Abstract
Given the presence of highly repetitive genomic regions such as subtelomeric regions, understanding human genomic evolution remains challenging. Recently, long-read sequencing technology has facilitated the identification of complex genetic variants, including structural variants (SVs), at the single-nucleotide level. Here, we resolved SVs and their underlying DNA damage–repair mechanisms in subtelomeric regions, which are among the most uncharted genomic regions. We generated ∼20 × high-fidelity long-read sequencing data from three Korean individuals and their partially phased high-quality de novo genome assemblies (contig N50: 6.3–58.2 Mb). We identified 131 138 deletion and 121 461 insertion SVs, 41.6% of which were prevalent in the East Asian population. The commonality of the SVs identified among the Korean population was examined by short-read sequencing data from 103 Korean individuals, providing the first comprehensive SV set representing the population based on the long-read assemblies. Manual investigation of 19 large subtelomeric SVs (≥5 kb) and their associated repair signatures revealed the potential repair mechanisms leading to the formation of these SVs. Our study provides mechanistic insight into human telomere evolution and can facilitate our understanding of human SV formation.
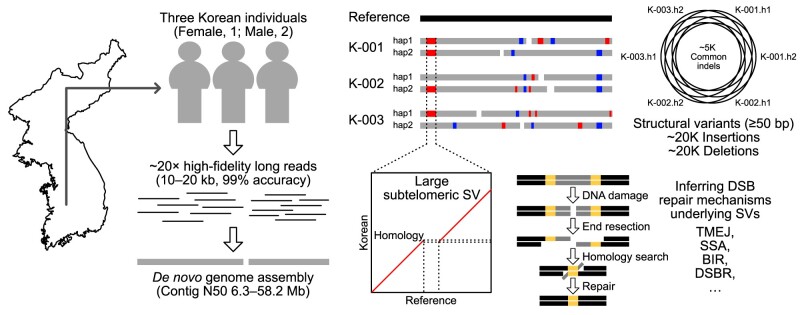
Graphical Abstract
Graphical Abstract.
Introduction
Understanding genetic variation is fundamental in the study of human evolution and genetic diseases. The first human reference genome, revealed by the Human Genome Project, allowed geneticists to study genetic variation between individuals at the genome level, leading to rapid advancement in targeted and whole-genome sequencing technologies (1–7). Moreover, the population-scale whole-genome sequencing data produced by several consortia have identified almost-saturated single-nucleotide polymorphisms (SNPs) and some structural variants (SVs) in the human population, along with matched individual phenotypes (8–11). Such efforts have improved our understanding of the human genome, including the tendency for mutations to accumulate in subtelomeric regions (9). Nonetheless, most of the telomeric and subtelomeric regions in the first human reference genome remain unavailable, which can be defined as distal clusters of telomeric-repeat motifs (TRMs) and the 500 kb distal ends without the TRM clusters, respectively. Such a limitation prevents the elucidation of human genetic variants in highly repetitive regions, since current population-scale whole-genome sequencing projects typically use short-read sequencing technology (12).
Advances in long-read sequencing technology have allowed for the assessment of genomic dark matter at the single-nucleotide level and the identification of the mechanisms that create genetic variants (12–18). The first complete human genome revealed centromeric and subtelomeric sequences, which are difficult to assemble because of their repetitive nature (19–22). Our knowledge of the genetic composition of the human population has been extended by a recently published draft human pangenome reference, providing 94 phased genome assemblies for 47 humans, as well as other long-read sequencing-based population-scale human genomes (23–27). These efforts have provided invaluable resources to identify genetic variants in any genomic region, not just region-specific SNPs. Moreover, it is now possible to examine in humans, at the single-nucleotide sequence level, >100 kb sized extreme subtelomeric SVs (≥50 bp), which could previously be elucidated only in model organisms with small genomes (18,20,28–34). This makes it possible to identify the mechanisms that generate subtelomeric SVs during telomere damage and repair, including single-strand annealing (SSA), polymerase theta-mediated end joining (TMEJ), break-induced replication (BIR) and double-strand break repair (DSBR).
In this study, given that Asian ancestry remains underrepresented in the current human pangenome reference, we present six phased high-quality genome assemblies from three Korean individuals. For each individual, we produced ∼20 × high-fidelity (HiFi) long-read sequencing data, comparable to the draft human pangenome HiFi data; we assembled these HiFi sequences and called genetic variants, including SVs, thereby expanding the human genetic variant catalog. Furthermore, we investigated human subtelomeric SVs and their DNA damage and repair signatures at the single-nucleotide sequence level to better understand human telomere evolution. This study provides valuable resources for further study of genetic variation and chromosomal evolution in humans.
Materials and methods
Sample preparation and DNA isolation
Blood samples were collected from three Korean individuals (K-001: female, aged 50 years; K-002 and K-003: males, aged 29 and 36 years, respectively) at the Chungnam National University Hospital (Daejeon, South Korea), with written informed consent from all participants and approval from the Institutional Review Board (IRB number: CNUH 2019-06-034). All methods were performed in accordance with the relevant guidelines and regulations and carried out in accordance with the Declaration of Helsinki. Genomic DNA was isolated from 5 ml blood samples using DNeasy Blood & Tissue Kit (Qiagen, Carlsbad, CA, USA), according to the manufacturer’s instructions. The quality and quantity of the extracted genomic DNA were analyzed using an ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Library preparation and sequencing
For long-read sequencing, we used the Sequel IIe HiFi system (Pacific Biosciences, Menlo Park, CA, USA). Briefly, HiFi sequencing libraries were prepared using a SMRTbell Express Template Prep Kit 2.0 (PN 101-853-100), followed by immediate treatment using a SMRTbell Enzyme Cleanup Kit (Pacific Biosciences). After pooling the fractions of the desired size (∼15–19 kb), the final libraries were further cleaned and concentrated using AMPure PB beads (Pacific Biosciences). Finally, library concentrations were assessed using a Qubit 1× dsDNA HS Assay Kit (Thermo Fisher Scientific), and the libraries were then sequenced using the Sequel IIe HiFi system (Pacific Biosciences).
We conducted an extensive analysis of SVs in a cohort of 103 Korean individuals, including the samples K-001, K-002 and K-003, using the MGI sequencing platform. For short-read sequencing, genomic DNA was isolated from 1 ml of blood from 103 normal individuals using a DNeasy Blood & Tissue Kit (Qiagen, Carlsbad, CA), according to the manufacturer’s instructions. The quality and quantity of the extracted genomic DNA were analyzed with an ND-1000 spectrophotometer. The MGIEasy FS DNA Prep kit (BGI, China) was used for short-read library construction according to the manufacturer’s instructions. Subsequently, raw sequence reads were obtained using the DNBSEQ-T7 sequencer (BGI) in the 150 bp paired-end sequencing mode.
Genome assembly
We assembled the raw HiFi reads de novo into contigs using hifiasm (version 0.16.0; default settings) and converted the GFA-formatted output to FASTA-formatted files (35). We used the CHM13 genome as the reference (version 1.1 for K-001, version 2.0, for K-002 and K-003) to scaffold our contigs into pseudo-chromosome-level genome assemblies using RagTag (version v2.0.1; ragtag.py scaffold -u) (36). To call SVs, we aligned our genome assemblies to the reference genome using Winnowmap2 (version 2.03; meryl count k=19, meryl print greater-than distinct=0.9998 and winnowmap -W -ax asm20 --cs -r2k), sorted and indexed the output alignment file using samtools (version 1.13; samtools sort -O BAM and samtools index) and ran SVIM-asm (version 1.0.2; SVIM-asm haploid) (37–39). Considering the accuracy of the identified SVs, we excluded the SVs located in chrY.
Genome annotation for called structural variants
The CHM13 genome annotation was obtained from publicly available databases (gene annotation: https://s3-us-west-2.amazonaws.com/human-pangenomics/T2T/CHM13/assemblies/annotation/chm13.draft_v2.0.gene_annotation.gff3; centromeric region annotation: https://s3-us-west-2.amazonaws.com/human-pangenomics/T2T/CHM13/assemblies/annotation/chm13v2.0_censat_v2.0.bed; RepeatMasker tracks: https://t2t.gi.ucsc.edu/chm13/dev/t2t-chm13-v2.0/rmsk/rmsk.bigBed). We defined the 500 kb distal ends of each chromosome as subtelomeric regions without TRMs. We did not analyze the p-arm subtelomeric regions of acrocentric chromosomes (chr13, chr14, chr15, chr21 and chr22).
Structural variant analysis using 94 public long-read assemblies
We downloaded 94 phased genome assemblies of 47 samples from the year 1 data of the Human Pangenome Reference Consortium (https://zenodo.org/record/5826274/files/HPRC-yr1.agc?download=1). We identified SVs of each assembly using the same SV-calling pipeline used for our Korean samples. The statistics of SVs from 100 assemblies including the six Korean assemblies from this study were obtained at the haplotype level. We merged these SVs using SURVIVOR (version 1.0.7; SURVIVOR merge 1000 1 1 1 0 50) (40) and set the merged SVs to one-hot vectors to perform the principal component analysis (PCA). Using this one-hot matrix, the PCs of 305 280 merged SVs were analyzed. The absolute loading value of each component was sorted, and 9160 SVs corresponding to the top 2% of the SVs for PC1 and top 1% of the SVs for PC2 loadings were selected for calculating average dissimilarity based on the Euclidean distances among the 100 assemblies. Moreover, by using the 9160 SVs predominantly contributing to the explained variances of the PCs, we calculated the ratio of SVs shared by three Korean assemblies over those identified from each superpopulation (i.e. KOR, EAS, AMR, SAS and AFR). We used only a superpopulation with more than six assemblies (i.e. three individuals).
Identification of subtelomeric structural variants
We manually trimmed the telomeric sequences (TTAGGG or its variants) from the end of the chromosome and used the 500 kb distal regions as the subtelomeric regions. We aligned our subtelomeric sequences to the locus-matched CHM13 subtelomeric sequence and visualized the alignment using the MUMmer package (version 4.0.0rc1; nucmer –maxmatch and mummerplot) (41).
We validated the subtelomeric SVs and their lengths (≥5 kb) using BLAST (version 2.12.0+; blastn -task megablast) (42), and manually removed SVs that were located near the scaffolded regions (filled with Ns) or that were too repetitive.
Short-read-based structural variant detection in all Korean samples
Following the short-read sequencing experiment, rigorous quality control measures were applied to ensure the integrity and reliability of the sequencing data. The sequenced reads were then aligned to the CHM13 human genome reference according to the best practices recommended by the GATK pipeline (v. 4.4.0) (43). Additionally, SVs were identified using Manta (44) (v. 1.6.0), a specialized tool designed for efficient and accurate detection of SVs in short-read sequencing data. SVs commonly detected from short- and long-read sequencing platforms were determined based on specific criteria: for deletions, SVs from two platforms were considered identical if >50% of their sequences were overlapped; for insertions, the genomic loci of the two SV breakpoints detected in CHM13 should be close enough (within 250 bp). Among the three individuals (K-001, K-002 and K-003), we identified the singleton SVs from short-read sequencing data as those detected from only one individual with heterozygous calls.
Detection of single nucleotide variants and short insertions and deletions from long-read sequencing data
We first downloaded 50 raw HiFi datasets, which include the samples previously used for phased genome assembly by the Human Pangenome Reference Consortium from the consortium website (https://github.com/Jeltje/HPRC_metadata). Including our three raw HiFi datasets, 53 long-read sequencing data were subjected to further variant analysis to identify single nucleotide variants (SNVs) and short insertions and deletions using DeepVariant (45). These small variants were used for PCA in a manner similar to the SVs detected from long-read sequencing data. The allele frequency (AF) of each variant was calculated across different superpopulations.
Polymerase chain reaction-based structural variant validation
To validate the deletions detected by our approach, we randomly selected the variants to (i) validate that the concordance of the SVs identified from the long- and short-read sequencing data is irrelevant with potential sequencing-prone biases; (ii) select the variants from diverse autosomal chromosomes; and (iii) focus on the repeat elements, which were abundant in telomeric and subtelomeric regions. To evaluate the concordance of the SVs between the two sequencing platforms, we performed a read-depth analysis. Briefly, for long- and short-read sequencing data, we calculated the read-depth ratios near the detected breakpoints of each deletion SV and determined the concordant SVs as those with their read-depth ratio of the deleted region to the non-deleted region (flanking region) >0.9 and randomly selected the 56 SVs with their length <5 kb, as well as 10 subtelomeric variants with their length ≥5 kb. Polymerase chain reaction (PCR) primers were designed for the individual breakpoints and their flanking sequences that do not include other deletions. Ten nanograms of the gDNA from K-001, K-002 and K-003 with a 20 μl PCR mixture containing primer sets and 2 × Master Mix (Doctor Protein, Seoul, Korea) was amplified using a GeneAmp PCR system 9 700 (Applied Biosystems, Waltham, MA). Detailed information on primer sequences and the experimental conditions used is provided in Supplementary Table S1. After PCR experiment, we confirmed the amplicon size for each target sequence by performing agarose gel electrophoresis.
Results
HiFi long-read sequencing enables assembly of highly contiguous Korean draft genomes
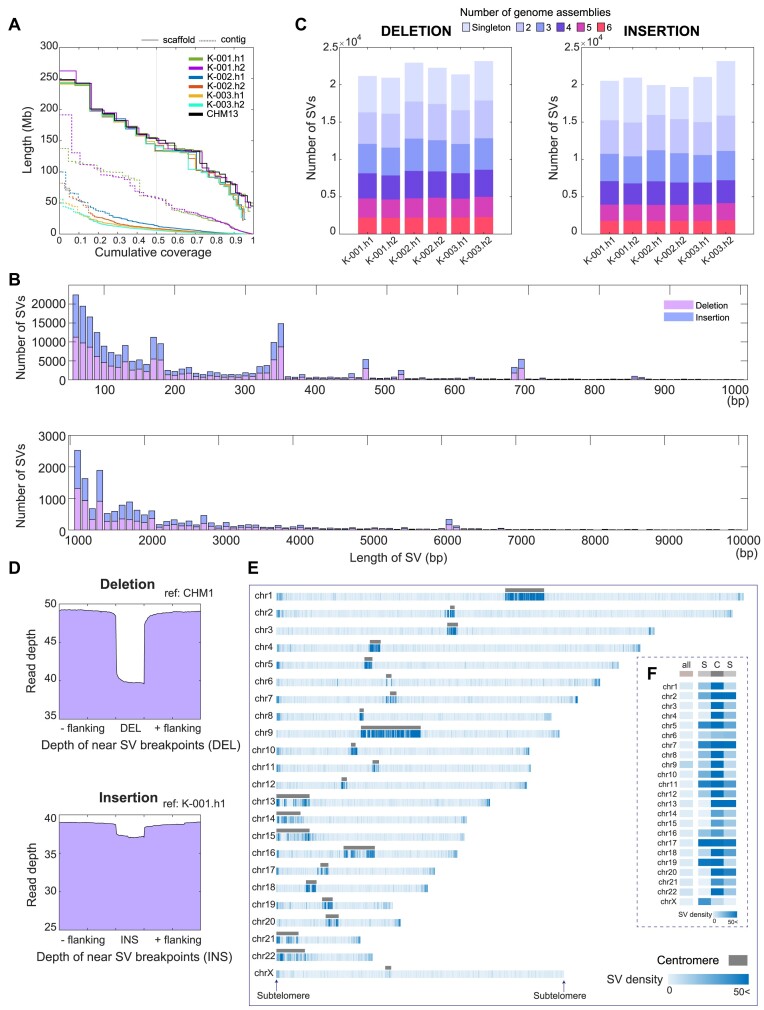
To understand genetic variation in Korean ancestry, we obtained ∼20 × HiFi long-read sequencing data from one female (hereafter, K-001; 31×) and two male (hereafter, samples K-002 and K-003; 21 × and 20×, respectively) individuals who did not have any significant genetic diseases or familial connections with each other (Supplementary Figure S1). Each raw HiFi read dataset covered 98% of the CHM13 genome, on average (Supplementary Figure S2). We de novo assembled the HiFi reads into contigs. The contig N50 lengths were 57.2 and 58.2 Mb for the K-001 assemblies, 13.8 and 9.3 Mb for the K-002 assemblies, and 8.1 and 6.3 Mb for the K-003 assemblies (Figure 1A; Supplementary Table S2), implying their high contiguity. The contigs were scaffolded into chromosome-level assemblies using the complete human CHM13 genome (Figure 1A; Supplementary Table S2).
Figure 1.
Total SVs of six haploid genomes compared to those of CHM13. (A) NGx plot for six genome assemblies of three Korean human samples. The de novo assembled contigs, scaffolds constructed using CHM13 genome and CHM13 genome are shown. Each color denotes a single assembly. Dotted lines represent cumulative coverage at the contig level, and solid lines represent coverage at the scaffold level. (B) Number of total SVs detected in each assembly and shared SVs among the six Korean assemblies. Colors depict the number of assemblies that share SVs with other assemblies. (C) The SV length distribution is presented from 50 bp to 1 kb in the upper panel and from 1 to 10 kb in the lower panel. (D) The read-depth distributions near SVs and their breakpoints. The upper panel shows the average read-depth near deletion breakpoints in the CHM13 genome; the lower panel shows the average read-depth of the insertion sequence regions in the K-001.h1 genome assembly. (Eand F) SV density distribution along the chromosomes including centromeric and subtelomeric regions. For each chromosome, the SV density was counted for every 100 kb bin. Maximal density is colored the same as 50 SVs per 100 kb. Subtelomeric regions are defined as 500 kb-long distal regions of each chromosome following manual removal of TRMs (TTAGGG or its variants). Centromeric regions are defined by CHM13 genome annotation.
Genomic structural variants are mostly located in centromeric and sub-telomeric regions
We identified ∼20 000 deletion and 20 000 insertion SVs (21 092, 20 856, 22 784, 22 153, 21 215 and 23 038 deletion SVs, and 20 466, 19 956, 20 798, 19 829, 19 512 and 20 900 insertion SVs, for K-001.h1, K-001.h2, K-002.h1, K-002.h2, K-003.h1 and K-003.h2, respectively; Supplementary Table S3) with ≥50 bp size in each of our six Korean genome assemblies compared with those in the CHM13 genome. For the 131 138 deletions and 121 461 insertions, the sum of the numbers from all the assemblies, the deletion and insertion SVs exhibited similar length distributions, and ∼82% of the SVs were <500 bp (Figure 1B and Supplementary Figure S3), consistent with a previous report (26).
We then analyzed the number of SVs shared between the genome assemblies. We obtained a similar number of deletion and insertion SVs for each assembly, with deletions being slightly more abundant. Among these SVs, ∼10% of the deletions and 9% of the insertions in each assembly were shared by all six assemblies, whereas ∼75% of the SVs were shared by at least two assemblies, and ∼25% of the SVs of each assembly were detected as singletons among the six assemblies (Figure 1C). Since the participants of this study did not have any familial connections, we estimated that 55% of the SVs (shared by three or more assemblies) may be common in the Korean population.
Notably, our variant sets identified by the long-read assembly-based method covered >91% (2 966, 3 137 and 3 013 out of 3 192, 3 477 and 3 285 for K-001, K-002 and K-003, respectively) and >95% (894, 1 069 and 921 out of 937, 1 109 and 962, respectively) of deletion and insertion SVs detected by short-read sequencing data of the same sample (Supplementary Figure S4A). Moreover, among the total SVs from short-read sequencing data, substantial numbers (597, 586 and 583 out of 4 129, 4 586 and 4 247 SVs for K-001, K-002 and K-003, respectively) were determined as the singleton SVs in long-read sequencing data (Supplementary Figure S4B). All these results support that the SVs identified from the long-read assemblies, even the singleton SVs, could be true positives. However, only 10% (2966, 3137 and 3013 out of 35 124, 36 687 and 36 272 for K-001, K-002 and K-003, respectively) and 3% (894, 1 069, and 921 out of 33 369, 32 788 and 32 762, respectively) of the deletion and insertion SVs from long-read sequencing data were detected from short-read sequencing data (Supplementary Figure S4A), suggesting that the long-read sequencing data contribute to the reduction of false negatives in SV detection.
Intriguingly, the comparative analysis of two variant calling methods showed that the long-read assembly-based variant calling method could provide significantly higher proportions of long (P < 0.05) deletion and insertion SVs for all three samples than the long-read mapping-based variant calling method (Supplementary Figure S5), which is similar to a previous report (46). In addition, the assembly-based variant sets covered most mapping-based variant sets across all three samples. For mapping-based SVs, 18 000–19 000 insertion and deletion SVs were detected per individual (Supplementary Table S4), with 63–69% covered by the assembly-based SVs (Supplementary Figure S5). Assembly-based variant calling may offer better precision than mapping-based methods for detecting large variants in complex genomic regions, such as subtelomeric regions, due to the longer contig lengths than raw reads (46–49). Therefore, we utilized the assembly-based variants for this study.
To investigate whether the assembly-based SVs are truly common in the Korean population, an orthogonal validation was performed by newly producing short-read sequencing data of an additional 100 Korean individuals, namely from K-004 to K-103. We analyzed these large sequencing data to obtain short-read-based SVs of each individual and compared them to the assembly-based SVs. We found that assembly-based SVs covered ∼76% of short-read-based SVs in each individual (Supplementary Figure S6), implying that these SVs are common in the Korean population. Furthermore, we showed that the three individuals (K-001, K-002 and K-003) represent the general genetic characteristics of the Korean population, by applying PCA on the small variants identified in the 103 Korean individuals. The PCA results confirmed that the three individuals used in the study were not outliers (Supplementary Figure S7), indicating that the SVs identified in these subjects are common and representative of the broader Korean population.
We next validated the presence of the detected deletion and insertion SVs by estimating read depths and performing PCR analysis. We calculated the read depths of the deletions detected in each assembly. Relative to their flanking sequences, the deleted SV regions in the reference genome (i.e. CHM13) were covered by fewer HiFi reads (Figure 1D and Supplementary Figure S8). Similarly, regarding the insertions detected in each assembly against the reference genome, we found that the loci of the insertions in each assembly (i.e. first or second assemblies of K-001, K-002 and K-003) were covered by a sufficient number of HiFi reads (Figure 1D and Supplementary Figure S8). Using read-depth analysis for long- and short-read sequencing data and comparing the read-depth distributions of SVs from the two sequencing platforms (see the details in ‘Materials and methods’ section; Supplementary Figure S9), we randomly selected 56 SVs with their length <5 kb among the ones showing the concordant read-depth distributions. We also designed primers for flanking sequences of these SVs, specifically targeting the unique sequences in repetitive regions, and conducted PCR analysis to validate their presence. Among a total of 56 tested SVs, 52 SVs (92.9%) were validated using PCR (Supplementary Figure S10 and Supplementary Table S5). The findings indicate the reliability of our SVs detected from the long-read assemblies.
These SVs were localized mainly in the centromeric region (defined via CHM13 annotation) or subtelomeric region (the 500 kb distal ends without distal clusters of TRMs). The subtelomeric regions exhibited ∼24 SVs per 100 kb while the centromeric regions exhibited 79 SVs per 100 kb, substantially higher than the overall SV density (<10 SVs per 100 kb; Figure 1E, F and Supplementary Figure S11). Since these centromeric and subtelomeric regions were recently assembled, most of the variants in the regions might not be thoroughly investigated yet in the human population.
Structural variants are predominantly localized in repetitive intergenic regions
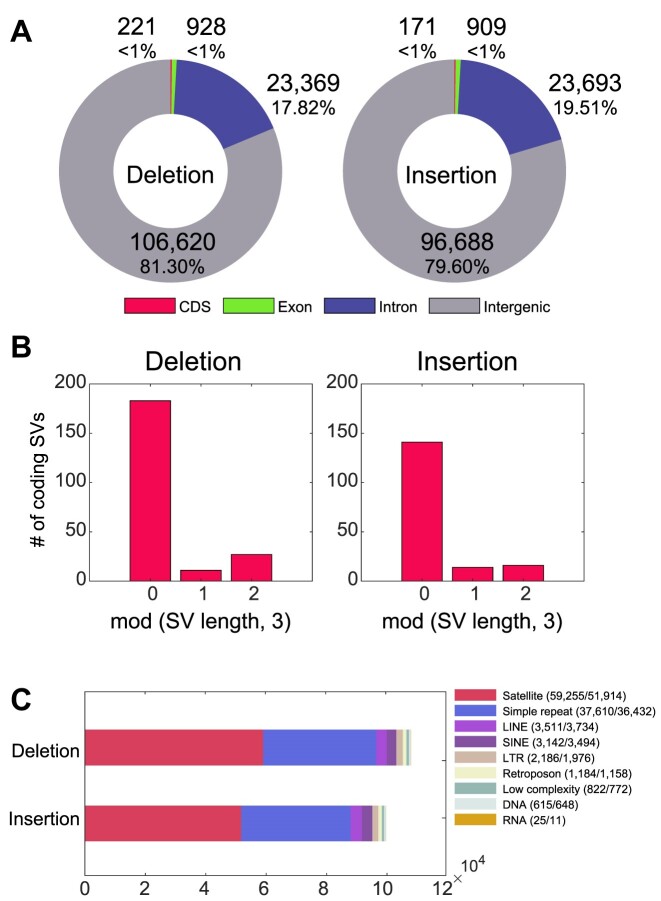
We calculated the number of SVs located in genic or repetitive regions. Among the 131 138 deletions and 121 461 insertions in the six Korean assemblies, only 1% of the deletion or insertion SVs were in coding or exonic regions (Figure 2A and Supplementary Figure S12A–C). This increased to 19–20% when including the SVs in the intronic regions. Moreover, 82% of the SVs in the coding regions (deletions: 82.8%; insertions: 82.5%) had lengths of multiples of three (Figure 2B and Supplementary Figure S12D–F). This implies that most of the coding SVs we detected were not frameshift variants, consistent with a previous report (26). Similarly, we confirmed that SVs of translational frameshifts would have been selected because of their phenotypic regulation.
Figure 2.
Functional characterization of the identified SVs. (A) Proportion of SVs in the genic regions. Each SV position is annotated as the coding sequence (CDS), exonic, intronic or intergenic region, according to the provided genome annotation. If an SV is commonly detected in intronic and exonic regions, it is considered an exonic SV. If an SV is commonly detected in both the exonic and coding regions, it is considered a CDS SV. (B) Number of coding region SVs according to the remainders after dividing each variant length by three. (C) Total number of SVs in eight different repetitive elements, provided by RepeatMasker tool, encompassing all SVs. The numbers within parentheses indicate deletion and insertion SV counts. LINE, long interspersed nuclear element; SINE, short interspersed nuclear elements; LTR, long terminal repeat; DNA, DNA repeat elements; and RNA, RNA repeats.
We next determined whether the remaining SVs were enriched in known repetitive genomic loci. This revealed that 61% of the SVs were present in known repetitive regions (79 908 deletion and 74 825 insertion SVs; 60.9% and 61.6%, respectively). Most of these sequences were detected in previously annotated repeats, such as satellites, simple repeats (single-nucleotide stretches or tandem repeats) and long and short interspersed nuclear elements (Figure 2C and Supplementary Figure S13). Satellite and simple repeat regions contained remarkably higher portions of the SVs (71.9% and 47.9%, respectively). This could be attributed to the fact that most SVs are localized at highly repetitive centromeric or subtelomeric regions.
Substantial proportions of Korean structural variants, including subtelomeric structural variants, are present in other ethnic groups
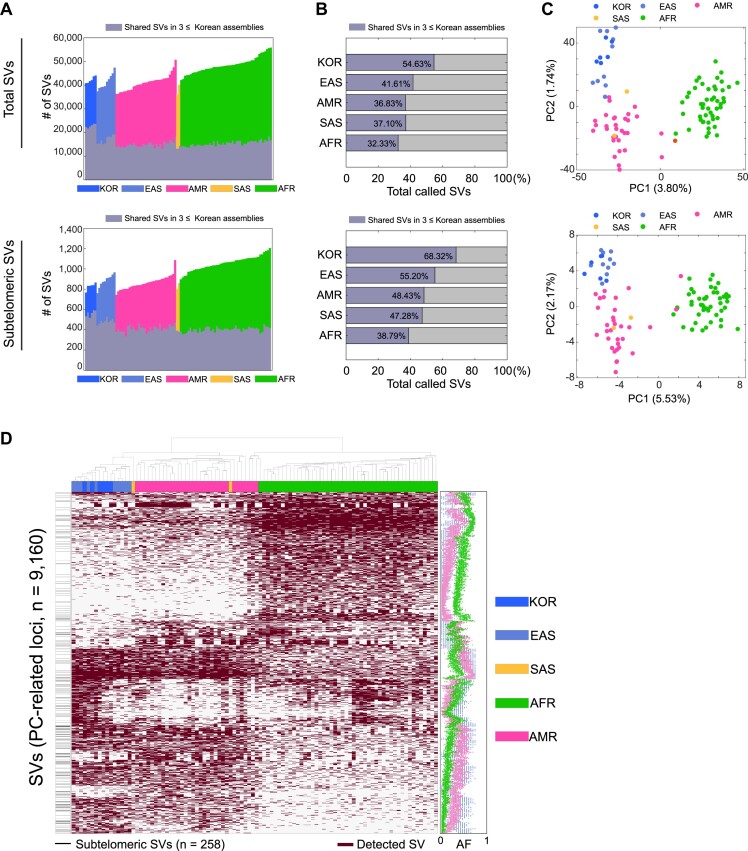
To investigate whether the SVs commonly identified from the six Korean genome assemblies were common to other populations, we identified deletion and insertion SVs from 94 phased genome assemblies in the publicly available human draft pangenome (25) by applying the same SV-calling pipeline. A total of 4 455 300 SVs were detected in 100 assemblies, including those from the six Korean assemblies in the present study. These were merged further into 305 343 SVs (154 281 deletions and 151 062 insertions). Among these SVs, we selected 25 595 SVs shared by three or more Korean assemblies, observing that substantial portions (41.6%, 36.8%, 37.1% and 32.3% of East Asians, admixed Americans, South Asians and Africans, respectively) were also common in other ethnic groups or superpopulations (Figure 3A and B; Supplementary Figure S14). This supports that the SVs from our assemblies were not artifacts. Interestingly, we also found comparable overlaps for subtelomeric SVs (i.e. 55.2%, 48.4%, 47.3% and 38.8% of East Asians, admixed Americans, South Asians and Africans, respectively).
Figure 3.
Commonality and uniqueness of SVs identified in diverse ethnic groups. (A) Number of deletion and insertion SVs identified in 100 pangenome assemblies and of SVs shared by Korean SVs common in at least three Korean assemblies. Each color represents a superpopulation, and gray-scaled bars represent the number of shared Korean SVs. (B) Proportion of shared Korean SVs. For each superpopulation, the ratio of the number of the shared Korean SVs over the union of SVs identified from the individual assemblies SVs was calculated. (C) PCA plot based on the merged SV one-hot matrix. For (A–C), the upper and lower panels present results of the total SVs and the subtelomeric SVs, respectively. (D) Clustering by PC1- and PC2-related SVs. For clustering, we selected the top 2% and 1% of SVs according to the top component coefficient values. The left horizontal gray lines in the white vertical box represent subtelomeric SVs. In the heatmap, each dark purple box represents an SV event. The dot graph (right) represents the population frequency for each superpopulation; we included the Korean and East Asian SVs (South Asian ancestry has only two assemblies and was removed). Blue, sky blue, magenta, orange and green represent data from the Korean (KOR), East Asian (EAS), admixed American (AMR), South Asian (SAS) and African (AFR) populations, respectively.
Different ancestries could be distinguished based on SVs, and subtelomeric regions contained population-specific SVs. We used the 305 280 merged SVs and 3 656 subtelomeric SVs to cluster 100 pangenome assemblies. Based on the PCA, the African, Asian and admixed American ancestries were separated, suggesting that SVs can be used to adequately distinguish populations (Figure 3C). Next, we extracted 9 160 SVs (3.0% of the merged SVs) that can be used to distinguish different ancestries according to the loadings of first PC (PC1) and second PC (PC2). Intriguingly, a higher portion of subtelomeric SVs (258 out of 3 656; 7.1%) were included in PC1- and PC2-related genes, suggesting the subtelomeric SVs as one of the distinguishing variant types (Figure 3C and D). After selecting the SVs according to the PC1 and PC2 loadings, we clustered the presence of these SVs, including subtelomeric SVs, and finally confirmed that each SV cluster represents distinct population frequencies (Figure 3D). Moreover, the patterns were similarly observed when their SNVs as well as small insertions and deletions were used (Supplementary Figure S15).
Overall, our HiFi-based genome assemblies showed high accuracy and resolution for SV detection. Next, we attempted to infer how such large variants emerge in the human population in terms of DNA damage and repair events. Given that subtelomeric SVs are among the most fragile genomic regions and have rarely been studied in the human population, we focused on them to understand their evolution by incorporating our knowledge of DNA damage–repair signatures.
Subtelomeric structural variants reveal ancestral telomere damage and repair events
Since telomeres are easily damaged during DNA replication, telomeric and subtelomeric regions may contain DNA double-strand breaks (DSBs), which leave behind SVs of >100 kb in subtelomeric regions when repaired (18,28,29,31,50). We assumed that our SV-calling pipeline might fail to identify these extremely large SVs; therefore, we manually curated large subtelomeric SVs (≥5 kb in size) in our Korean assemblies. In addition to the two large subtelomeric SVs detected using our SV-calling pipeline, this manual process identified 17 large subtelomeric SVs (Table 1). Among these 19 subtelomeric SVs, nine were shared by at least two assemblies and eight by at least two humans, supporting their prevalence in the Korean population. We could design primers only for 10 out of 19 subtelomeric SVs, and 5 out of the 10 subtelomeric SVs (50.0%) were validated using PCR (Supplementary Figure S10B and Supplementary Table S6).
Table 1.
Subtelomeric SVs (≥5 kb) in individual chromosomes, with their damage and repair signature types
| Type | Repair signature | Possible mechanism | Subtelomeric locus | Number of individuals | Number of genome assemblies |
|---|---|---|---|---|---|
| Deletion (9.3 kb) | Homology (933 bp) | SSA | chr1p | 1 | 1 |
| Deletion (5.4 kb) | Homology (267 bp) | SSA | chr3p | 3 | 4 |
| Deletion (19.4 kb) | Homology (171 bp) | SSA | chr11p | 2 | 2 |
| Deletion (43.8 kb) | Microhomology (9 bp) | TMEJ | chr1q | 3 | 3 |
| Deletion (6.3 kb) | Microhomology (20 bp) | TMEJ | chr3q | 1 | 1 |
| Deletion (7.5 kb) | Microhomology (3 bp) | TMEJ | chr5q | 1 | 1 |
| Deletion (13.7 kb) | Microhomology (3 bp) | TMEJ | chr5q | 1 | 1 |
| Deletion (9.4 kb) | Templated insertion (6 bp) | TMEJ | chr7p | 1 | 1 |
| Insertion (5.8 kb) | Templated insertion (182 bp) | TMEJ | chr5q | 1 | 1 |
| Insertion (84.3–158.7 kb) | Remaining TRM block (352, 480 and 549 bp) | BIR | chr6p | 3 | 5 |
| Insertion (141.0 kb) | Remaining TRM block (400 bp) | BIR | chr8p | 1 | 1 |
| Deletion (200.8 kb) | Remaining TRM block (1 363 bp) | BIR | chr11p | 2 | 2 |
| Deletion (117.9 kb) | Remaining TRM block (737 bp) | BIR | chr16q | 1 | 1 |
| Deletion (162.1 kb) | Remaining TRM block (585 bp) | BIR | chr18p | 3 | 4 |
| Substitution (37.0 kb–121.9 kb) | Large homology block (37.0 kb) | DSBR or BIR | chr9q | 1 | 1 |
| Substitution (32.8 kb to 24.5 kb) | Large homology block (32.8 kb) | DSBR or BIR | chr11p | 2 | 2 |
| Substitution (43.6 kb to 23.6 kb) | Large homology block (43.6 kb) | DSBR or BIR | chr19p | 1 | 2 |
| Deletion (142.6 kb) | Not applicable | Not specified | chr17q | 3 | 5 |
| Deletion (9.4 kb) | Not applicable | Not specified | chr20q | 1 | 1 |
SSA, single-strand annealing; TMEJ, polymerase theta-mediated end joining; BIR, break-induced replication; and DSBR, double-strand break repair.
DSB repair leaves specific signatures near the repaired sequences (51–55), and HiFi-based genome assemblies can precisely resolve these signatures. We categorized the 19 subtelomeric SVs by their DNA repair signatures as follows: deletion with homology (≥50 bp); deletion with microhomology (<50 bp); templated insertion; insertion with a short TRM block; and substitution between large homology blocks (Figure 4; Table 1). We detected a specific repair signature in 17 of the 19 subtelomeric SVs (Table 1). The remaining two SVs that were not categorized contained a substitution or insertion that could not be assigned to any of the five categories.
Figure 4.
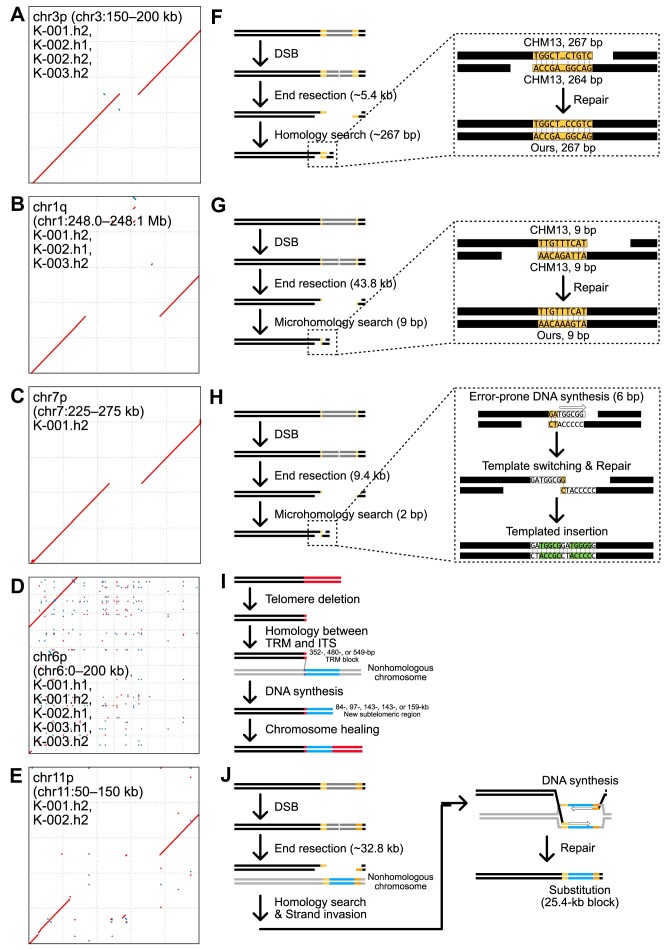
Subtelomeric SVs identified in six Korean genome assemblies contain subtelomere damage and repair signatures. (A–E) Dot plots representing large subtelomeric SVs. Red dots represent forward matches, and blue dots represent reverse matches. The x-axis represents the CHM13 genomic loci, and the y-axis represents the corresponding genomic loci in the Korean genome assemblies. Corresponding Korean genome assemblies and subtelomeric loci for each SV are shown in black-lined boxes. (F–J) Models representing several repair mechanisms and their corresponding clues found in the Korean genome assemblies. (F) A 5.4 kb deletion in chr3p shows 267 bp-long homology in the flanking sequences of the reference genome, suggesting its SSA-mediated repair in a Korean ancestor. (G) A 43.8 kb deletion in chr1q shows 9 bp-long microhomology in the flanking sequences, suggesting its TMEJ-mediated repair in a Korean ancestor. (H) A substitution from 9.4 kb to 5 bp in chr7p exhibits microhomology and templated insertion signatures, strongly supporting its TMEJ-mediated repair of CHM13 in an ancestor. (I) Five >84 kb insertions in the chr6p telomere comprise a short TRM cluster and new subtelomeric sequences, suggesting that the ancestral chr6p telomere was deleted and repaired via BIR in Korean ancestors. (J) A substitution between 32.8 and 25.4 kb blocks in chr11p has homology blocks in chr1, chr3, chr5, chr6 and chr15, possibly resulting from its DSBR-mediated repair via strand invasion to a nonhomologous chromosome in a Korean ancestor. SSA: single-strand annealing; TMEJ: polymerase theta-mediated end joining; TRM: telomeric-repeat motif, such as TTAGGG; BIR: break-induced replication; and DSBR: double-strand break repair.
Deletions with homology
A 5.4 kb deletion in the p arm of the chromosome 3 (chr3p) subtelomere was shared by four assemblies from the three individuals (Figure 4A; Table 1). Near the deletion, all four assemblies had the same 267 bp sequence, which exhibited >91% identity with the two flanking sequences in the CHM13 genome (Figure 4F). These long homology sequences may have resulted from DSB repair events via SSA, which typically excises both ends of the DSB site, anneals long (≥50 bp) homology sequences and repairs the remaining gaps (Figure 4F). The SVs in chr1p and chr11p had similar signatures, implying that these SVs were generated by SSA-mediated DSB repair (Table 1).
Deletions with microhomology
A 43.8 kb deletion in the q arm of chr1 (chr1q) was shared by the three individuals and three assemblies (Figure 4B; Table 1). All three assemblies had the same 43 818 bp deletion and 9 bp sequence near the deletion (Figure 4G). This 9 bp sequence was identical to a flanking sequence in the CHM13 genome, with only two mismatches in the other flanking sequence (Figure 4G). This microhomology implies that the SV was generated by TMEJ. The two additional SVs in chr3q and chr5q had the same signature (Table 1).
Templated insertions generated by polymerase theta-mediated end joining
A 9.4 kb deletion in chr7p had an additional 6 bp insertion (TGGCGG) that had a homology (TGGGGG) near the deletion (Figure 4C; Table 1). This templated insertion, which provides evidence of TMEJ (54), might have arisen via DSB followed by a 9.4 kb excision (Figure 4H). Exposed 2 bp (GA) microhomology sequences were used to anneal the ends of the DNA molecules (Figure 4H, right dotted box). Since the 6 bp sequence (TGGGGG) was located next to the 2 bp microhomology, it functioned as a template to synthesize a new complementary sequence, although not perfectly, owing to the error-prone replication of polymerase theta (Figure 4H).
However, the synthesized 6 bp sequence (TGGCGG) was released from the template, while polymerase theta searched for another microhomology (Figure 4H). Since the last nucleotide base (G) in the synthesized sequence was the same as the first base (G) of the 2 bp microhomology, this 1 bp microhomology was used to fill the remaining gaps and repair the DSB site (Figure 4H). A templated insertion was also detected in an SV in chr5q (Table 1).
Insertions with a short telomeric-repeat motif block
Insertions in chr6p are present in the telomeric region for all three humans and five assemblies (Figure 4D; Table 1). Although their lengths vary, ranging from 84 to 159 kb, they can be categorized into three groups according to the length of the remaining TRM blocks: a 352 bp TRM block in one assembly in K-002; a 480 bp TRM block in two assemblies in K-001 and K-003; and a 549 bp TRM block in two assemblies in K-001 and K-003 (Figure 4I). The independent acquisition of TRM blocks of the same length in two individuals suggests that each short TRM block was generated by the same telomere deletion and repair event. These remaining TRM blocks may have acted as homologous blocks enabling the invasion into another genomic locus with an interstitial telomeric sequence (ITS) (Figure 4I). Subsequently, a new sequence could have been synthesized using the adjacent sequence of ITS as a template (Figure 4I). This process might be mediated by BIR, a homologous recombination mechanism, generating an insertion of ∼100 kb in the telomeric region. Insertions in the Korean chr8p and CHM13 chr11p, chr16q and chr18p exhibited the same signatures (Table 1). Impressively, chr11p of K-001 and K-002 have the same insertion size (200 812 bp) and length as the remaining TRM block (1363 bp) in CHM13 (Table 1), implying that a single telomere damage and repair event may produce these large insertion SVs.
Substitutions between large homology blocks
A substitution of a 32.8 kb block by a 24.5 kb block in chr11p was shared by two individuals (Figure 4E; Table 1). Intriguingly, the substituted 24.5 kb block had homologous blocks in chr1, chr3, chr5, chr6 and chr15 subtelomeric or other genomic loci, but its flanking sequences were collinear with the reference sequences. We hypothesized that these were generated by homologous recombination-mediated repair, probably via DSBR or BIR. The subtelomeric region of chr11p may have undergone DSBs, with both ends being excised, and the 24.5 kb block was synthesized using a homologous block from another locus as a repair template (Figure 4J). If this synthesis was mediated by DSBR, the end of the synthesized block would have been ligated to the end of the original sequence block. If BIR was the main repair mechanism, this synthesized end switched its template to the original sequence to resume DNA replication. The two substitutions in chr9q and chr19p exhibited similar patterns (Table 1).
Uncategorized large subtelomeric structural variants
Two of the cases could not be explained by a single damage or repair event (Table 1). An SV in chr20q is a substitution of a 9.4 kb deletion and a 21 bp insertion (Table 1); however, we could not find any homologous block near the SV that was used as a template. While this SV could have been generated via templated insertion by TMEJ, using a different non-allelic locus as a template, we cannot determine this conclusively. Another SV in chr17q was shared by all three individuals and five assemblies (Table 1). This SV was also a substitution generated by a subtelomeric deletion in CHM13, followed by a 140 kb duplication of a nearby sequence block (Table 1). Although this block could have been duplicated via BIR, it contained an 11 bp insertion between the original and duplicated blocks. This 11 bp sequence is not present in the nearby sequences. Thus, we were unable to determine how the block was duplicated with the 11 bp insertion.
Discussion
Accurate detection of genetic variants helps us elucidate how they emerge and affect human evolution and cancer development (11,12,56–60). Unlike short-reads, which can be un- or mis-mappable, long-read sequencing technologies now allow for the resolution of all genetic variants in any individual, by providing high-quality genome assemblies (19,48,61). A limited number of previous research endeavors have focused on SVs by long-read sequencing within the Korean population (62–64). In this study, we used state-of-the-art long-read sequencing technology, HiFi sequencing, to identify SVs in the Korean genomes. Specifically, we identified and verified extremely large SVs in subtelomeric regions and postulated the mechanisms involved in their emergence. Although large subtelomeric SVs have been previously described in human populations, based on analysis of mapping data or parts of the sequences (28,65–70), our study is the first to report on full-length human subtelomeric SVs at the single-nucleotide level, to the best of our knowledge. These subtelomeric SVs were probably generated via telomere damage or subtelomeric DSB followed by repair processes. Therefore, analyzing these SVs may help us elucidate how genomic instability facilitates not only genomic changes in the human population but also cancer evolution.
Although half of the global human population is of Asian ancestry, Asian genetic diversity is relatively underrepresented in human population genetic studies (71). Korean ancestry exhibits a relatively uniform population structure compared with other East Asian populations (71,72). The modern Korean population has been shaped predominantly by two major ancestral components: East Siberian and Southeast Asian populations. This was elucidated through analyzing ancient and modern Korean genome sequencing data (73). Long-read sequencing-based high-quality genome assemblies provide the best and most extensive data for describing such underrepresented populations. Our results are inadequate for thorough characterization of even Korean ancestry, and some of our SVs could be CHM13-specific and, therefore, absent from the broader human population. Furthermore, our partially phased genome assemblies of the three individuals could not provide haplotype information, due to the lack of genome sequencing of their parents. Nonetheless, our data—six HiFi-based genome assemblies of three human individuals and their high-quality SVs—provide a foundation for future research. In the future, we intend to generate more comprehensive variant sets for people of Korean ancestry and produce phased genome assemblies of individuals by performing family trio-based sequencing, providing detailed haplotype information, given the impending national genome projects. This could resolve the origin of most rare or specific variants in people of Asian ancestry.
Subtelomeric variants should be thoroughly investigated to understand genome instability and the resulting variant formation. Telomeric and nearby subtelomeric regions can be easily broken by stochastic telomere deletion during DNA replication or via other telomere damage, which induces various DSB repair pathways (32,74,75). Nonhomologous end joining is the primary mechanism involved in this repair function. However, although nonhomologous end joining can result in chromosome fusion, it typically leaves only small and random insertions or deletions (∼5 bp) rather than SVs. In contrast, other DSB repair mechanisms, such as SSA, TMEJ, DSBR and BIR, may generate extreme SVs, as these mechanisms typically involve DSB end-excision. SSA or TMEJ can lead to large deletions. Moreover, human subtelomeric regions contain highly repetitive sequences, ITSs and segmentally duplicated blocks. Sequence blocks in these regions can be readily used as homologies for telomere or subtelomere repair after telomere deletions or subtelomeric DSBs (74–76). We identified BIR-mediated telomere damage–repair signatures that were produced using homology between short TRM blocks, in addition to DSBR-mediated substitutions of large homologous sequence blocks following subtelomeric DSBs. Whether such segmentally duplicated blocks in subtelomeric regions increase the stability of telomere maintenance or whether they are merely leftover sequences after telomere damage and repair events warrants further investigation (20). Moreover, all these findings in our study were identified by manual investigation. In the future, detecting large subtelomeric SVs (≥5 kb in size) and their associated repair signatures should be automated after establishing the general rules for our manual investigation. Centromeric SVs are also important; however, the centromeric region is typically too repetitive to infer what repair mechanisms have been involved in its SV formation (22,77). For example, if an SV has repetitive flanking sequences, these sequences could be overlapped. Such micro/homologous sequences can be inferred based on the signature of TMEJ or SSA, and they can also be created via independent mechanisms, such as replication slippage (78–80). Therefore, the repetitive nature of the centromeric region hinders not only the assembly of this region but also the inference of the underlying mechanisms of centromeric SVs.
Although our findings reveal that such mechanisms may participate in SV formation in normal human germlines, they may be much more important in cancer evolution. However, cancer cells are highly heterogeneous, and single-cell long-read DNA sequencing is currently immature (81–83). Thus, further advances in long-read sequencing technology are required to trace SVs in cancer cell populations. These efforts will facilitate the resolution of telomere damage-induced variant formation in cancer genomes. Furthermore, in-depth studies of the major mechanisms involved in telomere damage and repair during cancer evolution are necessary to understand the underlying mechanisms of cancer-specific SVs.
In this study, we developed high-quality genome assemblies and SV sets for three Korean individuals. Our results provide a valuable foundation to investigate SVs and extremely large subtelomeric SVs in the broader human population. Advances in sequencing technology and corresponding high-quality genome assembly production will shed light on genomic dark matter, thus helping elucidate all genetic variants in the human genome, including in subtelomeric regions. This will help explain the SV-level genetic architecture of human genetic disorders and the mechanisms of human genomic evolution.
Supplementary Material
Acknowledgements
Author contributions: Conceptualization: J.K., S.J. and J.-H.P.; Methodology: J.K., S.J., J.L.P., M.-J.C., S.L., J.-E.L., M.-K.Y., J.O.Y., S.K. and S.-Y.K.; Formal analysis: S.J., J.K., J.O.Y. and S.K.; Investigation: S.J., J.K. and J.-H.P.; Writing—original draft: J.K., S.J. and J.-H.P.; Writing—review & editing: J.K., S.J., G.P., T.H., J.-H.P., J.L.P., J.O.Y., S.K., M.-J.C. and S.-Y.K.; Funding acquisition: J.K., J.-H.P. and S.-Y.K.; Supervision, S.-Y. K, M.-K.Y. and J.-H.P.
Contributor Information
Jun Kim, Department of Convergent Bioscience and Informatics, College of Bioscience and Biotechnology, Chungnam National University, 99 Daehak-ro, Yuseong-gu, Daejeon 34134, Republic of Korea; Personalized Genomic Medicine Research Center, Korea Research Institute of Bioscience & Biotechnology, 125, Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Jong Lyul Park, Personalized Genomic Medicine Research Center, Korea Research Institute of Bioscience & Biotechnology, 125, Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea; Department of Bioscience, University of Science and Technology (UST), 217, Gajeong-ro, Yuseong-gu, Daejeon 34113, Republic of Korea.
Jin Ok Yang, Korea Bioinformation Center, Korea Research Institute of Bioscience & Biotechnology, 125, Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea; Department of Bio and Brain Engineering, Korea Advanced Institute of Science & Technology (KAIST), 291, Daehak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Sangok Kim, Korea Bioinformation Center, Korea Research Institute of Bioscience & Biotechnology, 125, Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea; Department of Bioscience, University of Science and Technology (UST), 217, Gajeong-ro, Yuseong-gu, Daejeon 34113, Republic of Korea.
Soobok Joe, Korea Bioinformation Center, Korea Research Institute of Bioscience & Biotechnology, 125, Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Gunwoo Park, Korea Bioinformation Center, Korea Research Institute of Bioscience & Biotechnology, 125, Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Taeyeon Hwang, Korea Bioinformation Center, Korea Research Institute of Bioscience & Biotechnology, 125, Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
Mun-Jeong Cho, Department of Bioscience, University of Science and Technology (UST), 217, Gajeong-ro, Yuseong-gu, Daejeon 34113, Republic of Korea.
Seungjae Lee, DNALink, Inc, 31, Magokjungang 8-ro 3-gil, Gangseo-gu, Seoul 07793, Republic of Korea.
Jong-Eun Lee, DNALink, Inc, 31, Magokjungang 8-ro 3-gil, Gangseo-gu, Seoul 07793, Republic of Korea.
Ji-Hwan Park, Korea Bioinformation Center, Korea Research Institute of Bioscience & Biotechnology, 125, Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea; Department of Biological Science, Ajou University, 206, World cup-ro, Yeongtong-gu, Suwon 16499, Republic of Korea.
Min-Kyung Yeo, Department of Pathology, Chungnam National University School of Medicine, 282, Munhwa-ro, Jung-gu, Daejeon 35015, Republic of Korea.
Seon-Young Kim, Korea Bioinformation Center, Korea Research Institute of Bioscience & Biotechnology, 125, Gwahak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea; Department of Bioscience, University of Science and Technology (UST), 217, Gajeong-ro, Yuseong-gu, Daejeon 34113, Republic of Korea.
Data availability
The Korean HiFi long-read sequencing datasets produced in this study are deposited in the Korean BioData Station (K-BDS) (84) (https://kbds.re.kr/; accession number KAP220172 and KAP241043) and the European Genome-Phenome Archive (EGA) (https://ega-archive.org/; accession number EGAS50000000375).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Ministry of Science and ICT (MSIT) [2020M3E5D708517212, 2023M3A9A7009442, 2020M3A9I6A0103605721; and RS-2023-00247499 and RS-2024-00415888 to J.K.]; Ministry of Health and Welfare (MOHW) [RS-2024-00438566]; Ministry of Trade, Industry and Energy (MOTIE) [P0009796]; KRIBB Research Initiative Program [KGM5192221 to J.K. and J.L.P.; and KGM5422221 to S.J., G.P., T.H., J.O.Y., S.K. and S.-Y.K.].
Conflict of interest statement. None declared.
References
- 1. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 2. International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004; 431:931–945. [DOI] [PubMed] [Google Scholar]
- 3. Church D.M., Schneider V.A., Graves T., Auger K., Cunningham F., Bouk N., Chen H.-C., Agarwala R., McLaren W.M., Ritchie G.R. Modernizing reference genome assemblies. PLoS Biol. 2011; 9:e1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klein R.J., Zeiss C., Chew E.Y., Tsai J.-Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T. Complement factor H polymorphism in age-related macular degeneration. Science. 2005; 308:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017; 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Loos R.J. 15 years of genome-wide association studies and no signs of slowing down. Nat. Commun. 2020; 11:5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Uffelmann E., Huang Q.Q., Munung N.S., De Vries J., Okada Y., Martin A.R., Martin H.C., Lappalainen T., Posthuma D. Genome-wide association studies. Nat. Rev. Methods Primers. 2021; 1:59. [Google Scholar]
- 8. Halldorsson B.V., Eggertsson H.P., Moore K.H., Hauswedell H., Eiriksson O., Ulfarsson M.O., Palsson G., Hardarson M.T., Oddsson A., Jensson B.O. The sequences of 150,119 genomes in the UK Biobank. Nature. 2022; 607:732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Collins R.L., Brand H., Karczewski K.J., Zhao X., Alföldi J., Francioli L.C., Khera A.V., Lowther C., Gauthier L.D., Wang H. A structural variation reference for medical and population genetics. Nature. 2020; 581:444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Byrska-Bishop M., Evani U.S., Zhao X., Basile A.O., Abel H.J., Regier A.A., Corvelo A., Clarke W.E., Musunuri R., Nagulapalli K. High-coverage whole-genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. Cell. 2022; 185:3426–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y., Roberts N.D., Wala J.A., Shapira O., Schumacher S.E., Kumar K., Khurana E., Waszak S., Korbel J.O., Haber J.E. Patterns of somatic structural variation in human cancer genomes. Nature. 2020; 578:112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Logsdon G.A., Vollger M.R., Eichler E.E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020; 21:597–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miga K.H., Koren S., Rhie A., Vollger M.R., Gershman A., Bzikadze A., Brooks S., Howe E., Porubsky D., Logsdon G.A. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020; 585:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bzikadze A.V., Pevzner P.A. Automated assembly of centromeres from ultra-long error-prone reads. Nat. Biotechnol. 2020; 38:1309–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garg S., Fungtammasan A., Carroll A., Chou M., Schmitt A., Zhou X., Mac S., Peluso P., Hatas E., Ghurye J. Chromosome-scale, haplotype-resolved assembly of human genomes. Nat. Biotechnol. 2021; 39:309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Porubsky D., Ebert P., Audano P.A., Vollger M.R., Harvey W.T., Marijon P., Ebler J., Munson K.M., Sorensen M., Sulovari A. Fully phased human genome assembly without parental data using single-cell strand sequencing and long reads. Nat. Biotechnol. 2021; 39:302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fujimoto A., Wong J.H., Yoshii Y., Akiyama S., Tanaka A., Yagi H., Shigemizu D., Nakagawa H., Mizokami M., Shimada M. Whole-genome sequencing with long reads reveals complex structure and origin of structural variation in human genetic variations and somatic mutations in cancer. Genome Med. 2021; 13:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim E., Kim J., Kim C., Lee J. Long-read sequencing and de novo genome assemblies reveal complex chromosome end structures caused by telomere dysfunction at the single nucleotide level. Nucleic Acids Res. 2021; 49:3338–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nurk S., Koren S., Rhie A., Rautiainen M., Bzikadze A.V., Mikheenko A., Vollger M.R., Altemose N., Uralsky L., Gershman A. The complete sequence of a human genome. Science. 2022; 376:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vollger M.R., Guitart X., Dishuck P.C., Mercuri L., Harvey W.T., Gershman A., Diekhans M., Sulovari A., Munson K.M., Lewis A.P. Segmental duplications and their variation in a complete human genome. Science. 2022; 376:eabj6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoyt S.J., Storer J.M., Hartley G.A., Grady P.G., Gershman A., de Lima L.G., Limouse C., Halabian R., Wojenski L., Rodriguez M. From telomere to telomere: the transcriptional and epigenetic state of human repeat elements. Science. 2022; 376:eabk3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Altemose N., Logsdon G.A., Bzikadze A.V., Sidhwani P., Langley S.A., Caldas G.V., Hoyt S.J., Uralsky L., Ryabov F.D., Shew C.J. Complete genomic and epigenetic maps of human centromeres. Science. 2022; 376:eabl4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. De Coster W., Weissensteiner M.H., Sedlazeck F.J. Towards population-scale long-read sequencing. Nat. Rev. Genet. 2021; 22:572–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang T., Antonacci-Fulton L., Howe K., Lawson H.A., Lucas J.K., Phillippy A.M., Popejoy A.B., Asri M., Carson C., Chaisson M.J. The Human Pangenome Project: a global resource to map genomic diversity. Nature. 2022; 604:437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liao W.-W., Asri M., Ebler J., Doerr D., Haukness M., Hickey G., Lu S., Lucas J.K., Monlong J., Abel H.J. A draft human pangenome reference. Nature. 2023; 617:312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beyter D., Ingimundardottir H., Oddsson A., Eggertsson H.P., Bjornsson E., Jonsson H., Atlason B.A., Kristmundsdottir S., Mehringer S., Hardarson M.T. Long-read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits. Nat. Genet. 2021; 53:779–786. [DOI] [PubMed] [Google Scholar]
- 27. Ebert P., Audano P.A., Zhu Q., Rodriguez-Martin B., Porubsky D., Bonder M.J., Sulovari A., Ebler J., Zhou W., Serra Mari R. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science. 2021; 372:eabf7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Riethman H. Human subtelomeric copy number variations. Cytogenet. Genome Res. 2009; 123:244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baird D.M. Telomeres and genomic evolution. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2018; 373:20160437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yue J.-X., Li J., Aigrain L., Hallin J., Persson K., Oliver K., Bergström A., Coupland P., Warringer J., Lagomarsino M.C. Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat. Genet. 2017; 49:913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim C., Kim J., Kim S., Cook D.E., Evans K.S., Andersen E.C., Lee J. Long-read sequencing reveals intra-species tolerance of substantial structural variations and new subtelomere formation in C. elegans. Genome Res. 2019; 29:1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim C., Sung S., Kim J., Lee J. Repair and reconstruction of telomeric and subtelomeric regions and genesis of new telomeres: implications for chromosome evolution. Bioessays. 2020; 42:e1900177. [DOI] [PubMed] [Google Scholar]
- 33. Chaux-Jukic F., O’Donnell S., Craig R.J., Eberhard S., Vallon O., Xu Z. Architecture and evolution of subtelomeres in the unicellular green alga Chlamydomonas reinhardtii. Nucleic Acids Res. 2021; 49:7571–7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee B.Y., Kim J., Lee J. Long-read sequencing infers a mechanism for copy number variation of template for alternative lengthening of telomeres in a wild C. elegans strain. MicroPubl. Biol. 2022; 2022:000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng H., Concepcion G.T., Feng X., Zhang H., Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods. 2021; 18:170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alonge M., Lebeigle L., Kirsche M., Jenike K., Ou S., Aganezov S., Wang X., Lippman Z.B., Schatz M.C., Soyk S. Automated assembly scaffolding using RagTag elevates a new tomato system for high-throughput genome editing. Genome Biol. 2022; 23:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jain C., Rhie A., Hansen N.F., Koren S., Phillippy A.M. Long-read mapping to repetitive reference sequences using Winnowmap2. Nat. Methods. 2022; 19:705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Subgroup G.P.D.P. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heller D., Vingron M. SVIM-asm: structural variant detection from haploid and diploid genome assemblies. Bioinformatics. 2020; 36:5519–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jeffares D.C., Jolly C., Hoti M., Speed D., Shaw L., Rallis C., Balloux F., Dessimoz C., Bähler J., Sedlazeck F.J. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun. 2017; 8:14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marçais G., Delcher A.L., Phillippy A.M., Coston R., Salzberg S.L., Zimin A. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 2018; 14:e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 43. Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013; 43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Källberg M., Cox A.J., Kruglyak S., Saunders C.T. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016; 32:1220–1222. [DOI] [PubMed] [Google Scholar]
- 45. Poplin R., Chang P.-C., Alexander D., Schwartz S., Colthurst T., Ku A., Newburger D., Dijamco J., Nguyen N., Afshar P.T. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018; 36:983–987. [DOI] [PubMed] [Google Scholar]
- 46. Lin J., Jia P., Wang S., Kosters W., Ye K. Comparison and benchmark of structural variants detected from long read and long-read assembly. Brief. Bioinform. 2023; 24:bbad188. [DOI] [PubMed] [Google Scholar]
- 47. Ahsan M.U., Liu Q., Perdomo J.E., Fang L., Wang K. A survey of algorithms for the detection of genomic structural variants from long-read sequencing data. Nat. Methods. 2023; 20:1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wenger A.M., Peluso P., Rowell W.J., Chang P.-C., Hall R.J., Concepcion G.T., Ebler J., Fungtammasan A., Kolesnikov A., Olson N.D. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019; 37:1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee H., Kim J., Lee J. Benchmarking datasets for assembly-based variant calling using high-fidelity long reads. BMC Genomics. 2023; 24:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 51. Bhargava R., Onyango D.O., Stark J.M. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 2016; 32:566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rodgers K., McVey M. Error-prone repair of DNA double-strand breaks. J. Cell. Physiol. 2016; 231:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen C.-C., Feng W., Lim P.X., Kass E.M., Jasin M. Homology-directed repair and the role of BRCA1, BRCA2, and related proteins in genome integrity and cancer. Annu. Rev. Cancer Biol. 2018; 2:313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schimmel J., van Schendel R., den Dunnen J.T., Tijsterman M. Templated insertions: a smoking gun for polymerase theta-mediated end joining. Trends Genet. 2019; 35:632–644. [DOI] [PubMed] [Google Scholar]
- 55. Al-Zain A.M., Symington L.S. The dark side of homology-directed repair. DNA Repari (Amst.). 2021; 106:103181. [DOI] [PubMed] [Google Scholar]
- 56. Hollox E.J., Zuccherato L.W., Tucci S. Genome structural variation in human evolution. Trends Genet. 2022; 38:45–58. [DOI] [PubMed] [Google Scholar]
- 57. Gerstung M., Jolly C., Leshchiner I., Dentro S.C., Gonzalez S., Rosebrock D., Mitchell T.J., Rubanova Y., Anur P., Yu K. The evolutionary history of 2,658 cancers. Nature. 2020; 578:122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Aganezov S., Goodwin S., Sherman R.M., Sedlazeck F.J., Arun G., Bhatia S., Lee I., Kirsche M., Wappel R., Kramer M. Comprehensive analysis of structural variants in breast cancer genomes using single-molecule sequencing. Genome Res. 2020; 30:1258–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Au K.F. The blooming of long-read sequencing reforms biomedical research. Genome Biol. 2022; 23:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Amarasinghe S.L., Su S., Dong X., Zappia L., Ritchie M.E., Gouil Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020; 21:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018; 36:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Seo J.S., Rhie A., Kim J., Lee S., Sohn M.H., Kim C.U., Hastie A., Cao H., Yun J.Y., Kim J. et al. De novo assembly and phasing of a Korean human genome. Nature. 2016; 538:243–247. [DOI] [PubMed] [Google Scholar]
- 63. Cho Y.S., Kim H., Kim H.-M., Jho S., Jun J., Lee Y.J., Chae K.S., Kim C.G., Kim S., Eriksson A. An ethnically relevant consensus Korean reference genome is a step towards personal reference genomes. Nat. Commun. 2016; 7:13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim H.-S., Jeon S., Kim Y., Kim C., Bhak J., Bhak J. KOREF_S1: phased, parental trio-binned Korean reference genome using long reads and hi-C sequencing methods. Gigascience. 2022; 11:giac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Riethman H., Ambrosini A., Paul S. Human subtelomere structure and variation. Chromosome Res. 2005; 13:505–515. [DOI] [PubMed] [Google Scholar]
- 66. Riethman H., Ambrosini A., Castaneda C., Finklestein J., Hu X.-L., Mudunuri U., Paul S., Wei J. Mapping and initial analysis of human subtelomeric sequence assemblies. Genome Res. 2004; 14:18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stong N., Deng Z., Gupta R., Hu S., Paul S., Weiner A.K., Eichler E.E., Graves T., Fronick C.C., Courtney L. Subtelomeric CTCF and cohesin binding site organization using improved subtelomere assemblies and a novel annotation pipeline. Genome Res. 2014; 24:1039–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Young E., Pastor S., Rajagopalan R., McCaffrey J., Sibert J., Mak A.C., Kwok P.-Y., Riethman H., Xiao M. High-throughput single-molecule mapping links subtelomeric variants and long-range haplotypes with specific telomeres. Nucleic Acids Res. 2017; 45:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Grigorev K., Foox J., Bezdan D., Butler D., Luxton J.J., Reed J., McKenna M.J., Taylor L., George K.A., Meydan C. Haplotype diversity and sequence heterogeneity of human telomeres. Genome Res. 2021; 31:1269–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Young E., Abid H.Z., Kwok P.-Y., Riethman H., Xiao M. Comprehensive analysis of human subtelomeres by whole genome mapping. PLoS Genet. 2020; 16:e1008347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pan Z., Xu S. Population genomics of East Asian ethnic groups. Hereditas. 2020; 157:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang C.-C., Yeh H.-Y., Popov A.N., Zhang H.-Q., Matsumura H., Sirak K., Cheronet O., Kovalev A., Rohland N., Kim A.M. Genomic insights into the formation of human populations in East Asia. Nature. 2021; 591:413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kim J., Jeon S., Choi J.-P., Blazyte A., Jeon Y., Kim J.-I., Ohashi J., Tokunaga K., Sugano S., Fucharoen S. The origin and composition of Korean ethnicity analyzed by ancient and present-day genome sequences. Genome Biol. Evol. 2020; 12:553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. McEachern M.J., Haber J.E. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006; 75:111–135. [DOI] [PubMed] [Google Scholar]
- 75. Zhang J.-M., Zou L. Alternative lengthening of telomeres: from molecular mechanisms to therapeutic outlooks. Cell Biosci. 2020; 10:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kramara J., Osia B., Malkova A. Break-induced replication: the where, the why, and the how. Trends Genet. 2018; 34:518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Eichler E.E. Repetitive conundrums of centromere structure and function. Hum. Mol. Genet. 1999; 8:151–155. [DOI] [PubMed] [Google Scholar]
- 78. Dieringer D., Schlötterer C. Two distinct modes of microsatellite mutation processes: evidence from the complete genomic sequences of nine species. Genome Res. 2003; 13:2242–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lovett S.T. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 2004; 52:1243–1253. [DOI] [PubMed] [Google Scholar]
- 80. Pumpernik D., Oblak B., Borštnik B. Replication slippage versus point mutation rates in short tandem repeats of the human genome. Mol. Genet. Genomics. 2008; 279:53–61. [DOI] [PubMed] [Google Scholar]
- 81. Fan X., Yang C., Li W., Bai X., Zhou X., Xie H., Wen L., Tang F. SMOOTH-seq: single-cell genome sequencing of human cells on a third-generation sequencing platform. Genome Biol. 2021; 22:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xie H., Li W., Hu Y., Yang C., Lu J., Guo Y., Wen L., Tang F. De novo assembly of human genome at single-cell levels. Nucleic Acids Res. 2022; 50:7479–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hård J., Mold J.E., Eisfeldt J., Tellgren-Roth C., Häggqvist S., Bunikis I., Contreras-Lopez O., Chin C.-S., Nordlund J., Rubin C.-J. Long-read whole-genome analysis of human single cells. Nat. Commun. 2023; 14:5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ko G., Lee J.H., Sim Y.M., Song W., Yoon B.-H., Byeon I., Lee B.H., Kim S.-O., Choi J., Jang I. et al. KoNA: Korean Nucleotide Archive as A New Data Repository for Nucleotide Sequence Data. Genomics Proteomics Bioinformatics. 2024; 22:qzae017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Korean HiFi long-read sequencing datasets produced in this study are deposited in the Korean BioData Station (K-BDS) (84) (https://kbds.re.kr/; accession number KAP220172 and KAP241043) and the European Genome-Phenome Archive (EGA) (https://ega-archive.org/; accession number EGAS50000000375).