Abstract
It has been shown that premature translation of the plasmid-mediated toxin in hok/sok of plasmid R1 and pnd/pndB of plasmid R483 is prevented during transcription of the hok and pnd mRNAs by the formation of metastable hairpins at the 5′-end of the mRNA. Here, an experimental approach is presented, which allows the accurate measurement of the refolding kinetics of the 5′-end RNA fragments in vitro without chemically modifying the RNA. The method is based on acid denaturation followed by a pH-jump to neutral pH as a novel way to trap kinetically favoured RNA secondary structures, allowing the measurement of a wide range of biologically relevant refolding rates, with or without the use of standard stopped-flow equipment. The refolding rates from the metastable to the stable conformation in both the hok74 and pnd58 5′-end RNA fragments were determined by using UV absorbance changes corresponding to the structural rearrangements. The measured energy barriers showed that the refolding path does not need complete unfolding of the metastable structures before the formation of the final structures. Two alternative models of such a pathway are discussed.
INTRODUCTION
Conformational switching between alternative secondary structures in RNA is thought to play a fundamental role in a number of biological processes (1–5). For instance, these alternative structures are involved in turning on or off of open-reading frames (6,7), in replication of RNA in viroids and viruses (2,8–11), in plasmid copy number control (12), and in rRNA during translation (5). The secondary structural elements in RNA are very stable as compared with proteins and tertiary RNA interactions (13–17). The high stability of these secondary structural elements could easily create such high energy barriers that the refolding rates could even become biologically irrelevant (18,19), since there is a high risk for the RNA to get captured into a kinetic trap (20,21). This is especially important, since the final structure of an RNA often depends on a specific folding pathway determined by the RNA itself (2–4,6,22,23) and because the folding pathway of an RNA molecule does not always lead to the conformation with the lowest free energy (6,12,18,23). First, RNA folds immediately when it leaves the RNA polymerase, and secondly, the early steps in RNA folding depend on the number of nucleation points available, which mostly lead to local hairpin formation (18,23). During the folding of the nascent RNA chain, immediately after synthesis, the formation of local hairpins is strongly favoured over long-range interactions, even if the long-range interactions are energetically more favourable. The sequential folding pathway can therefore lead to the formation of short-range hairpins, which can prevent the formation of possibly more stable long-range RNA interactions due to significant energy barriers (7,24,25). This trapping of short-range metastable RNA structures can either be further stabilised by tertiary interactions, or allowed to refold into the lower (global) minimum free energy structure (22,26,27).
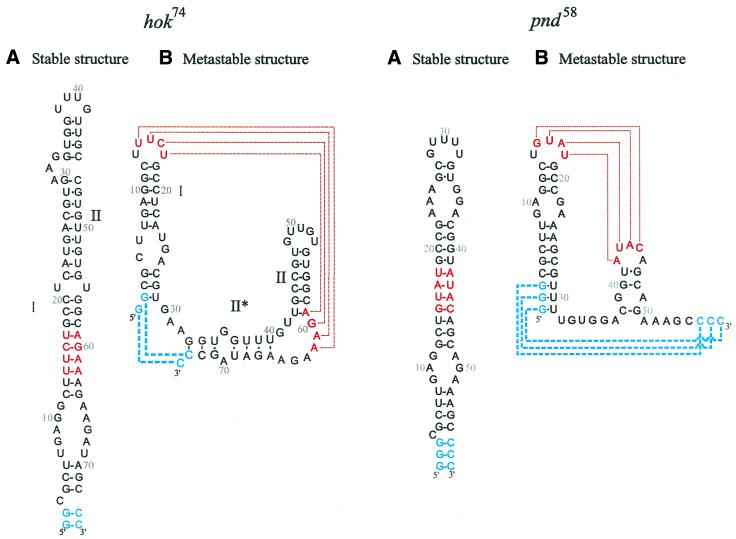
The formation of a relatively stable intermediate RNA structure in the folding pathway allows some RNAs to perform two biologically functional roles; one temporarily, when it is trapped into a metastable conformation, and another after refolding into its stable conformation (7,12). One of the systems, in which such a conformational switch plays an important role, is the hok/sok (host killing/suppression of killing) system in Escherichia coli. This hok/sok system mediates plasmid maintenance by killing of plasmid-free cells via a complex series of RNA-regulated mRNA rearrangements including antisense RNA binding (1,6,28). The conformational switch in the 5′-end of hok mRNA and in the family member pnd (promotion of nucleic acid degradation) mRNA, which has a shorter and simpler 5′-end switch, provides an excellent example to study the behaviour of the structural switches in RNA, because the main structural transition is located in a well defined separate domain in the mRNA. These hok and pnd domains (see Fig. 1) can be trapped in the metastable and stable structural states, which allowed us to utilise a variety of different experimental methods to examine the refolding process (25). Furthermore, proteins or RNA chaperones do not seem to be involved in the refolding process (K.Gerdes, unpublished data), indicating that the secondary structural refolding switch is entirely guided by the RNA itself (1).
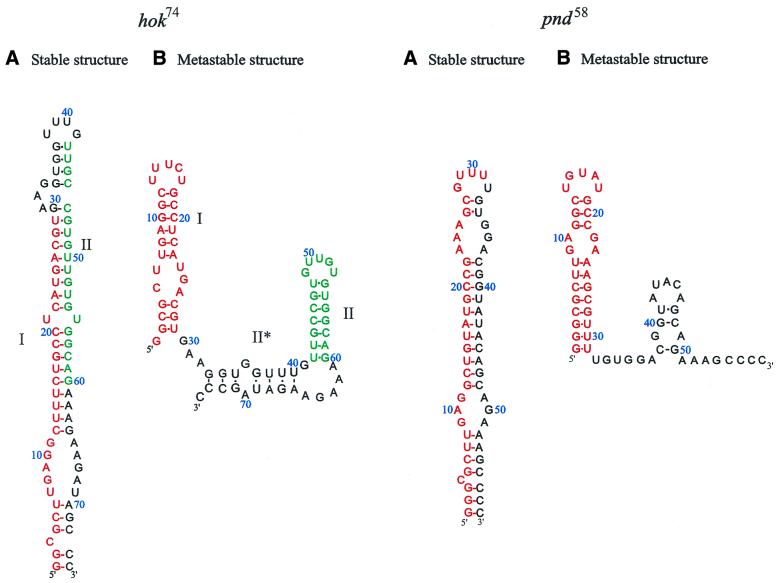
Figure 1.
The stable and metastable conformation of the hok74 and pnd58 5′-end RNA fragments. The position of metastable hairpin I is indicated in red and the position of metastable hairpin II is indicated in green. (A) Stable structure and (B) metastable structure.
Previously, we were able to determine the metastable structures and the kinetics of refolding into the stable conformation at the 5′-end of both the isolated hok and pnd mRNA fragments, by a heating and rapid cooling cycle followed by a structure probing analysis (25). However, determination of accurate kinetic parameters with enzymatic or chemical probing requires a laborious experimental set-up followed by electrophoretic separation and quantitation of the reaction products for each data point (29). Furthermore, the probing experiments are usually not suitable for detecting rapid conformational changes and are rarely used for kinetic refolding rates of <1 min (30). These limitations led us to search for a different experimental approach, which could allow us to determine kinetic refolding rates of the RNA fragments over a large time and temperature range. To achieve this goal, we used a pH-jump method, resembling the one described by Bina-Stein and Crothers (31,32). However, in our case the lowering of the pH was not done to form a new equilibrium structure but was meant to denature the RNA, followed by a rapid pH-jump to neutral pH, which allowed us to kinetically trap the metastable conformation in the RNA fragments. We could then follow the relaxation kinetics by measuring the change in absorbance at 260 nm. This experimental approach has several advantages over the previous method of capturing the metastable structures by a heating and rapid cooling cycle (25): (i) there is no need for probing or electrophoretic separation; (ii) it does not require chemical modification of the RNA fragments; (iii) the accuracy of the determined kinetic refolding rates is increased; (iv) it allows monitoring of real-time kinetics; (v) the use of standard stopped-flow equipment is possible. This last advantage is of great importance, because it provided us with real-time relaxation kinetics in time scales of <1 min, while maintaining a high experimental accuracy. Furthermore, the results of this approach could be cross-checked with the results of the previous structural probing analyses. Therefore, this method of acid denaturation, followed by a rapid pH-jump to neutral pH, should provide the possibility for a widespread application in studying refolding events in RNA domains.
MATERIALS AND METHODS
Bacterial strains and plasmids
For plasmid isolation and cloning, E.coli strain JM 101 was used in combination with the cloning vector pUC19. Isolation was done according to the Qiagen plasmid isolation protocol. The purified plasmids containing the clones of hok74 and pnd58 fragments, were used for the T7 RNA polymerase transcription (25).
Cloning of the RNA fragments
The DNA of the hok74 fragment for cloning was isolated with PCR, in which an extended 5′-end primer, containing a T7 promoter and an EcoRI site, was used. The template DNA used in the PCR was plasmid pBR322, which carries the 580-bp wt hok/sok system cloned between the EcoRI and BamHI sites [pPR633, (33,34)]. The DNA, isolated with the PCR, was cloned into the pUC19 plasmid between the EcoRI and SmaI sites. The DNA for cloning of the pnd58 fragment into the pUC19 plasmid was obtained by hybridising two primers (Gibco BRL) containing the desired sequence and ligating them to a T7 promoter sequence, which contained a partial HindIII site. These DNA fragments were cloned into pUC19 between the HindIII and SmaI site.
T7 RNA polymerase transcription
For the synthesis of the RNA fragments of hok74 and pnd58, purified pUC19 DNA was linearised with SmaI for both hok74 and pnd58. The blunt end plasmids were then used for a run off T7 RNA polymerase transcription. This transcription was done in a buffer containing 200 mM HEPES at pH 7.5, 30 mM MgCl2, 2 mM spermidine, 40 mM DTT, 24 mM of each NTP and 3.6 U/µl of T7 RNA polymerase (MBI Fermentas). The mixture was incubated for 4 h at 37°C. The plasmid DNA concentration in the T7 RNA polymerase reaction was 0.1 µg/µl.
5′-End 32P-labelling of RNA
The RNA was dephosphorylated with calf intestinal alkaline phosphatase (1 U/50 µl; Pharmacia) in an appropriate buffer (Pharmacia) for 30 min at 37°C, purified by phenol extraction and precipitated with ethanol. The 5′-end labelling was done in an appropriate buffer with 0.95 U T4 polynucleotide kinase (Pharmacia) and the mixture was incubated for 45 min at 37°C and purified by polyacrylamide gel electrophoresis.
Heating/cooling cycle experiment
The heating/cooling cycle experiment for hok74 and pnd58 tac stem fragment was done in 50 mM and 0.5 mM Na cacodylate buffer, pH 7.2, respectively. First, the sample was heated at 95°C for 2 min and put directly into liquid nitrogen. The sample was then carefully thawed by adding ice-cooled buffer, with either a concentration of 950 mM NaCl and 50 mM Na cacodylate or 100 mM NaCl, 10 mM MgCl2 and 50 mM Na cacodylate, pH 7.2 and used in the UV absorption experiments and kinetic probing studies.
Acid/alkaline cycle experiment
The acid denaturation of the RNA fragments was done in 50 mM HCl or 50 mM HCl, 10 mM MgCl2 solution at various temperatures, for both the hok74 and pnd58 tac stem fragments. This was done as fast as possible and the concentration of RNA was kept as low as possible when used in the stopped-flow apparatus to prevent unwanted precipitation of denatured RNA in the syringes. The renaturation of the RNA was achieved by adding either 50 mM NaOH, 900 mM NaCl and 50 mM Na cacodylate buffer, pH 7.2 (if only 50 mM HCl was used) or 50 mM NaOH, 50 mM NaCl and 50 mM Na cacodylate buffer, pH 7.2 (if Mg2+ was used in the denaturing buffer). After each experiment in the stopped-flow apparatus, UV spectrophotometer or kinetic structure probing experiments, the pH was checked.
Kinetic probing experiment
Kinetic probing experiments on the RNA fragments were performed in 50 mM Na cacodylate buffer at pH 7.2, 100 mM NaCl and 10 mM MgCl2. Incubation temperatures were at 15, 20 or 30°C in a PCR Mini Cycler (MJ Research) for various time periods. Samples were taken and added to a pre-cooled 0.2 M NaCl, 50 mM Na acetate pH 4.5, 1 mM ZnSO4, 0.5% glycerol buffer containing 8 U nuclease S1. Probing was done at 0°C on ice for 15 min and stopped by phenol extraction, followed by analysis with electrophoresis on a 20% polyacrylamide gel containing 8 M urea.
UV absorbance measurement
For the real-time UV absorbance kinetic experiments we used a thermostatted Perkin-Elmer Lambda 2 spectrophotometer, with a slit width of 2 nm, at a wavelength of 260 nm, with temperatures ranging from 15 to 47°C in a cell with an optical path length of 1 cm, which held a volume of 140 µl (minimum sample volume is 120 µl). For the heating and rapid cooling cycle kinetic experiments, the thawed samples of 140 µl were put into the pre-heated cell immediately. For the acid/alkaline cycle experiments, a 70 µl acidic RNA solution (containing 50 mM HCl with or without 10 mM MgCl2) was placed into the cell and pre-heated to the appropriate temperature. Then, the mildly cooled (to reduce the heat signal and/or prevent overheating of the sample) 70 µl alkaline solution (containing 50 mM NaOH, 1.8 M NaCl and 100 mM Na cacodylate buffer, pH 7.2 or 50 mM NaOH, 100 mM NaCl and 100 mM Na cacodylate buffer, pH 7.2, respectively) was added and quickly mixed, followed by measuring the UV absorbance. The number of measuring points varied between 500 and 4000 and the time interval varied between 0.5 and 6 s depending on the RNA fragment and temperature used. The RNA ranged between an absorbance of 0.1 and 0.9 at 260 nm and 1 cm path length.
Data analysis of the kinetic experiments
The data points of each individual kinetic curve were fitted to a first-order exponential to derive the kinetic parameters for that experiment (Fig. 3). The validity of the fitted curve was tested by changing the starting point of this curve, which did not change the calculated refolding rate nor did it increase or decrease the deviation from the measured data points significantly, strongly indicating that the measured curve show first-order kinetics. Changes in the experimental dead time and the concentration of the RNA at a given temperature did not alter the measured rates significantly, again indicating first-order kinetics. The error determined for the fitted curves compared with the experimental kinetic curves was significantly smaller than the error found between individual experiments at a given temperature indicating the quality of the fit due to the large number of data points in each experiment. This procedure was repeated for each experimentally determined kinetic curve at each temperature in both the acid/alkaline and heating/cooling cycle experiments. The averaged half-life and standard deviation at a given temperature was determined by using the kinetic parameters of at least three individual experiments at that temperature (Table 1).
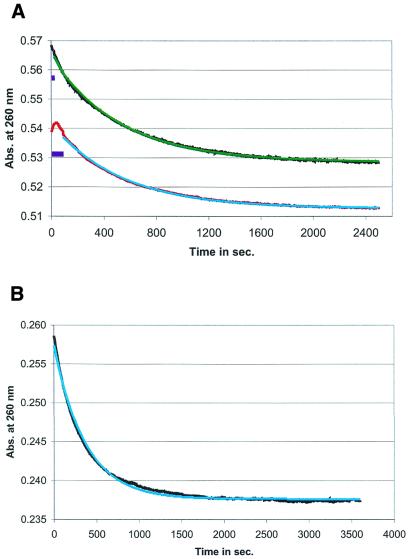
Figure 3.
Comparison of refolding kinetics, using the heating/cooling and acid/alkaline cycle. (A) The UV absorbance measured curve, showing the refolding kinetics from the metastable to the stable conformation in the hok74 RNA fragment at 37°C in 950 mM NaCl and 50 mM Na cacodylate buffer, pH 7.2, after the heating/cooling (in red) and acid/alkaline (in black) cycle and the fitted first-order kinetic curves [in light blue (fitted parameters t1/2 = 566 ± 1 s) and in green (fitted parameters t1/2 = 534 ± 2 s), respectively]. The dead times for the fitted curves are indicated with purple boxes (see also Fig. 2). (B) Measurement of refolding kinetics using the re-isolated (from the acid/alkaline cycle) hok74 RNA fragment and used again in another acid/alkaline cycle. The measured curve (in black) was obtained at 37°C in 950 mM NaCl and 50 mM Na cacodylate buffer, pH 7.2 and fitted with a first-order exponential curve [in light blue (with the fitted parameters t1/2= 521 ± 2 s)].
Table 1. The half-lives and energy barriers at different temperatures.
| Temperature (°C) | Half-life (s) | Standard deviation (s) | ΔG (kcal mol–1) | ΔH (kcal mol–1) |
|---|---|---|---|---|
| hok74 | ||||
| 27 | 42 × 102 | 2 × 102 | –16.0 | |
| 32 | 14 × 102 | 3 × 102 | –15.6 | |
| 35 | 7.7 × 102 | 0.2 × 102 | –15.4 | |
| 37 | 6.0 × 102 | 0.8 × 102 | –15.3 | –37.6 ± 1.1 |
| 40 | 2.67 × 102 | 0.06 × 102 | –15.0 | |
| 42 | 1.9 × 102 | 0.3 × 102 | –14.9 | |
| 47 | 0.84 × 102 | 0.07 × 102 | –14.6 | |
| pnd58 | ||||
| 15 | 24 × 102 | 2 × 102 | –15.0 | |
| 17 | 19 × 102 | 3 × 102 | –15.0 | |
| 20 | 10.6 × 102 | 0.7 × 102 | –14.8 | |
| 25 | 3.7 × 102 | 0.5 × 102 | –14.5 | –30.1 ± 1.3 |
| 30 | 1.4 × 102 | 0.2 × 102 | –14.1 | |
| 35 | 0.98 × 102 | 0.02 × 102 | –14.1 | |
| 37 | 0.61 × 102 | 0.03 × 102 | –13.9 | |
| 42 | 0.33 × 102 | 0.09 × 102 | –13.8 |
The half-life and standard deviation in seconds is derived from the experimentally determined fitted curves at each temperature (see Materials and Methods).
The stopped-flow experiments
The stopped-flow experiments were performed with an Applied Photophysics SX18MV apparatus. The temperature settings in the stopped-flow experiments ranged from 37 to 47°C and were controlled by a Nestlab RTE-111 cooler. The buffers used were the same as in the UV absorbance experiments, while the concentration for both the hok74 and pnd58 RNA fragments ranged between an absorbance of 0.1 and 0.2 at 260 nm. The cell width was 1 cm and it contained 120 µl of sample. The number of experiments (or shots) was limited to five in each experimental set-up to prevent precipitation of the denatured RNA in the acidic syringe.
All buffers used in the heating/cooling, acid/alkaline cycles and probing experiments contained 0.1 mM EDTA.
RESULTS
The hok/sok gene system of plasmid R1 and the pnd/pndB of plasmid R483 encode mRNAs with the same overall folding pathway (6,34). Both mRNAs form stable so-called tac (translation activation) stems at the extreme 5′-end. However, the tac stem of pnd is 58 nt instead of the 74 nt of hok (Fig. 1). The tac stem of pnd has a more simple structure in both the metastable and stable forms, making it an interesting candidate to study and compare the refolding kinetics and energy barrier with those found for the hok tac stem (25).
UV monitoring of kinetics
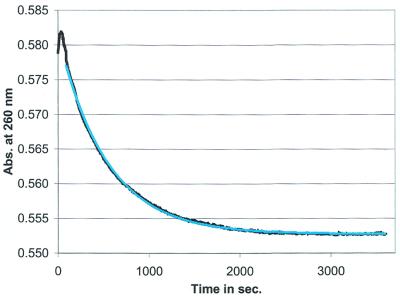
The two studied RNA fragments of 74 and 58 nt were synthesised by T7 RNA polymerase transcription and are indicated here as the hok74 and pnd58 RNA fragments. We have been able to determine the refolding of the tac stem fragments previously using structure probing experiments, after kinetically trapping the metastable structures, via a heating and rapid cooling cycle (25). Monitoring of the same refolding at 37°C, by hyperchromicity measurements at 260 nm instead of structural probing, yields a clear single exponential decay (Fig. 2). As compared with probing, the use of UV absorbance changes has the advantage to give more data points per experiment, thereby increasing the accuracy, in combination with measuring real-time kinetics. The observed refolding times of 10 ± 1 and 10 ± 4 min, in the hok74 RNA fragment, as determined by UV kinetic and structure probing experiments (see Materials and Methods), respectively, corresponded well to one another. Therefore, the UV hyperchromicity experiments did correspond to the real refolding process, having an absorbance change large enough to determine kinetic rates accurately for both the hok74 (Fig. 2) and pnd58 RNA fragments (data not shown). Furthermore, varying the concentration of the RNA fragments between A260 0.1 and 0.9 did not affect the measured kinetic curves and rates (data not shown), as expected for intra-molecular reactions (35).
Figure 2.
The kinetic refolding from the metastable to the stable conformation in the hok74 RNA fragment. The metastable conformation was trapped by the heating/cooling cycle and the kinetics monitored at 260 nm in a UV spectrophotometer at 37°C in 950 mM NaCl and 50 mM Na cacodylate buffer, pH 7.2. The measured real-time curve (in black) and the first-order exponentially fitted curve is indicated in light blue (fitted parameters t1/2 = 669 ± 1 s).
To check the integrity of the hok74 and pnd58 RNA fragments after the trapping of the metastable structures, by the heating/cooling cycle followed by a prolonged exposure to UV light at 260 nm, samples were tested by polyacrylamide gel electrophoresis, but no breakdown was observed. Moreover, the RNA fragments could be isolated and used again in the same type of experiments, without any change in kinetic behaviour (data not shown). However, trapping the metastable states by the heating/cooling cycle had several drawbacks, first of all the relatively large ‘dead-time’ of the experiment, which limits this approach (based on trapping the metastable conformations and determining subsequent refolding kinetics) to relatively longer time scales (Fig. 3A). The dead-time problem was determined by the time required for the heating of the sample to the appropriate experimental temperature after the heating/cooling cycle and subsequent thawing at 0°C. This was especially a problem for the pnd58 RNA fragment, since its refolding rate in 1 M NaCl at 37°C is faster than the dead-time of the experiment. In this case the change in hyperchromicity is lost to a great extent. Decreasing sample volumes did help to reduce dead-times, but only to a limited extent. The second drawback was the inability to use this approach in the standard stopped-flow experimental set-up, which did not have two separated thermostatted syringes to allow the reverse T-jump to trap the metastable conformation.
To overcome these problems, we opted for a different approach, similar to the one used for the unfolding of Crab satellite DNA (36) and the refolding of misfolded tRNATyr and tRNAfMet (31,32). In this approach, a pH-jump was used to unfold the DNA by adding an alkaline solution (36) or to refold the acid form of tRNATyr and tRNAfMet (31,32). However, in our system the acidic solution was used to denature the RNA fragments completely, followed by a quick renaturation by adding a neutralising alkaline solution (see Materials and Methods). This acid denaturation and rapid renaturation cycle was able to kinetically trap the RNA in the metastable state, so that the subsequent refolding to the stable conformation could be followed over time. Structure probing (data not shown) showed this refolding to have the same rate as the ones observed after the heating/cooling cycle (25). However, the pH-induced renaturing cycle does not have some of the essential drawbacks of the heating/cooling cycle. It does not have a relatively large dead-time (Fig. 3A) and this method is suitable for analyses in the stopped-flow apparatus. Furthermore, it preserved the attractive features of the heating/cooling cycle, because the RNA fragments were not degraded and can be re-isolated and used again (Fig. 3B). To test if the acid/alkaline cycle gives the same kinetics as the heating/cooling cycle, several experiments were done under similar conditions and the exponential change in UV absorbance was measured (Fig. 3A). The results show that the kinetic rates, as measured by each of the two approaches, are identical (within the experimental error), indicating that chemical denaturation does not influence the observed refolding kinetics in the hok74 (Fig. 3A) and pnd58 RNA fragments (data not shown). Furthermore, the exponential change in both cases gave clear first-order kinetics, which could be accurately fitted to theoretical first-order kinetic curves (see Materials and Methods). The observed deviations between the calculated and measured curves were smaller than the error between the individual kinetic experiments measured at the same temperature. Therefore, only the individual half-lives were used to calculate the average rates (Table 1).
The acid/alkaline cycle approach does allow the use of the stopped-flow method. However, this procedure has its own limitations in dead-time, because the renaturing reaction is exothermic. This causes a temperature increase during the mixing, accelerating the transition rate and thereby creating a loss of signal. Therefore, it is important to keep the acid concentration as low as possible to reduce the dead-time. The measured dead-time observed was ∼4 s, being approximately the time it takes to dissipate the heat from the cell to the surrounding thermostat. This dead-time was the experimental limit for determining the kinetic parameters of both the hok and pnd tac stem fragments (see Materials and Methods).
One can considerably (and possibly almost completely) reduce the heat-induced dead-time by lowering the acid concentration (see Materials and Methods). In our case, we were limited in lowering the HCl concentration to 50 mM when 10 mM MgCl2 was added together with the hok74 RNA fragment in the denaturing solution, since further lowering of the HCl concentration did not cause complete unfolding of the hok74 RNA fragment. To be able to compare the different buffer concentrations used in the various experiments, we kept the acid concentration at 50 mM despite its measurable heat signal in the stopped-flow of ∼4 s. Furthermore, a shorter dead-time was not necessary, since it was still considerably shorter than the observed refolding times and it did not give appreciable loss of signal at the given reaction conditions.
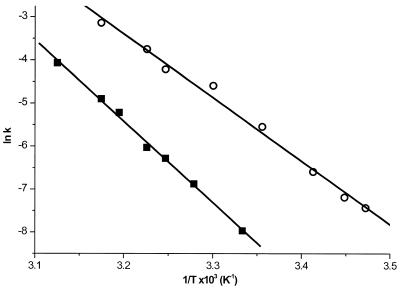
Kinetics at different temperatures
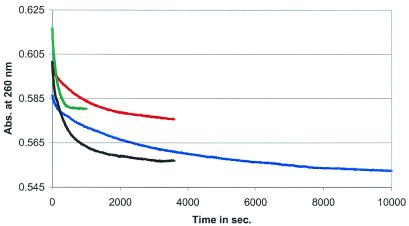
To determine the energy barrier of the conformational transition in the 5′-end hok74 and pnd58 stem fragments, we measured the refolding rates at different temperatures, using the acid/alkaline cycle method (Fig. 4). This was possible even though the hyperchromicity change decreased at low temperatures, probably due to increased stacking interactions, thereby decreasing the hyperchromicity effect of the refolding reaction. The temperatures used ranged from 27 to 47°C and from 15 to 37°C for the hok74 and pnd58 RNA fragment, respectively (Table 1). This allowed us to determine the Gibbs free energy of refolding at a given temperature and to calculate the enthalpy of the barrier of the refolding reaction via an Årrhenius plot (Fig. 5). The energy barriers calculated from this plot were 37.6 ± 1.0 and 30.1 ± 1.5 kcal mol–1 for hok74 and pnd58, respectively. These energy barriers correspond well to the previous rough estimate of ∼50 kcal mol–1 for both hok74 and pnd58 RNA fragments (25). The measured energy barriers of the refolding are less than required to disrupt the complete metastable structure, which is ∼100 kcal mol–1 for both RNA fragments (25). This indicates that there must be an alternative refolding pathway, which does not require a complete unfolding of the metastable structure before refolding into the stable tac stem (25).
Figure 4.
Refolding of the hok74 RNA fragment at different temperatures. After applying the acid/alkaline cycle, the UV absorbance was measured at 260 nm at 27 (in blue), 32 (in red), 37 (in black) and 42°C (in green) in 1 M NaCl, pH 7.2.
Figure 5.
The Årrhenius plots for the hok74 (squares) and pnd58 (circles) 5′-end RNA fragments.
Altogether, the acid/alkaline cycle technique in our case met all the experimental requirements needed, because it was able to kinetically trap the metastable states of both the hok74 and pnd58 5′-end tac stem fragments and because it allowed kinetic probing and measurements of real-time kinetics at different conditions without chemically changing the RNA fragments. This technique has also short dead-times and it can be used with standard stopped-flow equipment. Therefore, the acid denaturation and rapid base renaturation technique is likely to be a useful tool to study RNA kinetics in vitro.
DISCUSSION
In this paper, a new method of acid denaturation and rapid alkaline renaturation for trapping kinetically favourable metastable structures in RNA is described, allowing real-time measurement of RNA refolding kinetics involving alternative structures. This method provides the possibility to use a wide variety of reaction conditions and does not require modified RNA. As compared with the heating/cooling cycle, the acid/alkaline cycle has the advantage that it has a relatively short dead-time and that it can be used in a standard stopped-flow apparatus for measurements of relatively fast kinetics. Therefore, this technique is extremely useful for measuring kinetics in vitro, of independently folding RNA or DNA domains.
The values of the activation energies measured for the refolding reactions from the metastable to the stable conformation of the hok74 and pnd58 RNA fragments indicate that in the rate-limiting step several base pairs or structural elements need to be disrupted before the refolding takes place. However, complete disruption of all base pairs in the metastable hairpins of both the hok74 and pnd58 fragments would require ∼100 kcal mol–1 (25), whereas the measured activation energies are only 37.6 and 30.1 kcal mol–1, for the hok74 and pnd58 fragments, respectively. Therefore, a more gradual substitution of the base pairs in the metastable state by base pairs corresponding to the stable structure should take place, rather than complete disruption of the metastable hairpins, followed by the refolding into the stable conformation (25). We propose two alternative models here that can describe the refolding process. In the first model, the refolding starts by forming a pseudoknot between the loops of the metastable hairpins in the hok74 or pnd58 structures (Fig. 6), followed by a strand displacement into the stable hairpin. The second one is a branch migration, starting by disruption of the bottom of the metastable hairpin I and the formation of the terminal base pairs of the stable stem, followed by further helix propagation into the complete stable tac stem ‘bottom up’ (Fig. 6) (25). Both types of folding models were suggested for various switching RNAs (3,22,23,25,37,38).
Figure 6.
Models for the possible refolding pathways from the metastable to the stable conformation, of the hok74 and pnd58 5′-end RNA fragments. The bottom up or branch migration model is indicated in blue. The pseudoknot or strand displacement model is indicated in red. (A) Stable structure and (B) metastable structure.
The proposed pseudoknot as a starting point of the refolding pathways for the 5′-end of hok and pnd RNA fragments resembles the suggested pseudoknot as an intermediate structure in the refolding of the spliced leader of Leptomonas collosoma (3,37). The ‘bottom up’ or branch migration model seems to correspond to the P1/P(–1) refolding in the Tetrahymena group I intron, in which a branch migration was shown to exist, which was regulated by a third intermediate helix (22,39). This indicates that the refolding models proposed here might be representative for general refolding mechanisms found in other RNAs. However, we were not able to distinguish which refolding mechanism is dominant in the small hok74 and pnd58 fragments, on the basis of the kinetic results presented here.
Both models can explain the difference in refolding rates between the hok74 and the pnd58 fragment. In the case of the bottom up model, the initial refolding steps would require a more significant unfolding of the metastable structure of hok74, especially at the 3′-end of the fragment (Figs 1 and 6). However, the pseudoknot model could not yet be rejected either, because the difference in refolding half-lives (600 s for the hok74 and 60 s for the pnd58 fragment at 37°C) might also be explained by the possibility that the internal loop of hairpin II in the hok74 fragment is less accessible for the formation of the pseudoknot (Fig. 6). Nevertheless, we prefer the bottom up model, because it is more consistent with recent in vivo data (40).
The in vivo data seem to suggest a much slower refolding during transcription of the full-length mRNAs than measured here for the small fragments (40). This potentially slower refolding from the metastable to the stable and active conformation in the shorter active mRNAs (lacking the 3′-end nucleotides, which base-pair to the 5′-end of the full-length mRNAs) cannot be properly explained by the pseudoknot model, since the nucleotides for forming the pseudoknot are accessible in the metastable conformations of both the small and active mRNAs. In contrast, the bottom up model suggests that during the transcription of the full-length mRNAs the 3′-end nucleotides of the 5′-proximal domains are base-paired with the downstream Shine–Dalgarno region until the transcription is complete and therefore cannot trigger the refolding process. This is also consistent with the suggested biological role of the 5′-terminal metastable structures, to prevent premature refolding during transcription (1,6). Furthermore, the biology of the systems requires that in the complete hok and pnd mRNAs the metastable hairpins at the 5′-end, corresponding to the small fragments, have to be disrupted once the transcription of the mRNAs is complete (1,40). This is also consistent with the bottom up refolding model and it could explain the probably accelerated refolding of the full-length mRNAs by the pairing of the very 3′-end to the 5′-terminal region (40). Therefore, we prefer the bottom up model as the starting point of the refolding process from the metastable to the stable conformation in the small hok74 and pnd58 fragments as well, because it seems to fit the in vivo data and because it is consistent with the difference in refolding rates observed between the two fragments (Fig. 6 and Table 1), although more experimental work is needed to properly discriminate between these two refolding models.
The method of acid denaturation and rapid alkaline renaturation cycle is shown to be a powerful and flexible tool to determine real-time refolding kinetics in nucleic acids over a wide temperature and time range. It allows the use of RNA fragments of sizes that are both biologically and kinetically relevant. Experimental conditions provide the opportunity to use small volumes, low concentration of RNA and a wide variety of buffers. The acidic pH can be tuned in such a way that it still denatures the RNA without causing measurable breakdown of the sample. Furthermore, this method allows the trapping of kinetically favourable structures and the study of refolding events without the need to modify the RNA, and thus helps to broaden our understanding of folding events in nucleic acids in general.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Armand W. J. W. Tepper for his help with the stopped-flow equipment. We also want to thank one of the referees for drawing our attention to the work of Bina-Stein and Crothers on the pH-jump approach applied to tRNA. This work was supported in part by the European Commission (project BIO4-98-0189).
REFERENCES
- 1.Gerdes K., Gultyaev,A.P., Franch,T., Pedersen,K. and Mikkelsen,N.D. (1997) Antisense RNA-regulated programmed cell death. Annu. Rev. Genet., 31, 1–31. [DOI] [PubMed] [Google Scholar]
- 2.Loss P., Schmitz,M., Steger,G. and Riesner,D. (1991) Formation of a thermodynamically metastable structure containing hairpin II is critical for infectivity of potato spindle tuber viroid RNA. EMBO J., 10, 719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeCuyer K.A. and Crothers,D.M. (1994) Kinetics of an RNA conformational switch. Proc. Natl Acad. Sci. USA, 91, 3373–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poot R.A., Tsareva,N.V., Boni,I.V. and van Duin,J. (1997) RNA folding kinetics regulates translation of phage MS2 maturation gene. Proc. Natl Acad. Sci. USA, 94, 10110–10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lodmell J.S. and Dahlberg,A.E. (1997) A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science, 277, 1262–1267. [DOI] [PubMed] [Google Scholar]
- 6.Gultyaev A.P., Franch,T. and Gerdes,K. (1997) Programmed cell death by hok/sok of plasmid R1: coupled nucleotide covariations reveal a phylogenetically conserved folding pathway in the hok family of mRNAs. J. Mol. Biol., 273, 26–37. [DOI] [PubMed] [Google Scholar]
- 7.van Meerten D., Girard,G. and van Duin,J. (2001) Translational control by delayed RNA folding: identification of the kinetic trap. RNA, 7, 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gultyaev A.P., van Batenburg,F.H. and Pleij,C.W.A. (1998) Dynamic competition between alternative structures in viroid RNAs simulated by an RNA folding algorithm. J. Mol. Biol., 276, 43–55. [DOI] [PubMed] [Google Scholar]
- 9.Repsilber D., Wiese,S., Rachen,M., Schroder,A.W., Riesner,D. and Steger,G. (1999) Formation of metastable RNA structures by sequential folding during transcription: time-resolved structural analysis of potato spindle tuber viroid (–)-stranded RNA by temperature-gradient gel electrophoresis. RNA, 5, 574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsthoorn R.C., Mertens,S., Brederode,F.T. and Bol,J.F. (1999) A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J., 18, 4856–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biebricher C.K. and Luce,R. (1992) In vitro recombination and terminal elongation of RNA by Q beta replicase. EMBO J., 11, 5129–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gultyaev A.P., van Batenburg,F.H. and Pleij,C.W.A (1995) The influence of a metastable structure in plasmid primer RNA on antisense RNA binding kinetics. Nucleic Acids Res., 23, 3718–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee A.R., Jaeger,J.A. and Turner,D.H. (1993) Thermal unfolding of a group I ribozyme: the low-temperature transition is primarily disruption of tertiary structure. Biochemistry, 32, 153–163. [DOI] [PubMed] [Google Scholar]
- 14.Herschlag D. (1995) RNA chaperones and the RNA folding problem. J. Biol. Chem., 270, 20871–20874. [DOI] [PubMed] [Google Scholar]
- 15.Draper D.E. (1996) Parallel worlds. Nature Struct. Biol., 3, 397–400. [DOI] [PubMed] [Google Scholar]
- 16.Zarrinkar P.P. and Williamson,J.R. (1996) The kinetic folding pathway of the Tetrahymena ribozyme reveals possible similarities between RNA and protein folding. Nature Struct. Biol., 3, 432–438. [DOI] [PubMed] [Google Scholar]
- 17.Brion P. and Westhof,E. (1997) Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct., 26, 113–137. [DOI] [PubMed] [Google Scholar]
- 18.Nussinov R. and Tinoco,I.,Jr (1981) Sequential folding of a messenger RNA molecule. J. Mol. Biol., 151, 519–533. [DOI] [PubMed] [Google Scholar]
- 19.Thirumalai D. and Woodson,S.A. (2000) Maximizing RNA folding rates: a balancing act. RNA, 6, 790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlenbeck O.C. (1995) Keeping RNA happy. RNA, 1, 4–6. [PMC free article] [PubMed] [Google Scholar]
- 21.Woodson S.A. (2000) Compact but disordered states of RNA. Nature Struct. Biol., 7, 349–352. [DOI] [PubMed] [Google Scholar]
- 22.Cao Y. and Woodson,S.A. (1998) Destabilizing effect of an rRNA stem–loop on an attenuator hairpin in the 5′ exon of the Tetrahymena pre-rRNA. RNA, 4, 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flamm C., Fontana,W., Hofacker,I.L. and Schuster,P. (2000) RNA folding at elementary step resolution. RNA, 6, 325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan S.R. and Higgs,P.G. (1996) Evidence for kinetic effects in folding of large RNA molecules. J. Chem. Phys., 105, 7152–7157. [Google Scholar]
- 25.Nagel J.H.A., Gultyaev,A.P., Gerdes,K. and Pleij,C.W.A. (1999) Metastable structures and refolding kinetics in hok mRNA of plasmid R1. RNA, 5, 1408–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu M. and Tinoco,I.,Jr (1998) RNA folding causes secondary structure rearrangement. Proc. Natl Acad. Sci. USA, 95, 11555–11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clodi E., Semrad,K. and Schroeder,R. (1999) Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J., 18, 3776–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franch T., Gultyaev,A.P. and Gerdes,K. (1997) Programmed cell death by hok/sok of plasmid R1: processing at the hok mRNA 3′-end triggers structural rearrangements that allow translation and antisense RNA binding. J. Mol. Biol., 273, 38–51. [DOI] [PubMed] [Google Scholar]
- 29.Walter N.G. and Burke,J.M. (1997) Real-time monitoring of hairpin ribozyme kinetics through base-specific quenching of fluorescein-labeled substrates. RNA, 3, 392–404. [PMC free article] [PubMed] [Google Scholar]
- 30.Woodson S.A. (2000) Recent insights on RNA folding mechanisms from catalytic RNA. Cell Mol. Life Sci., 57, 796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bina-Stein M. and Crothers,D.M. (1974) Conformational changes of transfer ribonucleic acid. The pH phase diagram under acidic conditions. Biochemistry, 13, 2771–2775. [DOI] [PubMed] [Google Scholar]
- 32.Bina-Stein M. and Crothers,D.M. (1975) Localization of the structural change induced in tRNAfMET (Escherichia coli) by acidic pH. Biochemistry, 14, 4185–4191. [DOI] [PubMed] [Google Scholar]
- 33.Gerdes K., Poulsen,L.K., Thisted,T., Nielsen,A.K., Martinussen,J. and Andreasen,P.H. (1990) The hok killer gene family in gram-negative bacteria. New Biol., 2, 946–956. [PubMed] [Google Scholar]
- 34.Thisted T., Nielsen,A.K. and Gerdes,K. (1994) Mechanism of post-segregational killing: translation of Hok, SrnB and Pnd mRNAs of plasmids R1, F and R483 is activated by 3′-end processing. EMBO J., 13, 1950–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutfreund H. (1995) Kinetics for the Life Sciences: Receptors, Transmitters and Catalysts. Cambridge University Press, Cambridge, UK.
- 36.Hamori E., Iio,T., Senior,M.B. and Gutierrez,P.L. (1975) Kinetic investigation of unfolding and partial refolding of a crab satellite (dA–dT)n. Biochemistry, 14, 3618–3625. [DOI] [PubMed] [Google Scholar]
- 37.LeCuyer K.A. and Crothers,D.M. (1993) The Leptomonas collosoma spliced leader RNA can switch between two alternate structural forms. Biochemistry, 32, 5301–5311. [DOI] [PubMed] [Google Scholar]
- 38.Franch T. and Gerdes,K. (1996) Programmed cell death in bacteria: translational repression by mRNA end-pairing. Mol. Microbiol., 21, 1049–1060. [DOI] [PubMed] [Google Scholar]
- 39.Cao Y. and Woodson,S.A. (2000) Refolding of rRNA exons enhances dissociation of the Tetrahymena intron. RNA, 6, 1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moller-Jensen J., Franch,T. and Gerdes,K. (2001) Temporal translational control by a metastable RNA structure. J. Biol. Chem., 276, 35707–35713. [DOI] [PubMed] [Google Scholar]