Abstract
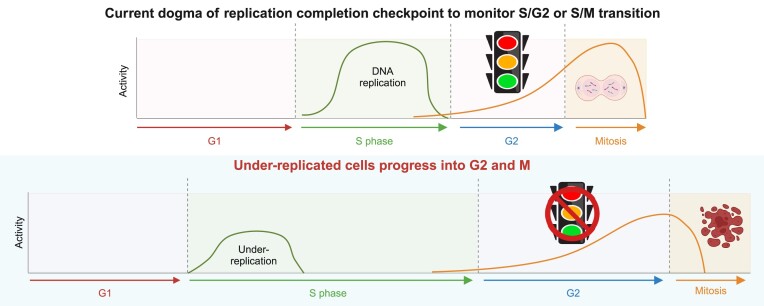
Cell cycle checkpoints are the regulatory mechanisms that secure the strict order of cellular events for cell division that ensure genome integrity. It has been proposed that mitosis initiation depends on the completion of DNA replication, which must be tightly controlled to guarantee genome duplication. Contrary to these conventional hypotheses, we showed here that cells were able to enter mitosis without completion of DNA replication. Although DNA replication was not completed in cells upon depletion of MCM2, CDC45 or GINS4, these under-replicated cells progressed into mitosis, which led to cell death. These unexpected results challenge current model and suggest the absence of a cell cycle checkpoint that monitors the completion of DNA replication.
Graphical Abstract
Graphical Abstract.
Introduction
To ensure that genetic information is precisely passed to daughter cells during cell division, cells must complete genome duplication in S phase before the initiation of mitosis (1,2). It is generally believed and well-accepted that mitosis initiation depends on the completion of DNA replication. Notably, some intriguing observations have been reported in yeast smc5-smc6 mutants (3) and cdc6 mutant (4), as well as zebrafish ticrr mutant (5). While these mutants fail to duplicate genome, these under-replicated cells still enter mitosis, leading to mitotic catastrophe and cell death. The key question is whether there is a checkpoint that ensures the completion of DNA replication before allowing cells to progress from S phase to G2 or M phase. This checkpoint was loosely defined as S/G2 or S/M checkpoint. We rename it here as replication completion checkpoint to clarify the nature of this checkpoint. If DNA replication completion checkpoint exists, how does it operate in cells with incomplete genome duplication?
To address this fundamental question in cell cycle regulation, we took advantage of the degradation tag (dTAG) system (6,7), which allows precise and well-controlled degradation of proteins of interest within 1 or 2 h and therefore enables detailed analysis of cell cycle progression in the absence of proteins that are essential for DNA replication. We showed that, as anticipated, cells could not complete DNA replication in the absence of MCM2, CDC45 or GINS4. Moreover, these cells exhibited robust activation of ATR/CHK1 and other DNA damage responsive (DDR) pathways, which is likely associated with DNA breaks induced by MCM2-, CDC45- or GINS4-depletion. However, the activation of ATR/CHK1 and other DDR pathways was transient. These cells with incomplete DNA replication eventually entered mitosis, which led to cell lethality. Taken together, these data indicate the absence of a replication completion checkpoint, which is supposed to monitor the completion of DNA replication before allowing cells transition to mitosis.
Materials and methods
Cell lines
HEK293A and HEK293T cells were purchased from the ATCC and cultured in Dulbecco’s modified Eagle’s medium (Corning) with 10% fetal bovine serum (Sigma). To generate HEK293A-MCM2/CDC45/GINS4-CKI-dTAG cell lines, HEK293A cells were co-transfected with sgRNA targeting the C-terminus of MCM2, CDC45 or GINS4 and donor vector. The donor vector contains dTAG that includes a linker, FKBPF36V-2HA, a P2A self-cleavage site and blasticidin resistance gene flanked by homology arms in a PUC19 backbone as reported previously (8). Forty-eight hours after transfection, cells were selected with 10 μg/ml blasticidin S (Gibco) for 5 days. The survived cells were diluted and seeded in 96-well plates for single cell cloning. Positive clones of dTAG knock-in were screened using genomic PCR and further validated by Western blotting.
To generate wild-type (WT) HEK293A and HEK293A-MCM2 (or -CDC45 or -GINS4)-CKI-dTAG cell lines with stable expression of tFucci (CA)5, super PiggyBac transposase expression vector (PB210PA-1; System Biosciences) and tFucci (CA)5 (Addgene, cat. no.153521) (9) were co-transfected into cells using PEI (polyethylenimine). Fucci (CA)-expressing single cells were selected by fluorescence-activated cell sorting (FACS) with both green and red channels and further diluted in 96-well plates for cloning (10). To generate WT HEK293A and HEK293A-MCM2 (or -CDC45 or -GINS4)-CKI-dTAG cell lines stably expressing GFP-H2B, pRRlsinPGK_H2BGFP_WPRE (Addgene, cat. no. 91788; a gift from Beverly Torok-Storb) was co-transfected with the packaging plasmid psPAX2 and the envelope vector pMD2.G using the X-tremeGENE HP DNA transfection reagent (Sigma-Aldrich, cat. no. 6366546001) for high-titer virus production in HEK293T cells. The viral supernatants were then used to infect cells, which were diluted and seeded in 96-well plates for cloning.
Constructs and cloning
CDC45 sgRNA (TGTCCTAGGGTGAGTTACAG), MCM2 sgRNA (GAGGCCCTATGCCAT CCATA) and GINS4 sgRNA (TGTCCAGCTAATTTAAAACT) were cloned into pX330 (Addgene, cat. no. 42230) (11) for knock-in of C-terminal dTAG tag at the endogenous CDC45, MCM2 and GINS4 locus respectively. The DNA fragments’ dTAG for CKI was amplified via PCR from pCRIS-PITChv2-dTAG-blasticidin (Addgene, cat. no. 91795). A donor vector for CDC45/MCM2/GINS4 knock-in was generated using Gibson assembly of the 5′ homolog arm, dTAG and 3′ homolog arm into a PUC19 vector. ETAA1-sgRNA1 (TCTATGCTGGACATGTGG AT), ETAA1-sgRNA2 (TTAAGCCAAGAACTGCCAG), TOPBP1-sgRNA1 (TCAAAGACAACGCCACTAAA) and TOPBP1-sgRNA2 (TATATCTGTGACCCTTTTAG) were cloned into lentiCRIPSRv2 (Addgene, cat. no. 52961) for respectively knockdown of ETAA1 or TOPBP1.
Antibodies and inhibitors
Antibodies against pKAP1 (S824; cat. no. 4127S), pATM (S1981; cat. no. 4526L), pChk2 (T68; cat. no. 2661S), MCM2 (cat. no. 4007S), CDC45 (cat. no. 3673S), HA (cat. no. 2999S), pChk1 (S345; cat. no. 2348S), pCDK1/2(Y15; cat. no. 9111S), pH3 (S10; cat. no. 9701S) and TOPBP1 (cat. no. 14 342) were purchased from Cell Signaling Technology. Antibodies against pRPA2 (S4/S8; cat. no. A300-245A) and pRPA2 (S33; cat. no. A300-246A) were purchased from Bethyl Laboratories. An antibody against vinculin (cat. no. V9264) was purchased from Sigma. An antibody against γH2AX was purchased from BioLegend (cat. no. 613402). An antibody against cyclin B1 was purchased from Santa Cruz Biotechnology (cat. no. sc-245). An antibody against GINS4/SLD5 was purchased from Novus Biologicals (cat. no. NBP2-16659). These antibodies were used for Western blot analysis. Antibodies against pH3(S10; cat. no. 06-570) and γH2AX (cat. no. 05-636) purchased from Millipore were used for immunofluorescent staining. Alexa Fluor 488 azide was purchased from Thermo Fisher Scientific (cat. no. A10266). Alexa Fluor 555-conjugated goat anti-rabbit IgG (H + L; cat. no. A-21428) was purchased from Thermo Scientific.
The CDK1 inhibitor RO-3306 (cat. no. S7747) was purchased from Selleck Chemicals. dTAGv-1-NEG (NEG; cat. no. 6915) and dTAGv-1 (cat. no. 6914) were purchased from Tocris Bioscience.
Western blot analysis
HEK293A-MCM2/CDC45/GINS4cki-dTAG cells were seeded in six-well plates overnight, followed by treatment with NEG or dTAGv-1 for the time as indicated. For cell synchronization at early S phase, HEK293A-MCM2/ CDC45/GINS4-dTAG cells were first treated with 2 mM thymidine for 18 h, followed by washing twice and cultured in thymidine-free media for 9 h, then again with second thymidine treatment for 16 h. The synchronized cells (at G1/S or early S) were released for 2 h before treatment with NEG or dTAGv-1 for the indicated time periods. Both adhesive and floated cells were collected and lysed using sodium dodecyl sulfate gel-loading buffer. Samples were boiled and saved for further analysis. Samples were then separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by immunoblotting with the indicated antibodies.
Immunofluorescent staining
HEK293A-MCM2-dTAG cells were seeded on cover glasses overnight. Cells were then treated with NEG or dTAGv-1 for different time points and pulse-labelled with 10 μM EdU for 30 min immediately before fixation with 3% paraformaldehyde. After fixation, cells were permeabilized with 0.5% Triton X-100 [in phosphate-buffered saline (PBS)] for 15 min and blocked with 3% bovine serum albumin in PBS for 30 min. Cells were further incubated with the antibody against pH3S10 overnight and then incubated with a secondary antibody the next day, followed by incubation with Click-iT cell reaction buffer (Invitrogen) for 30 min. Cells were later counterstained with DAPI, and images were acquired with a Nikon microscope.
Flow cytometry
HEK293A-MCM2/CDC45/GINS4-dTAG cells were treated with 1 μM NEG or dTAGv-1 for the indicated number of days. Both floated and adhesive cells were collected and fixed with pre-cold 70% ethanol. Cells were then permeabilized with 0.5% Triton X-100 (in PBS) for 15 min. The samples were washed with PBS and stained with FxCycle (Thermo Fisher Scientific) for 30 min, followed by flow cytometry analysis.
CellTiter-Glo cell viability assay
For short-term cell survival assays, HEK293A and HEK293A-MCM2/CDC45/GINS4-dTAG cells were seeded in 96-well plates with 1 μM NEG or dTAGv-1 in sextuplicate. Cell viability was evaluated via CellTiter-Glo luminescence assay (Promega) at the indicated days.
Colony formation assay
For the assay performed with asynchronized cells, HEK293A-MCM2/CDC45/GINS4-dTAG cells were seeded with 1 μM NEG or dTAGv-1 in six-well plates. For the assay performed with synchronized cells, HEK293A and HEK293A-MCM2/CDC45/GINS4-dTAG cells were seeded in six-well plates overnight, the cells first were incubated with 2 mM thymidine for 18 h, followed by washing twice and further cultured in thymidine-free media for 9 h. Cells were again incubated with 2 mM thymidine for 16 h, released by washing twice and cultured in thymidine-free media. Two hours after release from thymidine, cells were treated with 1 μM NEG or dTAGv-1. Cells were stained with crystal violet solution (Sigma-Aldrich) at the indicated days. The intensity of cell confluency was measured by ColonyArea ImageJ plugin (12).
Long-term live-cell imaging
Cells were grown in optically clear 96-well microplates (PhenoPlate, PerkinElmer, cat. no. 6055300) in DMEM containing 10% FBS. Time-lapse live-cell imaging was performed using a Yokogawa CV8000 spinning disk confocal microscope. Images were taken every 30 min until 48 h with cell cultivation environment (37°C and 5% CO2). Fluorescent and bright-field images were taken with 20×/0.75NA objectives using the following laser/filter combinations: 488 nm/525 nm (Ex/Em) and 561 nm/600 nm (Ex/Em). Laser power and exposure times were set to minimize photobleaching and kept constant throughout each imaging session. Image processing and export were performed using CellPathfinder software (Yokogawa). HEK293A and HEK293A-MCM2/CDC45/GINS4-dTAG cells transduced with either Fucci (CA) or GFP-H2B were used in Figures 4 and 5; Supplementary Videos S1–S16.
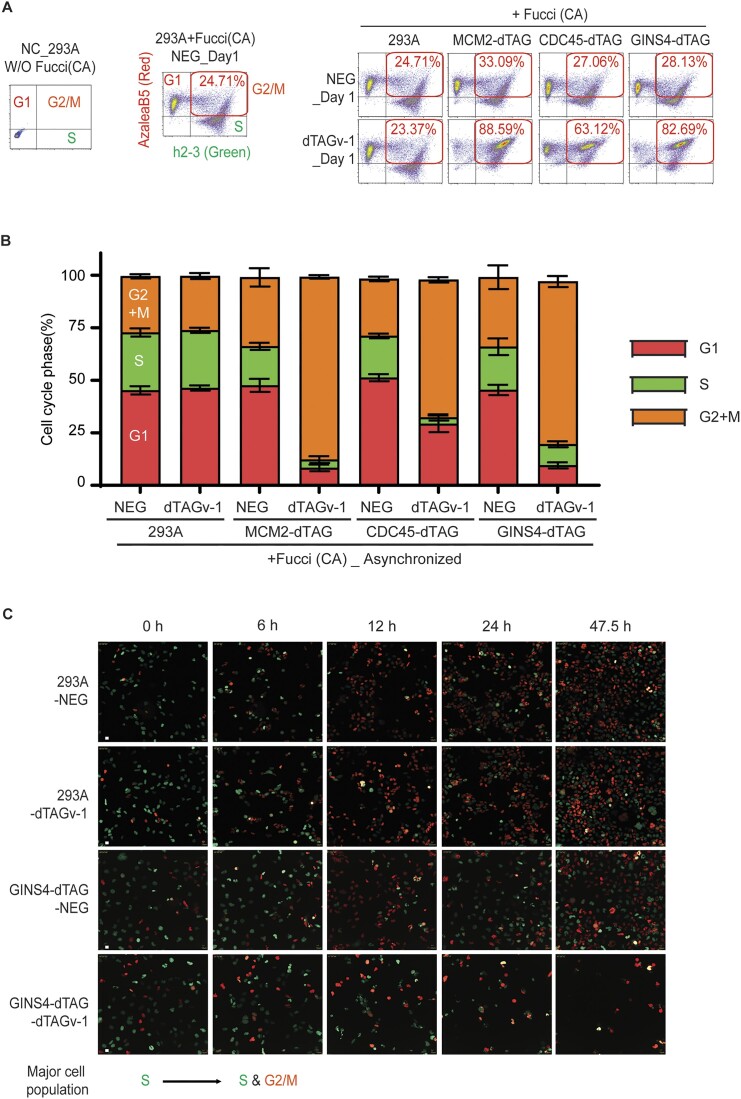
Figure 4.
MCM2-, CDC45- or GINS4-depleted cells enter and die in mitosis with incomplete DNA replication shown by Fucci (CA) assay. (A) Flow cytometry analyses of HEK293A-Fucci (CA) and HEK293A-MCM2/CDC45/GINS4-dTAG-Fucci (CA) cells. HEK293A cells without introducing Fucci (CA) were included as the negative control (NC) for gating G1, S and G2/M. NEG- or dTAGv-1-treated HEK293A-Fucci (CA) cells were further gated according to the expression pattern of fluorescent proteins. Cells only expressing AzaleaB5-hCdt1(1/100) (red) were gated as G1 cells, while cells only expressing h2-3-hGem(1/110) (green) were gated as S phase cells and cells expressing both proteins were gated as cells in G2/M. More than 10 000 cells were collected for analyses. (B) Flow cytometry analyses of asynchronized HEK293A-Fucci (CA) and HEK293A-MCM2/CDC45/GINS4-dTAG-Fucci (CA) cells using the same or similar samples in (A). HEK293A-Fucci (CA) and HEK293A-MCM2/CDC45/GINS4-dTAG-Fucci (CA) cells were treated with NEG- or dTAGv-1 for 24 h, followed by FACS analyses. More than 10 000 cells were collected for analyses. Bars represent the mean ± SEM (n = 3). (C) Live-cell imaging of HEK293A-Fucci (CA) and HEK293A-GINS4-dTAG-Fucci (CA) cells. Cells were synchronized with double thymidine block and then released for 2 h for most cells entering S phase, followed by treatment with NEG or dTAGv-1. Eight independent field of cell population for every sample was imaged, and images were taken every 30 min until 48 h. Representative images at indicated hours are shown (scale bar 100 px).
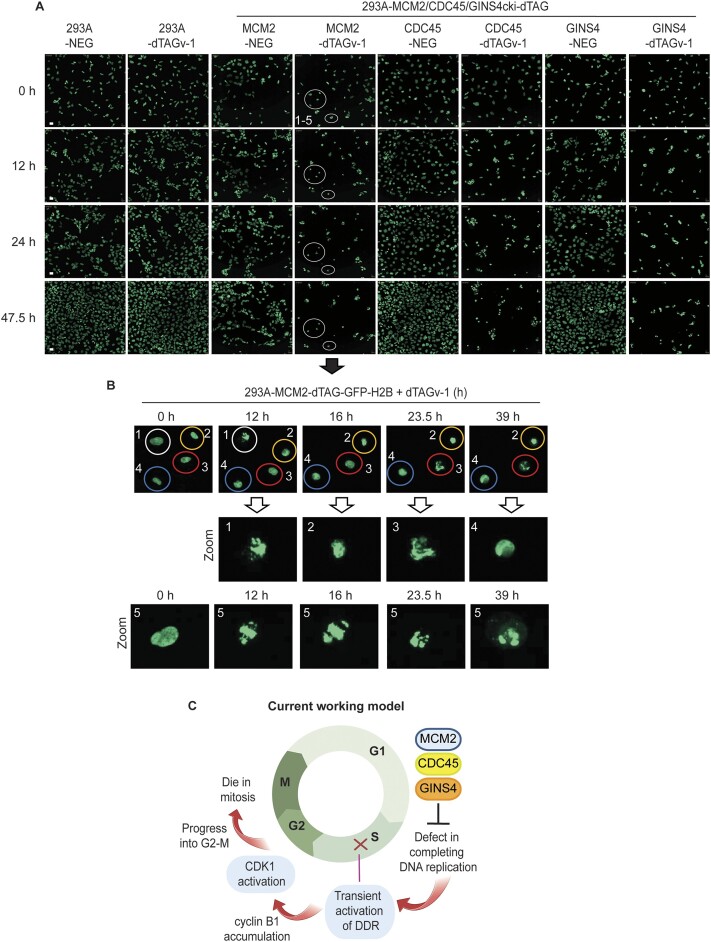
Figure 5.
MCM2-, CDC45- or GINS4-depleted cells enter and die in mitosis with incomplete DNA replication reflected by live cell imaging with GFP-H2B. (A) Live-cell imaging of HEK293A-GFP-H2B and HEK293A-MCM2/CDC45/GINS4-dTAG-GFP-H2B cells. Cells were synchronized in S phase and then released for 2 h followed by treatment with NEG or dTAGv-1, which was similar to that described in (B). Eight independent field of cell population for every sample was imaged, and images were taken every 30 min until 48 h. Representative images at indicated hours are shown (scale bar 100 px). (B) Live-cell imaging of dTAGv-1-treated HEK293A-MCM2-dTAG-GFP-H2B cells. These cells were the same cells shown by circle in Figure 4C and the zoomed in images were used for better representation. Representative images of abnormal nuclear condensation and chromosome segregation are shown. (C) Schematic representation of our current working hypothesis. Cells with MCM2, CDC45 or GINS4 depletion are not able to complete DNA replication. These cells transiently activate DNA damage responses for several hours. Although these cells never complete DNA replication, DNA damage responses in these cells diminish afterwards. Cells progress into G2/M phase and eventually die in mitosis, likely due to abnormal nuclear condensation and chromosome alignment as well as failed cytokinesis resulting from incomplete genome duplication. The image was created with BioRender.
Metaphase spread
HEK293A-MCM2-dTAG cells were treated with 1 μM NEG or dTAGv-1 for 48 h, then the samples were prepared as described previously (8). Briefly, NEG- or dTAGv-1-treated cells were synchronized with 100 ng/ml Nocodazole for 16 hand harvested. The harvested cells were subject to hypotonization with 0.075 M KCl for 15 min at 37°C. After centrifugation, cells were fixed with fresh and pre-chilled methanol: glacial acetic acid (3:1) solution. Then cells were resuspended with 0.5 ml methanol: glacial acetic acid (3:1) solution. A single drop of fixed cells was released vertically onto the slide from ∼20 cm height. Slides were mounted with DAPI after air dry. Images were captured by a Nikon microscope at ×60 magnification.
Neutral comet assay
The neutral comet assay was conducted with CometAssay kit (4250-050-K; R&D systems) following the instructions as described previously (8). Briefly, HEK293A-MCM2-dTAG cells were treated with 1 μM NEG or dTAGv-1 for indicated hours. Before collecting the samples, the comet LMAgarose was heated at 95°C for 5 min to melt and kept in 37°C for at least 20 min to cool. Cells were collected at the same time, washed with PBS once, and then diluted to the concentration of 4 × 105 cells/ml in PBS. The diluted cells (20 μl cells) were resuspended in 200 μl of pre-warmed LMAgarose and immediately spread 50 μl mixture onto CometSlide, followed by placing slides flat at 4°C for 30 min. Samples were lysed by immersing slides in lysis solution overnight at 4°C. Then slides were washed and immersed in 1× neutral electrophoresis buffer (50 nM Tris base and 150 mM sodium acetate) for 30 min, followed by subject to electrophoresis at 21 V for 45 min in 1× neutral electrophoresis buffer. Then slides were washed with water followed by 70% ethanol and dried overnight. Finally, slides were stained with SYBR-gold for 30 min, washed with water and dried for imaging. Images were obtained using a Nikon microscope at ×10 magnification. Collected images were analyzed using OpenComet (13), and the olive tails are shown and measured.
Results
Cells with MCM2 depletion transiently activate DNA damage responses
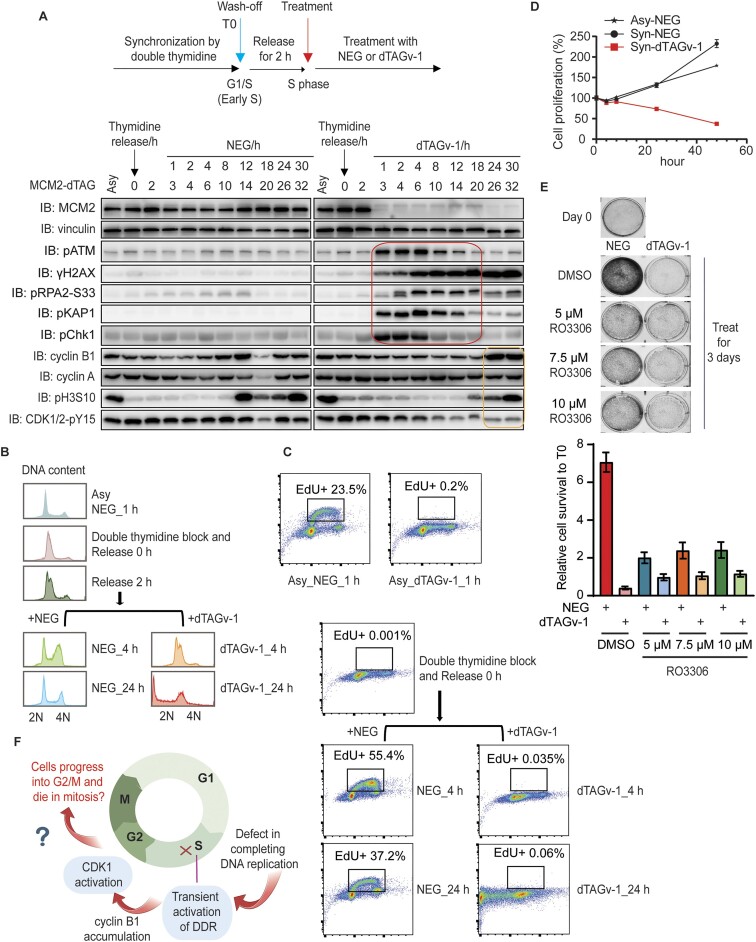
The cell cycle depicts a series of highly ordered critical events, such as DNA replication and chromosome segregation, which lead to the generation of daughter cells. According to the conventional hypothesis, there must be a S/G2 or S/M checkpoint that ensures the completion of DNA replication before mitosis. To determine what would happen if cells cannot complete DNA replication, we employed dTAG system, which allows inducible and rapid degradation of protein of interest within one or a few hours. Since MCM (2-7) is the helicase complex unwinding double-strand DNA and is essential for DNA replication, we generated homozygous C-terminal knock-in of dTAG at the MCM2 locus in HEK293A cells and validated the clones by PCR (Figure 1A). Our data show that the MCM2-dTAG protein rapidly decreased to strikingly low levels in the presence of dTAGv-1 after only 1 h compared to that of the same cells treated with dTAG-NEG (NEG) control (Figure 1B). Moreover, HEK293A-MCM2-dTAG cells treated with 5 nM or more dTAGv-1 exhibited dramatically deficient growth compared to that in NEG-treated cells (Figure 1C). Consistent with the dTAGv-1-induced significant degradation of MCM2 protein (Figure 1B), pulse-labeling of the dTAGv-1-treated MCM2-dTAG cells with EdU indicates that DNA replication declined to very low level after only 1 h of treatment and was almost undetectable after 2 h, compared to those in NEG-treated cells (Figure 1D). These results suggest that cells treated with dTAGv-1 for only 1 h would result in marked disruption of DNA replication. Thus, as expected, these data indicate that MCM2 is essential for DNA replication and cell proliferation. Moreover, MCM2-dTAG cells serve as a powerful tool to determine the cell fate of under-replicated cells.
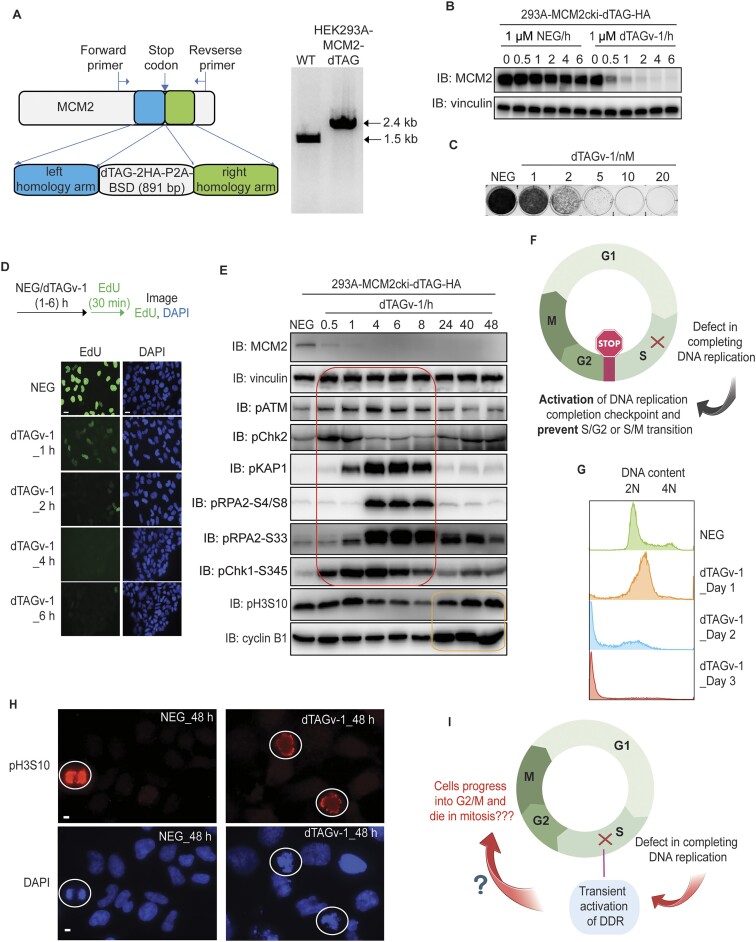
Figure 1.
MCM2 depletion results in transient activation of DNA damage responses. (A) Schematic design of the donor template used for the generation of HEK293A-MCM2-dTAG cells (left). The forward and reverse primers outside of respectively left and right homology arms were used for PCR validation of HEK293A-MCM2cki-dTAG clones (right). (B) Western blot validation of MCM2cki-dTAG cell line in the presence of 1 μM NEG or 1 μM dTAGv-1. (C) Clonogenic survival of MCM2-dTAG cells were determined in the presence of NEG or different concentrations of dTAGv-1. (D) Immunofluorescent staining of EdU-positive cells in MCM2-dTAG cells. Cells were treated 1 μM NEG or dTAGv-1 for the time indicated (hours), then pulse-labelled with 10 μM EdU for 30 min immediately before fixation. Representative images of EdU immunostaining are shown (scale bar 100 px). (E) Immunoblots of the indicated proteins prepared from MCM2-dTAG cells treated with NEG or dTAGv-1 for the indicated hours. Both floated and adhesive cells were collected for analysis. The red box shows the transient activation of DDR signaling, while the yellow box displays the increased levels of pH3S10 and cyclin B1 reflecting cells entering mitosis. (F) Conventional hypothesis for the activation of DNA replication completion checkpoint. The image was created with BioRender. (G) Flow cytometry analysis of DNA content of MCM2-dTAG cells that were treated with NEG or dTAGv-1 for the indicated time periods (days). More than 10 000 cells were collected for analyses. (H) Immunofluorescent staining of pH3S10 in MCM2-dTAG cells. Cells were treated 1 μM NEG or dTAGv-1 for 48 h. Representative images of pH3S10 are shown (scale bar 10 μm). (I) Schematic depiction of the observed outcomes of MCM2-depleted cells with incomplete DNA replication. The image was created with BioRender.
According to the principle of ordered cell cycle progression—which restricts DNA replication and chromosome segregation, respectively, to S and M phases—cells with incomplete DNA replication should be arrested and stopped at S phase (or S/G2 or S/M boundary) due to the activation of cell cycle checkpoints. We thus examined well-known DDR signaling pathways including ATR/CHK1, which are known to be activated in response to DNA breaks and/or replication stress in S phase cells. Indeed, we observed the activation of DDR signaling when MCM2-dTAG cells were treated with dTAGv-1 for 0.5–8 or more hours. Since TOPBP1 and ETAA1 are the major activators for ATR, we further examined whether ATR/CHK1 activation induced by MCM2 depletion would depend on TOPBP1 or ETAA1. The results showed that depletion of ETAA1 in MCM2-depleted cells mildly reduced pCHK1-S345 level when compared to that in MCM2-depleted cells (Supplementary Figure S1A). On the other hand, knockdown TOPBP1 in MCM2-depleted cells significantly decreased pCHK1-S345 level when compared to that in MCM2-depleted cells (Supplementary Figure S1B). Furthermore, the influence of TOPBP1 on pCHK1-S345 level in MCM2-depleted cells correlated with the knockdown of TOPBP1 protein levels (Supplementary Figure S1B). These results suggest that MCM2 depletion leads to ATR/CHK1 activation, which depends on TOPBP1, but relies minimally on ETAA1.
Intriguingly, DDR signaling diminished and followed by increased cyclin B1 and pH3S10 levels after 24 h (Figure 1E). We were wondering the possible reasons for the activation and termination of ATR/CHK1 and other DDR pathways in MCM2-depleted cells. We speculate that MCM2 depletion may cause DNA breaks at replication forks, which leads to the activation of ATR/CHK1 and other DDR pathways. These breaks may be repaired within several hours, which results in the termination of ATR/CHK1 and other DDR pathways. Therefore, we examined DNA breaks in MCM2-dTAG cells treated with dTAGv-1 at indicated hours. As shown in Supplementary Figure S1C, the neural comet tails increased in cells treated with dTAGv-1 for 1, 4 or 8 h, suggesting that DNA breaks induced by the rapid depletion of MCM2 are responsible for the activation of DDR signaling. Moreover, cells treated with dTAGv-1 for 24 or 48 h exhibited decreased comet tails, indicating that these breaks are being repaired, which may explain the termination of DDR signaling. Notably, there were tiny dots in samples treated with dTAGv-1 for 24 or 48 h (shown by arrow), which are likely dead cells with fragmented genome (Supplementary Figure S1C). Furthermore, we also employed γH2AX staining to document the induction and repair of DNA breaks in MCM2-depleted cells. Consistent with the results of neutral comet assay, cells treated with dTAGv-1 for 1, 4 or 8 h exhibited significantly increased γH2AX staining (in both percentage of cells and intensity), while its level rapidly decreased to much lower levels in cells treated with dTAGv-1 for 24 or 48 h (Supplementary Figure S1D), which correlated with the pattern of DDR signaling. Notably, partial γH2AX staining in cells treated with dTAGv-1 for 24 or 48 h were observed (Supplementary Figure S1D). These γH2AX staining appears to be associated with fragmented DNA and/or abnormal mitotic cells shown by bright DAPI staining (Supplementary Figure S1D), which probably due to abnormal chromosome segregation leading to chromosomal fragmentation and DNA breaks, therefore resulting in H2AX phosphorylation. Taken together, these results suggest that the activation and termination of ATR/CHK1 and other DDR pathways observed in MCM2-depleted cells is likely due to the induction and repair of DNA breaks caused by MCM2-depletion.
As mentioned above, the conventional hypothesis suggests that there should be a DNA replication completion checkpoint, which prevents S/G2 or S/M transition when DNA is not fully replicated (Figure 1F). The disappearance of DDR signaling in MCM2-depleted cells at 24 h could suggest that cells may somehow complete DNA replication even with very efficient MCM2 depletion. We thus examined DNA content by flow cytometry in MCM2-dTAG cells treated with dTAGv-1 for 1 to 3 days. As shown in Figure 1G, MCM2-dTAG cells treated with dTAGv-1 for 1 or 2 days did not complete DNA replication (<4N). Moreover, we observed abnormal pH3S10 signaling and chromosome alignment in cells treated with dTAGv-1 for 48 h compared to that in NEG-treated cells (Figure 1H). Furthermore, we observed abnormal metaphase spreads in MCM2-dTAG cells treated with dTAGv-1 for 48 h (Supplementary Figure S2A). These data suggest that under-replicated cells still progress to mitosis. Together, our results imply that although ongoing S phase progression may be monitored by ATR/CHK1 and other DDR pathways, cells do not appear to monitor the completion of DNA replication, since these under-replicated cells eventually enter mitosis with intact ATR/CHK1 and other DDR pathways (Figure 1I).
Cells with MCM2 depletion do not complete DNA replication but enter mitosis
As an essential replication helicase, MCM2 is critical for initiation as well as ongoing DNA replication. To focus on ongoing replication or replication progression in S phase cells, we synchronized MCM2-dTAG cells by double thymidine block and further released cells from thymidine for 2 h, which resulted in most cells in S phase (Figure 2A). We then treated these cells with 1 μM NEG or dTAGv-1 (Figure 2A). Under this situation, MCM2 was depleted only in cells that were already in S phase with active ongoing DNA synthesis.
Figure 2.
Cells with MCM2 depletion enter mitosis with incomplete DNA replication. (A) Immunoblots of the indicated proteins prepared from double-thymidine synchronized MCM2-dTAG cells. The synchronized cells were released for 2 h before treatment with NEG or dTAGv-1 for the indicated time periods (hours). The red box shows the transient activation of DDR signaling, while the yellow box displays the increased levels of pH3S10 and cyclin B1 as well as the decreased level of CDK1/2-pY15 that reflect cells entering mitosis. (B) Flow cytometry analyses of DNA content using asynchronized and double-thymidine synchronized MCM2-dTAG cells. More than 10 000 cells were collected for analyses. (C) Flow cytometry analyses of EdU intensity using the same or similar samples in (B). More than 10 000 cells were collected for analyses. The X and Y axes respectively represent DNA content and EdU intensity. (D) CellTiter-Glo luminescence assay was conducted to examine cell proliferation in double-thymidine synchronized MCM2-dTAG cells. (E) Clonogenic survival of MCM2-dTAG cells in the presence of 1 μM NEG or 1 μM dTAGv-1 combined with different concentrations of RO3306 (5, 7.5 or 10 μM) or DMSO for 3 days. Day 0 shows the cell confluency before any treatment. The representative images are shown. The intensity of cell confluency was measured by ColonyArea ImageJ plugin. Bars represent the mean ± SEM (n = 3). (F) Schematic illustration of the observed outcomes of MCM2-depleted cells with incomplete DNA replication. The image was created with BioRender.
Consistent with the data obtained in asynchronized cells (Figure 1), DDR was activated transiently but declined to baseline levels followed by the increased levels of cyclin B1 and pH3S10, when MCM2-dTAG cells were treated with dTAGv-1 for 24 h or longer (Figure 2A). In agreement with the increased level of cyclin B1 in MCM2-depleted cells, we also observed the decreased CDK1/2-pY15 at later time points, especially in cells treated with dTAGv-1 for 24 or 30 h (Figure 2A). These data indicate that the activity of CDK1 increases after the disappearance of DDR signaling.
Additionally, we examined pH3S10 level and DNA content by FACS in these synchronized cells (Supplementary Figure S2B). As expected, we observed normal mitotic cells (M) with much higher levels of pH3S10 and 4N DNA content in NEG-treated MCM2-dTAG cells (Supplementary Figure S2B). Moreover, we noted that the dTAGv-1-treated cells had <4N DNA content and increasing amount of sub-G1 (<2N) cells were observed over time. However, we observed that only some of these cells with <4N DNA content showed slightly increased intensity of pH3S10 staining, but we did not observe a distinct population of pH3S10 positive cells (Supplementary Figure S2B). These results suggest that the under-replicated cells do not accumulate very high levels of pH3S10 as that of normal mitotic cells.
Furthermore, we monitored DNA content as well as the percentage of cells that were able to replicate DNA (i.e. EdU positive cells; EdU+) in these asynchronized and synchronized populations treated with NEG or dTAGv-1 (Figure 2B and C). As expected, we observed cells with 2N or 4N or between 2N and 4N DNA content in asynchronized cells treated with NEG for 1 h, which we also observed in NEG-treated synchronized cells (Figure 2B). However, cells treated with dTAGv-1 had <4N DNA content (Figure 2B), suggesting these cells could not complete DNA replication. Consistent with the immunostaining results of EdU incorporation (Figure 1D), the percentage of EdU + cells (0.2%) in dTAGv-1-treated asynchronized MCM2-dTAG cells was much lower than that in NEG-treated cells (23.5%) determined by FACS analysis (Figure 2C). Similarly, there were almost no EdU + cells in synchronized MCM2-dTAG cells treated with dTAGv-1 for 4 or 24 h, while as expected the EdU + cells were the major population in NEG-treated cells (Figure 2C). These results revealed that these synchronized MCM2-depleted cells were not replicating and therefore had <4N DNA content (Figure 2B and C). Thus, as expected, cells could not complete DNA replication in the absence of MCM2. Notably, the synchronized MCM2-depleted cells showed significant cell death compared to that of the NEG-treated cells (Figure 2D).
We hypothesize that although MCM2-depleted cells cannot complete DNA replication, they yet enter mitosis and likely die due to failed mitosis caused by incomplete genome duplication. Thus, we wondered whether inhibiting CDK1 activity with RO-3306 could prevent cell lethality in these cells. As expected, when NEG-treated cells were treated with RO3306, the confluency of these cells was much lower than that of cells treated with DMSO, but still slightly higher than that of cells at Day 0 (Figure 2E). These results suggest that RO3306 inhibitor could efficiently arrest cells at G2/M phase. Moreover, the confluency of cells treated with dTAGv-1 + RO3306 was higher than that of cells treated with dTAGv-1 + DMSO (Figure 2E), suggesting that dTAGv-1 treatment induced cell death, but this phenotype could be rescued by RO3306 treatment. At Day 3, the density of cells treated with dTAGv-1 + RO3306 was slightly lower than that of cells treated with NEG + RO3306, but probably similar to those at Day 0 (Figure 2E), indicating that the dTAGv-1 + RO3306 treated cells may not die but they fail to proliferate. These results suggest that treatment with RO-3306 can rescue cell lethality, at least partially, in MCM2-depleted cells (Figure 2E). In addition, the partial, but not full, rescue of cell death was probably due to the fact that RO-3306 treatment can also induce cell lethality after cells were treated with RO-3306 for >24 h as previously reported (14). Taken together, our data indicate that cells with incomplete DNA replication only transiently activate DDR. These cells eventually progress to M phase and probably die in mitosis (Figure 2F).
Both CDC45 and GINS4 depletion recapitulate the phenotypes of MCM2 ablation
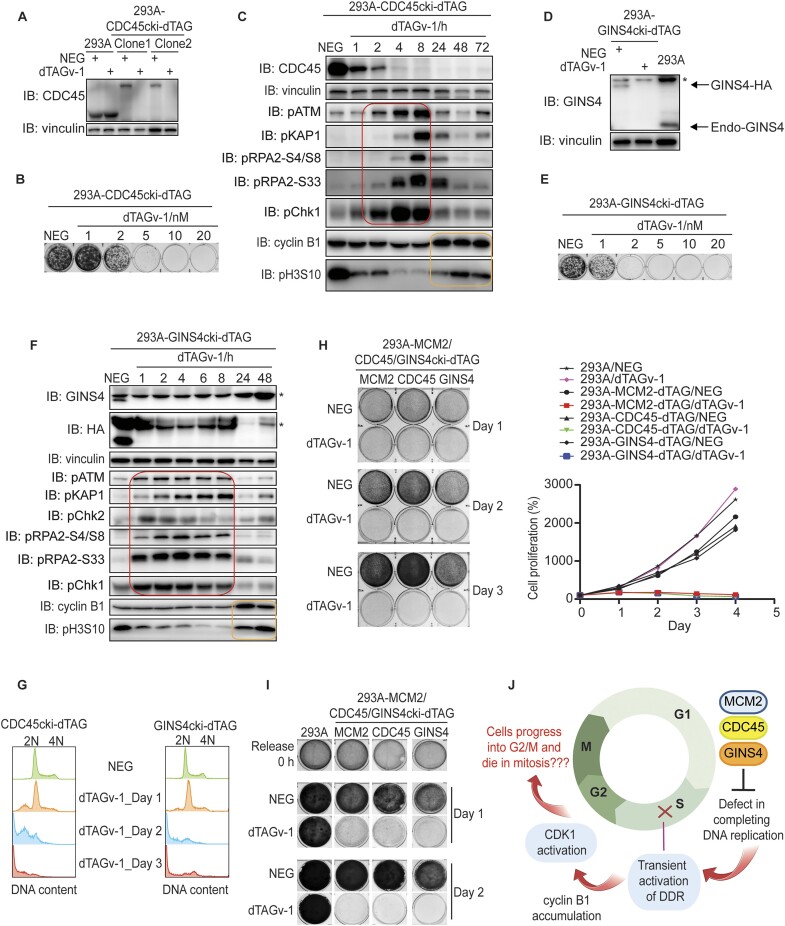
Our data demonstrate that under-replicated cells triggered by MCM2 depletion transiently activate DDR signaling. However, these cells are not arrested but instead enter and eventually die in mitosis. These surprising findings raise the question whether these phenotypes are unique in cells with MCM2 depletion. To address this question, we respectively generated cell lines with C-terminal dTAG knock-in of CDC45 and GINS4, which are also essential for DNA replication. The CDC45-dTAG cells showed marked protein degradation as well as cell proliferation defect following dTAGv-1 treatment compared to those treated with NEG (Figure 3A and B). Similar to the phenomena observed in MCM2-dTAG cells, CDC45-dTAG cells treated with dTAGv-1 displayed transient activation of DDR signaling followed by increased levels of cyclin B1 and pH3S10 (Figure 3C). GINS4-dTAG cells treated with dTAGv-1 also exhibited significant protein degradation and cell proliferation defect (Figure 3D and E). Moreover, these GINS4-dTAG cells also underwent transient DDR activation followed by increased levels of cyclin B1 and pH3S10 (Figure 3F).
Figure 3.
Both CDC45 and GINS4 depletion recapitulate phenotypes of MCM2 ablation. (A) Western blot validation of HEK293A-CDC45cki-dTAG cell lines in the presence of 1 μM NEG or 1 μM dTAGv-1. (B) Clonogenic survival of CDC45-dTAG cells in the presence of NEG or different concentrations of dTAGv-1. (C) Immunoblots of the indicated proteins prepared from CDC45-dTAG cells with treatment of NEG or dTAGv-1 for the indicated time periods (hours). Both floated and adhesive cells were collected for analyses. The red box shows the transient activation of DDR signaling, while the yellow box displays the increased levels of pH3S10 and cyclin B1 reflecting cells entering mitosis. (D) Western blot validation of HEK293A-GINS4cki-dTAG cell lines in the presence of 1 μM NEG or 1 μM dTAGv-1. The asterisk indicates the non-specific band. (E) Clonogenic survival of GINS4-dTAG cells in the presence of NEG or different concentrations of dTAGv-1. (F) Immunoblots of the indicated proteins prepared from GINS4-dTAG cells with treatment of NEG or dTAGv-1 for the indicated time periods (hours). Both floated and adhesive cells were collected for analyses. The asterisk indicates the non-specific band. The red box shows the transient activation of DDR signaling, while the yellow box displays the increased levels of pH3S10 and cyclin B1 reflecting cells entering mitosis. (G) Flow cytometry analyses of DNA content of CDC45- or GINS4-dTAG cells treated with NEG or dTAGv-1 for days as indicated. More than 10 000 cells were collected for analyses. (H) CellTiter-Glo luminescence assay as well as clonogenic assay were conducted to examine cell proliferation in HEK293A and HEK293A-MCM2/CDC45/ GINS4-dTAG cells. (I) Clonogenic assay for assessing cell proliferation in double-thymidine synchronized HEK293A and HEK293A-MCM2/CDC45/GINS4-dTAG cells. (J) Schematic illustration of similarities between MCM2 ablation and CDC45 or GINS4 depletion. The image was created with BioRender.
Again, we measured DNA content in either CDC45- or GINS4-depleted cells to examine whether DNA replication has been completed or not. Notably, CDC45- or GINS4-dTAG cells treated with dTAGv-1 for 2 or 3 days never reached 4N DNA content (Figure 3G), suggesting that CDC45- or GINS4-depleted cells are unable to fully replicate DNA within a cell cycle. Moreover, we observed 0–2N DNA content when cells were treated with dTAGv-1 for 2 or 3 days (Figure 3G), implying that DNA fragmentation occurs after cells enter mitosis, and such fragmentation is likely the product of abnormal or forced chromosome segregation of cells with incomplete DNA replication. We further examined the survival of MCM2-, CDC45- or GINS4-depleted cells. We showed that these cells began to die after 1 day (Figure 3H). Additionally, these cells following double-thymidine synchronization also displayed striking cell proliferation defect when treated with dTAGv-1 (Figure 1I). Again, these data suggest that cells with incomplete DNA replication enter mitosis and die likely due to abnormal chromosome segregation (Figure 3J). We did not observe cell cycle arrest due to incomplete DNA replication, which argues against the existence of a robust checkpoint that ensures the completion of DNA replication.
Cells with MCM2, CDC45 or GINS4 depletion enter and die in mitosis
To confirm our hypothesis that under-replicated cells with MCM2, CDC45 or GINS4 depletion die in mitosis, we transduced WT and MCM2-, CDC45- or GINS4-dTAG HEK293A cells with the cell cycle biosensor, Fucci (CA), i.e. fluorescent ubiquitination-based cell-cycle indicator, which can be used to reveal whether cells indeed exit S phase and enter G2 or M phase. We first performed cell cycle analysis of WT and MCM2-, CDC45- or GINS4-dTAG HEK293A cells stably expressing tFucci (CA)5 by flow cytometry after these cells were treated with NEG or dTAGv-1 for 24 h. The percentages of cells at G1, S and G2/M in NEG-treated WT and MCM2-, CDC45- or GINS4-dTAG cells were similar (Figure 4A and B). However, the percentage of cells at G2/M phases in dTAGv-1-treated MCM2-, CDC45- or GINS4-dTAG HEK293A cells showed a striking increase when compared to that in the NEG-treated cells (Figure 4A and B). These data are consistent with Western blotting data showing the increased level of cyclin B1 coupled with CDK1 activation, which probably drives these cells into G2/M. To clarify the fate of S phase cells that could not complete DNA replication, HEK293A-Fucci (CA) and HEK293A-MCM2-, CDC45- or GINS4-dTAG-Fucci (CA) cells were synchronized with double thymidine block and then released for 2 h to allow most cells to enter S phase. We then treated these cells with NEG or dTAGv-1. The NEG- and dTAGv-1-treated WT cells as well as NEG-treated MCM2-, CDC45-, or GINS4-dTAG cells divided repeatedly. However, the dTAGv-1-treated MCM2-, CDC45-, or GINS4-dTAG cells entered G2/M phase, and eventually died in middle or late mitosis (Figure 4C; Supplementary Videos S1–S8).
To further determine the fate of the majority of these under-replicated cells, i.e. whether they die in G2 or mitosis, we introduced GFP-H2B in our cell lines, which allows us to identify mitotic cells. Again, WT and HEK293A-MCM2-, CDC45- or GINS4-dTAG cells stably expressing GFP-H2B were synchronized in S phase, followed by releasing for 2 h and then treated with NEG or dTAGv-1. The NEG-treated MCM2-, CDC45- or GINS4-dTAG and NEG-/dTAGv-1-treated WT cells entered and exited mitosis (Figure 5A; Supplementary Videos S9–S16). However, in dTAGv-1-treated MCM2-, CDC45- or GINS4-dTAG cells, we observed chromosome condensation (as reflected by condensed GFP-H2B that exhibited high intensity) in most of the cells (Figure 5A; Supplementary Videos S9–S16), suggesting that these cells enter mitosis. Moreover, we observed abnormal chromosome alignment, failed cytokinesis and cell fusion in MCM2-, CDC45- or GINS4-depleted cells (Figure 5A; Supplementary Videos S9–S16), suggesting that these cells enter mitosis but cannot properly exit mitosis. Furthermore, some cells completed abnormal cytokinesis and generated fragmented chromosomes (Figure 5B; Supplementary Videos S9–S16), which we also observed when we measured DNA content (0–2N) in these cells. Taken together, these results indicate that MCM2-, CDC45- or GINS4-depleted cells with incomplete DNA replication transiently activate DDR signaling pathways, but these cells progress into G2/M and eventually die in mitosis (Figure 5C).
Discussion
In this study, we employed efficient dTAG system and high-throughput live-cell imaging to investigate the fate of under-replicated cells. As anticipated, we found that MCM2-, CDC45- or GINS4-depleted cells were not able to complete DNA replication. Unexpectedly, these cells still progressed into mitosis, which is contradictory to the conventional hypothesis that these cells should activate a checkpoint, which we called here as DNA replication completion checkpoint, and thus be arrested in S phase or at S/G2 or at S/M boundary. Notably, similar observations have been discovered in yeast smc5-smc6 mutants (3) and cdc6 mutant (4), as well as zebrafish ticrr mutant (5). These cells also progress into mitosis with severely defective genome duplication. Specifically, yeast smc5-smc6 mutants can lead to the activation of DNA damage/replication stress checkpoint response, but still enter mitosis with unfinished replication (3). These previous data agree with our observations and suggest that there is no checkpoint that ensures the completion of DNA replication.
Cell cycle checkpoints have been investigated by many groups. The general terms such as S/G2 or S/M checkpoints fail to capture the precise nature of these checkpoints. Specifically, do these checkpoints monitor ongoing DNA replication or the completion of DNA replication? Here we would like to distinguish these two conceptually distinct checkpoints. The checkpoint that monitors ongoing DNA replication is the intra-S phase checkpoint, which is likely controlled by ATR/CHK1 and other DDR pathways to ensure the fidelity of DNA replication. However, the question we want to address in this study is whether there is a checkpoint, which we dub as DNA replication completion checkpoint, that ensures genome duplication before cells exiting S phase and entering G2 or M phase. Our data and previous studies cited above argue against the existence of this DNA replication completion checkpoint.
In mammalian cells, mitotic DNA synthesis (MiDAS) occurs in cells with few incomplete replicated regions (15–17), which usually occur at common fragile sites (CFSs) especially upon mild replication stress (18,19). Since the very low level of stalled replication forks cannot lead to robust activation of ATR/CHK1 as well as other DDR pathways and thus may not be detected by any checkpoint, these cells progress into mitosis and continue DNA synthesis/repair in mitosis to minimize the under-replicated genome. In these scenarios, it is generally believed that cells with very few under-replicated regions are not detected by S/G2 or S/M checkpoint, i.e. replication completion checkpoint, since they fail below the detection threshold. Thus, these early studies do not directly challenge the existence or the concept of a replication completion checkpoint. In this study, we intentionally examined but failed to detect this checkpoint in cells with intact ATR/CHK1 and other DDR pathways.
In our study, MCM2-, CDC45- or GINS4-depleted mammalian cells, which contain largely under-replicated genome, progress and die in mitosis. Moreover, these largely under-replicated cells indeed activate the intra-S phase checkpoint, i.e. ATR/CHK1 and other DDR pathways, presumably for the timely repair of stalled replication forks. Since the activation of DDR pathways including ATR/CHK1 was observed after disruption of ongoing DNA replication by depleting MCM2, CDC45 or GINS4, we confirmed that these DDR pathways are likely activated due to DNA breaks induced by MCM2-, CDC45- or GINS4-depletion. Once DNA breaks are repaired, DDR signaling pathways are turned off and cells therefore progress into G2 or M. These data agree with the hypothesis that the ATR/CHK1 pathway mainly functions in S phase cells, probably by monitoring both the initiation and ongoing DNA replication (20,21). However, these DDR pathways are not employed to monitor the completion of DNA replication, since we showed that these pathways were only transiently activated and then turned off, although DNA replication was not completed during the same time period.
Notably, our data mentioned above agree that intra S phase checkpoint controlled by ATR/CHK1 pathway plays an important role in cell cycle progression. As a matter of fact, cells with MCM2-, CDC45- or GIN4-depletion transiently activate ATR/CHK1 and other DNA damage checkpoint pathways, which therefore enter mitosis much later than that of control cells. These observations agree with an early report suggesting that ATR/CHK1 pathway monitors ongoing DNA replication fork in S phase, which represses FOXM1-dependent expression of mitotic gene products until S/G2 transition to regulate mitotic entry (22). Moreover, a recent study showed that TRESLIN-MTBP monitors origin firing to manage S-phase progression for the timely progression to G2 phase (23). However, these intra-S phase checkpoints do not monitor and ensure the completion of DNA replication before allowing cells to enter G2 or M phase.
In summary, our results show that initiation of mitosis is not dependent on the completion of DNA replication. Our data favor a probability hypothesis to explain cell cycle progression. We envision that although cyclin B and other mitotic proteins start to accumulate in S phase, they are unlikely to reach levels sufficient for entering mitosis before the completion of DNA replication. Therefore, cells progress orderly from S to G2 and to M. In the presence of DNA damage or replication stress, DDR pathways, including the ATR/CHK1 pathway, are activated, which slow down cell cycle progression in part by inhibiting CDC25s and cyclin/CDKs (24,25). These DDR pathways not only deal with ongoing replication and replication-associated DNA damage, but also keep DNA replication progression in sync with the increase of cyclin/CDK activities. However, these pathways are not employed to ensure the completion of DNA replication. If cells cannot complete DNA replication, these cells will eventually accumulate enough proteins required for mitotic transition and enter mitosis. Of course, these under-replicated cells will die in mitosis, as we showed in this study. On the other hand, cells with minimal under-replicated regions cannot activate intra-S phase checkpoint as well as prevent cyclin/CDKs accumulation. Therefore, these cells enter mitosis and attempt to complete/continue DNA replication in mitosis, i.e. MiDAS and CFSs, which may still allow these cells to complete chromosome segregation and survive. Thus, our working hypothesis unifies our and those early observations and allow us to understand what is monitored and more importantly what is not monitored during cell cycle progression.
Indeed, several recent studies suggest that we should reconsider our hypothesis of cell cycle regulation and/or cell cycle checkpoints. For example, a recent study found that the decision to proliferate is actually reversible at various cell cycle phases in the absence of mitogens (26), which is contrary to the current model that this decision is irreversibly made at the restriction point of the cell cycle. Another study reported that the time it takes for cells to transit from G2 to G1 varies widely, suggesting that some or all of the feedback loops involved in the switch-like activation of APC/C at metaphase to anaphase transition may not be fully operational during a normal cell cycle (27). Moreover, another recent study concludes that S-CDK (S-phase related) and M-CDK (mitosis-related) regulate cell cycle progression by increasing the overall CDK activity, instead of functioning specifically for controlling different and unique substrates (28). All these findings imply buffering and/or relatively flexible regulation, but not the conventional view of precise and irreversible regulation, of the cell cycle, which encourages us to rethink whether cells employ any irreversible and/or stringent checkpoints to control cell cycle progression.
We would like to point out that the current study has some limitations. We generated independent cell lines to deplete respectively three essential components of replication machinery to prevent completion of DNA replication to test our hypothesis. However, these dTAG cells were generated using the same cellular background, i.e. HEK293A. Future experiments are warranted to further test our hypothesis in multiple cellular backgrounds. Additionally, we have tested all the well-known DDR signaling molecules that may be involved in monitoring ongoing and completion of DNA replication. DDR signaling could only be activated transiently, but do not appear to prevent these under-replicated cells from entering mitosis. Interestingly, γH2AX signaling appears to be relatively high over the course of our experiments, which may due to abnormal chromosome segregation, leading to chromosomal fragmentation and DNA breaks that result in another wave of H2AX phosphorylation. However, we could not definitively rule out the possibility that some kind of DDR checkpoint slippage, signified by γH2AX, may lead to the escape of some of these cells to mitosis. Moreover, the replication completion checkpoint may still exist, which could act somehow by monitoring the removal of CDC45:MCM2-7:GINS (CMG) complex from chromatin. If this was the case, we may have missed this checkpoint due to our experimental setting. Future studies are needed by depleting other essential replication proteins to test whether CMG removal is a part of this enigmatic replication completion checkpoint.
Supplementary Material
Acknowledgements
Author contributions: M.H. and J.C. conceived the project. M.H., C.Y., L.N., H.Z., D.Z., X.F., M.T., C.W., M.S., E.M., S.K., S.L. and F.S. performed the experiments. M.H. and J.C. wrote the manuscript with input from all authors. We thank all members of the Chen laboratory for their help and constructive discussion and Dr. Jingkun Zeng for his suggestion on data analysis. We thank Drs. Atsushi Miyawaki and Asako Sakaue-Sawano for constructive discussion about Fucci (CA) system and data analysis. We thank Dr. David Cortez for kindly sharing antibody against ETAA1. The graphical abstract was created with BioRender.com. Editorial support was provided by Bryan Tutt, Scientific Editor, Research Medical Library at MD Anderson Cancer Center.
Contributor Information
Min Huang, Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030, USA.
Chang Yang, Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030, USA.
Litong Nie, Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030, USA.
Huimin Zhang, Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030, USA.
Dandan Zhu, Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030, USA.
Chao Wang, Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030, USA.
Jeong-Min Park, Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030, USA.
Mrinal Srivastava, Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030, USA.
Elina Mosa, Integrated Microscopy Core, Advanced Technology Cores, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA.
Siting Li, Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030, USA.
Mengfan Tang, Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030, USA.
Xu Feng, Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030, USA.
Sarah J Keast, Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030, USA.
Fabio Stossi, Department of Molecular and Cellular Biology, Integrated Microscopy Core, Advanced Technology Cores, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA.
Junjie Chen, Department of Experimental Radiation Oncology, The University of Texas MD Anderson Cancer Center, 6565 MD Anderson Blvd, Houston, TX 77030, USA.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Cancer Prevention & Research Institute of Texas [RP160667 and RP180813 to J.C.; RP150578 and RP170719 to F.S.]; National Cancer Institute [CA193124, CA210929, CA216911, CA216437, CA274234, CA275712 to J.C.; P30CA016672 to MD Anderson Cancer Center/Cancer Center Support Grant]; National Institutes of Health [DK56338, CA125123, ES030285, S10OD030414 to F.S.].
Conflict of interest statement. None declared.
References
- 1. Hartwell L.H., Weinert T.A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989; 246:629–634. [DOI] [PubMed] [Google Scholar]
- 2. Elledge S.J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996; 274:1664–1672. [DOI] [PubMed] [Google Scholar]
- 3. Torres-Rosell J., De Piccoli G., Cordon-Preciado V., Farmer S., Jarmuz A., Machin F., Pasero P., Lisby M., Haber J.E., Aragon L. Anaphase onset before complete DNA replication with intact checkpoint responses. Science. 2007; 315:1411–1415. [DOI] [PubMed] [Google Scholar]
- 4. Piatti S., Lengauer C., Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995; 14:3788–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sansam C.L., Cruz N.M., Danielian P.S., Amsterdam A., Lau M.L., Hopkins N., Lees J.A. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev. 2010; 24:183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nabet B., Roberts J.M., Buckley D.L., Paulk J., Dastjerdi S., Yang A., Leggett A.L., Erb M.A., Lawlor M.A., Souza A. et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 2018; 14:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nabet B., Ferguson F.M., Seong B.K.A., Kuljanin M., Leggett A.L., Mohardt M.L., Robichaud A., Conway A.S., Buckley D.L., Mancias J.D. et al. Rapid and direct control of target protein levels with VHL-recruiting dTAG molecules. Nat. Commun. 2020; 11:4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang M., Yao F., Nie L., Wang C., Su D., Zhang H., Li S., Tang M., Feng X., Yu B. et al. FACS-based genome-wide CRISPR screens define key regulators of DNA damage signaling pathways. Mol. Cell. 2023; 83:2810–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ando R., Sakaue-Sawano A., Shoda K., Miyawaki A. Two coral fluorescent proteins of distinct colors for sharp visualization of cell-cycle progression. Cell Struct. Funct. 2023; 48:135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sakaue-Sawano A., Yo M., Komatsu N., Hiratsuka T., Kogure T., Hoshida T., Goshima N., Matsuda M., Miyoshi H., Miyawaki A. Genetically encoded tools for optical dissection of the mammalian cell cycle. Mol. Cell. 2017; 68:626–640.e5. [DOI] [PubMed] [Google Scholar]
- 11. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guzman C., Bagga M., Kaur A., Westermarck J., Abankwa D. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One. 2014; 9:e92444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gyori B.M., Venkatachalam G., Thiagarajan P.S., Hsu D., Clement M.V. OpenComet: an automated tool for comet assay image analysis. Redox. Biol. 2014; 2:457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vassilev L.T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D.C., Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl Acad. Sci. U.S.A. 2006; 103:10660–10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mocanu C., Karanika E., Fernandez-Casanas M., Herbert A., Olukoga T., Ozgurses M.E., Chan K.L. DNA replication is highly resilient and persistent under the challenge of mild replication stress. Cell Rep. 2022; 39:110701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Minocherhomji S., Ying S., Bjerregaard V.A., Bursomanno S., Aleliunaite A., Wu W., Mankouri H.W., Shen H., Liu Y., Hickson I.D. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015; 528:286–290. [DOI] [PubMed] [Google Scholar]
- 17. Spies J., Lukas C., Somyajit K., Rask M.B., Lukas J., Neelsen K.J. 53BP1 nuclear bodies enforce replication timing at under-replicated DNA to limit heritable DNA damage. Nat. Cell Biol. 2019; 21:487–497. [DOI] [PubMed] [Google Scholar]
- 18. Debatisse M., Le Tallec B., Letessier A., Dutrillaux B., Brison O. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012; 28:22–32. [DOI] [PubMed] [Google Scholar]
- 19. Ozeri-Galai E., Bester A.C., Kerem B. The complex basis underlying common fragile site instability in cancer. Trends Genet. 2012; 28:295–302. [DOI] [PubMed] [Google Scholar]
- 20. Menolfi D., Lee B.J., Zhang H., Jiang W., Bowen N.E., Wang Y., Zhao J., Holmes A., Gershik S., Rabadan R. et al. ATR kinase supports normal proliferation in the early S phase by preventing replication resource exhaustion. Nat. Commun. 2023; 14:3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saldivar J.C., Cortez D., Cimprich K.A. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017; 18:622–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saldivar J.C., Hamperl S., Bocek M.J., Chung M., Bass T.E., Cisneros-Soberanis F., Samejima K., Xie L., Paulson J.R., Earnshaw W.C. et al. An intrinsic S/G2 checkpoint enforced by ATR. Science. 2018; 361:806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zonderland G., Vanzo R., Amitash S., Martin-Doncel E., Coscia F., Mund A., Lerdrup M., Benada J., Boos D., Toledo L. The TRESLIN-MTBP complex couples completion of DNA replication with S/G2 transition. Mol. Cell. 2022; 82:3350–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rudolph J. Inhibiting transient protein–protein interactions: lessons from the Cdc25 protein tyrosine phosphatases. Nat. Rev. Cancer. 2007; 7:202–211. [DOI] [PubMed] [Google Scholar]
- 25. Bartek J., Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003; 3:421–429. [DOI] [PubMed] [Google Scholar]
- 26. Cornwell J.A., Crncec A., Afifi M.M., Tang K., Amin R., Cappell S.D. Loss of CDK4/6 activity in S/G2 phase leads to cell cycle reversal. Nature. 2023; 619:370–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zeng J., Hills S.A., Ozono E., Diffley J.F.X. Cyclin E-induced replicative stress drives p53-dependent whole-genome duplication. Cell. 2023; 186:528–542. [DOI] [PubMed] [Google Scholar]
- 28. Basu S., Greenwood J., Jones A.W., Nurse P. Core control principles of the eukaryotic cell cycle. Nature. 2022; 607:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.