Abstract
Homologous recombination is a meiotic process that generates diversity along the genome and interacts with all evolutionary forces. Despite its importance, studies of recombination landscapes are lacking due to methodological limitations and limited data. Frequently used approaches include linkage mapping based on familial data that provides sex-specific broad-scale estimates of realized recombination and inferences based on population linkage disequilibrium that reveal a more fine-scale resolution of the recombination landscape, albeit dependent on the effective population size and the selective forces acting on the population. In this study, we use a combination of these 2 methods to elucidate the recombination landscape for the Afro-European barn owl (Tyto alba). We find subtle differences in crossover placement between sexes that lead to differential effective shuffling of alleles. Linkage disequilibrium-based estimates of recombination are concordant with family-based estimates and identify large variation in recombination rates within and among linkage groups. Larger chromosomes show variation in recombination rates, while smaller chromosomes have a universally high rate that shapes the diversity landscape. We find that recombination rates are correlated with gene content, genetic diversity, and GC content. We find no conclusive differences in the recombination landscapes between populations. Overall, this comprehensive analysis enhances our understanding of recombination dynamics, genomic architecture, and sex-specific variation in the barn owl, contributing valuable insights to the broader field of avian genomics.
Keywords: linkage map, linkage disequilibrium, meiotic recombination, heterochiasmy, hotspots
Introduction
Homologous meiotic recombination (hereafter recombination) is the reciprocal exchange of genetic material between homologous chromosomes during the first meiotic division. The physical exchange, called a crossover (CO), generates the necessary tension between homologous chromosomes to ensure their proper segregation in the daughter cells. In most species, in the absence of a CO, the daughter cells have an aberrant number of chromosome pairs (aneuploidy) leading to deleterious effects (Koehler et al. 1996; Hassold and Hunt 2001; Hassold et al. 2007; Zickler and Kleckner 2015; Zelkowski et al. 2019). Hence, at least 1 CO is expected per chromosome (or chromosome arm), termed an “obligate CO.”
Beyond contributing to the integrity of functional meiotic division, recombination has evolutionary consequences. It shuffles alleles between haplotypes, affecting the genomic composition of a population, which, in turn, can incur evolutionary benefits, including faster adaptation to a changing environment and more efficient selection (Hill and Robertson 1966; Otto and Lenormand 2002). However, recombination can increase the rate of mutations and chromosomal rearrangements or impede adaptation by breaking up beneficial combinations of alleles (Barton and Charlesworth 1998; Arbeithuber et al. 2015; Halldorsson et al. 2019). In fact, it can often be beneficial to link haplotypes together by suppressing recombination between loci, for example during the formation of a sex chromosome (Wright et al. 2016; Charlesworth 2017) or through the maintenance of inversions capturing ecologically relevant phenotypes (Küpper et al. 2016; Rowan et al. 2019; Todesco et al. 2020). As a result, the evolutionary net outcome of recombination is context dependent and may differ between populations, sexes, and genomic coordinates (Stapley et al. 2017).
The variation in recombination rates along the genomic sequence affects almost all genome-wide processes. For instance, in some, but not all species, genetic diversity along the genome correlates with recombination rate. Multiple forces can explain this pattern such as linked selection, a mutagenic effect of recombination, and GC-biased gene conversion (gBGC) (Begun and Aquadro 1992; Duret and Galtier 2009; Flowers et al. 2012; Cutter and Payseur 2013; Campos et al. 2014; Halldorsson et al. 2019). Importantly, the strength of these forces can vary among species, and their relative contribution in shaping genetic diversity is still unclear. In addition, regions with high recombination rates are often enriched in foreign DNA from past introgressions both in plants and animals (Schumer et al. 2018; Martin et al. 2019; Dreissig et al. 2020; Edelman and Mallet 2021). Beyond influencing these processes, variable recombination rates can confound inferences, like scans for selection based on genetic differentiation (Booker et al. 2020). Thus, in order to advance our understanding of the evolutionary consequences of recombination and distinguish it from other forces that vary along the genome, we need to quantify recombination and understand the sources of its variation in different species and under different evolutionary backgrounds.
However, quantifying recombination is a challenging task. One approach is linkage mapping, the positioning of markers along the sequence with a distance proportional to the recombination rate between them. This approach requires family data (or controlled crosses, when available) and uses the cosegregation of alleles from one generation to the next, to identify the contemporary COs in the sample. Linkage mapping has been applied in several species thus far, providing a reliable measure of CO frequency (Kong et al. 2002; Stapley et al. 2017; Brazier and Glémin 2022). Linkage mapping enables quantification of the differences in recombination rates between sexes (i.e. heterochiasmy; Kong et al. 2010; Johnston et al. 2017; Brekke et al. 2022). However, historically, linkage mapping studies used a small set of genomic markers (Stapley et al. 2017) and the resolution of the method is limited by the number of observed meioses, making it impossible to quantify recombination accurately at a fine scale with the sample sizes available in many nonmodel species (Halldorsson et al. 2019).
Another approach used to estimate recombination rates uses whole genome sequences or reduced representation genotyping from tens of unrelated individuals and models the observed linkage disequilibrium (LD) between markers (Auton and McVean 2007; Chan et al. 2012; Spence and Song 2019). This method (hereafter referred to as LD-based inference) assesses ancestral recombination events that occurred in the coalescent history of the samples (Li and Stephens 2003; Stumpf and McVean 2003) and has enabled the quantification of fine-scale recombination variation (at the kilobase scale). These fine-scale inferences have identified that in many species COs occur in “hotspots,” defined as regions with elevated recombination rate scattered along the genome (Myers et al. 2005; Mézard 2006; Paape et al. 2012; Choi and Henderson 2015). This observation is true for most species studied to date but not all (e.g. Caenorhabditis elegans and Drosophila; Kaur and Rockman 2014; Smukowski Heil et al. 2015). While initial discoveries in humans and mice implicated the fast-evolving PRDM9 gene for the localization of hotspots along the sequence through motif matching, most species, notably birds, amphibians, most arthropods, and plants, do not have a functioning copy of the gene (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010; Baker et al. 2017). In these organisms, hotspots are found in accessible chromatin, near transcription start sites (TSSs) and CpG islands (CGIs), and are usually conserved even after millions of years of species’ divergence (Auton et al. 2013; Lam and Keeney 2015; Singhal et al. 2015).
Despite the ability of LD-based inference to quantify fine-scale variation using a small number of genomes, the method has certain limitations. It infers the population recombination rate (rho), the product of the effective population size (Ne), and the recombination rate, rather than estimating the recombination rate directly. This has 2 major implications for the use of LD-based inference: (1) it does not distinguish between crossing over in male and female meioses and (2) it is affected by forces that change Ne and not the recombination rate itself. The consequences of the latter are that forces that modify Ne, such as selection and fluctuating population sizes, can confound estimates of recombination (O’Reilly et al. 2008). Even if recent methods account for demography (Spence and Song 2019), estimates of recombination based on LD often need to be validated with a different method, such as linkage mapping (McVean et al. 2004; Axelsson et al. 2012; Shanfelter et al. 2019; Wall et al. 2022). While a combination of approaches is the preferred route to accurately infer the recombination rates, this has rarely been used in practice beyond in model species.
Birds have been the target of multiple evolutionary studies on speciation, hybridization, phenotypic microevolution, and biogeography (e.g. Poelstra et al. 2014; Vijay et al. 2016; Bosse et al. 2017; Enbody et al. 2023) but we still know very little about recombination variation in this class. Most of the current information on recombination in birds is from linkage mapping and cytological studies, which show that recombination exhibits broad-scale among-species variation and inconclusive patterns of sex differences (Groenen et al. 2009; Backström et al. 2010; Kawakami et al. 2014; van Oers et al. 2014; Malinovskaya et al. 2018, 2020; Hagen et al. 2020; Peñalba et al. 2020; Robledo-Ruiz et al. 2022; McAuley et al. 2024; Tan et al. 2024). Rates of recombination inferred from linkage mapping tend to differ between species, despite a rather conserved avian karyotype (Ellegren 2010; Bravo et al. 2021; Waters et al. 2021). For example, 2 members of the passerine order [collared flycatcher (Ficedula albicollis) and the superb fairy-wren (Malurus cyaneus)] show a 2-fold difference in genetic length for their largest syntenic chromosome despite a similar physical length (Kawakami et al. 2014; Peñalba et al. 2020). Further, birds show no consistent patterns of sex differences in recombination with the direction and strength of heterochiasmy varying between species (Malinovskaya et al. 2020; Sardell and Kirkpatrick 2020; McAuley et al. 2024; Tan et al. 2024). However, until recently, conclusions on heterochiasmy were only based on the total recombination frequency (genetic length) summed over all chromosomes in each sex. Recent studies have found evidence for differences in the placement of COs between sexes suggesting that heterochiasmy could be more subtle (McAuley et al. 2024; Tan et al. 2024; Zhang et al. 2024). Considering that the reasons behind heterochiasmy are not well understood, and that multiple adaptive hypotheses have been proposed (Lenormand and Dutheil 2005; Mank 2009; Brandvain and Coop 2012; Sardell and Kirkpatrick 2020), the avian class provides fertile ground for research on the forces driving heterochiasmy. Finally, since all avian species studied so far—with the exception of chickens (Gallus gallus)—belong to the passerine order, the sampled diversity may not be truly representative of the complete avian class.
Here, using both linkage mapping and LD information, we present the first recombination landscape for a species of the Strigiformes order, the barn owl (Tyto alba). This species has a high-quality genome assembly (Ducrest et al. 2020; Machado, Cumer, et al. 2022), a set of whole genome sequences available from previous studies (Cumer, Machado, Dumont, et al. 2022; Cumer, Machado, Siverio, et al. 2022; Machado, Cumer, et al. 2022; Machado, Topaloudis, et al. 2022; Cumer et al. 2024), and a long-term pedigreed population (Roulin 1999) with an untapped genomic potential (Charmantier et al. 2014; Sheldon et al. 2022). We capitalize on 176 genomes previously published, along with 326 newly sequenced individuals to build a high-confidence variant set of polymorphisms spanning the diversity of the species across the Western Palearctic. For recombination inference, we use linkage mapping on a subset of our data set, 250 owls belonging to 28 families, to (1) identify linkage groups (LGs) in the barn owl sequence assembly, (2) estimate the sex-averaged linkage map length, and (3) quantify sex differences in recombination. Additionally, we employ an LD-based approach on 102 unrelated individuals from 3 populations to infer fine-scale recombination rate variation and scale our results using the estimates from the linkage map. With these complementary resources, we quantify variation in recombination between sexes as well as identify substantial differences in fine-scale patterns among chromosomes and populations.
Methods
Samples and sequencing
A total of 333 barn owl samples from Switzerland were sequenced for this study. In Western Switzerland, a subset of breeding barn owls has been monitored for over 30 years. During the breeding season, installed nest boxes are controlled for occupancy every 4 weeks. Individuals are ringed and measured, and a blood sample is taken to sex and genotype each individual (Py et al. 2006; Roulin et al. 2007; Antoniazza et al. 2010). Adult parents are also captured and sampled when possible. Given that barn owls rarely show extrapair paternity (Roulin et al. 2004), the unique ring identifiers of parents and offspring can be used to construct an observational pedigree.
Based on pedigree information, 285 individuals belonging to families between 1994 and 2020 were sequenced in 2020 and 2021. Families with more than 4 offspring and grandparent information were prioritized whenever possible. Sample DNA was extracted from blood using DNeasy Blood & Tissue Kit (QIAGEN) following manufacturer's instructions, quantified with dsDNA HS Qubit Kit (Thermo Fisher), and diluted to 6.3 ng/μL with 10 mM Tris-HCl, pH 8.0, in 40 μL. Libraries were prepared with Nextera DNA Flex (Illumina) and sequenced in Illumina HiSeq 4000 at the Lausanne Genomic Technologies Facility (GTF) giving an average depth of 12X (8–37X). This initial data set was increased using 6 samples from Georgia and 48 from Switzerland. The additional samples from Switzerland are from the same long-term study population, chosen so that they had the maximum number of descendants based on the field pedigree. These samples were sequenced in 2021. Sample preparation was as above, and sequencing was performed using Illumina NovaSeq 6000 with a produced depth of 30X (22–40X). All sequencing took place at the Lausanne GTF (University of Lausanne, Switzerland).
Variant discovery and filtering
All available barn owl sequences were used for variant discovery. This included individuals mentioned above and samples from previous sequencing efforts (Cumer, Machado, Dumont, et al. 2022; Cumer, Machado, Siverio, et al. 2022; Machado, Cumer, et al. 2022; Machado, Topaloudis, et al. 2022; Cumer et al. 2024) along with 6 samples from Georgia and 3 from the island of Corsica (Supplementary File 2). In total, 502 samples were processed through the variant discovery pipeline described below. Raw reads were processed with Trimmomatic v0.39 (Bolger et al. 2014). Sequence adapters were removed, and reads with a length less than 70 bp were excluded. Mapping was performed with BWA-MEM v0.7.17 (Li 2013) on the barn owl genome assembly v.4.0 (https://www.ncbi.nlm.nih.gov/nuccore/JAEUGV000000000) (Machado, Cumer, et al. 2022), and read groups were added with samtools v1.15.1 (Li et al. 2009). Since the GATK v4.2.6 (Auwera et al. 2013) pipeline was used for variant discovery, base quality score recalibration was performed using a previously published variant “truth set” (Cumer, Machado, Dumont, et al. 2022). GATK's Haplotype caller was run with default parameters for each individual separately to generate individual gvcf files. These files were merged, and joint calling was performed with all individuals together using GenotypeGVCFs. We initially identified 30,620,917 variants in the data set. Filtering focused on biallelic SNPs and consisted of the core technical filters suggested in the GATK pipeline, a “mappability” mask and a manual individual depth filtering. Specifically, technical filters included the following criteria: QD < 2.0, QUAL < 30, SOR > 3.0, FS > 60.0, MQ < 40.0, MQRankSum < −12.5, and ReadPosRankSum < −8.0. A further filtering was the exclusion of regions of the genome where our ability to confidently map reads is limited (i.e. a “mappability” mask) (Corval et al. 2023). Briefly, the reference genome was split into reads of 150 base pairs (bp) with a sliding of 1 bp. These artificial reads were mapped back to the reference using BWA-MEM v0.7.17. Regions of the reference sequence where less than 90% of the reads mapped perfectly and uniquely were discarded. This step masked 118 Mb of sequence that corresponds to 10% of the assembly, half of which belongs to scaffolds that did not make it into the linkage map, probably representing misassembled repeats. Variants were also filtered based on individual depth. A minimum and a maximum cutoff were applied. For the minimum cutoff, any genotype with less than 5 reads supporting it was set to missing (Benjelloun et al. 2019). For the maximum, a distribution of autosomal read depth per individual was extracted for a sample region (Super-Scaffold_1 and Super-Scaffold_2) with a length of 133.5 Mb. The mean and SD of depth was estimated, and any genotype with a read depth of more than 3 SD from the mean was set to missing to avoid the effect of repeated regions. After filtering, 26,933,469 variants were kept in 1,080 Mb of callable sequence, corresponding to 1 SNP per 40 bp. This is the raw variant data set used in all downstream analyses. Analysis-specific filters and number of SNPs used can be found below and in Supplementary Table 1 in Supplementary File 1.
Pedigree and relatedness
The pedigree from observational data was confirmed with genomic information from a subset of the genome. SNPs from a sample subset of the genome, specifically 3 scaffolds (Super-Scaffold_11, 12, and 14), were filtered for minor allele count (>5) and missing data (<10%) and were pruned for LD using plink v.1.9 (Chang et al. 2015) with the command --indep-pairwise 100 10 0.1. This filtering created a data set with 91,874 SNPs. A genomic kinship matrix was calculated using the Weir and Goudet (2017) method as implemented in hierfstat v0.5-11 (Goudet 2005) R package. The kinship from genomic data was compared with the pedigree kinship, calculated using the kinship2 v1.9.6 (Sinnwell et al. 2014) R package, and the pedigree was completed by manually fixing the first- and second-degree links when those could be resolved, for example identifying full siblings and parent–offspring links. Both k1 and k2 statistics, the probabilities of sharing 1 or 2 alleles, respectively, were calculated with the SNPRelate v.1.34.1 (Zheng et al. 2012) R library and were used to discern between relationships with the same kinship value (e.g. parent–offspring and full siblings). A set of unrelated individuals was selected automatically by pruning the genomic kinship table to only include individuals with a kinship of less than 0.03125. When selecting for unrelated individuals, we removed individuals with multiple high-kinship links and then we prioritized individuals with higher depth of coverage. This method left a subset of 187 unrelated individuals of which 76 were from the Swiss population.
Linkage mapping
Lep-MAP3 v0.2 (LM3) (Rastas 2017) was used to create a linkage map. A set of 250 individuals in 28 families was used, where a family in LM3 is defined as a set of individuals around a unique mating pair. Most families had 4 offspring and 4 grandparents but numbers differed, ranging from 2 to 8 offspring and from 0 to 4 grandparents. To run LM3, a stringently filtered data set of biallelic SNPs was used. Specifically, we removed Mendelian incompatibilities using bcftools’ mendelian plugin (Danecek et al. 2021) and retained a minimum of 5% MAF and a maximum of 5% missing data per site. We also filtered out SNPs that were less than 1,000 bp (1 kb) apart using VCFtools v.0.1.16 (Danecek et al. 2011). The first step of the LM3 pipeline ParentCall was used to transform the data into the appropriate LM3 format, and the options halfSibs and removeNonInformative were included. Data were filtered in LM3 using the Filtering2 command, to shrink the size of the data set. Specifically, dataTolerance was set to 0.01 as suggested by the author, and missingLimit and familyInformativeLimit were set to 28. This meant that only variants that were nonmissing and informative in all families were kept. After filtering the data set, we retained 163,950 variants. We used SeparateChromosomes to identify the putative LGs based on a user-defined logarithm of odds (LOD) score cutoff. We selected a LOD score of 15 (for a justification see Supplementary Text 1 and Supplementary Fig. 1 in Supplementary File 1). Finally, OrderMarkers with the usePhysical option was executed. Ordering was repeated 3 times, and the output with the best Likelihood was selected for each LG. All 3 runs were compared to test for variation in estimated genetic maps.
After confirming that certain Super-Scaffolds correspond to the LGs, we used a larger set of variants to build the linkage map from. Specifically, we filtered on 10% missingness and 10% minor allele frequency, instead of the 5% filter used before. For all SNPs in each LG, we used ParentCall and Filtering2 as before but ran SeparateChromosomes on each LG with a LOD cutoff of 5. Finally, we ran OrderMarkers by enforcing the physical map of the genome assembly and estimating the genetic map without de novo ordering by using the parameters evaluateOrder and improveOrder=0. This allowed us to use most variants on the scaffold (a total of 4,889,667 SNPs) and detect possible missing COs not identified by the previously filtered data set used in de novo ordering. A comparison of the 2 runs of Lep-MAP3, 1 with de novo marker ordering and 1 using the physical order, can be found in Supplementary Fig. 2 in Supplementary File 1.
In linkage mapping, certain markers might be erroneously mapped especially at the extremities of LGs. Thus, all markers with a separation larger than an arbitrary threshold of 2 cM in a region of 100 markers concentrated toward the ends of LGs were filtered out using a custom script adapted from LepWrap (Dimens 2022).
LD recombination
To run SMC++ v1.15.2 (Terhorst et al. 2017), we followed the authors’ instructions as presented on the software's GitHub page (https://github.com/popgenmethods/smcpp). In summary, SNPs were filtered on 10% missingness, HWE (using a Fisher's exact test P-value cutoff of 0.05) and 5% minor allele frequency using VCFtools v.0.1.16. In addition, missing data were recoded using plink v1.9 (Chang et al. 2015), and the 5 samples with the highest coverage were selected as individuals to be provided to SMC++. The command vcf2smc was run for each of these 5 individuals. When executing “vcf2smc,” the mappability mask was excluded by using the -m option. The model was estimated using all output files from the previous step and with a mutation rate of 1.93e−9 estimated from family data of a snowy owl Bubo scandiacus (Bergeron et al. 2023). The csv-formatted estimate of piecewise-constant Ne in past generation intervals was used in subsequent pyrho (Spence and Song 2019) analyses.
We ran pyrho v0.1.7 with an unphased set of markers for 76 unrelated Swiss individuals. We used the data set of SNPs we used in SMC++, but we further filtered variants to be at least 10 bp apart using VCFTools v.0.1.16. The first step in the pyrho implementation was the precalculation of a 2-locus likelihood look-up table. This step takes into account the Ne estimates from SMC++ (Supplementary Fig. 3 in Supplementary File 1). For the Swiss samples, the number of diploid individuals was 76 and we used the Moran approximation parameter with a size of 200. The “hyperparameter” command was run to estimate metrics on the performance of different window sizes and block penalties. The authors’ guidelines were followed on how to select the best combination of parameters. Briefly, we summed the Pearson correlation statistics outputted by pyrho and plotted their total sum against the L2 values. The authors suggest (https://github.com/popgenmethods/pyrho#hyperparam) that depending on the implementation, one might opt to choose the parameter combination that maximizes the correlation measures or minimizes L2. In our case, both conditions were satisfied with one combination of parameters and we run pyrho with that set of parameters. A table of the hyperparameter values for all populations can be found in Supplementary Table 2 in Supplementary File 1. With the inferred hyperparameters, the recombination rate was estimated using the “optimize” command on vcfs containing individual scaffolds that were previously filtered for singletons and a minimum distance of 10 bp between variants as in Wall et al. (2022).
Downstream analyses
Putative centromeres were identified using a repeat annotation through a combination of 2 software, RepeatObserver v1 (Elphinstone et al. 2023) and TRASH v1.2 (Wlodzimierz et al. 2023), run with default parameters on the barn owl assembly v.4.0 (Machado, Cumer, et al. 2022) (more details in Supplementary Text 2 in Supplementary File 1). After identifying LGs, synteny with the chicken reference genome was inferred with the method presented in Waters et al. (2021). Synteny matches can be found in Supplementary Fig. 4 and Table 3 in Supplementary File 1. The rate of intrachromosomal shuffling was calculated from the genetic map distances following Supplementary information S5 in Veller et al. (2019). For every chromosome, there is a set of sequenced markers A = (a1, a2,…, an) and a cumulative genetic distance up to each marker (in cM) of Da = Σi<adi, but these markers are not placed at even distance intervals. Thus, we created a set of equidistant (10 kb) pseudomarkers B, starting from the beginning and including the end of the sequence, and calculated their centimorgan position by using adjacent true markers. For every b marker, the distance (Db) is:
where sa(b) is the genomic position of the nearest marker below marker b, sb the genomic position of marker b, and sa(b) + 1 is the genomic position of the nearest marker above b. Similarly Da(b) and Da(b) + 1 are the cumulative genetic positions of the nearest true markers below and above pseudomarker b, respectively. Then, we averaged all pairwise distances of the pseudomarkers, weighing the result by the proportion of length belonging to each LG (L2). The end value corresponds to the total shuffling attributable to each chromosome (intra).
Recombination rate estimates from pyrho were averaged across nonoverlapping windows of different lengths using a custom script. Windows of sizes 1 kb, 10 kb, 100 kb, and 1 Mb were created from the reference sequence using bedtools makewindows from bedtools v2.3 (Quinlan 2014). These windows were overlapped with the pyrho windows, and the recombination rate in centimorgans was calculated by multiplying the recombination probability estimate with the length of each interval and then translating this to centimorgans using Haldane's function (Haldane 1919). For each window, nucleotide diversity was calculated using VCFtools and the --window-pi command. Estimates were corrected for masked nucleotides in each window. Sequence GC content was calculated using the reference sequence and the bedtools nuc command. We annotated CGIs using the UCSC genome browser CGI annotation tool cpg_hl (Kent et al. 2002) with default parameters. The Gini coefficient was calculated using Desctools v.0.99 (Signorell 2023). Transcription start and end sites were annotated using the genome annotation from NCBI as the first and last positions of the genomic sequence for each gene. The intersection of different bed files was performed using bedtools. Local hotspots were annotated by dividing the estimate of recombination rate in each focal window with the average recombination in 80 kb around (40 kb upstream and 40 kb downstream). In addition, because power to infer hotspots is lower in very low and high recombination rates (Singhal et al. 2015), we restricted hotspots to windows between 1 and 10 cM/Mb. Lastly, we removed any hotspots found in windows with more than 500 bp annotated as repeats. Global hotspots were annotated as windows with at least 10 times the genome-average recombination rate.
Owl images are from PhyloPic (https://www.phylopic.org). Map was made using tmap v3.3-4 (Tennekes 2018) and the Natural Earth high-resolution data set. Corrplot v.0.92 was used for correlation (Wei and Simko 2021). Vioplot v0.4.0 was used to create the violin plots (Adler et al. 2022). All analyses were executed in R v.4.3.1 (R Core Team 2023) using the Rstudio IDE (Posit team 2022). Scripts with commands used for data generation and downstream analyses can be found in https://github.com/topalw/Recombination_barn_owl.
Results
In order to build the most comprehensive set of polymorphism to date in the barn owl, we performed variant identification on a total of 502 whole genome sequences of medium to high coverage (mean 16X, range = 8X–43X). Samples originated from 19 distinct localities spanning the Western Palearctic distribution of the species (with 3–13 samples from 19 localities, see Supplementary File 2 for details). The Swiss population in particular included 346 individuals with a family structure originating from an observational pedigree. After filtering, we retained 26,933,469 single nucleotide variants (SNPs) and used subsets of those individuals and variants for each analysis below (Supplementary Table 1 in Supplementary File 1).
LGs and recombination rate of the barn owl
After additional filtering on the variant set for technical errors, allele frequency, distance, and missingness (see Methods), we ordered 154,706 SNPs along the 41 largest scaffolds of the barn owl genome assembly v.4.0 (Machado, Cumer, et al. 2022) to create a linkage map for the species. Based on segregation of these markers in 350 meioses of 250 individuals from 28 families, we identified 40 LGs spanning 1,196.47 million base pairs (Mb) covering 95.7% of the genome assembly. All the LGs we identified correspond to scaffolds in the genome assembly, except for Super-Scaffold 2, which was split into 2 LGs (see Supplementary Text 1 and Table 3 in Supplementary File 1). In addition, we merged Super-Scaffold_3 and Super-Scaffold_49 into LG 20 and Super-Scaffold_13 and Super-Scaffold_42 into LG 40, corresponding to the sex chromosome Z. The genome assembly of the barn owl therefore contains the sequence of 39 LGs out of 45 expected pairs of autosomal chromosomes and 1 LG corresponding to the Z chromosome. Of the 39 autosomal LGs identified, 17 correspond to microchromosomes—chromosomes with a size less than 24 Mb (Waters et al. 2021). After we were confident with the correspondence between scaffolds and LGs, we inferred the genetic map positions of a set of 4,889,667 SNPs, which were filtered with less stringent parameters, with an average inter-SNP nucleotide distance of 266 bp. This way we inferred the genetic map for each LG independently by capturing COs over most of the assembled sequence. We observed 3,972 male COs and 4,250 female COs, which correspond to 23 COs per meiosis on average. For the 39 autosomal LGs, the final sex-averaged linkage map length spanned 2,580 cM resulting in an autosomal genome-wide average estimate of recombination rate of ∼2.33 cM/Mb for this species.
The genetic length of LGs increased slightly with their physical length (Fig. 1a). As a result of the obligate CO, the genetic map of a chromosome is expected to be at least 50 cM long. The LGs of the barn owl showed an average genetic length of 66 cM (range: 35–97 cM), and all LGs recombined on average less than twice per meiosis (<100 cM). The slope of the regression of the genetic length on the physical length was significantly positive [β = 0.378, 95% confidence interval (CI): 0.18–0.57], and the intercept was not different from the expected minimum of 50 cM (α = 55, 95% CI: 48–62) under 1 obligate CO per chromosome. Three LGs around 30 Mb of physical length had an inferred genetic length less than 50 cM (LG13: 40 cM, LG18: 49.2 cM, and LG31: 34.8 cM, Supplementary Table 3 in Supplementary File 1).
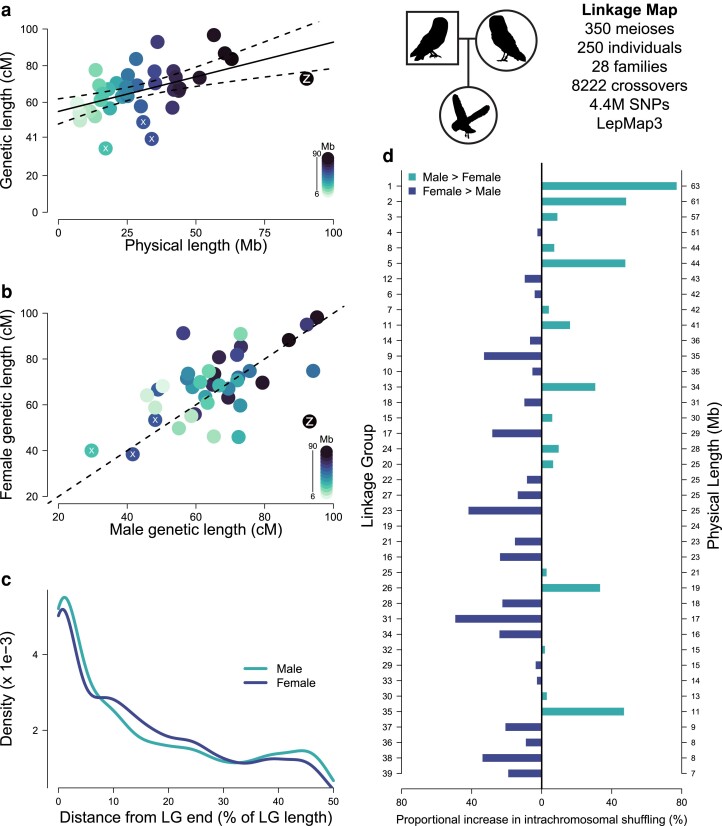
Fig. 1.
Linkage map for the barn owl (T. alba). All plots in this figure are products of the linkage map data set consisting of 350 meioses of 250 individuals in 28 families, illustrated with the pedigree of owl symbols on the top right. a) Estimates of sex-averaged genetic lengths for the LGs plotted against their physical length. Regression line is shown with 95% CIs (α = 55, β = 0.378, t = 3.73, P < 0.001). Color intensity scales with LG physical lengths. The Z LG is marked with a Z, and LGs with less than 50 cM are marked with a white X. b) Female map length (cM) of LGs against the male map length. Dashed line is the identity (y = x) line. c) Density plot of male (aquamarine) and female (blue) CO counts plotted along the distance from the LGs’ end. X-axis is in percentages of total LG sequence. Density values are scaled so that they sum to 1. d) Differences between sexes in rates of intrachromosomal shuffling (intra) presented as a ratio (male intra/female intra) for different LGs. Bars to the right of the black line colored green signify higher intrachromosomal shuffling in males, and bars to the left of the black line colored in blue correspond to LGs with higher shuffling in females. LGs are ordered by decreasing physical length as on the right-hand axis.
We found that the Z chromosome is the largest chromosome, with an assembled physical length of 90.3 Mb (Fig. 1a; Supplementary Fig. 5, final panel, and Table 3 in Supplementary File 1). The identified pseudoautosomal region (PAR) spans 4.4 Mb at the end of the LG (Supplementary Fig. 5 in Supplementary File 1). Males, with 2 copies of the Z chromosome, had a genetic length of 69.28 cM (approximately 10 cM in the PAR), which corresponds to a rate of approximately 0.77 cM/Mb (Supplementary Table 3 in Supplementary File 1). Females, having a single copy of the Z chromosome, harbored 53 cM just in the PAR (Supplementary Table 3 and Fig. 5 in Supplementary File 1).
Subtle heterochiasmy
To infer heterochiasmy, we compared the sex-specific linkage map estimates (Supplementary Fig. 5 and Table 3 in Supplementary File 1). Females, with a map length of 2,672 cM, had a 4.7% larger genetic map than males (2,560 cM). There appeared to be no consistent pattern of heterochiasmy among LGs at the chromosome scale (Fig. 1b). To investigate potential differences in localization of recombination events in males and females, we looked at the positioning of COs along the length of all LGs (Fig. 1c; Supplementary Figs. 5 and 6 in Supplementary File 1). Overall, COs occurred closer to the LG extremities than in their center. However, the distribution of COs differed between the sexes (two-sample Kolmogorov–Smirnov test D = 0.30; P < 0.001). Notably, males appeared to recombine more at the extremities and the middle of the LGs since their COs were less evenly distributed.
To test if pericentromeric rates differ between males and females, we attempted to annotate centromeres in silico in the barn owl assembly. Annotated tandem repeats were often concentrated at the ends of chromosomes as expected by the presence of a centromere in acrocentric chromosomes. We retained only annotated centromeres in 18 LGs, where the centromere was inferred in the distal 20% of the sequence. However, most annotated centromeres overlapped recombination peaks, and we were not fully confident in the end result to draw any conclusions (Supplementary Text 2 and Figs. 7 and 8 in Supplementary File 1).
Because sexes showed different locations of COs and not all COs are as effective at shuffling alleles between haplotypes, we quantified the rate of intrachromosomal shuffling (intra) as defined in Veller et al. (2019). Briefly, this quantity measures the relative shuffling of alleles due to a CO along the length of the chromosome. For example, a CO in the middle of the chromosome shuffles more alleles than a distal one. We estimated rates of intrachromosomal shuffling in males and females (Fig. 1d). Despite an overall lower recombination frequency (Fig. 1b), males showed up to 50% higher intrachromosomal shuffling for larger LGs (Fig. 1d). On the other hand, females showed higher rates of shuffling in intermediate to smaller LGs.
Fine-scale variation among LGs
To investigate fine-scale variation in recombination rates, we turned to recombination rates estimated from patterns of LD (Supplementary Fig. 9 in Supplementary File 1). We estimated recombination rates using pyrho v0.1.7 (Spence and Song 2019) in a set of 5.8 million variants (number of variants per population is presented in Supplementary Table 1 in Supplementary File 1) identified along the whole genome of 76 unrelated birds from Switzerland (CH). The total genetic length for the autosomal part of the assembly was estimated from LD to be ∼3,100 cM, 25% larger than the linkage map estimate for the same population. We scaled the total length inferred from LD to be the same as the linkage mapping estimate for each scaffold separately, to account for the confounding effect of Ne and compared the estimates in non-overlapping 1 Mb windows. The correlation of recombination rate estimated from the linkage map and from pyrho at the 1 Mb scale was high (r = 0.83, 95% CI: 0.813–0.85), but pyrho showed larger estimates in regions of low recombination compared to linkage mapping (Fig. 2a).
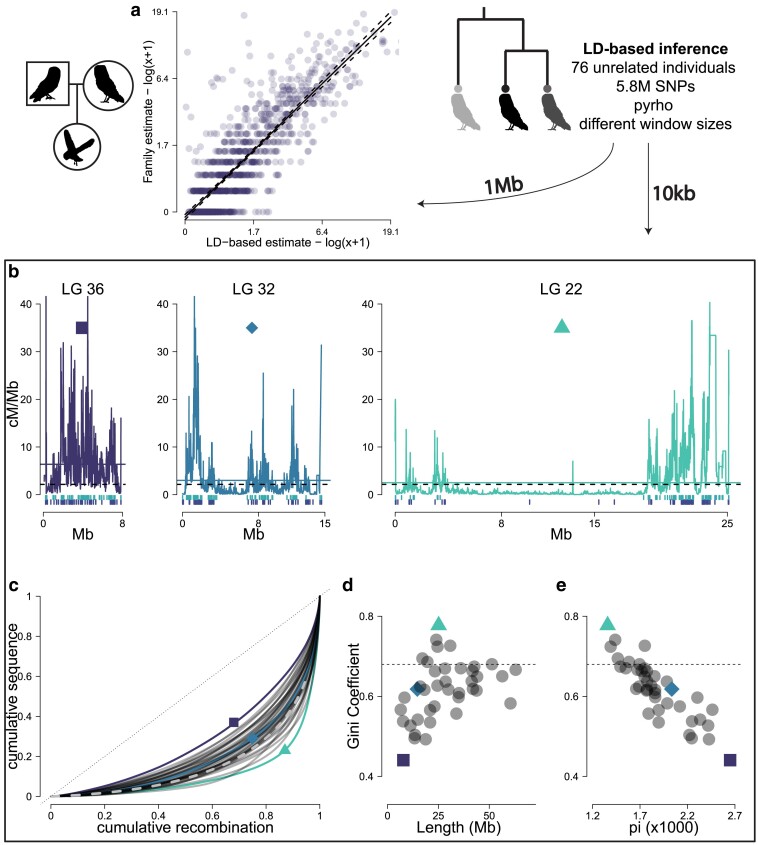
Fig. 2.
Variation of recombination among LGs. a) Comparison of recombination rate estimates from linkage mapping and LD inference for the Swiss population. The comparison is made in 1 Mb windows. Regression line is shown with prediction intervals as dashed lines (α = −0.08, β = 0.999, t = 53.15, P < 0.001). b) The recombination frequency (cM/Mb) in 10 kb windows along the physical map of 3 example LGs: LG 36 (purple square), LG 32 (aquamarine diamond), and LG 22 (green triangle), respectively. The dashed horizontal line represents the genome-average recombination rate and the full line the LG average recombination rate. These LGs represent LGs with different recombination landscapes. Vertical lines below indicate locations of male (top row) and female (bottom row) COs from the family estimates. c) Cumulative sequence plotted against cumulative ordered recombination length for each LG (dark gray curves) and genome-wide (gray dashed curve). The black dotted line is the identity (y = x) line. The Gini coefficient corresponds to the area delimited by each curve and the identity line. d) The Gini coefficient of recombination rates for each LG plotted against its physical length. Dashed gray line is the genome-average Gini coefficient. e) The Gini coefficient of recombination plotted against the average nucleotide diversity of each LG.
The recombination landscape differed among chromosomes. To further quantify this variation in recombination rates, we calculated the proportion of genomic sequence where recombination occurs. We ordered all 10 kb windows for each LG by decreasing recombination rate and quantified the cumulative recombination percentage against the cumulative percentage of sequence (Fig. 2c). Overall, 80% of recombination occurs in approximately 30% of the sequence (dashed gray line in Fig. 2c). However, there was substantial variation in the distribution of recombination among LGs. To further measure this skewness, we used the Gini coefficient of recombination rates for each chromosome, which is a measure of inequality among the values of a frequency distribution. Graphically, this corresponds to the area between each curve in Fig. 2c and the identity (y = x) line and ranges from 0 to 1. Smaller values indicate an evenly spread landscape (every window has the same recombination rate) and higher values a more variable one (windows show large differences in recombination rates). The genome-wide average Gini coefficient was 0.67, and the LG-specific estimates varied between 0.44 and 0.77 (LG 36 and LG 22, marked with a blue square and a green triangle, respectively, in Fig. 2). Along with a LG of an intermediate Gini coefficient of 0.61, LG 32 (aquamarine diamond in Fig. 2), their recombination landscapes are presented in Fig. 2b. We found that the Gini coefficient depended on the physical length of the LG, with more evenly spread (and elevated) recombination rates in smaller LGs and more concentrated landscapes in larger ones (Fig. 2d), although the effect reached a plateau as the length increased above 25 Mb. Further, the Gini coefficients were strongly negatively correlated with the average nucleotide diversity of the LG, with more concentrated recombination peaks associated with lower average nucleotide diversity (Pearson's r = −0.87, 95% CI: −0.92, −0.76) (Fig. 2e). Overall, recombination rates varied substantially among the different LGs of the barn owl assembly.
Correlates of recombination
To identify genomic correlates of recombination rates, we annotated 10 kb windows with more than 5 times the genome-wide average recombination rate as recombination “hotspots” windows (Fig. 3a). We identified 3,805 such windows containing 30% of the total genetic length. We also annotated 4,677 windows with less than 5 times the average recombination rate, i.e. recombination “coldspots.” We compared both hotspot and coldspot with a set of 4,000 randomly chosen windows that were not annotated as either hotspots or coldspots. An example of hotspots and coldspots across a particular LG is shown in Fig. 3b. We found that windows with high recombination rates were most often identified at the ends of LGs, had higher GC content and nucleotide diversity, and colocalized with genes (Fig. 3c–f). On the contrary, coldspots were found in the middle of the LGs and were depleted in GC content, nucleotide diversity, and genes.
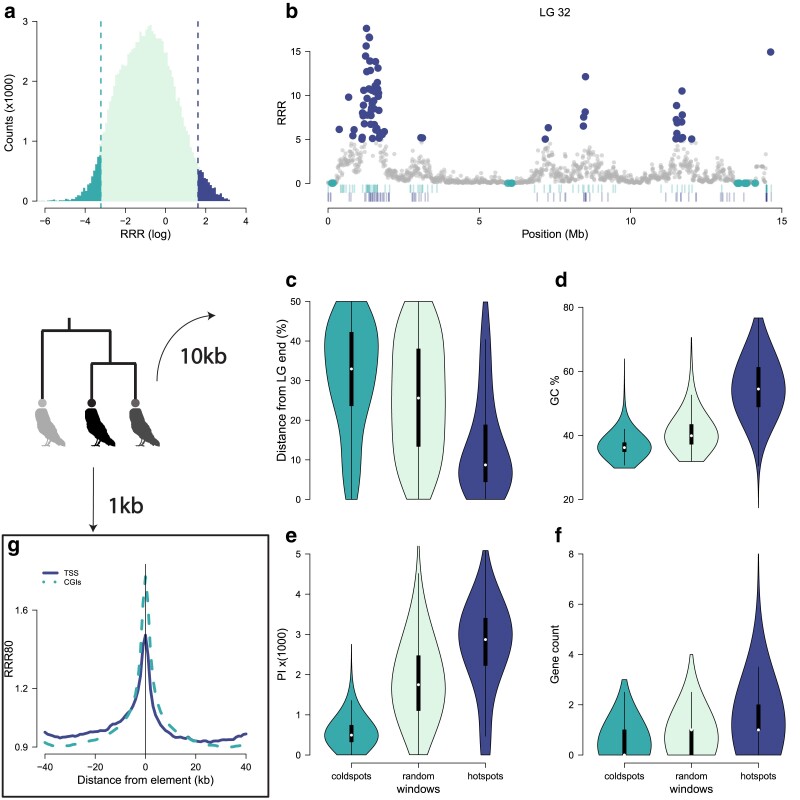
Fig. 3.
Correlates of recombination. a) The distribution of recombination rates in 10 kb windows. We define hotspots as windows with more than 5 times the genome-average recombination rate and coldspots as windows with a recombination rate less than a fifth of the average. b) Example of annotated coldspots and hotspots in LG 32. Vertical lines below are locations of male (top row) and female (bottom row) COs from the family estimates. Violin plots of characteristics of hotspots, coldspots, and a similar sized set of random windows that belong in neither category. c) Position along LG. d) GC content. e) Nucleotide diversity. f) Number of annotated genes. g) Recombination is increased around annotated TSSs (full line) as well as CGIs (dashed line). RRR80 is the recombination rate of each 1 kb window divided by the average in 80 kb around. The lines show the average across all identified elements.
In addition, because birds lack the PRDM9 gene, recombination hotspots are expected to localize to TSSs, as well as CGIs (Singhal et al. 2015; Baker et al. 2017). To verify this, we used estimates of recombination frequency in non-overlapping windows of 1,000 bp (1 kb) along the genome. Windows that were annotated to contain a TSS (n = 17,191, 1.5% of windows) or contained a CGI spanning the whole window (n = 13,971, 1.3% of windows) were identified, and their recombination rate was divided by the average recombination rate in 40 kb upstream and 40 kb downstream of the focal window (relative recombination rate in 80 kb—RRR80). The focal windows showed elevated recombination rates compared to other windows in their vicinity (Fig. 3g).
Recombination landscapes across populations
To quantify the change of the recombination landscape across European populations of the species, we used 3 populations from the Western Palearctic: Portugal (PT, n = 13), Great Britain (GB, n = 13), and Switzerland (CH, n = 76) (Fig. 4a). For Portugal, we pooled together 3 samples from Morocco and 10 from Portugal since they have very high genetic similarity (Cumer, Machado, Dumont, et al. 2022). Because the sample size in Switzerland was far larger than the other 2 populations and to test the robustness of our results, we randomly subsampled 5 sets of 13 individuals from Switzerland, creating pseudoreplicate populations. Genetic length estimates differed between populations. Portugal showed a 5-fold increase in genetic length while GB showed a 2-fold increase compared to the family linkage map. The Swiss subset data sets underestimated the total genetic length up to 2.4 times. For all populations, we scaled the results so that each scaffold would match the corresponding linkage map estimate. After scaling, we compared all populations with the linkage map in 1 Mb windows along the genome and found that genome-average correlations were higher than 0.69 (Supplementary Fig. 10 in Supplementary File 1). At a finer scale, however, recombination landscapes varied among populations (an example landscape for all populations is presented at the 1 kb scale for the first 5 Mb of LG 32 in Fig. 4b; all landscapes in Supplementary Fig. 11 in Supplementary File 1). Estimates in GB showed reduced resolution at the finer scales (aquamarine line in Fig. 4b), likely due to reduced genomic diversity and historically smaller Ne in the population (Supplementary Table 1 and Fig. 3 in Supplementary File 1; see also Machado, Cumer, et al. 2022).
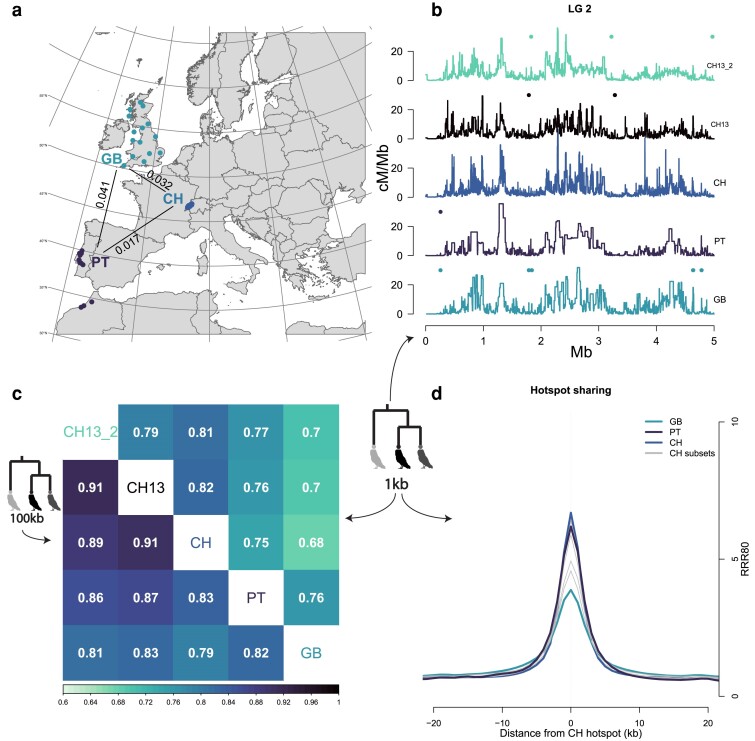
Fig. 4.
Comparison of recombination landscapes between populations. a) Map of sampled populations. Numbers correspond to genome-wide pairwise FST values (Machado, Cumer, et al. 2022). b) Example of recombination landscape for 1 kb windows in all data sets for the first 5 Mb of LG 2. Points above each plot show local hotspots for each population. Local hotspots are identified as at least two consecutive windows with a recombination rate of at least five times the average in a region of 80kb around them (RRR80 > 5). c) Correlation matrix of recombination rates for 1 kb windows (above diagonal) or 100 kb windows (below diagonal). d) Relative recombination rates in 80 kb around (only 40 kb plotted) 1 kb windows annotated as hotspots in the full Swiss data set. Each line is the relative recombination rate in all other populations. Gray lines are the subsets from Switzerland with n = 13. CH: full Swiss data set (n = 76); PT: Portuguese data set (n = 13); GB: Great Britain data set (n = 13); CH13: undersampled first Swiss data set (n = 13); CH13_2: undersampled second Swiss data set (n = 13).
We quantified the divergence of the recombination landscapes as the correlation of recombination rates on different window sizes (1 and 100 kb) among all pairs of populations (Fig. 4c). The recombination landscapes between populations were concordant at the broad scale with the correlation steadily decreasing as the window size got smaller. The subsampled sets from Switzerland showed that correlations between CH and PT were slightly lower than between CH and the subsets. It also showed that broad-scale correlations in GB were not substantially no different between the CH subsets and PT while the fine scale (1 kb) was not as well correlated.
Figure 2b and c provide evidence for the existence of fine-scale hotspots in the barn owl recombination landscape. Thus, we looked for at least 2 consecutive 1 kb windows with a recombination rate higher than 5 times the average in 80 kb around them (relative recombination rate in 80 kb—RRR80), following definitions of a local hotspot in the literature (Myers et al. 2005; Singhal et al. 2015; Kawakami et al. 2017), and careful filtering based on recombination rates and repeat content. After filtering, a set of 1,566 windows were identified as local hotspots, harboring 8,171 variants. We then quantified the RRR80 of the other populations on those windows (Fig. 4d). The 5 Swiss subsampled data sets with n = 13 provided a measure of estimation error and showed strong to moderate increase on the hotspot windows. Recombination rates in the Portuguese sample showed a marked increase in the windows, indistinguishable from the Swiss subsampled data sets, while the signal in the samples from Great Britain was outside the range of the subsampled data sets.
Discussion
Recombination is an evolutionary force with direct and indirect implications for reproduction and evolution, and while it varies on different scales, our knowledge and ability to quantify this variation are often limited. In this study, using an extensive whole genome sequencing data set of barn owls, we inferred recombination with 2 different methods to describe broad and fine-scale variation in recombination variation: linkage mapping on a pedigreed population and a LD-based approach on 3 different populations. Using both methods allowed us to identify crossing over in contemporary families and infer haplotype shuffling in the coalescent history of our data set. We first identified 40 LGs in the barn owl genome assembly and estimated the recombination rate to be ∼2.3 cM/Mb. We showed that in the barn owl, the overall length of genetic maps shows no clear distinction between males and females, but sexes had fine-scale differences in CO placement and shuffling proportions. Despite few (1 to 2) COs per chromosome, we found large variation in the location of these COs among different LGs, with some LGs harboring large stretches of reduced recombination. We showed that this variation in the distribution of recombination comes in tandem with variation in the genetic diversity of the different LGs. At a more fine-scale resolution, recombination rates were increased in windows that contain TSSs and CGIs. Equally, recombination hotspots at the 10 kb scale showed an elevated GC ratio, diversity, and gene density and were most often found at the ends of LGs. Lastly, population comparisons showed local recombination hotspot conservation despite high statistical noise. We discuss these results and their implications below.
LGs confirm the near completeness of the barn owl assembly
Complete genome assemblies are a useful resource that requires multiple sources of information. The karyotype of the barn owl contains 45 autosomal pairs with a large Z chromosome and degraded W (Belterman and De Boer 1984; Rebholz et al. 1993; Peona et al. 2018). The latest barn owl assembly v4.0 (Machado, Cumer, et al. 2022) was assembled into superscaffolds using optical genome mapping (BioNano; Lam et al. 2012). In the present study, we verified and improved the barn owl assembly by anchoring the largest 41 scaffolds into 40 LGs and revealed that the genome assembly of the barn owl is of chromosome-level quality (though not telomere to telomere). However, there are still 6 autosomes unaccounted for in the linkage map, and the W chromosome is also not present because the assembly uses a male individual (Ducrest et al. 2020; Machado, Cumer, et al. 2022). These small autosomes might be partially present in the physical assembly, since smaller scaffolds with a few tens of identified markers that passed filtering could not be confidently allocated to LGs. Regardless, the “missing” chromosomes are likely the smallest 6 microchromosomes, or dot chromosomes, notoriously difficult to sequence and assemble due to their high GC content and reduced chromatin accessibility (Burt 2002; Bravo et al. 2021; Waters et al. 2021). Notably, the dot chromosomes were only recently assembled in the chicken genome (Huang et al. 2023) and are missing from most available bird reference genomes (Peona et al. 2018; Baalsrud et al. 2024). It is likely that most microchromosomes will remain elusive until future studies make use of advances in long-read technologies (Marx 2023) to complete the reference genomes of birds. In this endeavor, linkage mapping, when available, can be a valuable tool (e.g. Peñalba et al. 2020; Robledo-Ruiz et al. 2022).
Differential shuffling between sexes due to CO placement
Our results suggest the presence of variation in CO placement between sexes that is not immediately apparent when investigating sex differences at overall genetic map length of each LG. This variation generates a differential shuffling of markers in each sex among different chromosomes with the overall pattern showing that males recombine less evenly than females along the length of the LGs. In turn, this leads to a greater shuffling of markers, as observed in larger chromosomes, where male COs situated more toward the middle of the sequence shuffle most of the variants in the chromosome. In avian studies, results are inconclusive for a general pattern of heterochiasmy in the class. For example, male collared flycatchers and male hihis (Notiomystis cincta) exhibit higher genetic lengths than females and recombine more toward the ends of the chromosomes (Kawakami et al. 2014; Smeds et al. 2016; Tan et al. 2024). On the other hand, sparrows (Passer domesticus) and great tits (Parus major) show higher female recombination (van Oers et al. 2014; McAuley et al. 2024), and other species such as the great reed warbler (Acrocephalus arundinaceus) show no pattern on the broad scale but a clear male bias toward the telomeres (Zhang et al. 2024). Recently, there has been an effort to recharacterize heterochiasmy in CO placement with studies showing that there are heritable fine-scale sex differences in recombination in birds (McAuley et al. 2024; Tan et al. 2024; Zhang et al. 2024). Although fine-scale information on sex differences is not available for other bird species, the emerging pattern is that descriptions of recombination patterns at the LG level might not reveal the whole picture of heterochiasmy in birds and a more thorough quantification of sex-specific recombination is required.
Heterochiasmy is pervasive in eukaryotes (Burt et al. 1991; Sardell and Kirkpatrick 2020). In the most extreme version of sex differences, one sex does not recombine at all (achiasmy), almost always the heterogametic sex [for example XY Drosophila males and ZW females in Lepidoptera or the common silk worm (Bombyx mori); Morgan 1914; Burt et al. 1991; Goldsmith et al. 2005; Jiggins et al. 2005]. For most species, the reality is more nuanced and is independent of which sex is heterogametic. A prominent pattern of heterochiasmy is that males often recombine more toward the telomeres of a chromosome (Kong et al. 2010; Giraut et al. 2011; Johnston et al. 2016; Brekke et al. 2023; Venu et al. 2024), although there are exceptions (Kianian et al. 2018; Rifkin et al. 2022). While the evolutionary reasons behind the existence of heterochiasmy remain unexplained (Burt et al. 1991; Mank 2009; Sardell and Kirkpatrick 2020), hypotheses have been proposed such as differences in haploid selection intensity (Lenormand and Dutheil 2005) or female meiotic drive (Brandvain and Coop 2012). Haploid selection predicts that the sex with more intense selection during the haploid phase (e.g. sperm competition) will show reduced recombination while the meiotic drive hypothesis predicts increased rates of recombination close to the centromeres in females. Barn owls show very low rates of extrapair copulation (Roulin et al. 2004) and therefore low sperm competition, and in birds and mammals, the egg completes meiosis II only after fertilization leaving little room for haploid selection (Mira 1998). Further, the emerging patterns of heterochiasmy, and its frequent absence, do not lend support to the haploid selection theory in birds, which was also not associated with sperm competition intensity in eutherian mammals (Mank 2009). Concerning female meiotic drive, because most avian assemblies lack annotation of centromeres, thus far no studies have been published on this topic in birds. While we attempted to annotate centromeres in our assembly, the lack of assembled repeats (often the last part of a reference to be assembled) and the placement of centromeres at the distal ends of the LGs confounded results (Belterman and De Boer 1984; Rebholz et al. 1993; Benham et al. 2024). Thus, to reach conclusions about heterochiasmy in birds, the use of more complete reference genomes with assembled repeats will be invaluable (Peona et al. 2018; Robledo-Ruiz et al. 2022; Huang et al. 2023).
Heterochiasmy is most apparent on the sex-linked chromosome. Females, the heterogametic sex in birds, recombine exclusively on the PAR of the Z chromosome where sequence homology between the Z and W chromosomes is maintained. Beyond the PAR region, the 2 chromosomes have diverged so drastically in the >100 million years since their evolution from a pair of autosomes, that in karyotypic studies they appear like different elements (i.e. they are heteromorphic) (Belterman and De Boer 1984; Rebholz et al. 1993; Handley et al. 2004). While this is true for most birds (Neognathae), ratite birds (Palaeognathae) maintain homomorphic sex chromosomes with large PAR, which spans 50 Mb in the ostrich (Mank and Ellegren 2007; Yazdi et al. 2023). The variable degradation of sex chromosome pairs has been linked to multiple factors, like age of the sex chromosomes and strength of sexual conflict, in animals and especially in plants that have evolved ZW and XY systems independently multiple times (Bergero and Charlesworth 2009; Charlesworth 2013; Wright et al. 2016; Charlesworth 2019). Future work in the barn owl such as assembling the W chromosome and identifying the sex-determining region and its contents will help shed more light on the place of the Tytonidae on the sex chromosome evolution landscape.
Variation in recombination rate among and within LGs
Our results show that barn owl LGs recombine at most twice per meiosis. This result is in line with an expectation of 1 CO per chromosome (or chromosome arm) and the generally small (<70 Mb) acrocentric (or telocentric) chromosomes in the barn owl karyotype (Belterman and De Boer 1984; Rebholz et al. 1993; Coop and Przeworski 2007). However, these recombination frequencies contrast with results from other bird species. For example, the syntenic chromosome 2 of chickens and flycatchers with an approximate length of 150 Mb shows an average of 6 COs per meiosis (300 cM) (Groenen et al. 2009; Kawakami et al. 2014). The same chromosome has a genetic length of 175 cM in sparrows and 100 cM in great tits and superb fairy-wrens (van Oers et al. 2014; Peñalba et al. 2020; McAuley et al. 2024). The source of this variation in the order is unknown. Reasonable hypotheses include the evolution of the recombination landscape, localized suppression of recombination in some species (for example through segregating structural variations like inversions), or interspecific variation in the strength of CO interference (Kirkpatrick 2010; Otto and Payseur 2019). Higher quality linkage map data on more bird species can help identify the breadth of recombination variation in the class, and a meta-analysis of the available data sets can provide a much needed rigorous comparison.
In the barn owl, some LGs appear to recombine less than once per meiosis (genetic length < 50 cM). However, since the absence of an obligate CO can lead to aneuploidy, which, coupled with the LGs’ intermediate size, should generate severely deleterious consequences, this is unlikely to be the true recombination frequency of these LGs (Hassold et al. 2007). A more likely explanation is that in these chromosomes, the genetic length is less than 50 cM due to missing markers at the distal parts of the chromosome. Because our marker data set used is relatively extensive, this discrepancy likely originates at the assembly stage, where parts of the sequence might have been misassembled or be present in small scaffolds. Another notable outlier in our study is the Z chromosome that in males recombines almost 3 times less than the autosomes. One explanation is a lack of marker coverage, especially for scaffold 13, that might have been filtered out during linkage mapping. On the other hand, the Z chromosome harbors a low-recombination region in the middle and is the only metacentric chromosome in the barn owl karyotype and the reduction could be due to pericentromeric suppression or segregating structural variations that are found on the Z chromosome in other bird species (Knief et al. 2016, 2017; Yazdi and Ellegren 2018).
We find recombination rates to vary substantially between and within chromosomes. As expected from an obligate CO per chromosome, smaller chromosomes tend to have higher rates (per bp) of recombination compared to longer chromosomes. In addition, smaller chromosomes show a more uniform distribution of recombination rates along their length. Longer chromosomes exhibit a U-shaped pattern, with reduced recombination in their center sometimes spanning large parts of the chromosome. This relationship between and within chromosomes has been found in other species (e.g. Backström et al. 2010; Bascón-Cardozo et al. 2024; Castellani et al. 2024), and Haenel et al. (2018), in a meta-analysis of recombination rates of different species, proposed a model where the length of a chromosome and the distance from the telomere are the major factors impacting recombination rates. This model was recently extended by Brazier and Glémin (2022) based on a large data set of plant linkage maps, to include centromeric position and the placement of a single CO per chromosome. Our results follow this general pattern, although most LGs have interspersed signals of reduced recombination that could be due to segregating structural variants and/or CO interference (Kirkpatrick 2010; Otto and Payseur 2019). Regardless, the placement of COs on distal parts of the chromosomes is an important aspect of recombination that can inform different genetic analyses, from calculation of summary statistics to selection inference (Knief et al. 2017; Booker et al. 2020).
In the barn owl, these broad-scale patterns of recombination variation are associated with varying levels of nucleotide diversity, a correlation which we observe on 2 scales. Small LGs with uniform and large recombination rates show higher genetic diversity than large LGs with punctuated landscapes. At the same time, at a finer scale (10 kb), hotspot windows coinciding with family COs show higher nucleotide diversity than coldspot windows where family COs are rare. A similar correlation of genetic diversity and recombination is observed in bird species and across the tree of life, although there are exceptions and its magnitude varies (Nordborg et al. 2005; Webster and Hurst 2012; Cutter and Payseur 2013; Kawakami et al. 2014; Rowan et al. 2019; Peñalba et al. 2020). A proposed explanation for this association is the mutagenic effect of recombination that has been shown to exist in multiple species (Arbeithuber et al. 2015; Rattray et al. 2015; Halldorsson et al. 2019). Another explanation is the interplay of selection and recombination. If recombination is spread throughout the length of the sequence, neutral alleles are uncoupled faster from selected ones that tend to drag them to extinction or fixation, thus allowing an increase of standing variation. On the contrary, long stretches of reduced recombination, through the action of linked selection, lead to reduced diversity (Charlesworth et al. 1993; Charlesworth and Jensen 2021). While both effects can be acting at the same time, it can be hard to distinguish their relative contributions especially because recombination-induced mutation is hard to quantify in nonmodel species and the relative effects might be species specific (Cutter and Payseur 2013).
A recurring observation is that recombination rates correlate with multiple genomic features without a clear direction of causality. For example, as illustrated here, windows with increased recombination rates are found in gene-rich regions, a finding prevalent in both plants and animals (e.g. Paape et al. 2012; Kawakami et al. 2014; Rifkin et al. 2022). An attractive explanation for this pattern is an adaptive one. In gene-rich regions, targets for selection (positive or negative) are increased, and as a consequence, so is the potential for interference between selected alleles [Hill–Robertson interference (HRI); Hill and Robertson 1966]. Higher recombination rates in these regions can mitigate this effect. However, indirect selection for increased recombination might be too weak to drive this phenomenon (Roze 2021), since only a few COs are required to overcome HRI. Instead genes and COs might colocalize because of underlying factors like accessible chromatin, as suggested here by the increase of recombination around TSSs and CGIs or through modulating effects of transposable element density (Baker et al. 2017; Kent et al. 2017; Kianian et al. 2018; Venu et al. 2024). Similarly, GC content can correlate with increased recombination both through colocalization of COs and CGIs and through gBGC (Eyre-Walker 1993; Duret and Galtier 2009). As recombination landscapes of more species accumulate, comparative studies will be more equipped to answer questions regarding the directionality and strength of these forces. For example, characterizing recombination patterns in species with varying magnitudes of these forces (e.g. different GC conversion bias or different strength of recombination-induced mutagenesis) might help pick apart their relative contribution. To this end, exploring the recombination landscapes of different species is a valuable first step.
Population differences
The populations we use in our study show a shallow genetic differentiation resulting from an out of refugium expansion following the last glacial maximum and facilitated by moderate dispersal over the Western European continent (Cumer, Machado, Dumont, et al. 2022; Machado, Topaloudis, et al. 2022). Inspired by Talbi et al. (2024), we attempted to measure statistical noise from the sampling of genealogies in the inference of recombination landscapes through subsampling the Swiss data set 5 times down to n = 13. This showed that while correlations between Switzerland and Portugal were slightly lower than between Switzerland and the subsets, hotspot sharing was indistinguishable. We interpret this as evidence of divergence in parts of the recombination landscape. While this can be a signal of between-population differences in segregating structural variants, transposable elements, or variation in some part of the recombination machinery, it can also be confounded through the effect of LD. Inferring recombination rates through LD means that selection and population size fluctuations can impact the inferred result (O’Reilly et al. 2008; Johnston and Cutler 2012; Dapper and Payseur 2018). Especially at the scale of a few kilobases, methods are known to show large statistical noise that depends on sample size, demography, and sequencing artifacts (Raynaud et al. 2023; Talbi et al. 2024). In our study, estimates were especially problematic with the samples from Great Britain where low genetic diversity coupled with a small sample size led to inconclusive results. While LD-based approaches provide an easy way to quantify the recombination landscape in multiple populations or species, ruling out confounding factors can be challenging, unless a more robust method of inferring recombination is used, such as linkage mapping.
Concerning evolving recombination landscapes, past investigations have focused on the effect of the presence and absence of the PRDM9 gene. Species where the gene is active seem to have fast-evolving hotspots, possibly through erosion of identified motif sites (Coop and Myers 2007; Myers et al. 2010; Baker et al. 2017; Raynaud et al. 2024). On the contrary, in species without PRMD9, recombination hotspots tend to be preserved over longer evolutionary scales and colocalize with open chromatin (Auton et al. 2013; Lam and Keeney 2015; Singhal et al. 2015; Kawakami et al. 2017). Recently, both the relevance of PRDM9-driven hotspots (Hoge et al. 2024; Joseph et al. 2024) and the evolutionary stability of recombination landscapes in species without the gene have been questioned (Talbi et al. 2024). Beyond mammals, the lack of a fast-evolving hotspot gene does not imply the lack of recombination divergence that has been observed even in closely related taxa (Stapley et al. 2017). Multiple genes have been shown to harbor heritable variation in modifying the number and placement of CO events and have thus the potential to evolve under direct and indirect selective pressures (Reynolds et al. 2013; Johnston et al. 2018; Dreissig et al. 2020; Arter and Keeney 2024 ; Johnston 2024). Yet, how often differences in recombination landscapes are adaptive is less well known.
Conclusion
To conclude, we present the recombination landscape of the barn owl using both linkage mapping and LD-based inference. The barn owl genome is now equipped with an assembly comprised of 40 identified distinct LGs and a detailed recombination map. It is thus the first species in the Strigiformes order with significant genomic resources, paving the way for further analyses like genome-wide association studies and haplotype phasing. Concerning recombination, the 2 methods applied allow us to quantify variation in recombination between populations, sexes, chromosomes, and fine-scale genomic windows. We verify that observations in passerine species, like fine-scale heterochiasmy and large regions of no recombination, are found outside of this clade. We also highlight the complex interplay of recombination and genetic diversity. Overall, these results contribute to our growing understanding of recombination in eukaryotes and birds specifically, providing a more comprehensive overview of the changing recombination landscape and divergence between sexes.
Supplementary Material
Acknowledgments
The authors would like to thank Museum national d’Histoire naturelle in Paris for the Corsican samples used in the variant discovery. They would also like to thank Julien Marquis and Melanie Dupasquier of the GTF facility for assistance in library preparation and sequencing of the samples used in the study. The authors also thank Milan Malinsky for helpful discussions on analysis and interpretation and Anna Hewett for comments and careful proofreading of the final version. Lastly, the work and manuscript greatly benefited from the insightful comments of two anonymous reviewers.
Contributor Information
Alexandros Topaloudis, Department of Ecology and Evolution, University of Lausanne, Lausanne 1015, Switzerland; Swiss Institute of Bioinformatics, Lausanne 1015, Switzerland.
Tristan Cumer, Department of Ecology and Evolution, University of Lausanne, Lausanne 1015, Switzerland; Swiss Institute of Bioinformatics, Lausanne 1015, Switzerland.
Eléonore Lavanchy, Department of Ecology and Evolution, University of Lausanne, Lausanne 1015, Switzerland; Swiss Institute of Bioinformatics, Lausanne 1015, Switzerland.
Anne-Lyse Ducrest, Department of Ecology and Evolution, University of Lausanne, Lausanne 1015, Switzerland.
Celine Simon, Department of Ecology and Evolution, University of Lausanne, Lausanne 1015, Switzerland.
Ana Paula Machado, Department of Ecology and Evolution, University of Lausanne, Lausanne 1015, Switzerland.
Nika Paposhvili, Institute of Ecology, Ilia State University, Tbilisi 0162, Georgia.
Alexandre Roulin, Department of Ecology and Evolution, University of Lausanne, Lausanne 1015, Switzerland.
Jérôme Goudet, Department of Ecology and Evolution, University of Lausanne, Lausanne 1015, Switzerland; Swiss Institute of Bioinformatics, Lausanne 1015, Switzerland.
Data availability
All sample information is found in Supplementary File 2. Sequence data used in the study from previous publications are available on NCBI under BioProject codes PRJNA700797, PRJNA727915, PRJNA727977, PRJNA774943, and PRJNA925445. Sequence data generated for this study are available on NCBI under BioProject code PRJNA1172395. Code to reproduce the figures can be found on github: https://github.com/topalw/Recombination_barn_owl. Downstream data to be used with aforementioned scripts can be found on Zenodo under the following DOI: 10.5281/zenodo.13982582.
Supplemental material available at GENETICS online.
Funding
This work was supported by the Swiss National Science Foundation (SNF, grant 310030_215709) to JG.
Author contributions
AT and JG devised the project. AR and NP provided the samples. A-LD, CS, and APM carried out the DNA extraction and library preparation. AT, EL, and TC generated and filtered the variant data set. AT performed analyses and wrote the manuscript with input from all coauthors.
Literature cited
- Adler D, Kelly ST, Elliott T, Adamson J. 2022. vioplot: violin plot. [accessed 2023 Dec 13]. https://github.com/TomKellyGenetics/vioplot.
- Antoniazza S, Burri R, Fumagalli L, Goudet J, Roulin A. 2010. Local adaptation maintains clinal variation in melanin-based coloration of European barn owls (Tyto alba). Evolution. 64(7):1944–1954. doi: 10.1111/j.1558-5646.2010.00969.x. [DOI] [PubMed] [Google Scholar]
- Arbeithuber B, Betancourt AJ, Ebner T, Tiemann-Boege I. 2015. Crossovers are associated with mutation and biased gene conversion at recombination hotspots. Proceedings of the National Academy of Sciences. 112(7):2109–2114. doi: 10.1073/pnas.1416622112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arter M, Keeney S. 2024. Divergence and conservation of the meiotic recombination machinery. Nat Rev Genet. 25(5):309–325. doi: 10.1038/s41576-023-00669-8. [DOI] [PubMed] [Google Scholar]
- Auton A, Li YR, Kidd J, Oliveira K, Nadel J, Holloway JK, Hayward JJ, Cohen PE, Greally JM, Wang J, et al. 2013. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 9(12):e1003984. doi: 10.1371/journal.pgen.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, McVean G. 2007. Recombination rate estimation in the presence of hotspots. Genome Res. 17(8):1219–1227. doi: 10.1101/gr.6386707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2013. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 43(1110):11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E, Webster MT, Ratnakumar A, Consortium TL, Ponting CP, Lindblad-Toh K. 2012. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 22(1):51–63. doi: 10.1101/gr.124123.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baalsrud HT, Garmann-Aarhus B, Enevoldsen ELG, Krabberød AK, Fischer D, Tooming-Klunderud A, Skage M, Árnyasi M, Sandve SR, Jakobsen KS, et al. 2024. Evolutionary new centromeres in the snowy owl genome putatively seeded from a transposable element. bioRxiv 602039. 10.1101/2024.07.05.602039, preprint: not peer reviewed. [DOI]
- Backström N, Forstmeier W, Schielzeth H, Mellenius H, Nam K, Bolund E, Webster MT, Öst T, Schneider M, Kempenaers B, et al. 2010. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 20(4):485–495. doi: 10.1101/gr.101410.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker Z, Schumer M, Haba Y, Bashkirova L, Holland C, Rosenthal GG, Przeworski M. 2017. Repeated losses of PRDM9-directed recombination despite the conservation of PRDM9 across vertebrates. Elife. 6:e24133. doi: 10.7554/eLife.24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Charlesworth B. 1998. Why sex and recombination? Science. 281(5385):1986–1990. doi: 10.1126/science.281.5385.1986. [DOI] [PubMed] [Google Scholar]
- Bascón-Cardozo K, Bours A, Manthey G, Durieux G, Dutheil JY, Pruisscher P, Odenthal-Hesse L, Liedvogel M. 2024. Fine-scale map reveals highly variable recombination rates associated with genomic features in the Eurasian blackcap. Genome Biol Evol. 16(1):evad233. doi: 10.1093/gbe/evad233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 327(5967):836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 356(6369):519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Belterman R, De Boer L. 1984. A karyological study of 55 species of birds, including karyotypes of 39 species new to cytology. Genetica. 65(1):39–82. doi: 10.1007/BF00056765. [DOI] [Google Scholar]
- Benham PM, Cicero C, Escalona M, Beraut E, Fairbairn C, Marimuthu MPA, Nguyen O, Sahasrabudhe R, King BL, Thomas WK, et al. 2024. Remarkably high repeat content in the genomes of sparrows: the importance of genome assembly completeness for transposable element discovery. Genome Biol Evol. 16(4):evae067. doi: 10.1093/gbe/evae067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjelloun B, Boyer F, Streeter I, Zamani W, Engelen S, Alberti A, Alberto FJ, BenBati M, Ibnelbachyr M, Chentouf M, et al. 2019. An evaluation of sequencing coverage and genotyping strategies to assess neutral and adaptive diversity. Mol Ecol Resour. 19(6):1497–1515. doi: 10.1111/1755-0998.13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D. 2009. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 24(2):94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Bergeron LA, Besenbacher S, Zheng J, Li P, Bertelsen MF, Quintard B, Hoffman JI, Li Z, St. Leger J, Shao C, et al. 2023. Evolution of the germline mutation rate across vertebrates. Nature. 615(7951):285–291. doi: 10.1038/s41586-023-05752-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker TR, Yeaman S, Whitlock MC. 2020. Variation in recombination rate affects detection of outliers in genome scans under neutrality. Mol Ecol. 29(22):4274–4279. doi: 10.1111/mec.15501. [DOI] [PubMed] [Google Scholar]
- Bosse M, Spurgin LG, Laine VN, Cole EF, Firth JA, Gienapp P, Gosler AG, McMahon K, Poissant J, Verhagen I, et al. 2017. Recent natural selection causes adaptive evolution of an avian polygenic trait. Science. 358(6361):365–368. doi: 10.1126/science.aal3298. [DOI] [PubMed] [Google Scholar]
- Brandvain Y, Coop G. 2012. Scrambling eggs: meiotic drive and the evolution of female recombination rates. Genetics. 190(2):709–723. doi: 10.1534/genetics.111.136721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo GA, Schmitt CJ, Edwards SV. 2021. What have we learned from the first 500 avian genomes? Annu Rev Ecol Evol Syst. 52(1):611–639. doi: 10.1146/annurev-ecolsys-012121-085928. [DOI] [Google Scholar]
- Brazier T, Glémin S. 2022. Diversity and determinants of recombination landscapes in flowering plants. PLoS Genet. 18(8):e1010141. doi: 10.1371/journal.pgen.1010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekke C, Berg P, Gjuvsland AB, Johnston SE. 2022. Recombination rates in pigs differ between breeds, sexes and individuals, and are associated with the RNF212, SYCP2, PRDM7, MEI1 and MSH4 loci. Genetics Selection Evolution. 54(1):33. doi: 10.1186/s12711-022-00723-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekke C, Johnston SE, Knutsen TM, Berg P. 2023. Genetic architecture of individual meiotic crossover rate and distribution in Atlantic Salmon. Sci Rep. 13(1):20481. doi: 10.1038/s41598-023-47208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt DW. 2002. Origin and evolution of avian microchromosomes. Cytogenet Genome Res. 96(1–4):97–112. doi: 10.1159/000063018. [DOI] [PubMed] [Google Scholar]
- Burt A, Bell G, Harvey PH. 1991. Sex differences in recombination. J of Evolutionary Biology. 4(2):259–277. doi: 10.1046/j.1420-9101.1991.4020259.x. [DOI] [Google Scholar]
- Campos JL, Halligan DL, Haddrill PR, Charlesworth B. 2014. The relation between recombination rate and patterns of molecular evolution and variation in Drosophila melanogaster. Mol Biol Evol. 31(4):1010–1028. doi: 10.1093/molbev/msu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani M, Zhang M, Thangavel G, Mata-Sucre Y, Lux T, Campoy JA, Marek M, Huettel B, Sun H, Mayer KFX, et al. 2024. Meiotic recombination dynamics in plants with repeat-based holocentromeres shed light on the primary drivers of crossover patterning. Nat Plants. 10(3):423–438. doi: 10.1038/s41477-024-01625-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AH, Jenkins PA, Song YS. 2012. Genome-wide fine-scale recombination rate variation in Drosophila melanogaster. PLoS Genet. 8(12):e1003090. doi: 10.1371/journal.pgen.1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 4(1):7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. 2013. Plant sex chromosome evolution. J Exp Bot. 64(2):405–420. doi: 10.1093/jxb/ers322. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. 2017. Evolution of recombination rates between sex chromosomes. Philosophical Transactions of the Royal Society B: Biological Sciences. 372(1736):20160456.. doi: 10.1098/rstb.2016.0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. 2019. Young sex chromosomes in plants and animals. New Phytol. 224(3):1095–1107. doi: 10.1111/nph.16002. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Jensen JD. 2021. Effects of selection at linked sites on patterns of genetic variability. Annu Rev Ecol Evol Syst. 52(1):177–197. doi: 10.1146/annurev-ecolsys-010621-044528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. 1993. The effect of deleterious mutations on neutral molecular variation. Genetics. 134(4):1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmantier A, Garant D, Kruuk LEB, editors. 2014. Quantitative Genetics in the Wild. Oxford University Press. [Google Scholar]
- Choi K, Henderson IR. 2015. Meiotic recombination hotspots—a comparative view. Plant J. 83(1):52–61. doi: 10.1111/tpj.12870. [DOI] [PubMed] [Google Scholar]
- Coop G, Myers SR. 2007. Live hot, die young: transmission distortion in recombination hotspots. PLoS Genet. 3(3):e35. doi: 10.1371/journal.pgen.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Przeworski M. 2007. An evolutionary view of human recombination. Nat Rev Genet. 8(1):23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- Corval H, Cumer T, Topaloudis A, Roulin A, Goudet J. 2023. Where and when local adaptation happens: lessons from the European barn owl (Tyto alba). bioRxiv 533108. 10.1101/2023.03.17.533108, preprint: not peer reviewed. [DOI]
- Cumer T, Machado AP, Dumont G, Bontzorlos V, Ceccherelli R, Charter M, Dichmann K, Kassinis N, Lourenço R, Manzia F, et al. 2022. Landscape and climatic variations shaped secondary contacts amid barn owls of the Western Palearctic. Mol Biol Evol. 39(1):msab343. doi: 10.1093/molbev/msab343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumer T, Machado AP, San-Jose LM, Ducrest A-L, Simon C, Roulin A, Goudet J. 2024. The genomic architecture of continuous plumage colour variation in the European barn owl (Tyto alba). Proc R Soc Lond B Biol Sci. 291(2014):20231995. doi: 10.1098/rspb.2023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumer T, Machado AP, Siverio F, Cherkaoui SI, Roque I, Lourenço R, Charter M, Roulin A, Goudet J. 2022. Genomic basis of insularity and ecological divergence in barn owls (Tyto alba) of the Canary Islands. Heredity (Edinb). 129(5):281–294. doi: 10.1038/s41437-022-00562-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Payseur BA. 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat Rev Genet. 14(4):262–274. doi: 10.1038/nrg3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics. 27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. Gigascience. 10(2):giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapper AL, Payseur BA. 2018. Effects of demographic history on the detection of recombination hotspots from linkage disequilibrium. Mol Biol Evol. 35(2):335–353. doi: 10.1093/molbev/msx272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimens PV. 2022. pdimens/LepWrap: 4.0.1. doi: 10.5281/zenodo.6326228. [accessed 2023 Feb 4]. https://zenodo.org/record/6326228. [DOI]
- Dreissig S, Maurer A, Sharma R, Milne L, Flavell AJ, Schmutzer T, Pillen K. 2020. Natural variation in meiotic recombination rate shapes introgression patterns in intraspecific hybrids between wild and domesticated barley. New Phytol. 228(6):1852–1863. doi: 10.1111/nph.16810. [DOI] [PubMed] [Google Scholar]
- Ducrest A-L, Neuenschwander S, Schmid-Siegert E, Pagni M, Train C, Dylus D, Nevers Y, Vesztrocy AW, San-Jose LM, Dupasquier M, et al. 2020. New genome assembly of the barn owl (Tyto alba alba). Ecol Evol. 10(5):2284–2298. doi: 10.1002/ece3.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Galtier N. 2009. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 10(1):285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- Edelman NB, Mallet J. 2021. Prevalence and adaptive impact of introgression. Annu Rev Genet. 55(1):265–283. doi: 10.1146/annurev-genet-021821-020805. [DOI] [PubMed] [Google Scholar]
- Ellegren H. 2010. Evolutionary stasis: the stable chromosomes of birds. Trends Ecol Evol. 25(5):283–291. doi: 10.1016/j.tree.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Elphinstone C, Elphinstone R, Todesco M, Rieseberg L. 2023. RepeatOBserver: tandem repeat visualization and centromere detection. bioRxiv 573697. 10.1101/2023.12.30.573697, preprint: not peer reviewed. [DOI]
- Enbody ED, Sendell-Price AT, Sprehn CG, Rubin C-J, Visscher PM, Grant BR, Grant PR, Andersson L. 2023. Community-wide genome sequencing reveals 30 years of Darwin's finch evolution. Science. 381(6665):eadf6218. doi: 10.1126/science.adf6218. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. 1993. Recombination and mammalian genome evolution. Proc R Soc Lond B Biol Sci. 252(1335):237–243. doi: 10.1098/rspb.1993.0071. [DOI] [PubMed] [Google Scholar]
- Flowers JM, Molina J, Rubinstein S, Huang P, Schaal BA, Purugganan MD. 2012. Natural selection in gene-dense regions shapes the genomic pattern of polymorphism in wild and domesticated rice. Mol Biol Evol. 29(2):675–687. doi: 10.1093/molbev/msr225. [DOI] [PubMed] [Google Scholar]
- Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C. 2011. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 7(11):e1002354. doi: 10.1371/journal.pgen.1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith MR, Shimada T, Abe H. 2005. The genetics and genomics of the silkworm, Bombyx mori. Annu Rev Entomol. 50(1):71–100. doi: 10.1146/annurev.ento.50.071803.130456. [DOI] [PubMed] [Google Scholar]
- Goudet J. 2005. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes. 5(1):184–186. doi: 10.1111/j.1471-8286.2004.00828.x. [DOI] [Google Scholar]
- Groenen MAM, Wahlberg P, Foglio M, Cheng HH, Megens H-J, Crooijmans RPMA, Besnier F, Lathrop M, Muir WM, Wong GK-S, et al. 2009. A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Res. 19(3):510–519. doi: 10.1101/gr.086538.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenel Q, Laurentino TG, Roesti M, Berner D. 2018. Meta-analysis of chromosome-scale crossover rate variation in eukaryotes and its significance to evolutionary genomics. Mol Ecol. 27(11):2477–2497. doi: 10.1111/mec.14699. [DOI] [PubMed] [Google Scholar]
- Hagen IJ, Lien S, Billing AM, Elgvin TO, Trier C, Niskanen AK, Tarka M, Slate J, Sætre G-P, Jensen H. 2020. A genome-wide linkage map for the house sparrow (Passer domesticus) provides insights into the evolutionary history of the avian genome. Mol Ecol Resour. 20(2):544–559. doi:doi: 10.1111/1755-0998.13134. [DOI] [PubMed] [Google Scholar]
- Haldane JB. 1919. The combination of linkage values and the calculation of distances between the loci of linked factors. J Genet. 8(29):299–309. [Google Scholar]
- Halldorsson BV, Palsson G, Stefansson OA, Jonsson H, Hardarson MT, Eggertsson HP, Gunnarsson B, Oddsson A, Halldorsson GH, Zink F, et al. 2019. Characterizing mutagenic effects of recombination through a sequence-level genetic map. Science. 363(6425):eaau1043. doi: 10.1126/science.aau1043. [DOI] [PubMed] [Google Scholar]
- Handley L-JL, Ceplitis H, Ellegren H. 2004. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics. 167(1):367–376. doi: 10.1534/genetics.167.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Hall H, Hunt P. 2007. The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet. 16(R2):R203–R208. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P. 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2(4):280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Hill WG, Robertson A. 1966. The effect of linkage on limits to artificial selection. Genet Res (Camb). 8(3):269–294. doi: 10.1017/S0016672300010156. [DOI] [PubMed] [Google Scholar]
- Hoge C, de Manuel M, Mahgoub M, Okami N, Fuller Z, Banerjee S, Baker Z, McNulty M, Andolfatto P, Macfarlan TS, et al. 2024. Patterns of recombination in snakes reveal a tug-of-war between PRDM9 and promoter-like features. Science. 383(6685):eadj7026. doi: 10.1126/science.adj7026. [DOI] [PubMed] [Google Scholar]
- Huang Z, Xu Z, Bai H, Huang Y, Kang N, Ding X, Liu J, Luo H, Yang C, Chen W, et al. 2023. Evolutionary analysis of a complete chicken genome. Proc Natl Acad Sci U S A. 120(8):e2216641120. doi: 10.1073/pnas.2216641120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins CD, Mavarez J, Beltrán M, McMillan WO, Johnston JS, Bermingham E. 2005. A genetic linkage map of the mimetic butterfly Heliconius melpomene. Genetics. 171(2):557–570. doi: 10.1534/genetics.104.034686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE. 2024. Understanding the genetic basis of variation in meiotic recombination: past, present, and future. Mol Biol Evol. 41(7):msae112. doi: 10.1093/molbev/msae112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE, Bérénos C, Slate J, Pemberton JM. 2016. Conserved genetic architecture underlying individual recombination rate variation in a wild population of soay sheep (Ovis aries). Genetics. 203(1):583–598. doi: 10.1534/genetics.115.185553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston HR, Cutler DJ. 2012. Population demographic history can cause the appearance of recombination hotspots. Am J Hum Genet. 90(5):774–783. doi: 10.1016/j.ajhg.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE, Huisman J, Ellis PA, Pemberton JM. 2017. A high-density linkage map reveals sexual dimorphism in recombination landscapes in red deer (Cervus elaphus). G3 (Bethesda). 7(8):2859–2870. doi: 10.1534/g3.117.044198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE, Huisman J, Pemberton JM. 2018. A genomic region containing REC8 and RNF212B is associated with individual recombination rate variation in a wild population of red deer (Cervus elaphus). G3 (Bethesda). 8(7):2265–2276. doi: 10.1534/g3.118.200063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J, Prentout D, Laverré A, Tricou T, Duret L. 2024. High prevalence of PRDM9-independent recombination hotspots in placental mammals. Proc Natl Acad Sci U S A. 121(23):e2401973121. doi: 10.1073/pnas.2401973121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur T, Rockman MV. 2014. Crossover heterogeneity in the absence of hotspots in Caenorhabditis elegans. Genetics. 196(1):137–148. doi: 10.1534/genetics.113.158857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Mugal CF, Suh A, Nater A, Burri R, Smeds L, Ellegren H. 2017. Whole-genome patterns of linkage disequilibrium across flycatcher populations clarify the causes and consequences of fine-scale recombination rate variation in birds. Mol Ecol. 26(16):4158–4172. doi: 10.1111/mec.14197. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Smeds L, Backström N, Husby A, Qvarnström A, Mugal CF, Olason P, Ellegren H. 2014. A high-density linkage map enables a second-generation collared flycatcher genome assembly and reveals the patterns of avian recombination rate variation and chromosomal evolution. Mol Ecol. 23(16):4035–4058. doi:doi: 10.1111/mec.12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res. 12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent TV, Uzunović J, Wright SI. 2017. Coevolution between transposable elements and recombination. Philos Trans R Soc Lond B Biol Sci. 372(1736):20160458. doi: 10.1098/rstb.2016.0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kianian PMA, Wang M, Simons K, Ghavami F, He Y, Dukowic-Schulze S, Sundararajan A, Sun Q, Pillardy J, Mudge J, et al. 2018. High-resolution crossover mapping reveals similarities and differences of male and female recombination in maize. Nat Commun. 9(1):2370. doi: 10.1038/s41467-018-04562-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M. 2010. How and why chromosome inversions evolve. PLoS Biol. 8(9):e1000501. doi: 10.1371/journal.pbio.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knief U, Forstmeier W, Pei Y, Ihle M, Wang D, Martin K, Opatová P, Albrechtová J, Wittig M, Franke A, et al. 2017. A sex-chromosome inversion causes strong overdominance for sperm traits that affect siring success. Nat Ecol Evol. 1(8):1177–1184. doi: 10.1038/s41559-017-0236-1. [DOI] [PubMed] [Google Scholar]
- Knief U, Hemmrich-Stanisak G, Wittig M, Franke A, Griffith SC, Kempenaers B, Forstmeier W. 2016. Fitness consequences of polymorphic inversions in the zebra finch genome. Genome Biol. 17(1):199. doi: 10.1186/s13059-016-1056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knief U, Kempenaers B, Forstmeier W. 2017. Meiotic recombination shapes precision of pedigree- and marker-based estimates of inbreeding. Heredity (Edinb). 118(3):239–248. doi: 10.1038/hdy.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler KE, Hawley RS, Sherman S, Hassold T. 1996. Recombination and nondisjunction in humans and flies. Hum Mol Genet. 5(Suppl 1):1495–1504. doi: 10.1093/hmg/5.supplement_1.1495. [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. 2002. A high-resolution recombination map of the human genome. Nat Genet. 31(3):241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Aslaug J, Walters GB, Adalbjorg J, Gylfason A, Kristinsson KT, et al. 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 467(7319):1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- Küpper C, Stocks M, Risse JE, dos Remedios N, Farrell LL, McRae SB, Morgan TC, Karlionova N, Pinchuk P, Verkuil YI, et al. 2016. A supergene determines highly divergent male reproductive morphs in the ruff. Nat Genet. 48(1):79–83. doi: 10.1038/ng.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, Austin MD, Deshpande P, Cao H, Nagarajan N, Xiao M, et al. 2012. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol. 30(8):771–776. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam I, Keeney S. 2015. Nonparadoxical evolutionary stability of the recombination initiation landscape in yeast. Science. 350(6263):932–937. doi: 10.1126/science.aad0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T, Dutheil J. 2005. Recombination difference between sexes: a role for haploid selection. PLoS Biol. 3(3):e63. doi: 10.1371/journal.pbio.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:13033997. 10.48550/arXiv.1303.3997, preprint: not peer reviewed. [DOI]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Stephens M. 2003. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 165(4):2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado AP, Cumer T, Iseli C, Beaudoing E, Ducrest A-L, Dupasquier M, Guex N, Dichmann K, Lourenço R, Lusby J, et al. 2022. Unexpected post-glacial colonisation route explains the white colour of barn owls (Tyto alba) from the British Isles. Mol Ecol. 31(2):482–497. doi: 10.1111/mec.16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado AP, Topaloudis A, Cumer T, Lavanchy E, Bontzorlos V, Ceccherelli R, Charter M, Kassinis N, Lymberakis P, Manzia F, et al. 2022. Genomic consequences of colonisation, migration and genetic drift in barn owl insular populations of the eastern Mediterranean. Mol Ecol. 31(5):1375–1388. doi: 10.1111/mec.16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovskaya L, Shnaider E, Borodin P, Torgasheva A. 2018. Karyotypes and recombination patterns of the Common Swift (Apus apus Linnaeus, 1758) and Eurasian Hobby (Falco subbuteo Linnaeus, 1758). Avian Res. 9(1):4. doi: 10.1186/s40657-018-0096-7. [DOI] [Google Scholar]
- Malinovskaya LP, Tishakova K, Shnaider EP, Borodin PM, Torgasheva AA. 2020. Heterochiasmy and sexual dimorphism: the case of the barn swallow (Hirundo rustica, Hirundinidae, Aves). Genes (Basel). 11(10):1119. doi: 10.3390/genes11101119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. 2009. The evolution of heterochiasmy: the role of sexual selection and sperm competition in determining sex-specific recombination rates in eutherian mammals. Genet Res (Camb). 91(5):355–363. doi: 10.1017/S0016672309990255. [DOI] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. 2007. Parallel divergence and degradation of the avian W sex chromosome. Trends Ecol Evol. 22(8):389–391. doi: 10.1016/j.tree.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Salazar C, Jiggins CD. 2019. Recombination rate variation shapes barriers to introgression across butterfly genomes. PLoS Biol. 17(2):e2006288. doi: 10.1371/journal.pbio.2006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. 2023. Method of the year: long-read sequencing. Nat Methods. 20(1):6–11. doi: 10.1038/s41592-022-01730-w. [DOI] [PubMed] [Google Scholar]
- McAuley JB, Servin B, Burnett HA, Brekke C, Peters L, Hagen IJ, Niskanen AK, Ringsby TH, Husby A, Jensen H, et al. 2024. The genetic architecture of recombination rates is polygenic and differs between the sexes in wild house sparrows (Passer domesticus). Mol Biol Evol. 41(9):msae179. doi: 10.1093/molbev/msae179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GAT, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P. 2004. The fine-scale structure of recombination rate variation in the human genome. Science. 304(5670):581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- Mézard C. 2006. Meiotic recombination hotspots in plants. Biochem Soc Trans. 34(4):531–534. doi: 10.1042/BST0340531. [DOI] [PubMed] [Google Scholar]
- Mira A. 1998. Why is meiosis arrested? J Theor Biol. 194(2):275–287. doi: 10.1006/jtbi.1998.0761. [DOI] [PubMed] [Google Scholar]
- Morgan TH. 1914. No crossing over in the male of Drosophila of genes in the second and third pairs of chromosomes. Biol Bull. 26(4):195–204. doi: 10.2307/1536193. [DOI] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science. 310(5746):321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 327(5967):876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3(7):e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly PF, Birney E, Balding DJ. 2008. Confounding between recombination and selection, and the Ped/Pop method for detecting selection. Genome Res. 18(8):1304–1313. doi: 10.1101/gr.067181.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Lenormand T. 2002. Resolving the paradox of sex and recombination. Nat Rev Genet. 3(4):252–261. doi: 10.1038/nrg761. [DOI] [PubMed] [Google Scholar]
- Otto SP, Payseur BA. 2019. Crossover interference: shedding light on the evolution of recombination. Annu Rev Genet. 53(1):19–44. doi: 10.1146/annurev-genet-040119-093957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paape T, Zhou P, Branca A, Briskine R, Young N, Tiffin P. 2012. Fine-scale population recombination rates, hotspots, and correlates of recombination in the Medicago truncatula genome. Genome Biol Evol. 4(5):726–737. doi: 10.1093/gbe/evs046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. 2010. Prdm9 controls activation of mammalian recombination hotspots. Science. 327(5967):835–835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalba JV, Deng Y, Fang Q, Joseph L, Moritz C, Cockburn A. 2020. Genome of an iconic Australian bird: high-quality assembly and linkage map of the superb fairy-wren (Malurus cyaneus). Mol Ecol Resour. 20(2):560–578. doi: 10.1111/1755-0998.13124. [DOI] [PubMed] [Google Scholar]
- Peona V, Weissensteiner MH, Suh A. 2018. How complete are “complete” genome assemblies?—an avian perspective. Mol Ecol Resour. 18(6):1188–1195. doi: 10.1111/1755-0998.12933. [DOI] [PubMed] [Google Scholar]
- Poelstra JW, Vijay N, Bossu CM, Lantz H, Ryll B, Müller I, Baglione V, Unneberg P, Wikelski M, Grabherr MG, et al. 2014. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science. 344(6190):1410–1414. doi: 10.1126/science.1253226. [DOI] [PubMed] [Google Scholar]
- Posit Team . 2022. RStudio: integrated development environment for R. Posit Software, PBC. [accessed 2023 Dec 13]. http://www.posit.co/.
- Py I, Ducrest A, Duvoisin N, Fumagalli L, Roulin A.. 2006. Ultraviolet reflectance in a melanin-based plumage trait is heritable. Evol Ecol Res. 8(3):483–491. [Google Scholar]
- Quinlan AR. 2014. BEDTools: the Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics. 47(1):11.12.1–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastas P. 2017. Lep-MAP3: robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics. 33(23):3726–3732. doi: 10.1093/bioinformatics/btx494. [DOI] [PubMed] [Google Scholar]
- Rattray A, Santoyo G, Shafer B, Strathern JN. 2015. Elevated mutation rate during meiosis in Saccharomyces cerevisiae. PLoS Genet. 11(1):e1004910. doi: 10.1371/journal.pgen.1004910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud M, Gagnaire P-A, Galtier N. 2023. Performance and limitations of linkage-disequilibrium-based methods for inferring the genomic landscape of recombination and detecting hotspots: a simulation study. Peer Community J. 3:e27. doi: 10.24072/pcjournal.254. [DOI] [Google Scholar]
- Raynaud M, Sanna P, Joseph J, Clément J, Imai Y, Lareyre J-J, Laurent A, Galtier N, Baudat F, Duret L, et al. 2024. PRDM9 drives the location and rapid evolution of recombination hotspots in salmonids. bioRxiv 583651. 10.1101/2024.03.06.583651, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- R Core Team . 2023. R: a language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R-project.org/.
- Rebholz WER, Boer LEMD, Sasaki M, Belterman RHR, Nishida-Umehara C. 1993. The chromosomal phylogeny of owls (Strigiformes) and new karyotypes of seven species. Cytologia (Tokyo). 58(4):403–416. doi: 10.1508/cytologia.58.403. [DOI] [Google Scholar]
- Reynolds A, Qiao H, Yang Y, Chen JK, Jackson N, Biswas K, Holloway JK, Baudat F, de Massy B, Wang J, et al. 2013. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat Genet. 45(3):269–278. doi: 10.1038/ng.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin JL, Hnatovska S, Yuan M, Sacchi BM, Choudhury BI, Gong Y, Rastas P, Barrett SCH, Wright SI. 2022. Recombination landscape dimorphism and sex chromosome evolution in the dioecious plant Rumex hastatulus. Philos Trans R Soc Lond B Biol Sci. 377(1850):20210226. doi: 10.1098/rstb.2021.0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo-Ruiz DA, Gan HM, Kaur P, Dudchenko O, Weisz D, Khan R, Lieberman Aiden E, Osipova E, Hiller M, Morales HE, et al. 2022. Chromosome-length genome assembly and linkage map of a critically endangered Australian bird: the helmeted honeyeater. Gigascience. 11:giac025. doi: 10.1093/gigascience/giac025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin A. 1999. Nonrandom pairing by male barn owls (Tyto alba) with respect to a female plumage trait. Behav Ecol. 10(6):688–695. doi: 10.1093/beheco/10.6.688. [DOI] [Google Scholar]
- Roulin A, Christe P, Dijkstra C, Ducrest A-L, Jungi TW. 2007. Origin-related, environmental, sex, and age determinants of immunocompetence, susceptibility to ectoparasites, and disease symptoms in the barn owl. Biol J Linn Soc Lond. 90(4):703–718. doi: 10.1111/j.1095-8312.2007.00759.x. [DOI] [Google Scholar]
- Roulin A, Müller W, Sasvári L, Dijkstra C, Ducrest A-L, Riols C, Wink M, Lubjuhn T. 2004. Extra-pair paternity, testes size and testosterone level in relation to colour polymorphism in the barn owl Tyto alba. J Avian Biol. 35(6):492–500. doi: 10.1111/j.0908-8857.2004.03294.x. [DOI] [Google Scholar]
- Rowan BA, Heavens D, Feuerborn TR, Tock AJ, Henderson IR, Weigel D. 2019. An ultra high-density Arabidopsis thaliana crossover map that refines the influences of structural variation and epigenetic features. Genetics. 213(3):771–787. doi: 10.1534/genetics.119.302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze D. 2021. A simple expression for the strength of selection on recombination generated by interference among mutations. Proc Natl Acad Sci U S A. 118(19):e2022805118. doi: 10.1073/pnas.2022805118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardell JM, Kirkpatrick M. 2020. Sex differences in the recombination landscape. Am Nat. 195(2):361–379. doi: 10.1086/704943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumer M, Xu C, Powell DL, Durvasula A, Skov L, Holland C, Blazier JC, Sankararaman S, Andolfatto P, Rosenthal GG, et al. 2018. Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science. 360(6389):656–660. doi: 10.1126/science.aar3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanfelter AF, Archambeault SL, White MA. 2019. Divergent fine-scale recombination landscapes between a freshwater and marine population of threespine stickleback fish. Genome Biol Evol. 11(6):1552–1572. doi: 10.1093/gbe/evz090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon BC, Kruuk LEB, Alberts SC. 2022. The expanding value of long-term studies of individuals in the wild. Nat Ecol Evol. 6(12):1799–1801. doi: 10.1038/s41559-022-01940-7. [DOI] [PubMed] [Google Scholar]
- Signorell A. 2023. DescTools: tools for descriptive statistics. [accessed 2023 Dec 13]. https://andrisignorell.github.io/DescTools/.
- Singhal S, Leffler EM, Sannareddy K, Turner I, Venn O, Hooper DM, Strand AI, Li Q, Raney B, Balakrishnan CN, et al. 2015. Stable recombination hotspots in birds. Science. 350(6263):928–932. doi: 10.1126/science.aad0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnwell JP, Therneau TM, Schaid DJ. 2014. The kinship2 R package for pedigree data. Hum Hered. 78(2):91–93. doi: 10.1159/000363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeds L, Mugal CF, Qvarnström A, Ellegren H. 2016. High-resolution mapping of crossover and non-crossover recombination events by whole-genome re-sequencing of an avian pedigree. PLoS Genet. 12(5):e1006044. doi: 10.1371/journal.pgen.1006044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukowski Heil CS, Ellison C, Dubin M, Noor MA. 2015. Recombining without hotspots: a comprehensive evolutionary portrait of recombination in two closely related species of Drosophila. Genome Biol Evol. 7(10):2829–2842. doi: 10.1093/gbe/evv182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JP, Song YS. 2019. Inference and analysis of population-specific fine-scale recombination maps across 26 diverse human populations. Sci Adv. 5(10):eaaw9206. doi: 10.1126/sciadv.aaw9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J, Feulner PGD, Johnston SE, Santure AW, Smadja CM. 2017. Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Philos Trans R Soc Lond B Biol Sci. 372(1736):20160455. doi: 10.1098/rstb.2016.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf MPH, McVean GAT. 2003. Estimating recombination rates from population-genetic data. Nat Rev Genet. 4(12):959–968. doi: 10.1038/nrg1227. [DOI] [PubMed] [Google Scholar]
- Talbi M, Turner GF, Malinsky M.. 2024. Rapid evolution of recombination landscapes during the divergence of cichlid ecotypes in Lake Masoko. bioRxiv 585960. 10.1101/2024.03.20.585960, preprint: not peer reviewed. [DOI] [PubMed]
- Tan HZ, Scherer P, Stuart KC, Bailey S, Lee KD, Brekke P, Ewen JG, Whibley A, Santure AW. 2024. A high-density linkage map reveals broad- and fine-scale sex differences in recombination in the hihi (stitchbird; Notiomystis cincta). Heredity (Edinb). 133(4):262–275. doi: 10.1038/s41437-024-00711-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennekes M. 2018. Tmap: thematic maps in R. J Stat Softw. 84(6):1–39. doi: 10.18637/jss.v084.i06.30450020 [DOI] [Google Scholar]
- Terhorst J, Kamm JA, Song YS. 2017. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet. 49(2):303–309. doi: 10.1038/ng.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, Owens GL, Bercovich N, Légaré J-S, Soudi S, Burge DO, Huang K, Ostevik KL, Drummond EBM, Imerovski I, et al. 2020. Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature. 584(7822):602–607. doi: 10.1038/s41586-020-2467-6. [DOI] [PubMed] [Google Scholar]
- van Oers K, Santure AW, De Cauwer I, van Bers NE, Crooijmans RP, Sheldon BC, Visser ME, Slate J, Groenen MA. 2014. Replicated high-density genetic maps of two great tit populations reveal fine-scale genomic departures from sex-equal recombination rates. Heredity (Edinb). 112(3):307–316. doi: 10.1038/hdy.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veller C, Kleckner N, Nowak MA. 2019. A rigorous measure of genome-wide genetic shuffling that takes into account crossover positions and Mendel's second law. Proc Natl Acad Sci U S A. 116(5):1659–1668. doi: 10.1073/pnas.1817482116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venu V, Harjunmaa E, Dreau A, Brady S, Absher D, Kingsley DM, Jones FC. 2024. Fine-scale contemporary recombination variation and its fitness consequences in adaptively diverging stickleback fish. Nat Ecol Evol. 8(7):1337–1352. doi: 10.1038/s41559-024-02434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay N, Bossu CM, Poelstra JW, Weissensteiner MH, Suh A, Kryukov AP, Wolf JBW. 2016. Evolution of heterogeneous genome differentiation across multiple contact zones in a crow species complex. Nat Commun. 7(1):13195. doi: 10.1038/ncomms13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Robinson JA, Cox LA. 2022. High-resolution estimates of crossover and noncrossover recombination from a captive baboon colony. Genome Biol Evol. 14(4):evac040. doi: 10.1093/gbe/evac040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters PD, Patel HR, Ruiz-Herrera A, Álvarez-González L, Lister NC, Simakov O, Ezaz T, Kaur P, Frere C, Grützner F, et al. 2021. Microchromosomes are building blocks of bird, reptile, and mammal chromosomes. Proc Natl Acad Sci U S A. 118(45):e2112494118. doi: 10.1073/pnas.2112494118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MT, Hurst LD. 2012. Direct and indirect consequences of meiotic recombination: implications for genome evolution. Trends Genet. 28(3):101–109. doi: 10.1016/j.tig.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Wei T, Simko V.. 2021. R package “corrplot”: visualization of a correlation matrix. [accessed 2023 Dec 13]. https://github.com/taiyun/corrplot.
- Weir BS, Goudet J. 2017. A unified characterization of population structure and relatedness. Genetics. 206(4):2085–2103. doi: 10.1534/genetics.116.198424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodzimierz P, Hong M, Henderson IR. 2023. TRASH: tandem repeat annotation and structural hierarchy. Bioinformatics. 39(5):btad308. doi: 10.1093/bioinformatics/btad308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AE, Dean R, Zimmer F, Mank JE. 2016. How to make a sex chromosome. Nat Commun. 7(1):12087. doi: 10.1038/ncomms12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdi HP, Ellegren H. 2018. A genetic map of ostrich Z chromosome and the role of inversions in avian sex chromosome evolution. Genome Biol Evol. 10(8):2049–2060. doi: 10.1093/gbe/evy163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdi HP, Olito C, Kawakami T, Unneberg P, Schou MF, Cloete SWP, Hansson B, Cornwallis CK. 2023. The evolutionary maintenance of ancient recombining sex chromosomes in the ostrich. PLoS Genet. 19(6):e1010801. doi: 10.1371/journal.pgen.1010801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelkowski M, Olson MA, Wang M, Pawlowski W. 2019. Diversity and determinants of meiotic recombination landscapes. Trends Genet. 35(5):359–370. doi: 10.1016/j.tig.2019.02.002. [DOI] [PubMed] [Google Scholar]
- Zhang H, Lundberg M, Ponnikas S, Hasselquist D, Hansson B. 2024. Male-biased recombination at chromosome ends in a songbird revealed by precisely mapping crossover positions. G3 (Bethesda). 14(9):jkae150. doi: 10.1093/g3journal/jkae150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 28(24):3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 2015. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol. 7(6):a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Dimens PV. 2022. pdimens/LepWrap: 4.0.1. doi: 10.5281/zenodo.6326228. [accessed 2023 Feb 4]. https://zenodo.org/record/6326228. [DOI]
Supplementary Materials
Data Availability Statement
All sample information is found in Supplementary File 2. Sequence data used in the study from previous publications are available on NCBI under BioProject codes PRJNA700797, PRJNA727915, PRJNA727977, PRJNA774943, and PRJNA925445. Sequence data generated for this study are available on NCBI under BioProject code PRJNA1172395. Code to reproduce the figures can be found on github: https://github.com/topalw/Recombination_barn_owl. Downstream data to be used with aforementioned scripts can be found on Zenodo under the following DOI: 10.5281/zenodo.13982582.
Supplemental material available at GENETICS online.