs-derived exosomes accelerated macrophage M1 polarization, oxLDL uptake and foam cell formation by ubiquitinating IκBα/PPARγ and phosphorylating Smad1/2. These data support targeted therapies to reduce NEDD4L levels in endothelial cells to alleviate the development of DM-related AS by altering macrophage activity.
Supplementary information
Supplementary Fig. 1 LPS induced macrophage M1 polarization. (A-B) qRT-PCR for detecting CD80, iNOS, IL-6, CD206, Arg-1 and IL-10 mRNA levels in LPS-induced THP1-M0 cells. (C-D) Flow cytometry for analyzing CD80 and CD206 positive cells in LPS-induced THP1-M0 cells. (E-H) ELISA was used to test the levels of IL-6, TNF-α, IL-1β and IL-10 in LPS-induced THP1-M0 cells. All experiments were performed in triplicate and P<0.05 was denoted statistically significant. (PNG 402 kb)
(PDF 2272 kb)
Acknowledgements
None.
Author contribution
Guozhu Chen conducted the experiments and wrote the draft, designed and supervised the study. Yisong Pei and Peng Jiang collected and analyzed the data. Qiaoling Ye plotted the figures and arrange the images. Zulong Xie and Laxman Gyawali contributed the methodology, operated the software and edited the manuscript. All authors reviewed and approved the final version.
Funding
This study was supported by the mechanism of metformin protection against ischemia-reperfusion myocardial injury based on AMPK (2019-02).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Conflicts of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bai S, Yin Q, Dong T, Dai F, Qin Y, Ye L, et al. Endothelial progenitor cell-derived exosomes ameliorate endothelial dysfunction in a mouse model of diabetes. Biomed Pharmacother. 2020;131:110756. [DOI] [PubMed] [Google Scholar]
- Cai H, Pang Y, Wang Q, Qin W, Wei C, Li Y, et al. Proteomic profiling of circulating plasma exosomes reveals novel biomarkers of Alzheimer's disease. Alzheimers Res Ther. 2022;14(1):181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Li K, Guo J, Chen HN, Ming Y, Jin Y, et al. circNEIL3 inhibits tumor metastasis through recruiting the E3 ubiquitin ligase Nedd4L to degrade YBX1. Proc Natl Acad Sci USA. 2023;120(13):e2215132120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Xie X, Hu L, Zhou W, Mi B, Xiong Y, et al. Hypoxia endothelial cells-derived exosomes facilitate diabetic wound healing through improving endothelial cell function and promoting M2 macrophages polarization. Bioact Mater. 2024;33:157–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Ma J. NEDD4L Promotes IkappaBalpha Ubiquitination and Degradation in the Pathogenesis of Diabetic Retinopathy. Curr Eye Res. 2024;49(1):62–72. [DOI] [PubMed] [Google Scholar]
- Ding X, Qiu Y, Wu G, Li S, Cai M, Liang Y, et al. l-thyroxine attenuates extracellular Hsp90alpha-induced vascular endothelial calcification in diabetes mellitus, as revealed by parallel metabolic profiles. Atherosclerosis. 2024;392:117527. [DOI] [PubMed] [Google Scholar]
- Goel P, Manning JA, Kumar S. NEDD4-2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene. 2015;557(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Wu S, Dong Y, Liu Y, Sun B, Chen L. Aberrant expression of NEDD4L disrupts mitochondrial homeostasis by downregulating CaMKKbeta in diabetic kidney disease. J Transl Med. 2024;22(1):465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heather LC, Hafstad AD, Halade GV, Harmancey R, Mellor KM, Mishra PK, et al. Guidelines on models of diabetic heart disease. Am J Physiol Heart Circ Physiol. 2022;323(1):H176–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Gao W, Zhong X, Wu H, Zhou Y, Ma Y, et al. Epigenetically altered macrophages promote development of diabetes-associated atherosclerosis. Front Immunol. 2023;14:1196704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac R, Reis FCG, Ying W, Olefsky JM. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021;33(9):1744–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylova SV, Feng D. The Machinery of Exosomes: Biogenesis, Release, and Uptake. Int J Mol Sci. 2023;24(2):1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Song F, Zhou X, Wu C, Huang H, Wu W, et al. NEDD4L is a promoter for angiogenesis and cell proliferation in human umbilical vein endothelial cells. J Cell Mol Med. 2024a;28(8):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang N, Yang R, Luan J, Cao M, Zhai C, et al. E3 Ubiquitin Ligase NEDD4L Negatively Regulates Skin Tumorigenesis by Inhibiting IL-6/GP130 Signaling Pathway. J Invest Dermatol. 2024b; [DOI] [PubMed]
- Liu Z, Wang H, Yang Z, Lu Y, Zou C. Causal associations between type 1 diabetes mellitus and cardiovascular diseases: a Mendelian randomization study. Cardiovasc Diabetol. 2023;22(1):236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes DE, Zhu J, Tsai NP. E3 ubiquitin ligase Nedd4-2 exerts neuroprotective effects during endoplasmic reticulum stress. J Neurochem. 2022;160(6):613–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malahfji M, Mahmarian JJ. Imaging to Stratify Coronary Artery Disease Risk in Asymptomatic Patients with Diabetes. Methodist Debakey Cardiovasc J. 2018;14(4):266–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamudu HM, Alamian A, Paul T, Subedi P, Wang L, Jones A, et al. Diabetes, subclinical atherosclerosis and multiple cardiovascular risk factors in hard-to-reach asymptomatic patients. Diab Vasc Dis Res. 2018;15(6):519–27. [DOI] [PubMed] [Google Scholar]
- Nila IS, Sumsuzzman DM, Khan ZA, Jung JH, Kazema AS, Kim SJ, et al. Identification of exosomal biomarkers and its optimal isolation and detection method for the diagnosis of Parkinson's disease: A systematic review and meta-analysis. Ageing Res Rev. 2022;82:101764. [DOI] [PubMed] [Google Scholar]
- Park K, Li Q, Evcimen ND, Rask-Madsen C, Maeda Y, Maddaloni E, et al. Exogenous Insulin Infusion Can Decrease Atherosclerosis in Diabetic Rodents by Improving Lipids, Inflammation, and Endothelial Function. Arterioscler Thromb Vasc Biol. 2018;38(1):92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman P, Khanal S. Leptin in Atherosclerosis: Focus on Macrophages, Endothelial and Smooth Muscle Cells. Int J Mol Sci. 2021;22(11):5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattar N, McMurray J, Boren J, Rawshani A, Omerovic E, Berg N, et al. Twenty Years of Cardiovascular Complications and Risk Factors in Patients With Type 2 Diabetes: A Nationwide Swedish Cohort Study. Circulation. 2023;147(25):1872–86. [DOI] [PubMed] [Google Scholar]
- Shen J, San W, Zheng Y, Zhang S, Cao D, Chen Y, et al. Different types of cell death in diabetic endothelial dysfunction. Biomed Pharmacother. 2023;168:115802. [DOI] [PubMed] [Google Scholar]
- Song F, Li JZ, Wu Y, Wu WY, Wang Y, Li G. Ubiquitinated ligation protein NEDD4L participates in MiR-30a-5p attenuated atherosclerosis by regulating macrophage polarization and lipid metabolism. Mol Ther Nucleic Acids. 2021;26:1303–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li Z, Liu Y, Yuan L. Exosomes in atherosclerosis: performers, bystanders, biomarkers, and therapeutic targets. Theranostics. 2021;11(8):3996–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MG, Li G, Hao M. Apoptotic beta-cells induce macrophage reprogramming under diabetic conditions. J Biol Chem. 2018;293(42):16160–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Fan Z. VSMCs-derived exosomes: A novel therapeutic target for diabetic atherosclerosis. Int J Cardiol. 2021;343:14. [DOI] [PubMed] [Google Scholar]
- Yang DR, Wang MY, Zhang CL, Wang Y. Endothelial dysfunction in vascular complications of diabetes: a comprehensive review of mechanisms and implications. Front Endocrinol. 2024;15:1359255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Li L, Wang M, Ma Q, Tian Y, Zhang Q, et al. Diabetes Mellitus Promotes the Development of Atherosclerosis: The Role of NLRP3. Front Immunol. 2022;13:900254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Douglas HF, Wathieu D, Braun RA, Edomwande C, Lightell DJ Jr, et al. Diabetes is accompanied by secretion of pro-atherosclerotic exosomes from vascular smooth muscle cells. Cardiovasc Diabetol. 2023;22(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li L, Li Y, Jiang H, Sun Z, Zang G, et al. Disruption of COMMD1 accelerates diabetic atherosclerosis by promoting glycolysis. Diab Vasc Dis Res. 2023;20(1):14791641231159009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang S, Chong N, Chen D, Shu J, Sun J, et al. GDF-15 alleviates diabetic nephropathy via inhibiting NEDD4L-mediated IKK/NF-kappaB signalling pathways. Int Immunopharmacol. 2024;128:111427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 LPS induced macrophage M1 polarization. (A-B) qRT-PCR for detecting CD80, iNOS, IL-6, CD206, Arg-1 and IL-10 mRNA levels in LPS-induced THP1-M0 cells. (C-D) Flow cytometry for analyzing CD80 and CD206 positive cells in LPS-induced THP1-M0 cells. (E-H) ELISA was used to test the levels of IL-6, TNF-α, IL-1β and IL-10 in LPS-induced THP1-M0 cells. All experiments were performed in triplicate and P<0.05 was denoted statistically significant. (PNG 402 kb)
(PDF 2272 kb)
Data Availability Statement
No datasets were generated or analysed during the current study.