Abstract
The emergence of insecticide resistance has increased the need for alternative pest management tools. Numerous genetic biocontrol approaches, which involve the release of genetically modified organisms to control pest populations, are in various stages of development to provide highly targeted pest control. However, all current mating-based genetic biocontrol technologies function by releasing engineered males which skew sex-ratios or reduce offspring viability in subsequent generations which leaves mated females to continue to cause harm (e.g. transmit disease). Here, we demonstrate intragenerational genetic biocontrol, wherein mating with engineered males reduces female lifespan. The toxic male technique (TMT) involves the heterologous expression of insecticidal proteins within the male reproductive tract that are transferred to females via mating. In this study, we demonstrate TMT in Drosophila melanogaster males, which reduce the median lifespan of mated females by 37 − 64% compared to controls mated to wild type males. Agent-based models of Aedes aegypti predict that TMT could reduce rates of blood feeding by a further 40 – 60% during release periods compared to leading biocontrol technologies like fsRIDL. TMT is a promising approach for combatting outbreaks of disease vectors and agricultural pests.
Subject terms: Genetic engineering, Environmental biotechnology, Drosophila, Transgenic organisms
Current methods for genetic biocontrol of insect pests (e.g. gene drives) take generations to reduce harm. Here, authors engineered male fruit flies to express venom proteins in their seminal fluid that reduce female lifespan after mating, demonstrating a rapid approach to sustainable pest control.
Introduction
Insect pests pose a major challenge to human and environmental health. Malaria, spread by several species of Anopheles mosquitoes, causes 608,000 deaths per year1 and rates of arboviral diseases spread primarily by the Aedes aegypti mosquito, including dengue, Zika, chikungunya, and yellow fever, are reaching unprecedented levels due to increased global trade and warming climates2. Dengue virus alone causes 390 million human infections each year and is now considered the most common vector-borne viral infection worldwide3. Recent studies have calculated that the cumulative cost of biological invasions is increasing four-fold every decade4, with a conservative estimate of US$162.7 billion for damages caused in 20175. Invertebrate pests, primarily insects, are responsible for ~40% of these damages5,6, and estimates of annual losses for all major food crops ranges between 20 and 30% due to damages by pests and pathogens7. Arboviral diseases such as avian malaria also pose a significant ecological threat to many native species, and are believed to be a key driver of population decline in New Zealand and Hawaiian forest birds8,9.
Pesticides are the first line of defence against many invasive species, particularly mosquitoes10. However, over-reliance on insecticides has resulted in the widespread emergence of resistance11. Over-application of insecticides has also resulted in declining populations of non-target species, and are responsible for many other environmental and human-health concerns12. Modern integrated pest management is trending towards reducing reliance on chemical insecticides in favour of other environmentally friendly management techniques to reduce the emergence of resistances10,13.
Genetic biocontrol, defined as the release of organisms which have been genetically altered to reduce the spread and harm caused by a target species14, is one such alternative that’s been gaining broader public acceptance15. Early and well established genetic biocontrol technologies (GBT) such as the sterile insect technique (SIT) involve the release of mass numbers of males which have been radiologically sterilised16, thereby reducing the reproductive potential of the females. More modern GBT, such as the release of insects carrying a dominant lethal gene (RIDL)17 and gene drives18 function by propagating transgenes which reduce the fitness of future generations as the transgenes are inherited. Cage and field trials of these second-generation GBT have begun to demonstrate their improved efficacy19–21, requiring fewer released males compared to conventional approaches to achieve similar population control, albeit with a potential trade-off between enhanced efficacy and reduced control over the degree of transgene introgression14.
Currently, all GBT require a minimum of a generation to take effect on the target population and often much longer until the harm from a pest outbreak is mitigated19. While this generational lag period is less problematic for species where immature life stages are the primary cause of harm, it poses a significant challenge for many disease vectors and agricultural pests where adult females are the main concern. For example, wild female Ae. aegypti have a median adult lifespan of 2–3 weeks22,23, will typically mate within 24–48 h of emerging24, and on average will take 0.63–0.76 blood meals per day25. Females that mate with GBT males may not produce viable offspring, but they can continue to spread disease or damage crops. Although current GBT are highly effective at intergenerational population control, a more rapid response to outbreaks of disease vectors is critical to mitigate the spread of arboviruses and avert the risk of epidemics, particularly in regions with seasonal population dynamics26.
An alternative approach to GBT that has been speculated on is utilising seminal fluid proteins (SFP) as a target to affect the fitness or fertility of mated females27–30. SFPs are produced within the male accessory glands (MAG) and stored within the lumen until they are transferred to females along with sperm and other compounds. SFPs such as ovulin (Acp26Aa) and sex peptides are known to generate various post-mating responses and physiological changes in females, such as reduced mating receptivity and median lifespan31,32. It has been demonstrated in Drosophila melanogaster that several low molecular weight SFP can pass through the female reproductive tract and enter the circulatory system (haemolymph), and even to act on receptors in the central nervous system (CNS)33,34. Evidence is mounting that mosquito SFP also act on receptors found within the CNS, with 30% of radiolabelled Culex SFP localising to the female head and thorax35, and intra-thoracic but not intra-abdominal SFP injections resulting in mating refractoriness in Anopheline and Aedes females36–39.
Here we describe the toxic male technique (TMT), which is—to the best of our knowledge—the first example of an intragenerational GBT. By genetically engineering pest species to express toxic, low molecular weight compounds within the MAG, the survival of mated females can be reduced (Fig. 1). Using D. melanogaster as a proof of concept, we engineer males to express one of seven insect-specific venom proteins in a MAG-specific pattern. Of the >1600 characterised venom compounds, 60% of these are cysteine-knotted mini-proteins40, which range from 2.6 to 14.8 kDa and are highly stable, taxonomically selective and less susceptible to pore region mutations associated with insecticide resistances41. Like endogenous SFP, the recombinant venom peptides should remain safely sequestered within the lumen of the MAG and not enter the male’s haemolymph. If the recombinant venom peptides pass through the mated female’s reproductive tract to affect the target ion channel receptors in her CNS, the neurotoxic effects are likely to be fast-acting and dose-dependent.
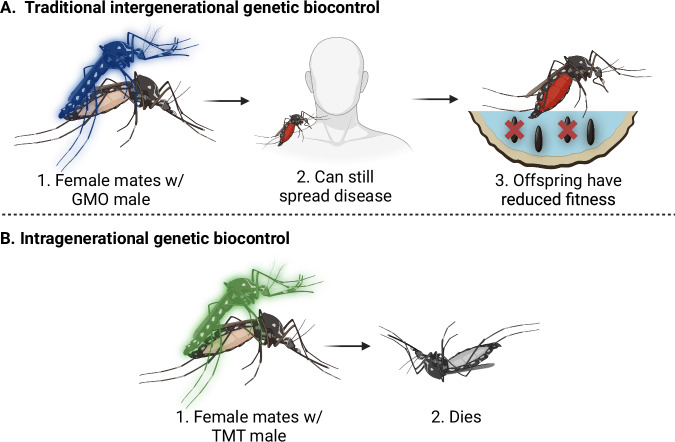
Fig. 1. Intergenerational vs intragenerational genetic biocontrol of pest insects.
Current mating-based methods of genetic biocontrol (A) function by affecting the viability or sex ratio of the offspring of subsequent generations. However, mated females persist in the target area and can continue to cause harm (e.g. spread disease). Intragenerational biocontrol (B), such as TMT, directly affects the fitness of mated females, thereby rapidly reducing the harm caused by the target population. Created in BioRender. Maselko, M. (2024) https://BioRender.com/r25q201.
The median lifespans of wild-type females mated to two of the TMT strains are reduced by 37–64% after initial exposure compared to control females mated to wild-type males. Single-pair mating assays find that TMT males are able to court wild-type females as effectively as wild-type males. MAG-specific expression of a 5.2 kDa venom peptide has no significant effect on the lifespan of adult TMT males, but expression of a 2.9 kDa venom peptide reduces lifespan by 59% compared to wildtype and transgenic controls. To determine the theoretical capability of TMT to suppress a pest population in comparison to female-specific RIDL (fsRIDL), the current state-of-the-art GBT, we develop an agent-based model to simulate an idealised Ae. aegypti release programme19. Sensitivity analysis of the model found that the improved performance of TMT compared to fsRIDL is consistent across a broad range of values of polyandry, density-dependent mortality, and release ratios, with TMT reducing blood feeding rates by a further 40–60% in most simulations. These results demonstrate the potential of TMT as the next generation of genetic biocontrol, which is especially suited to rapidly respond to outbreaks of disease vectors and agricultural pests.
Results
Heterologous expression of insecticidal venom proteins
We first identified a list of candidate venom proteins based on several criteria. Critically, the target receptor of the venom could not be present in the male reproductive tract, which was determined by tissue-specific gene enrichment values of the target ion channel given by FlyAtlas 242. The venom proteins must also only interact with invertebrate ion channels, with no measurable LD50 found for mammals. Candidates were preferred if they had lower LD50 for Drosophila or another Dipteran. Candidates of lower molecular weight were also preferred, as we expect that to increase the likelihood of the venom proteins entering the haemolymph of mated females. To keep the number of candidate proteins manageable, we limited the list to the most promising candidate for a given ion channel target or class of neurotoxin (agatoxin, ctenitoxin, etc.) given our criteria (Table 1). Venom protein sequences were cloned into a UAS expression vector along with the signal peptide of an endogenous SFP (Acp26Aa) to generate transgenic D. melanogaster strains.
Table 1.
Candidate venom proteins investigated for accessory gland-specific recombinant expression
| Venom protein | Native host | Target | kDA |
|---|---|---|---|
| μ-AGTX-Aa1d | Agelenopsis aperta | NaV channel (site 4) | 4.2 |
| Γ-CNTX-Pn1a | Phoneutria nigriventer | NMDA glutamate receptors | 5.2 |
| δ-CNTX-Pn1a | Phoneutria nigriventer | NaV channel (site 3) | 5.3 |
| κ-HXTX-Hv1c | Hadronyche versuta | BKCa channels | 3.8 |
| ω-HXTX-Hv1a | Hadronyche versuta | CaV1 channels | 4.1 |
| ω-HXTX-Hv2a | Hadronyche versuta | CaV2 channels | 4.5 |
| δ-AITX-Avd2a | Anemonia sulcata | NaV channel (site 3) | 2.9 |
Testing functional expression of recombinant venoms
To assess the functionality of the recombinant venom proteins in a heterologous host, homozygous UAS:venom strains were crossed to hemizygous Act5C-GAL4 flies43. This GAL4 parental line will transmit one of two homologous chromosomes to its offspring: one containing the gene for the GAL4 transcriptional activator under regulation of the strong Actin-5C promoter; or the balancer chromosome containing a CyO marker resulting in a curled-wing phenotype. Progeny from this cross which inherit the Act5C-GAL4 chromosome would be non-viable if the venom is functionally expressed by the flies, as ubiquitous expression of the venom peptide should be immediately lethal, so 100% of the viable offspring ought to have inherited the CyO chromosome and display curled-wing phenotype. Conversely, if the venom is non-functional, then inheritance of either the Act5C-GAL4 or CyO chromosome would have no impact on the viability of the offspring, so 50% of the offspring would display the curled-wing phenotype. From these crosses (N = 3), progeny from 5/7 of the UAS:venoms strains displayed 100% curled-wing phenotype, including μ-AGTX-Aa1d, Γ-CNTX-Pn1a, κ-HXTX-Hv1c, ω-HXTX-Hv1a and δ-AITX-Avd2a. Progeny from UAS:venom strains expressing δ-CNTX-Pn1a exhibited 46 ± 2% curled-wing phenotype, and ω-HXTX-Hv2a exhibited 44 ± 10%, so these venom transgenes were presumed to be non-functional and not investigated further.
Accessory gland-specific expression of transgenes
The success of this approach hinges on our ability to get high levels of MAG-specific expression of the venom proteins, while reducing off-target expression as much as possible. We could not express the venom proteins directly by a MAG-specific promoter, as the strains would not be stable and would require backcrossing transgenic females to wild-type males each generation to maintain them. As such, a bipartite approach such as UAS:GAL4 was required to facilitate separating the regulation of venom expression between the driver (GAL4) and responder (UAS) lines. To determine the most suitable expression pattern, we tested three different MAG-specific GAL4 drivers: antr-GAL4; prd-GAL4; and Acp26Aa-GAL444–46. These drivers were crossed to a UAS:mCherry reporter47, and the degree of on and off-target expression for each cross was determined by fluorescence microscopy of dissected male progeny (Fig. 2). prd-GAL4 produced the strongest accessory gland-specific RFP expression, but RFP was also observed in the digestive tracts of adult males (Fig. 2b, e). Acp26Aa-GAL4 was highly specific to the accessory glands, but the expression was relatively weak (Fig. 2c, f) Only antr-GAL4 produced a strong fluorescent signal in the accessory glands without also driving expression in earlier developmental stages or off-target tissue (Fig. 2a, d).
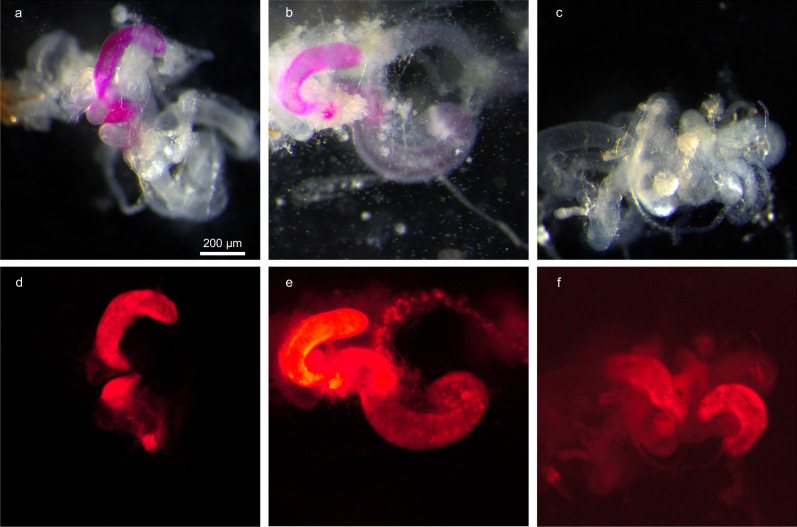
Fig. 2. Accessory gland-specific expression of RFP in transgenic D. melanogaster males.
Dissected reproductive and digestive tracts of adult male offspring from UAS:mCherry reporter and accessory gland-specific GAL4 drivers (antr-GAL4 (a, d), prd-GAL4 (b, e), and Acp26Aa-GAL4 (c, f)) observed under bright-field (a–c) and red fluorescence (d–f) microscopy. Dissections were performed on ≥5 males per cross. Scale bar in a: 200 µm, applies to all panels.
TMT males reduce the lifespan of mated females
Lifespan assays of mated females were performed to determine whether exposure to TMT males reduces the lifespan of wild-type females. TMT males were prepared by crossing UAS:venom strains (μ-AGTX-Aa1d, Γ-CNTX-Pn1a, κ-HXTX-Hv1c, ω-HXTX-Hv1a or δ-AITX-Avd2a) to the antr-GAL4 driver, and transheterozygous male offspring were isolated soon after eclosion. As female D. melanogaster are poorly receptive to mating for 24–48 h post-eclosion48, virgin wild-type females were isolated for 2–3 days to ensure that they had reached sexual maturity. Females were then housed in groups of eight with an equal number of TMT or wild-type males and were transferred to fresh media every 3 days. As female D. melanogaster are refractory to mating for at least 5 days after fertilisation49, to ensure that females had access to mates with fully replenished SFP when they became receptive, as well as to maintain the 1:1 sex ratio, males were replaced during each transfer. Females lived for a median of 27 days post-initial exposure to wild-type males (95% CI: 25–31 days, N = 40, Fig. 3a). Females mated to Γ-CNTX-Pn1a TMT males had a median post-exposure (MPE) lifespan of 17 days (95% CI: 12–19 days, N = 40, Fig. 3a), 37% lower than controls (log-rank test, χ2 (1, N = 80) = 20.7, P < 0.005). Females mated to δ-AITX-Avd2a TMT males had an MPE lifespan of 15 days (95% CI: 11–18 days, N = 40, Fig. 3a), 44% lower than controls (log-rank test, χ2 (1, N = 80) = 17.7, P < 0.005).
Fig. 3. Female D. melanogaster exposed to TMT males have reduced lifespan.
Females were exposed to males at an equal sex ratio (a), or with males at a three-fold excess (b). Data were presented as Kaplan–Meier survival probability estimates with 95% Greenwood confidence intervals (indicated by the shaded area). The dashed lines indicate a median time of 50% survival. Source data are provided as a Source Data file.
Females mated to μ-AGTX-Aa1d or κ-HXTX-Hv1c TMT males did not have significantly reduced lifespans compared to wild-type controls, indicating that the reduction in lifespan observed in the Γ-CNTX-Pn1a and δ-AITX-Avd2a treatment groups is the result of specific venom expression rather than exposure to transgenic males. Females mated to ω-HXTX-Hv1a TMT males had an MPE lifespan of 20 days (95% CI: 17–23 days, N = 40), 26% lower than controls (log-rank test, χ2 (1, N = 80) = 9.51, P < 0.005). However, most ω-HXTX-Hv1a TMT males were not surviving the 3 days until their replacement. Upon further investigation, we found that while expression of the primary subunit (cac) of the calcium channel targeted by ω-HXTX-Hv1a is not enriched in the MAG, the pore-forming subunit (Ca-α1D) is. As such, expression of ω-HXTX-Hv1a was not investigated further.
Release programmes of currently availably GBT such as SIT and RIDL utilise overflooding ratios of modified males to wild individuals19,50,51. However, female D. melanogaster are known to have reduced fitness at higher rates of male exposure52. To determine if an increase in the sex ratio of TMT males results in a greater reduction in median lifespan than would be expected from a simple increase in male exposure, we performed another female lifespan assay with a 3:1 male-to-female sex ratio. In this assay, control females mated to wild-type males had an MPE lifespan of 22 days (95% CI: 15–30 days, N = 39, Fig. 3b). Comparatively, females mated to Γ-CNTX-Pn1a TMT males had an MPE lifespan of 11 days (95% CI: 8–14 days, N = 40, Fig. 3b), 50% lower than controls (log-rank test, χ2 (1, N = 79) = 24.4, P < 0.005), and females mated to δ-AITX-Avd2a TMT males had a MPE lifespan of 8 days (95% CI: 6–13 days, N = 40, Fig. 3b), 64% lower than controls (log-rank test, χ2 (1, N = 79) = 28.1, P < 0.005). The MPE lifespans of females mated 3:1 to wild-type males was not significantly reduced compared to females mated 1:1 to wild-type males (log-rank test, χ2 (1, N = 79) = 0.2, P = 0.6), while females mated to 3:1 Γ-CNTX-Pn1a and δ-AITX-Avd2a TMT males had a 35% (log-rank test, χ2 (1, N = 80) = 6.8, P = 0.01) and 47% (log-rank test, χ2 (1, N = 80) = 7.4, P = 0.01) respectively reduced MPE lifespans relative to their equal ratio counterparts. Therefore, the negative effect of exposure to TMT males on the survival of mated females is greater than the negative effect of increased male exposure in general.
Single-pair courtship assay
To determine whether the expression of venom proteins in the accessory glands impaired the males’ ability to court females, single-pair courtship assays were performed as previously described53. A single wild-type virgin female was paired with a virgin male in a mating arena, which were observed for up to two hours to identify males which successfully courted a female. To control for off-target effects of accessory gland-specific expression of heterologous proteins, the competitiveness of negative control males expressing an unrelated low molecular weight protein (NbVHH0554, 13 kDa) in the same expression pattern (antr-GAL4/ UAS:NbVHH05) was also measured. The percentage of wild-type males who successfully courted a female was 30% (95% CI: 19–44%, N = 50), Γ-CNTX-Pn1a TMT males was 30% (95% CI: 19–44%, N = 50), δ-AITX-Avd2a TMT males was 22% (95% CI: 13–35%, N = 50), and negative control males was 24% (95% CI: 14–37%, N = 50). No significant difference was observed in the ability of the males in any treatment group to court females (χ2 (3, N = 200) = 1.3, P = 0.73).
Competitive mating assay
To determine if the intrinsic competitiveness of the GAL4-UAS transgenic males was negatively affected by variations in genetic background, we performed a control competitive mating assay in which equal numbers of virgin wild-type females, wild-type males, and antr-GAL4/UAS:NbVHH05 males were housed together overnight and allowed to mate freely. As transgenic offspring lacked a distinct phenotypic marker, 10 offspring from each female were genotyped to determine paternity. Male competitiveness (C) was calculated as C = PW/O(1 – P)19, where P is the proportion of females which produced transgenic offspring, W and O are the number of wild and transgenic males, respectively, with C = 1 indicating equal competitiveness to wild-type males. The competitiveness of the control males was calculated to be just 0.14 (95% CI: 0.05–0.25, N = 42), indicating that the GAL4-UAS genetic background of the transgenic males significantly affected their mating competitiveness.
TMT male longevity
Longevity assays were performed to determine what effect accessory gland-specific venom protein expression had on the longevity of TMT males relative to wild-type and negative control NbVHH05 males. Males were isolated soon after eclosion and housed in groups of 8, being transferred onto fresh media every 3 days. The median lifespan of wild-type males was 46 days (95% CI: 38–50 days, N = 40, Fig. 4), negative control males was 43 days (95% CI: 41–46 days, N = 40), Γ-CNTX-Pn1a males was 46 days (95% CI: 38–47 days, N = 40), and δ-AITX-Avd2a males was 19 days (95% CI: 11–27 days, N = 40). Γ-CNTX-Pn1a TMT males had no significant difference in longevity compared to wild-type males (log-rank test, χ2 (1, N = 80) = 2.8, P = 0.09) or negative control males (log-rank test, χ2 (1, N = 80) = 0.8, P = 0.36). However, δ-AITX-Avd2a TMT males had a 59% reduced median lifespan compared to wild-type males (log-rank test, χ2 (1, N = 80) = 50.9, P < 0.005).
Fig. 4.

Longevity of male D. melanogaster expressing Γ-CNTX-Pn1a or δ-AITX-Avd2a venom proteins in their accessory glands compared to wild type and negative control (NbVHH05) strains. Data were presented as Kaplan–Meier survival probability estimates with 95% Greenwood confidence intervals (indicated by the shaded area). The dashed lines indicate time to 50% survival. Source data are provided as a Source Data file.
Modelling of Ae. aegypti population control
To evaluate how a TMT release programme might compare to currently deployed GBT in its ability to suppress a mosquito population, we developed an agent-based model simulating a hypothetical Ae. aegypti release programme of SIT, fsRIDL and TMT mosquitoes using GAMA 1.9 (https://gama-platform.org). This model simulates the full life cycle of each mosquito within the population in hourly intervals (Supplementary Fig. 1), with adult females having more complex behaviour in response to various internal and external factors such as age, time of day, mate choice, etc. (Supplementary Fig. 2). Models were instantiated with a seed population that is left for a year of simulation time to reach equilibrium, at which point adult transgenic males are released every 3 days until the end of the simulation at a release ratio of 12:1 males per wild adult. The frequency of polyandry was set to ~20%55, and density-dependent mortality (DDM) of the immature stages was modelled as a logistic function (see Methods for further information). The fitness and lifespan of all transgenic males were presumed to be equal to wild males, but the mating competitiveness estimated from Brazil field trials of RIDL mosquitoes19 (3.1%) was used. Each technology was likewise ‘idealised’, with all SIT offspring and female fsRIDL pupae being completely non-viable, and wild females that mated with TMT males dying within the hour (which functionally may also be acute incapacitation, eventually leading to their death). A negative control (no release) simulation was also performed for comparison.
The target female populations in SIT and fsRIDL simulations were reduced to 50% of their initial size (PR50) within 56.1 and 36.3 days, respectively, and reduced by 95% (PR95) within 109.6 and 87.8 days (Fig. 5a). Qualitatively, these results follow a similar trend to the rate of population reduction observed during the suppression phase of the Brazil field trials. The PR50 and PR95 for TMT simulations were 15 and 62.2 days, 58.7 and 29.2% faster than fsRIDL, respectively. The model also tracked key epidemiological factors such as cumulative blood feeding events, which on average were 8183 for SIT simulations, and 5659 for fsRIDL simulations (Fig. 5b). The average number of blood feeding events in TMT simulations was just 2804, 67% lower than SIT and 50% lower than fsRIDL simulations. The number of gonotrophic cycles undergone by a female mosquito correlates closely with two important factors, multiple feeding rates and the extrinsic incubation period (EIP) of arboviruses56,57, with the earliest that a female is likely to transmit a virus being her second gonotrophic cycle. The median number of gonotrophic cycles per female was 5 for the negative control (IQR: 3–7), SIT (IQR: 3–7) and fsRIDL (IQR: 4–7) simulations, whereas the median for TMT simulations was just 1 (IQR: 1–5).
Fig. 5. Agent-based models simulating Ae. aegypti population control programmes with SIT, fsRIDL, TMT, or no transgenic male releases.

a The size of the female population during the release period relative to initial conditions. 50% reduction in the female population indicated by the dashed line. b The cumulative number of biting events which occurred throughout the releases. Data were presented as the mean of 10 simulations ± the maximum and minimum values, respectively, observed at a given time point (indicated by the shaded area). Source data are available through Zenodo under accession code 13927099.
Simulating reduced mating lethality
We then explored what performance could be expected if the mating lethality (i.e. likelihood that a female will die after mating with a TMT male) is less than 100%. We repeated the TMT simulations with mating lethality (hereafter ML) between 10 and 100% (in increments of 10%) and presumed that females that had survived the mating event had no other adverse effects and may even go on to produce offspring. Due to the non-linear response to venom dosage, and the uncertainty in the difference in venom production between homozygotes and heterozygotes, we decided that modelling heritability of the TMT construct was impractical at this stage. We, therefore, decided to model these simulations as a ‘worst-case’ scenario with TMT males producing fertile, wild-type offspring. However, it would be unlikely that no attempt to mitigate the risk of transgene escape would be made in the development of any TMT application. As such, we also modelled a scenario with TMT males that had reduced ML but would produce no viable offspring, which could be achieved through a combination with some form of SIT, IIT, or RIDL. We found that the cumulative blood feeding rates for fertile TMT male scenarios were only significantly lower than fsRIDL simulations when the ML was ≥80% (P < 0.001, Supplementary Fig. S3), but sterile males only required ≥40% ML to outperform fsRIDL (P < 0.001, Supplementary Fig. S4). While the PR50 of fertile TMT males was lower than fsRIDL at ML ≥70%, the target population was not eliminated when ML was <80% (Supplementary Fig. S3). However, sterile TMT males had a PR50 lower than fsRIDL at ML ≥50% (Supplementary Fig. S4) and had a PR95 lower than fsRIDL at ≥80%.
Sensitivity analysis
We conducted sensitivity analysis58 to quantify the uncertainty in the model’s outputs and to determine the relative influence of key parameters on the comparative efficacy of TMT and fsRIDL. Our analysis focused on three critical parameters: density-dependent mortality (DDM), polyandry, and release ratio. We systematically varied these parameters across defined ranges (see Methods) and assessed their impact on the percentage difference in cumulative blood-feeding events between TMT and fsRIDL simulations, serving as a proxy for their capacity to rapidly reduce the spread of disease (Fig. 6). We performed linear regression with polynomial and interaction terms by utilising a robust linear model (RLM) to account for non-normality and heteroscedasticity in the data, followed by global and local sensitivity analyses. We also calculated standardised regression coefficients (SRC) to compare the magnitude and direction of the parameters’ effects, and variance inflation factors (VIF) to identify multicollinearity between effects.
Fig. 6. Heatmap of the percentage difference in cumulative bite rates between fsRIDL and TMT simulations under different input parameter combinations.
The influence of key parameters (polyandry, release ratio, and density-dependent mortality) on the comparative ability of fsRIDL or TMT to reduce blood feeding rates was determined by sensitivity analysis. The data shown are the average of ten simulations performed per technology and parameter value combination. Positive values (blue) indicate relatively lower bite rates for fsRIDL simulations, and negative values (red) indicate lower bite rates for TMT simulations. Source data are available through Zenodo under accession code 13927099.
No significant multicollinearity was observed (all VIF <2.5), indicating that each predictor variable provides unique information to the model. The mean absolute error (MAE) of 15.7% reflects the average deviation in the model’s predictions, indicating a reasonable level of uncertainty in its outputs. This, combined with the pseudo R-squared of 0.857, suggests that while the model captures a significant portion of the variability in the data, there remains some uncertainty in individual predictions. First-order Sobol indices indicate that the variance in the model’s output can primarily be attributed to variation in polyandry (45.0 ± 6.2%) and release ratio (38.2 ± 4.8%), while DDM contributed less to output uncertainty (14.8 ± 3.5%). One-at-a-time sensitivity analysis revealed that polyandry and release ratio induced output variations of −45.0 to 60.1% and −46.0 to 46.8%, respectively, while DDM’s impact ranged from −36.3 to 28.4%.
The regression model indicates that polyandry (β = 1.51, P < 0.001, SRC (95% CI) = 40.4–47.0) and release ratio (β = 17.4, P < 0.001, SRC (95% CI) = 100.3–110.5) have significant positive effects on the performance of TMT relative to fsRIDL, while DDM (β = −89.0, P < 0.001, SRC (95% CI) = −67.1 to −56.6) has significant negative effects. The SRC suggest that the release ratio has the strongest relative influence, followed by DDM and polyandry. However, the significant quadratic terms for all three parameters (all P < 0.001) indicate there may be non-linear effects, which may explain the discrepancy between Sobol indices and SRC rankings. Partial dependence plots (PDP) were used to further investigate these non-linear effects. While the effect of DDM on the relative performance of TMT and fsRIDL is negative, this trend reverses as the influence of DDM increases (SRC (95% CI) = 37.4–47.5, Supplementary Fig. S5). The positive effect of increased release ratios also appears to plateau around the highest value observed in the model (SRC (95% CI) = −74.9 to −65.0, Supplementary Fig. S6). However, the positive effect of polyandry on TMT’s relative performance continues to increase up to the highest value observed in the model (SRC (95% CI) = 5.3–11.1, Supplementary Fig. S7). The interaction term between release ratio and polyandry (β = −0.09, P < 0.001, SRC (95% CI) = −24.5 to −21.2) suggests that the positive effect of release ratios may be mitigated in populations with high polyandry. Conversely, the negative effect of DDM may be mitigated by the interaction of increased release ratios (β = 1.58, P < 0.001, SRC (95% CI) = 7.1–10.5).
Discussion
Here, we describe a genetic biocontrol technology which can reduce the lifespan of mated females, called the toxic male technique, or TMT. The heterologous expression of insect-specific venom proteins, Γ-CNTX-Pn1a from the P. nigriventer spider and δ-AITX-Avd2a from the A. sulcata sea anemone, within the seminal fluid of male D. melanogaster resulted in a 37–64% reduction in the median lifespan of mated wild-type females after initial exposure to TMT males. The development of intragenerational genetic biocontrol technologies such as TMT represents a paradigmatic shift in pest management. Relative to current biocontrol technologies, the speed of population suppression achievable by intragenerational biocontrol may make these techniques a viable alternative to chemical pesticides as a first-line response to outbreaks while retaining taxonomic specificity and reducing off-target effects on local ecologies.
Over 1000 unique spider venom proteins have been characterised41, and many more from hosts such as scorpions and centipedes59. Many insecticidal venom proteins are highly taxonomically selective, with toxicities that vary by orders of magnitude between different classes of insect60, which allows for targeted application of particular use cases. Other insecticidal proteins from plants or bacteria may also make good candidates for TMT, though further investigation is required to determine their suitability. Co-expression of multiple toxins could help to mitigate the emergence of resistance alleles, as well as potentially resulting in synergistic toxicity from affecting multiple ion channel targets. It needs to be determined whether heterologous venom protein expression may result in TMT males becoming toxic to natural predators. However, the oral toxicity of venom proteins is typically between 1 and 2 orders of magnitude lower than when they are directly injected61, and venom proteins can be selected which have greater toxicity for the target species relative to natural predators.
To determine the theoretical capabilities of TMT, we developed an agent-based model of an Ae. aegypti release programme to compare its performance to fsRIDL under different assumptions of density-dependent mortality, polyandry, and release ratios of transgenic males. These results suggest that compared to fsRIDL, TMT may reduce the incidence of blood feeding during the release period by a further 40–60% under most scenarios, which has significant epidemiological implications. Our model indicates for TMT to outperform fsRIDL in this regard, it would require ~80% of the females to die during mating. However, this presumes that the TMT males are fertile, and any application of TMT will likely also require sterilisation of the males to mitigate the risk of transgene escape. If the TMT males cannot produce viable offspring, then only 40% mating lethality is required to outperform fsRIDL.
Sensitivity analysis revealed that the model has a mean absolute error of 17%, with most of the variation being explained by the direct effects of these key parameters. Release ratio had the most significant effect on the relative performance of TMT simulations, with TMT benefitting from higher ratios more so than fsRIDL, up to a point of diminishing returns where the wild population is fully saturated by transgenic males. As expected, DDM had a negative effect on TMT’s performance relative to fsRIDL, though increases in release ratio may help to overcome this limitation. Higher rates of polyandry increased TMT’s performance, and the positive effect continues up to the highest rates tested, suggesting that TMT would likely be even more effective in disease vectors with higher polyandry, such as Brown Planthoppers62,63. As Ae. aegypti females typically mate and attempt to blood feed within 24–48 h of eclosion24, and arboviruses such as Dengue require a 3–10-day incubation period57, most females killed by TMT males are not likely to transmit these illnesses. However, further studies will be required to understand the effects of epidemiology and other extrinsic factors (e.g. migration) on TMT’s ability to mitigate epidemics.
The ideal phenotype for TMT would be immediate and irreversible incapacitation of the mated female. While the observed reduction in median female lifespan of 37–64% demonstrates the concept of TMT, mating lethality will inevitably need to be increased for TMT to approach the performance observed in the modelling. Anecdotally, we observed that mated females appeared lethargic and perhaps even mildly paralysed prior to their death, which might suggest that even if the TMT males do not kill the females directly, they may increase their mortality indirectly by making them more susceptible to predation. The expression of Γ-CNTX-Pn1a in male accessory glands did not affect the lifespan of TMT males, but expression of δ-AITX-Avd2a reduced median lifespan by 59% compared to wild-type males. We believe that this discrepancy is likely due to the difference in molecular weights of these venoms (5.2 and 2.9 kDa, respectively). The smaller size of δ-AITX-Avd2a may allow it to pass through the accessory gland epithelium into the haemolymph via paracellular routes, as has been described for some orally toxic insecticidal proteins64. The performance of TMT can, therefore, potentially be improved by identifying venoms or other insecticidal proteins within some optimal molecular weight range, and by characterising accessory gland-specific regulatory elements to increase their expression.
The expression of either Γ-CNTX-Pn1a or δ-AITX-Avd2a did not reduce TMT male’s ability to court females in single-pair settings, though due to differences in genetic background, we were unable to perform a meaningful assay to determine their ability to compete with wild-type males for mates. Competitive mating assays, which assess the ability of genetically engineered males to compete with wild-type males for mates, predominately utilise transgenic males prepared from wild-type genetic background65–67. This is because males prepared using the GAL4-UAS system68, as well as ectopic expression of the white (mini-w+) marker, can confound the results of competitive mating assays due to differences in body size and colour69 or by increased rates of male-male courtship70. Due to the stringent spatiotemporal regulation necessary to characterise TMT, the use of antr-GAL4 drivers were required to regulate the MAG-specific expression of the venom peptides. While it may have been possible to isogenize the antr-GAL4 and UAS strains by multigenerational backcrossing, characteristics unique to TMT (i.e. increased female mortality) would skew the paternity share to wild males regardless, further confounding indirect estimations of male competitiveness.
Determining the mating competitiveness of TMT males in target species will, therefore, require these strains to be engineered in wild-type genetic backgrounds, as well as utilising phenotypic markers such as fluorescent sperm so that female preference can be determined directly rather than through offspring. D. melanogaster is useful as a model for characterising systems like TMT, but limitations inherent in model systems hinder our ability to conclude how effective TMT may be if it can be implemented in other species, even closely related Dipterans like Ae. aegypti. However, current evidence indicates that successful translation of TMT into mosquitoes appears probable, as SFP are known to act on targets in the CNS of both Drosophila and mosquito females33–39, males of both species can mate three to five times before depleting their SFP27,71, and accessory gland-specific promoters have been characterised in Ae. aegypti27. However, female Ae. aegypti are more monandrous than D. melanogaster females, and it is unclear whether this difference in mating strategies may affect the quantity or proportion of SFP transferred by males.
Optimisations in the expression patterns of insecticidal proteins, such as changes to regulatory elements or co-expression of multiple venoms, will likely result in further decreases in mated female lifespan. Dose-dependent responses to venoms tend to follow a sigmoidal curve, so depending on the current levels of expression, it’s possible that minor improvements could result in a significant increase in female mortality. In order to develop a stable TMT strain for mass rearing that does not produce venom peptides until the males are released, it will be necessary to engineer conditional expression or post-translational activation of venom proteins through mechanisms such as tetracycline-repression. As UAS:venom/antr-GAL4 crosses produced viable male and female offspring, it may also be beneficial to engineer TMT strains with genetic sorting of females to increase the efficiency of large-scale production of male-only cohorts.
Intragenerational biocontrol techniques such as TMT present an approach to pest management which addresses many limitations in current insecticide and intergenerational biocontrol practices. TMT is inherently self-limiting due to strong selection pressure against venom expression, which may be advantageous from a regulatory perspective as, unlike GBT such as gene drives, there is little risk of substantial transgene introgression into non-target populations, and TMT transgenes are predicted to be rapidly lost from all populations in the absence of ongoing releases. TMT could theoretically be implemented in any species with internal fertilisation, provided that heterologous expression of the target protein can be constrained to the seminal fluid. Compared to current GBT such as SIT and fsRIDL, TMT will likely be most effective in species where the primary harm-causing life stage is the adult female, particularly anautogenous disease vectors like Black Flies and Horse Flies. TMT may also be highly effective for species which are already candidates for SIT that have relatively long lived and highly polyandrous females, including many fruit flies and Lepidoptera. In species where the adults are short lived, have low rates of female remating, and the immature stages are the primary concern, TMT will likely function comparably to SIT in its ability to supress the population. As an alternative to chemical insecticides, TMT may be uniquely well suited for rapidly responding to outbreaks of human, animal, and agricultural disease vectors.
Methods
Ethical statement
Biosafety approval was granted by the Macquarie University Institutional Biosafety Committee (#5202).
Design and construction of UAS expression plasmids
The coding sequences for mature venom proteins (μ-AGTX-Aa1d, δ-CNTX-Pn1a, Γ-CNTX-Pn1a, κ-HXTX-Hv1c, ω-HXTX-Hv1a, ω-HXTX-Hv2a and δ-AITX-Avd2a), NbVHH05, as well as the Acp26Aa signal peptide, were codon optimised for expression in D. melanogaster using Geneious Prime 2023.2.1. pJFRC81-10XUAS:IVS-Syn21-GFP-p10 (hereafter pJFRC81) was a gift from Gerald Rubin (Addgene plasmid # 36432; http://n2t.net/addgene:36432; RRID:Addgene_36432). The region 67 bp upstream of the start codon of the GFP CDS of pJFRC81 was used as an extended upstream homology arm to re-introduce the Syn21 5’UTR during assembly. About 30 bp of the p10 3′ UTR was used as the downstream homology arm, with a SapI recognition site introduced between the stop codon and terminator for future mutagenesis. Sequences comprised of the Acp26Aa signal peptide, a coding sequence, and both homology arms were ordered as gBlocks from Integrated DNA Technologies (IDT) and assembled (NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs (NEB))) into pJFRC81 after linearisation with NotI and XbaI. All plasmids were sequence verified by Plasmidsaurus (Eugene, OR) whole plasmid sequencing. Annotated plasmid DNA sequences used in this study have been deposited in Zenodo under accession code 13910640 (10.5281/zenodo.13910639)72.
Fly strains, transgenesis, and husbandry
Canton S wild-type flies were sourced from the Bloomington Drosophila Stock Centre (RRID: BDSC #64349). Act5C-GAL4/CyO flies were sourced from the Bloomington Drosophila Stock Centre (RRID: BDSC #4414). UAS:mCherry flies were sourced from the Bloomington Drosophila Stock Centre (RRID: BDSC #52268). prd-GAL4, antr-GAL4 and Acp26Aa-GAL4 flies were a kind gift from Mariana F. Wolfner from Cornell University.
UAS expression plasmids were sent to BestGene Inc (Chino Hills, CA) for plasmid DNA midipreps, microinjections into a strain with a 2nd chromosome attP docking site for φC31 mediated genomic integration (RRID: BDSC #9752) and outcrossing of transgenic lines to balancer strains. Homozygous UAS stocks were prepared by selecting against the CyO marker on the balancer chromosome.
Flies were maintained on a cornmeal diet based on the Bloomington Drosophila Stock Centre (BDSC) standard Nutri-Fly formulation (catalogue number 66-113; Genesee Scientific). Flies were reared in a controlled environment room at 25 °C, 75% humidity, and a 12 h light-dark cycle with a 30 min transition period.
Recombinant venom functionality assay
To test the functionality of recombinant venom expression, hemizygous Act5C-GAL4/CyO virgin females were crossed to homozygous UAS:(μ-AGTX-Aa1d, δ-CNTX-Pn1a, Γ-CNTX-Pn1a, κ-HXTX-Hv1c, ω-HXTX-Hv1a, ω-HXTX-Hv2a and δ-AITX-Avd2a) males. Three crosses were prepared for each of the venoms, with each cross comprising ten females and ten males, which mated freely for 7 days before the parents were discarded. Offspring were collected for up to 10 days after the parents were discarded, and the presence or absence of the CyO marker was scored for each offspring.
Characterising accessory gland-specific GAL4 drivers
The strength of on and off-target heterologous gene regulation by various accessory gland GAL4 drivers was determined by crossing UAS:mCherry reporter strains to prd-GAL4, antr-GAL4 and Acp26Aa -GAL4 driver strains. Offspring were observed throughout their development, and five to ten adult male offspring from each cross were dissected to isolate their reproductive tracts. The strength of off-target and accessory gland-specific expression of mCherry was identified by fluorescence microscopy (Olympus SZX16 stereomicroscope, 6.3x magnification, Excitation: 530–550 nm, Emission: 575 nm longpass).
Mated female lifespan assays
Wild-type females were isolated soon after eclosion and housed together in groups of eight for 2–3 days prior to male exposure to ensure that they would be receptive to male courtship. Transhemizygous antr-GAL4/UAS:(μ-AGTX-Aa1d, Γ-CNTX-Pn1a, κ-HXTX-Hv1c, ω-HXTX-Hv1a and δ-AITX-Avd2a) males were prepared continuously throughout the experiment by crossing homozygous UAS males to homozygous antr-GAL4 virgin females. Male offspring, including wild-type males, were isolated soon after eclosion and housed in groups of eight for 3–5 days prior to female exposure. Five groups of eight females were allocated to each treatment group. Females were briefly anaesthetised with CO2 to be transferred to fresh vials every 3 days, and deaths were scored every 1–2 days. Males were initially added at this first transfer (2–3 days post-female eclosion) and were replaced during subsequent transfers to maintain the desired sex ratio, as well as to ensure that females had constant access to robust males with full reserves of SFP.
Male lifespan assays
Transhemizygous antr-GAL4/UAS:(Γ-CNTX-Pn1a, δ-AITX-Avd2a and NbVHH05) males were prepared by crossing homozygous UAS males to homozygous antr-GAL4 virgin females. Forty male offspring, including wild-type males, were isolated soon after eclosion and housed in five groups of eight. Males were briefly anaesthetised with CO2 to be transferred to fresh vials every 3 days, and deaths were scored every 1–2 days.
Single-pair courtship assays
About 200 virgin wild-type females, 50 virgin wild-type males and 50 virgin transhemizygous antr-GAL4/UAS:(Γ-CNTX-Pn1a, δ-AITX-Avd2a and NbVHH05) males were isolated soon after eclosion and housed in groups of 10–15. Males were aged for 3–5 days, and females for 2–3 days prior to the experiment. Mating arenas were prepared by adding 6.2 g of sucrose diet (5% sucrose, 2% agar) to 35-mm petri dishes, leaving enough headroom for the flies to comfortably move around. Experiments were performed at the same time of day, starting 30 min after dawn. Flies were briefly anaesthetised on ice, and single pairs were transferred to mating arenas. The pairs were observed for up to 2 h, with courtship considered to be successful if the male remained firmly mounted on the female for >1–2 min, and the fraction of successful courtship was calculated to be the percentage courtship of a given treatment group.
Competitive mating assay
About 50 virgin wild-type females, 50 virgin wild-type males and 50 virgin antr-GAL4/UAS:NbVHH05 males were isolated soon after eclosion and housed in groups of 5. Males were aged for 3–5 days, and females for 2–3 days prior to the experiment. Assays were prepared just before dusk to reduce visual courtship stimuli and increase reliance on chemosensory cues73. Flies were briefly anaesthetised, and ten groups of five wild-type females, five wild-type males and five transgenic males were transferred to fresh media. The flies were immediately placed into darkened environment chambers and were allowed to mate freely until the next morning, at which point the males were discarded, and the females were isolated on fresh media. Ten offspring from each female were collected, and genomic DNA was extracted from pooled offspring as previously described74. The flies were homogenised in 20 µL PBS, after which 20 µL extraction buffer (10 Mm Tris HCl, 10 Mm KCl, 10 Mm MgCl2, 2 mM EDTA and 0.4 M NaCl) and 10 µL SDS (10%) were added. The homogenate was vortexed for 1 min, briefly centrifuged, incubated for 25 min at 60 °C, and 10 µL of NaCl (5 M) added. Finally, the samples were cooled to 4 °C, centrifuged for 30 min at 4 °C, 16,000 × g. The clarified supernatant containing the genomic DNA was extracted, diluted 1:10 to reduce the salt concentration, then used as a template for 20 µL PCR reactions with OneTaq® DNA Polymerase (New England Biolabs (NEB)). The quality of the extracted genomic DNA was determined by using primers against the endogenous white locus using the following primers: white-F (GCCAACTAGCCGAGAACCTC), and white-R (GCCGTTAGGGAGCCGATAAA). Paternity of the offspring was determined by the presence or absence of the UAS:NbVHH05 locus using the following primers: UAS-F (TCAACCACTGATGCCTAGGC), and UAS-R (CACGTTCCTGACTGCGTACT).
Agent-based modelling
To model the expected performance of TMT relative to existing GBT such as fsRIDL, we developed an agent-based model of Ae. aegypti using GAMA 1.9 (https://gama-platform.org). The model is not spatially explicit, but instead assumes a homogenous, randomly mixing population of a size for which the major limiting factor (in the absence of extrinsic effects such as predation or fluctuations in temperature and precipitation) is density-dependent mortality (DDM) of the early life stages (eggs and first and second instar larvae)75. We incorporated empirically derived parameters such as density-independent survivorship, durations of immature stages, and polyandry (model functionality and parameters detailed in Supplementary Figs. S1,S2 and Supplementary Data S1A–E)76–79. DDM was modelled as a non-linear logistic (sigmoidal) function of the larval population75, which was calibrated so that the population equilibrium and proportion of life stages fit observations from field studies22. DDM was calibrated such that the midpoint of the sigmoidal curve was centred on the hourly survival rate (0.97) and number of larvae (300) which produced a stable population of 500–600 adults, manually determining reasonable constants which produced the strongest and weakest response to larval density while maintaining the population equilibrium, then using the midpoint between these extremes for the analysis (see Supplementary Data S1A). Female remating was modelled as a logarithmic decay function which was calibrated to result in the desired rate of polyandry while producing a similar pattern of refractoriness as described in the literature55. Females that mate with fsRIDL males will produce carrier offspring, with all female carriers dying during pupation and male carriers passing the fsRIDL transgene on to 50% of their offspring. Females that mate with SIT males will produce no viable offspring, and females that mate with TMT males will die immediately after mating.
Simulating reduced mating lethality
GAMA’s factorial sampling method was used to run batches of 10 simulations for each value for mating lethality to be tested (i.e. 0.1–1.0), in combination with either fertile TMT males (surviving females produced offspring of a ‘wild’ genotype) or sterile males (surviving females produced non-viable ‘SIT’ genotype offspring). The differences between the cumulative biting rates between fsRIDL simulations and each of the reduced mating lethality simulations were assessed by performing a two-sample independent t-test.
Sensitivity analysis
We performed a sensitivity analysis to assess the robustness of our results and the model’s sensitivity to changes in three key parameters: density-dependent mortality (DDM) of the immature stages, polyandry, and release ratio. Female Ae. aegypti become receptive to mating within 24–48 h of eclosion24, and upon mating for the first time, will become increasingly refractory to remating over the course of 24 h55, after which she will never remate. The fraction of Ae. aegypti females which mate with more than a single male is estimated to be between 14 and 24%38,55, so the medium value for polyandry was set as 20%, with 0% remating as low, and the highest rates of polyandry reported for Ae. aegypti (23% of females remating two or more times, 53% remating at least once)80 as high. The values for release ratio were set at 3, 6 and 12, respectively, roughly corresponding to the per-hectare release ratios in the rangefinder, scale-up, and suppression phases of the Brazil field trials19. The high and low values for DDM were derivations of the logistic function, which produced the strongest and weakest response to larval density, respectively while maintaining the population equilibrium, with the medium value set as the midpoint between these (the same function as used in the initial analysis). GAMA’s factorial sampling method was used to run batches of 10 simulations for each parameter and technology combination, with the number of successful blood meals taken between the start of the release period and the final cycle of the simulation used as the cumulative bite rate.
The statsmodels (v0.14.1)81 package for Python 3.12 was used to perform the regression analysis using a robust linear model (RLM) with polynomial and interaction terms, as well as to evaluate multicollinearity using variance inflation factors (VIF). To perform global sensitivity analysis, the SALib (v1.4.5)82 package was used to calculate first-order, total-order, and second-order Sobol indices with 95% confidence intervals. One-at-a-time (OAT) sensitivity analysis was also used to examine the individual effects of each parameter. The Scikit-learn (v1.5.1)83 package was used to calculate standardised regression coefficients (SRC) with 95% confidence intervals and assess model fit using pseudo R-squared and mean absolute error (MAE).
Statistics and reproducibility
Lifespans of mated females and transheterozygous males were analysed with the lifelines (v0.28.0)84 Python package. Survival functions were calculated using the Kaplan–Meier estimate, differences in the survival of groups was determined by log-rank analysis, and 95% confidence intervals were calculated using Greenwood’s method. To test for differences in single-pair male courtship, a one-way chi-square test for contingency tables were used to calculate P values. Confidence intervals for the single-pair male courtship, as well as the mating competitiveness of antr-GAL4/UAS:NbVHH05 males, were calculated using the Wilson method. No statistical method was used to predetermine sample sizes, and no data were excluded from the analyses. The Investigators were not blinded to allocation during experiments and outcome assessment.
Figures
Kaplan–Meier survival curves and cumulative biting rates of model Ae. aegypti populations were plotted using the Matplotlib (v3.9.0)85 Python package. The model sensitivity heatmap and partial dependence plots were plotted using the Matplotlib and seaborn (v0.13.2)86 packages.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Information
Source data
Acknowledgements
The authors are grateful to Prof. Mariana F. Wolfner from Cornell University for providing many of the GAL4 strains used in this project, and to Prof. Glenn King from the University of Queensland for sharing his expertise on venom proteins and their heterologous expression. This material is based upon work supported by Revive and Restore Catalyst Science Fund Grant. Contract No. 2021-034: M.M. and S.J.B.
Author contributions
S.J.B. and M.M. conceived this study, designed experiments and analysed data. S.J.B. performed the experiments and developed the model. All authors contributed to writing the manuscript.
Peer review
Peer review information
Nature Communications thanks Luke Alphey, Jackson Champer, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Source data are provided with this paper or have been made available through Zenodo under accession code 13927099 (10.5281/zenodo.11465042)87. Source data are provided with this paper.
Code availability
GAMA 1.9 model files have been made available through Zenodo under accession code 13927094 (10.5281/zenodo.11439089)88. All codes used to perform statistical analysis and generate plots used in this study have been made available through Zenodo under accession code 13927099 (10.5281/zenodo.11465042)87.
Competing interests
M.M. and S.J.B. have submitted a patent application (AU2023903662A0) to the Australian patent office pertaining to the enablement of the Toxic Male Technique.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54863-1.
References
- 1.World Health Organization. World malaria report 2023. (2023)
- 2.Roiz, D. et al. Integrated Aedes management for the control of Aedes-borne diseases. PLoS Negl. Trop. Dis.12, e0006845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilder-Smith, A. et al. Epidemic arboviral diseases: priorities for research and public health. Lancet Infect. Dis.17, e101–e106 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Diagne, C. et al. High and rising economic costs of biological invasions worldwide. Nature592, 571–576 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Bacher, S. et al. Impacts of invasive alien species on nature, nature's contributions to people, and good quality of life. In Thematic Assessment Report on Invasive Alien Species and their Control of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services. (eds. Roy, H. E., Pauchard, A., Stoett, P. & Renard Truong, T.) Ch. 4 (Zenodo, 2023). 10.5281/zenodo.7430731.
- 6.Leroy, B. et al. Analysing economic costs of invasive alien species with the invacost r package. Methods Ecol. Evol.13, 1930–1937 (2022). [Google Scholar]
- 7.Savary, S. et al. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol.3, 430–439 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Howe, L. et al. Malaria parasites (Plasmodium spp.) infecting introduced, native and endemic New Zealand birds. Parasitol. Res.110, 913–923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Riper, III C., van Riper, S. G., Goff, M. L. & Laird, M. The epizootiology and ecological significance of malaria in Hawaiian land birds. Ecol. Monogr.56, 327–344 (1986). [Google Scholar]
- 10.World Health Organization. Test Procedures for Insecticide Resistance Monitoring in Malaria Vector Mosquitoes (World Health Organization, 2013).
- 11.Liu, N. Insecticide resistance in mosquitoes: impact, mechanisms, and research directions. Annu. Rev. Entomol.60, 537–559 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Ansari, M. S., Moraiet, M. A. & Ahmad, S. In Environmental Deterioration and Human Health (eds. Malik, A., Grohmann, E. & Akhtar, R.) Ch. 6 (Springer, 2014).
- 13.Bueno, A. F. et al. Challenges for adoption of integrated pest management (IPM): the soybean example. Neotrop. Entomol.50, 5–20 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Teem, J. L. et al. Genetic biocontrol for invasive species. Front. Bioeng. Biotechnol.8, 452 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winneg, K. M., Stryker, J. E., Romer, D. & Jamieson, K. H. Differences between Florida and the rest of the United States in response to local transmission of the zika virus: implications for future communication campaigns. Risk Anal.38, 2546–2560 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Knipling, E. F. Possibilities of insect control or eradication through the use of sexually sterile males. J. Econ. Entomol.48, 459–462 (1955). [Google Scholar]
- 17.Fu, G. et al. Female-specific insect lethality engineered using alternative splicing. Nat. Biotechnol.25, 353–357 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Esvelt, K. M., Smidler, A. L., Catteruccia, F. & Church, G. M. Concerning RNA-guided gene drives for the alteration of wild populations. eLife3, e03401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvalho, D. O. et al. Suppression of a field population of Aedes aegypti in Brazil by sustained release of transgenic male mosquitoes. PLoS Negl. Trop. Dis.9, e0003864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shelton, A. M. et al. First field release of a genetically engineered, self-limiting agricultural pest insect: evaluating its potential for future crop protection. Front. Bioeng. Biotechnol.7, 482 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammond, A. et al. Gene-drive suppression of mosquito populations in large cages as a bridge between lab and field. Nat. Commun.12, 4589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheppard, P. M., Macdonald, W. W., Tonn, R. J. & Grab, B. The dynamics of an adult population of Aedes aegypti in relation to dengue haemorrhagic fever in Bangkok. J. Anim. Ecol.38, 661–702 (1969). [PMC free article] [PubMed] [Google Scholar]
- 23.Sowilem, M. M., Kamal, H. A. & Khater, E. I. Life table characteristics of Aedes aegypti (Diptera: Culicidae) from Saudi Arabia. Trop. Biomed. 30, 301–314 (2013). [PubMed]
- 24.Ramírez-Sánchez, L. F., Hernández, B. J., Guzmán, P. A., Alfonso-Parra, C. & Avila, F. W. The effects of female age on blood-feeding, insemination, sperm storage, and fertility in the dengue vector mosquito Aedes aegypti (Diptera: Culicidae). J. Insect Physiol.150, 104570 (2023). [DOI] [PubMed] [Google Scholar]
- 25.Scott, T. W. et al. Longitudinal Studies of Aedes aegypti (Diptera: Culicidae) in Thailand and Puerto Rico: blood feeding frequency. J. Med. Entomol.37, 89–101 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Weaver, S. C. Prediction and prevention of urban arbovirus epidemics: a challenge for the global virology community. Antivir. Res.156, 80–84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alfonso-Parra, C. et al. Synthesis, depletion and cell-type expression of a protein from the male accessory glands of the dengue vector mosquito Aedes aegypti. J. Insect Physiol.70, 117–124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall, J. M., Pittman, G. W., Buchman, A. B. & Hay, B. A. Semele: a killer-male, rescue-female system for suppression and replacement of insect disease vector populations. Genetics187, 535–551 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurtado, J., Revale, S. & Matzkin, L. M. Propagation of seminal toxins through binary expression gene drives could suppress populations. Sci. Rep.12, 6332 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sirot, L. K. et al. Identity and transfer of male reproductive gland proteins of the dengue vector mosquito, Aedes aegypti: potential tools for control of female feeding and reproduction. Insect Biochem. Mol. Biol.38, 176–189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapman, T., Liddle, L. F., Kalb, J. M., Wolfner, M. F. & Partridge, L. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature373, 241–244 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Avila, F. W., Sirot, L. K., LaFlamme, B. A., Rubinstein, C. D. & Wolfner, M. F. Insect seminal fluid proteins: identification and function. Annu. Rev. Entomol.56, 21–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lung, O. & Wolfner, M. F. Drosophila seminal fluid proteins enter the circulatory system of the mated female fly by crossing the posterior vaginal wall. Insect Biochem. Mol. Biol.29, 1043–1052 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Yapici, N., Kim, Y.-J., Ribeiro, C. & Dickson, B. J. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature451, 33–37 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Young, A. D. M. & Downe, A. E. R. Male accessory gland substances and the control of sexual receptivity in female Culex tarsalis. Physiol. Entomol.12, 233–239 (1987). [Google Scholar]
- 36.Klowden, M. J. Sexual receptivity in Anopheles gambiae mosquitoes: absence of control by male accessory gland substances. J. Insect Physiol.47, 661–666 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Shutt, B., Stables, L., Aboagye-Antwi, F., Moran, J. & Tripet, F. Male accessory gland proteins induce female monogamy in anopheline mosquitoes. Med. Vet. Entomol.24, 91–94 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Helinski, M. E. H., Deewatthanawong, P., Sirot, L. K., Wolfner, M. F. & Harrington, L. C. Duration and dose-dependency of female sexual receptivity responses to seminal fluid proteins in Aedes albopictus and Ae. aegypti mosquitoes. J. Insect Physiol.58, 1307–1313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duvall, L. B., Basrur, N. S., Molina, H., McMeniman, C. J. & Vosshall, L. B. A peptide signaling system that rapidly enforces paternity in the Aedes aegypti mosquito. Curr. Biol.27, 3734–3742.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhn-Nentwig, L., Stöcklin, R. & Nentwig, W. Venom composition and strategies in spiders. Adv. Insect Physiol. 40, 1–86 (2011).
- 41.King, G. F. Tying pest insects in knots: the deployment of spider‐venom‐derived knottins as bioinsecticides. Pest Manag. Sci.75, 2437–2445 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Leader, D. P., Krause, S. A., Pandit, A., Davies, S. A. & Dow, J. A. T. FlyAtlas 2: a new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Res.46, D809–D815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishikawa, T. et al. DCRY is a Drosophila photoreceptor protein implicated in light entrainment of circadian rhythm. Genes Cells4, 57–65 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Lee, P.-T. et al. A gene-specific T2A-GAL4 library for Drosophila. eLife7, e35574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoffe, K. B., Manoukian, A. S., Wilder, E. L., Brand, A. H. & Perrimon, N. Evidence for engrailed-independent wingless autoregulation in Drosophila. Dev. Biol.170, 636–650 (1995). [DOI] [PubMed] [Google Scholar]
- 46.Chapman, T. et al. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc. Natl Acad. Sci. USA100, 9923–9928 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shearin, H. K., Macdonald, I. S., Spector, L. P. & Stowers, R. S. Hexameric GFP and mCherry reporters for the Drosophila GAL4, Q, and LexA transcription systems. Genetics196, 951–960 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manning, A. The control of sexual receptivity in female Drosophila. Anim. Behav.15, 239–250 (1967). [DOI] [PubMed] [Google Scholar]
- 49.Manning, A. A sperm factor affecting the receptivity of Drosophila melanogaster females. Nature194, 252–253 (1962). [Google Scholar]
- 50.Proverbs, M. D., Newton, J. R. & Campbell, C. J. Codling moth: a pilot program of control by sterile insect release in British Columbia. Can. Entomol.114, 363–376 (1982). [Google Scholar]
- 51.Wee, S. L. et al. Effects of substerilizing doses of gamma radiation on adult longevity and level of inherited sterility in Teia anartoides (Lepidoptera: Lymantriidae). J. Econ. Entomol.98, 732–738 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Edward, D. A., Fricke, C., Gerrard, D. T. & Chapman, T. Quantifying the life‐history response to increased male exposure in female Drosophila melanogaster. Evolution65, 564–573 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Von Philipsborn, A. C., Shohat-Ophir, G. & Rezaval, C. Single-pair courtship and competition assays in Drosophila. Cold Spring Harb. Protoc.2023, pdb.prot108105 (2023). [DOI] [PubMed] [Google Scholar]
- 54.Ling, J. et al. A nanobody that recognizes a 14-residue peptide epitope in the E2 ubiquitin-conjugating enzyme UBC6e modulates its activity. Mol. Immunol.114, 513–523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Degner, E. C. & Harrington, L. C. Polyandry depends on postmating time interval in the dengue vector Aedes aegypti. Am. J. Trop. Med. Hyg.94, 780–785 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrington, L. C. et al. Heterogeneous feeding patterns of the dengue vector, Aedes aegypti, on individual human hosts in rural Thailand. PLoS Negl. Trop. Dis.8, e3048 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan, M. & Johansson, M. A. The incubation periods of dengue viruses. PLoS ONE7, e50972 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.ten Broeke, G., van Voorn, G. & Ligtenberg, A. Which sensitivity analysis method should I use for my agent-based model? JASSS19, 5 (2016). [Google Scholar]
- 59.Quistad, G. B., Dennis, P. A. & Skinner, W. S. Insecticidal activity of spider (Araneae), centipede (Chilopoda), scorpion (Scorpionida), and snake (Serpentes) venoms. J. Econ. Entomol.85, 33–39 (1992). [Google Scholar]
- 60.Schwartz, E. F., Mourão, C. B. F., Moreira, K. G., Camargos, T. S. & Mortari, M. R. Arthropod venoms: a vast arsenal of insecticidal neuropeptides. Pept. Sci.98, 385–405 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Guo, S., Herzig, V. & King, G. F. Dipteran toxicity assays for determining the oral insecticidal activity of venoms and toxins. Toxicon150, 297–303 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Oh, R. J. Repeated copulation in the brown planthopper, Nilaparvata lugens Stál (Homoptera; Delphacidae). Ecol. Entomol.4, 345–353 (1979). [Google Scholar]
- 63.Cabauatan, P. Q., Cabunagan, R. C. & Choi, I.-R. In Planthoppers: New Threats to the Sustainability of Intensive Rice Production Systems in Asia (eds. Heong, K. L. & Hardy, B.) 357–368 (International Rice Research Institute, 2009).
- 64.Denecke, S., Swevers, L., Douris, V. & Vontas, J. How do oral insecticidal compounds cross the insect midgut epithelium. Insect Biochem. Mol. Biol.103, 22–35 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Kandul, N. P. et al. Transforming insect population control with precision guided sterile males with demonstration in flies. Nat. Commun.10, 84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Massonnet-Bruneel, B. et al. Fitness of transgenic mosquito Aedes aegypti males carrying a dominant lethal genetic system. PLoS ONE8, e62711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Upadhyay, A. et al. Genetically engineered insects with sex-selection and genetic incompatibility enable population suppression. eLife11, e71230 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ejima, A. & Griffith, L. C. Measurement of courtship behavior in Drosophila melanogaster. Cold Spring Harb. Protoc.2007, pdb.prot4847 (2007). [DOI] [PubMed]
- 69.Mueller, J. L., Linklater, J. R., Ravi Ram, K., Chapman, T. & Wolfner, M. F. Targeted gene deletion and phenotypic analysis of the Drosophila melanogaster seminal fluid protease inhibitor Acp62F. Genetics178, 1605–1614 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, S. D. & Odenwald, W. F. Misexpression of the white (w) gene triggers male-male courtship in Drosophila. Proc. Natl Acad. Sci. USA92, 5525–5529 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sirot, L. K., Buehner, N. A., Fiumera, A. C. & Wolfner, M. F. Seminal fluid protein depletion and replenishment in the fruit fly, Drosophila melanogaster: an ELISA-based method for tracking individual ejaculates. Behav. Ecol. Sociobiol.63, 1505–1513 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maselko, M. & Beach, S. Plasmid sequences for: Recombinant venom proteins in insect seminal fluid reduces female lifespan. Zenodo 10.5281/zenodo.13910640 (2024).
- 73.Joiner, M. A. & Griffith, L. C. Visual input regulates circuit configuration in courtship conditioning of Drosophila melanogaster. Learn. Mem.7, 32–42 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hegde, R., Hegde, S., Gataraddi, S., Kulkarni, S. S. & Gai, P. B. Novel and PCR ready rapid DNA isolation from Drosophila. Nucleosides Nucleotides Nucleic Acids41, 1162–1173 (2022). [DOI] [PubMed] [Google Scholar]
- 75.Sauers, L. A., Hawes, K. E. & Juliano, S. A. Non-linear relationships between density and demographic traits in three Aedes species. Sci. Rep.12, 8075 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goindin, D., Delannay, C., Ramdini, C., Gustave, J. & Fouque, F. Parity and longevity of Aedes aegypti according to temperatures in controlled conditions and consequences on dengue transmission risks. PLoS ONE10, e0135489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Christophers, A. A. The yellow fever mosquito, its life history, bionomics and structure. J. Natl Med. Assoc.54, 132 (1960). [Google Scholar]
- 78.Almeida, S. J. D., Martins Ferreira, R. P., Eiras, Á. E., Obermayr, R. P. & Geier, M. Multi-agent modeling and simulation of an Aedes aegypti mosquito population. Environ. Model. Softw.25, 1490–1507 (2010). [Google Scholar]
- 79.Maneerat, S. & Daudé, E. A spatial agent-based simulation model of the dengue vector Aedes aegypti to explore its population dynamics in urban areas. Ecol. Model.333, 66–78 (2016). [Google Scholar]
- 80.Pimid, M. et al. Parentage assignment using microsatellites reveals multiple mating in Aedes aegypti (Diptera: Culicidae): implications for mating dynamics. J. Med. Entomol.59, 1525–1533 (2022). [DOI] [PubMed] [Google Scholar]
- 81.Seabold, S. & Perktold, J. Statsmodels: econometric and statistical modeling with Python. In Proc. 9th Python in Science Conference 92–96 (2010).
- 82.Herman, J. & Usher, W. SALib: an open-source Python library for sensitivity analysis. J. Open Source Softw.2, 97 (2017). [Google Scholar]
- 83.Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res.12, 2825–2830 (2011). [Google Scholar]
- 84.Davidson-Pilon, C. lifelines: survival analysis in Python. J. Open Source Softw.4, 1317 (2019). [Google Scholar]
- 85.Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng.9, 90–95 (2007). [Google Scholar]
- 86.Waskom, M. L. seaborn: statistical data visualization. J. Open Source Softw.6, 3021 (2021). [Google Scholar]
- 87.Maselko, M. & Beach, S. J. Data and code for: recombinant venom proteins in insect seminal fluid reduces female lifespan. Zenodo10.5281/zenodo.13927099 (2024).
- 88.MaselkoLab. TMT-Aedes-model. GitHub (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Information
Data Availability Statement
Source data are provided with this paper or have been made available through Zenodo under accession code 13927099 (10.5281/zenodo.11465042)87. Source data are provided with this paper.
GAMA 1.9 model files have been made available through Zenodo under accession code 13927094 (10.5281/zenodo.11439089)88. All codes used to perform statistical analysis and generate plots used in this study have been made available through Zenodo under accession code 13927099 (10.5281/zenodo.11465042)87.