Abstract
Lytic cell death culminates in plasma membrane rupture, which releases large intracellular molecules to augment the inflammatory response. Plasma membrane rupture is mediated by the effector membrane protein ninjurin-1 (NINJ1)1, which polymerizes and ruptures the membrane via its hydrophilic face1–4. How NINJ1 is restrained under steady-state conditions to ensure cell survival remains unknown. Here we describe the molecular underpinnings of NINJ1 inhibition. Using cryogenic electron microscopy, we determined the structure of inactive-state mouse NINJ1 bound to the newly developed nanobody Nb538. Inactive NINJ1 forms a face-to-face homodimer by adopting a three-helix conformation with unkinked transmembrane helix 1 (TM1), in contrast to the four-helix TM1-kinked active conformation2–4. Accordingly, endogenous NINJ1 from primary macrophages is a dimer under steady-state conditions. Inactive dimers sequester the membrane rupture-inducing hydrophilic face of NINJ1 and occlude the binding site for kinked TM1 from neighbouring activated NINJ1 molecules. Mutagenesis studies in cells show that destabilization of inactive face-to-face dimers leads to NINJ1-mediated cell death, whereas stabilization of face-to-face dimers inhibits NINJ1 activity. Moreover, destabilizing mutations prompt spontaneous TM1 kink formation, a hallmark of NINJ1 activation. Collectively, our data demonstrate that dimeric NINJ1 is autoinhibited in trans to prevent unprovoked plasma membrane rupture and cell death.
Subject terms: Cryoelectron microscopy, Apoptosis
Structure-function studies reveal that the plasma membrane rupture protein NINJ1 homodimerizes through its hydrophilic membrane-rupturing face, thereby rendering NINJ1 inactive in the resting state.
Main
Various programmed cell death pathways culminate in plasma membrane rupture (PMR) and the concomitant release of intracellular proinflammatory molecules called damage-associated molecular patterns5. PMR is mediated by the 16-kDa membrane protein NINJ11. In response to cell death stimuli, NINJ1 orchestrates PMR by forming megadalton-sized multimers at the plasma membrane1. Recent cryogenic electron microscopy (cryo-EM) structures of NINJ1 multimers in their active state2–4 display two key features: (1) a pronounced kink between helices α1 and α2; and (2) a large hydrophilic face exposed to the plane of the membrane, which presumably repels lipids and triggers PMR. The presence of a PMR-inducing hydrophilic face in a membrane protein raises the fundamental mechanistic question of how NINJ1 PMR activity is restrained under steady-state conditions. We hypothesized that the hydrophilic face of NINJ1 in its inactive state must be neutralized to prevent unprovoked PMR and cell death.
Stabilization of inactive-state NINJ1
To understand how NINJ1 activity is kept in check, we sought to determine the structure of inactive NINJ1. However, overexpressed wild-type NINJ1 is constitutively active and leads to PMR and cell death in HEK293T cells1. Recombinant purified wild-type NINJ1 yields higher-order multimers approximately 1 MDa in size that permeabilize liposomes2,3,6. Capturing wild-type NINJ1 in the inactive state presents a challenge. By contrast, overexpressed mutant NINJ1(K45Q) causes less PMR and cell death1,2, despite being properly localized to the plasma membrane7. Furthermore, whereas activated wild-type NINJ1 in macrophages forms approximately 1-MDa oligomers1,8, NINJ1(K45Q) resembles inactive wild-type NINJ1 and migrates at 40–200 kDa in native PAGE1. These data prompted us to use the K45Q mutant to capture NINJ1 in its biochemically elusive inactive state.
Purified mouse NINJ1(K45Q) was less active than wild-type NINJ1 in a liposome permeabilization assay (Extended Data Fig. 1a). Size-exclusion chromatography (SEC) analysis of NINJ1(K45Q) yielded three distinct peaks (Extended Data Fig. 1b). 2D classification analysis from cryo-EM of peak 1 revealed a filament composed of several copies of a repeating unit (Extended Data Fig. 1c,d). Some classes showed stacking between two protofilaments, probably owing to hydrophobic interactions, as seen previously for NINJ12–4. 2D classes of peak 2 showed two copies of the same repeating unit, whereas peak 1 contained one copy of the repeating unit (Extended Data Fig. 1c,d). Native PAGE of peaks 1, 2, and 3 yielded bands at 500–1,000 kDa, 200 kDa and 70 kDa, respectively, reflecting the number of repeating units in 2D classes (Extended Data Fig. 1e). Despite extensive efforts, we were limited to a severely anisotropic low-resolution map of peak 2, owing to poor particle alignment and strong orientation bias (Extended Data Fig. 1f–h).
Extended Data Fig. 1. Biochemical and structural analysis of NINJ1 K45Q.
a, Liposome cargo release by NINJ1 WT or K45Q. Data are means (bars) of nine individual replicates (circles) from three independent replicates. Significance was determined using Welch one-way ANOVA test. ***P < 0.001. 100% cargo release is the total cargo release following addition of 1% cholamidopropyl(dimethylammonio)-2-hydroxy-1-propanesulfonate (CHAPSO). Melittin is used as a positive control. b, SEC trace of NINJ1 K45Q purified in LMNG/CHS. Peak 1 at 9 ml, peak 2 at 11.5 ml and peak 3 at 12.3 ml. Arrows indicate molecular weight standard marker positions. mAU: milli-absorbance units. c, Representative micrographs of NINJ1 K45Q peaks 1, 2 and 3. (n = 7,114, 4,233 and 9,316 micrographs). White arrow indicates a filament. d, Representative 2D class averages of NINJ1 K45Q peaks 1, 2 and 3, showing particles are composed of similar repeating subunits. e, Immunoblot of NINJ1 in SEC fractions corresponding to peaks 1, 2 and 3. (n = 3 independent immunoblots). f, Example 2D class averages including a close-up view of four class averages showing different orientations of apo NINJ1 K45Q peak 2. Scale bar, 110 Å. g, Flowchart of cryoEM image processing. h, Low resolution anisotropic cryoEM map of apo NINJ1 K45Q peak 2.
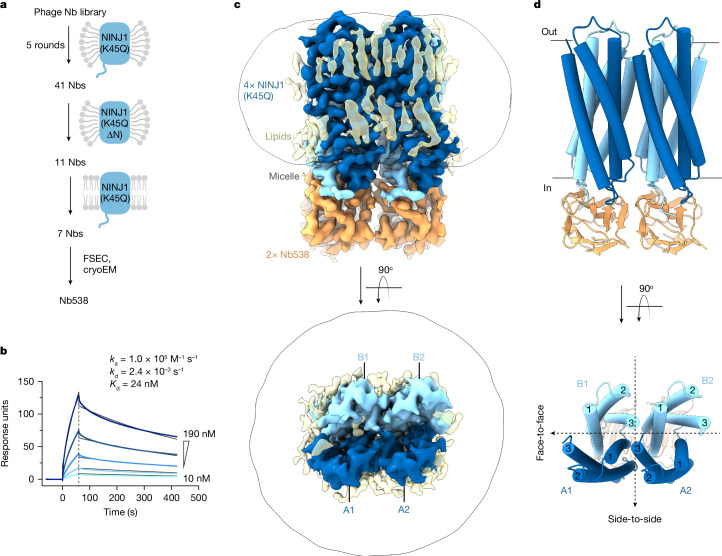
To overcome these limitations, we sought a structure chaperone and screened synthetic single-domain antibodies (nanobodies) for binding to full-length NINJ1(K45Q) solubilized in lauryl maltose neopentyl glycol (LMNG) and cholesteryl hemisuccinate (CHS), as well as NINJ1(K45Q) reconstituted in lipid nanodiscs (Fig. 1a). We prioritized nanobodies that retained binding to NINJ1(K45Q) residues 30–152 (NINJ1(K45Q ΔN)) to exclude binders of the unstructured NINJ1 N terminus. This strategy yielded seven nanobodies, five of which co-eluted with NINJ1(K45Q) by fluorescence SEC (FSEC) when incubated with peak 2 fractions (Extended Data Fig. 2a). Among these, Nb538 bound inactive NINJ1(K45Q), but not active wild-type NINJ1 by ELISA (Extended Data Fig. 2b). Similarly, Nb538 bound NINJ1(K45Q) by surface plasmon resonance (SPR) with a dissociation constant (Kd) of 24 nM (Fig. 1b), but showed no detectable binding to wild-type NINJ1 even at higher analyte concentrations (Extended Data Fig. 2c). Therefore, we selected Nb538 as a NINJ1 inactive-state-specific structure chaperone.
Fig. 1. Structure of inactive NINJ1(K45Q) bound to Nb538.
a, Schematic outline of nanobody (Nb) selection strategy. Nanobodies were selected using full-length mouse NINJ1(K45Q) or NINJ1(K45Q ∆N) (residues 30–152) in LMNG/CHS, and full-length NINJ1(K45Q) in lipid nanodiscs. b, SPR sensorgram, showing binding of NINJ1(K45Q) to immobilized Nb538. Raw traces are coloured blue and global kinetic fits are coloured black. Data are representative of three independent experiments. c, Cryo-EM density map of NINJ1(K45Q) in complex with Nb538. The map reveals four NINJ1 subunits (blue) bound to two Nb538 molecules (orange) and several probable unmodelled detergents or lipids (yellow). The map is contoured at 0.27σ. d, Ribbon diagram of NINJ1(K45Q) in complex with Nb538. NINJ1 A1 and B1 form a face-to-face dimer. NINJ1 A1:B1 and A2:B2 form a side-to-side dimer of dimers. Transmembrane helices are numbered 1–3. Black dotted lines indicate symmetry between face-to-face dimers and side-to-side dimer of dimers.
Extended Data Fig. 2. Nb538 specifically binds inactive-state NINJ1.
a, FSEC to screen for co-elution of NINJ1 K45Q peak 2 fractions with several His-tagged nanobodies. Five nanobodies co-elute with NINJ1 K45Q (light orange), one nanobody does not (grey). Nb538 is highlighted in orange. AU: arbitrary units. (n = 2 independent experiments). b, ELISA shows binding of Nb538-displaying phage particles (orange) to immobilised full-length NINJ1 K45Q or NINJ1 K45Q ∆N (residues 30–152) or WT NINJ1 in LMNG/CHS or in lipid nanodiscs (ND). A non-binding nanobody was used as a specificity control (grey). Bars represent the mean and circles represent data from three independent replicates. OD: optical density. c, Surface plasmon resonance (SPR) sensorgram showing WT NINJ1 fails to bind immobilised Nb538. Raw traces are coloured blue. Data representative of three independent experiments. d, SEC trace and coomassie-stained SDS-PAGE gel of the NINJ1 K45Q Nb538 complex used for cryoEM. Peak fraction at 11.3 ml was used for grid preparation. Representative of 5 independent purifications.
Structure of inactive-state NINJ1
We generated a complex of NINJ1(K45Q) SEC peak 2 with Nb538 (Extended Data Fig. 2d) and determined its structure at 3 Å resolution by single-particle cryo-EM (Extended Data Figs. 3 and 4 and Extended Data Table 1). The map showed well-defined density for all transmembrane helices and enabled us to unambiguously build atomic models for four subunits of NINJ1(K45Q) and two Nb538 molecules (Fig. 1c,d). NINJ1(K45Q) forms a face-to-face symmetric dimer (subunits A1:B1), which packs in a side-to-side orientation with another face-to-face dimer (subunits A2:B2) to form a dimer of dimers. Each face-to-face A:B dimer is bound to one Nb538 molecule. NINJ1(K45Q) dimerization is not induced by Nb538 binding because the NINJ1 face-to-face dimer is present even in the absence of Nb538 (Extended Data Fig. 5a–d). Accordingly, NINJ1(K45Q) dimers without Nb538 remained stable in 3 independent 1-µs all-atom molecular dynamics simulations (Extended Data Fig. 5e,f).
Extended Data Fig. 3. cryoEM data processing for NINJ1 K45Q bound to Nb538.
a, A representative cryoEM micrograph (n = 9,838 micrographs). b, Example 2D class averages including a close-up view of four class averages showing different orientations of NINJ1 K45Q bound to Nb538. Scale bar, 90 Å. c, Flowchart of cryoEM image processing.
Extended Data Fig. 4. cryoEM map of NINJ1 K45Q bound to Nb538.
a, Final EM map used for structural modelling. Surface colour indicates estimated local resolution. b, Final Fourier Shell Correlation (FSC) curve. c, Final viewing direction distribution. Representative cryoEM densities for all transmembrane helices in NINJ1 K45Q protomer d, A1 e, B1 f, A2 and g, B2. All maps are contoured at 0.6 σ. Residues discussed in the manuscript are labelled.
Extended Data Table 1.
cryoEM data collection, refinement and validation statistics
Extended Data Fig. 5. NINJ1 forms face-to-face dimers in the absence of Nb538.
a, Representative 2D class averages of NINJ1 K45Q in the presence and absence of Nb538. Density contributed by Nb538 is identified with orange arrows. b, The anisotropic cryoEM density map of the apo NINJ1 K45Q peak 2 sample. The map is consistent with four NINJ1 subunits (blue) surrounded by a detergent micelle (yellow). c, Ribbon diagram of NINJ1 K45Q (blue) in complex with Nb538 (orange). d, Rigid body fitting of NINJ1 K45Q with Nb538 removed into the cryoEM density map of apo NINJ1 K45Q. e-h, All-atom molecular dynamics simulations to assess stability of inactive conformation. e, f, NINJ1 K45Q in the absence of Nb538. g, h, NINJ1 WT in the absence of Nb538. e, g, Starting model for simulations shown in blue (NINJ1). Simulation frames sampled every 100 ns from a representative 1 µs long simulation shown in grey. f, h, Root mean square deviation (RMSD) of NINJ1 TM backbone atoms over the course of three independent simulation replicates. Dark blue and light blue traces indicate mean and standard deviation respectively from three simulation replicates.
Endogenous NINJ1 exists as a dimer
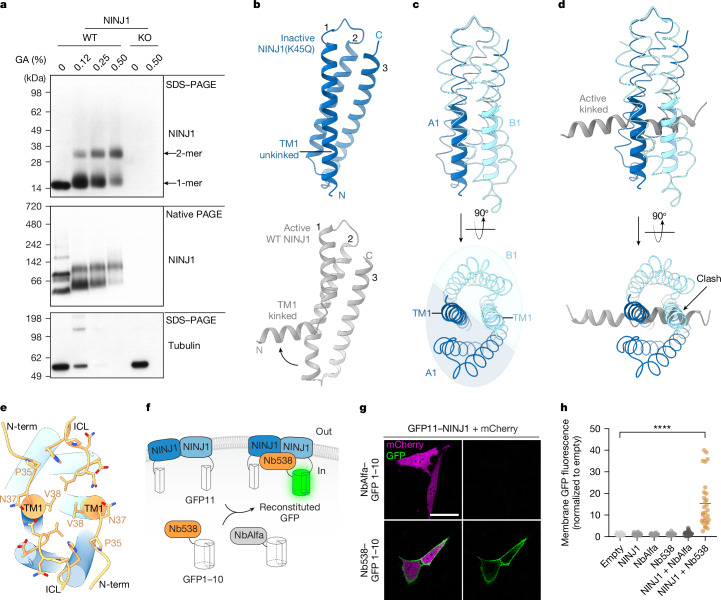
To assess the oligomeric state of NINJ1 in a native membrane under steady-state conditions, we treated mouse bone marrow-derived macrophages (BMDMs) with the crosslinker glutaraldehyde prior to detergent solubilization and immunoblot analysis. Endogenous NINJ1 migrated exclusively as a monomer in denaturing SDS–PAGE at the expected molecular weight of 16.5 kDa. Increasing amounts of glutaraldehyde decreased the amount of monomeric NINJ1 and elicited a larger NINJ1 species around 33 kDa, consistent with a crosslinked NINJ1 dimer. In agreement with previous reports, uncrosslinked NINJ1 migrated primarily as two species by non-denaturing native PAGE1,8 (Fig. 2a). Glutaraldehyde addition did not introduce any new NINJ1 species, suggesting that the observed crosslinking is specific to physiological NINJ1 dimers.
Fig. 2. Inactive face-to-face NINJ1 dimers are stabilized by unkinked TM1.
a, Immunoblot of NINJ1 in extract from primed BMDMs following treatment with indicated amounts of glutaraldehyde (GA) crosslinker (n = 3 independent immunoblots). KO, knockout; WT, wild type. b, Ribbon diagrams comparing the TM1 conformation in inactive NINJ1(K45Q) (blue, this study) and active wild-type NINJ1 (grey, Protein Data Bank (PDB) ID: 8SZA). Transmembrane helices are numbered 1–3. c, NINJ1 A1:B1 dimerization is driven by TM1–TM1 packing. Unkinked TM1 residues N33–A55 are shown as thick ribbons. Residues L56–Q142 are shown as thin ribbons. d, Inactive NINJ1 dimers occlude the binding site for active-state TM1 from hypothetical neighbouring NINJ1 molecules. Residues N33–A55 of active-state TM1 are coloured grey. e, Cytoplasmic view of the NINJ1 dimer displayed as cylinders. Nb538 (not shown) engages a composite epitope (orange) formed by the NINJ1 A1:B1 dimer in the TM1-unkinked conformation. Key NINJ1 residues that mediate contacts with Nb538 are shown as sticks. ICL, intracellular loop; N-term, residues N33–V38. f, Schematic of SplitGFP reconstitution system. g, Live-cell imaging of Ninj1-deficient BALB/3T3 cells transfected with mCherry, N-terminally tagged GFP11–NINJ1 and C-terminally tagged NbAlfa–GFP1–10 or Nb538–GFP1–10. Scale bar, 40 µm. h, Quantification of membrane GFP fluorescence in Ninj1-deficient BALB/3T3 cells transfected with mCherry and the indicated constructs. Lines represent the mean and circles represent 30–34 cells quantified over three independent experiments. Welch’s one-way ANOVA test. ****P < 0.0001.
In agreement with our cryo-EM and native PAGE data, AlphaFold2-predicted dimers showed high confidence in face-to-face A1:B1 dimerization9,10 (Extended Data Fig. 6). Of note, tetrameric NINJ1 AlphaFold2 predictions resulted in an A1:B1–A2:B2 dimer of dimers assembly with high confidence, whereas monomeric and trimeric NINJ1 models showed low confidence. Similarly, molecular dynamics simulations starting with our cryo-EM structure, but with Q45 mutated back to K45, showed that wild-type NINJ1 dimers without Nb538 remain stable in molecular dynamics simulations (Extended Data Fig. 5g,h).
Extended Data Fig. 6. AlphaFold2 predicts lower order multimerization of WT NINJ1.
Top-ranked AlphaFold2-predicted models of WT NINJ1 shown as ribbons. a, monomer; b, dimer; d, trimer; or e, tetramer models are coloured by pLDDT (predicted local distance difference test) scores. c, cryoEM structure of NINJ1 K45Q dimer in blue superimposed on AlphaFold2-predicted WT NINJ1 dimer in grey.
Inactive NINJ1 features an unkinked TM1
NINJ1 TM1 is highly conserved and mutations within TM1 render NINJ1 incapable of inducing cell death and PMR1. Active-state NINJ1 features a pronounced kink in the middle of TM1, which results in a four-helix fold2–4 (Fig. 2b, bottom). The TM1 kink binds the neighbouring NINJ1 protomer to stabilize an active-state multimer resembling a riverdance line. In marked contrast, inactive-state NINJ1 TM1 is unkinked and continuous, resulting in a three-helix fold (Fig. 2b, top). Unkinked TM1 of the NINJ1 A1 subunit packs against the unkinked TM1 of NINJ1 B1 at the core of the face-to-face A:B dimer (Fig. 2c). TM1 of A1 forms additional contacts with TM2 and TM3 of B1 and drives NINJ1 face-to-face dimerization. Notably, the inactive NINJ1 dimer occludes the binding site observed in the active state, which accommodates a kinked TM1 from a neighbouring activated NINJ1 molecule2–4 (Fig. 2d). Thus NINJ1 face-to-face dimerization probably avoids spurious NINJ1 activation by preventing TM1 kink formation and kinked TM1 invasion from neighbouring NINJ1 molecules.
Nb538 binds to inactive TM1-unkinked NINJ1(K45Q), but not active TM1-kinked wild-type NINJ1 (Extended Data Fig. 2b,c), by recognizing a composite epitope formed by both subunits of the face-to-face NINJ1 dimer at the start of unkinked TM1 helices (residues 33–38) and cytoplasmic loops (residues 105–112) (Fig. 2e). We therefore used Nb538 as a biosensor to interrogate whether dimeric wild-type NINJ1 has the TM1-unkinked three-helix conformation in live cells. We expressed split-GFP fusions of NINJ1, Nb538 or control nanobody NbAlfa in Ninj1-deficient BALB/3T3 cells11,12 (Fig. 2f). Both NbAlfa and Nb538 were expressed at equivalent levels (Extended Data Fig. 7a). NINJ1 specifically interacted with Nb538, but not NbAlfa, as measured by GFP fluorescence at the plasma membrane in live cells (Fig. 2g,h). These results coupled with our structural insights illustrate that wild-type NINJ1 forms face-to-face dimers with an unkinked TM1 in unstimulated cells.
Extended Data Fig. 7. Analysis of NINJ1 constructs and activity mutants.
a, Immunoblot of WT and Ninj1-deficient BALB/3T3 cells transfected with indicated plasmids used in Fig. 2g,h. (n = 2 independent immunoblots). b, Left, cytoplasmic view of a representative dimer-stabilised mutation A48S (blue sticks), predicted to interact with E49 (grey sticks). Right, cytoplasmic view of a representative dimer-destabilised mutation K45R (green sticks). Clashes are highlighted in magenta. c, Immunoblot of HEK293T cells transfected with indicated plasmids. Left, dimer-stabilised mutants; and Right, dimer-destabilised mutants corresponding to Fig. 3g. (n = 3 independent immunoblots). d, Liposome cargo release by WT NINJ1 or indicated mutants. Bars show the mean of three independent replicates performed in triplicate. Each circle represents the mean of one replicate performed in triplicate. 100% cargo release is the total cargo release following addition of 1% CHAPSO. e-h, Nb538 shows reduced binding to NINJ1 dimer-destabilised mutants. e, Immunoblot of WT and Ninj1-deficient BALB/3T3 cells transfected with WT or dimer-destabilising NINJ1 mutants fused to GFP11. (n = 2 independent immunoblots). f, Live imaging of Ninj1-deficient BALB/3T3 cells transfected with either NbAlfa-GFP 1-10 or Nb538-GFP 1-10 and indicated WT or dimer-destabilising NINJ1 mutants fused to GFP11. mCherry indicates transfected cells. Scale bar, 40 µm. g, Quantification of membrane GFP fluorescence in Ninj1-deficient BALB/3T3 cells transfected with mCherry and indicated constructs. mCherry was used to threshold for cells with similar expression levels. Lines represent the mean and circles represent individual cells quantified over two independent experiments. h, Immunofluorescence imaging of Ninj1−/− BALB/3T3 cells expressing either WT Ninj1 or indicated mutants. Cells are stained for NINJ1 and DAPI (nuclei). Images are representative of 3 independent experiments. Scale bar, 40 µm.
Trans-autoinhibition of dimeric NINJ1
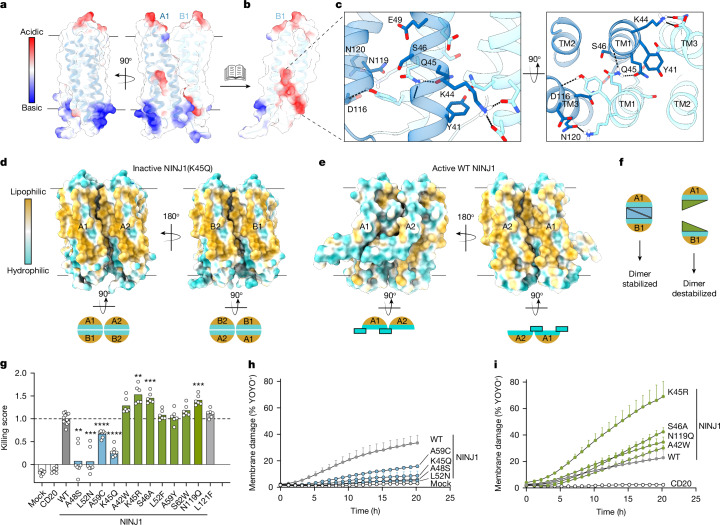
The inactive face-to-face NINJ1 dimer is stabilized by polar contacts between the two copies of amphipathic TM1 (Fig. 3a–c). S46, E49, D116, N119 and N120 from one protomer form a polar pocket at the centre of the dimer, which accommodates Y41, K44 and Q45 from the opposite protomer (Fig. 3c). The inactivating K45Q mutation likely further stabilizes face-to-face dimerization by forming strong polar contacts between two adjacent Q45 residues (Fig. 3c). This dimerization mechanism sequesters the hydrophilic face of NINJ1 inside the dimer core, enabling the exposed hydrophobic regions of NINJ1 to pack in membranes (Fig. 3d). By contrast, active-state NINJ1 filaments feature a membrane-exposed hydrophilic face, which is responsible for membrane rupture2–4 (Fig. 3e). These structural insights prompted us to posit that NINJ1 face-to-face dimer subunits autoinhibit in trans.
Fig. 3. Dimerization sequesters the hydrophilic face to trans-autoinhibit NINJ1.
a, NINJ1(K45Q) face-to-face dimer displayed as a semi-transparent surface coloured by electrostatic potential and as a ribbon diagram. b, NINJ1 A1 is hidden to reveal a polar pocket at the core of the dimer. c, Close-up view of the polar interactions between NINJ1 A1 (dark blue) and B1 (light blue). Interacting residues are labelled and shown as sticks. Dotted black lines denote hydrogen bonds identified by ChimeraX. d, Top, inactive NINJ1(K45Q) dimer of dimers coloured by lipophilicity to highlight sequestration of the hydrophilic face. Bottom, cytoplasmic view of inactive NINJ1 in cartoon representation. e, A side-to-side dimer of wild-type NINJ1 (PDB ID: 8SZA) highlights membrane exposure of the hydrophilic face. Bottom, cytoplasmic view of active NINJ1 in cartoon representation. Cyan rectangle with a black outline highlights kinked TM1. Surface lipophilicity was calculated in ChimeraX. f, Cartoon representation of point mutants designed to stabilize the inactive face-to-face dimer or to destabilize the inactive face-to-face dimer. g, Cytotoxicity of wild-type NINJ1, and dimer-stabilizing (blue) and dimer-destabilizing (green) NINJ1 in HEK293T cells. CD20 is an unrelated membrane protein control. A previously reported gain-of-function mutant, L121F, is included for comparison. Killing score is the cytotoxicity normalized to wild-type NINJ1. Data are mean (bars) of 12, 7 or 6 individual replicates (circles) for wild-type, dimer-stabilizing mutants or dimer-destabilizing mutants, respectively from three individual experiments. Welch’s one-way ANOVA test. A48S, **P = 0.0015; K45R, **P = 0.005; L52N, ***P = 0.0006; S46A, N119Q, ***P = 0.0002; A59C, K45Q, ****P < 0.0001. h,i, Membrane damage measured by YOYO-1 dye (1.2 kDa) incorporation in NINJ1-expressing HEK293T cells. Data are mean ± s.d.; three independent replicates.
To test this, we generated structure-based point mutants engineered to stabilize or destabilize the face-to-face dimer (Fig. 3f and Extended Data Fig. 7b) without modifying amino acids involved in the active-state NINJ1 filament interface (PDB: 8SZA4). Consistent with previous reports1,2, expression of wild-type NINJ1 in HEK293T cells induced cell death (Fig. 3g). Similar to NINJ1(K45Q), dimer-stabilizing mutants A48S, L52N or A59C were less toxic than wild-type NINJ1 (Fig. 3g and Extended Data Fig. 7c). In addition, these loss-of-function mutants caused less membrane damage in cells (Fig. 3h) and less liposome permeabilization (Extended Data Fig. 7d) than wild-type NINJ1. Remarkably, four out of the seven mutations predicted to destabilize the inactive face-to-face dimer were more toxic than wild-type NINJ1 (Fig. 3g and Extended Data Fig. 7c), caused more membrane damage (Fig. 3i), and showed more robust liposome permeabilization (Extended Data Fig. 7d).
Collectively, our structure–function studies demonstrate that the NINJ1 face-to-face dimer is autoinhibited in trans under basal conditions and that disrupting dimerization relieves this autoinhibition.
Proposed model for NINJ1 activation
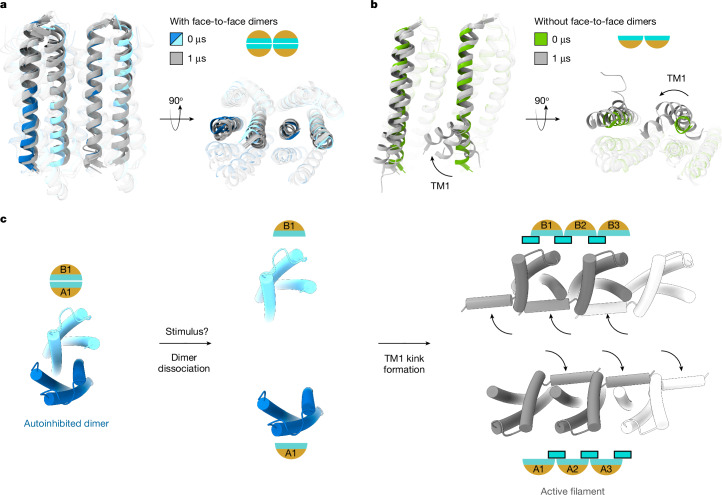
Kinked TM1 is a key structural feature of active-state NINJ12–4 (Fig. 2b). Since destabilizing autoinhibited face-to-face dimers enhanced NINJ1 cell killing and membrane damage function (Fig. 3g,i), we interrogated whether dissociating face-to-face dimers results in TM1 kinking. We used Nb538 as a biosensor for the TM1-kinked conformation in live cells, since it binds specifically to the inactive TM1-unkinked NINJ1 dimer. Consistent with our model, all four dimer-destabilizing NINJ1 mutants showed less binding to Nb538 than wild-type NINJ1 (Extended Data Fig. 7e–h). Next, we performed molecular dynamics simulations to measure TM1 displacement. Starting from our NINJ1(K45Q) cryo-EM structure, autoinhibited face-to-face dimers (A1:B1–A2:B2) showed minimal TM1 displacement over the course of simulations (Fig. 4a and Extended Data Fig. 8a,b). By contrast, without face-to-face dimerization, NINJ1(K45Q) in side-to-side orientation (A1:A2) showed increased TM1 movement in all simulations (Fig. 4b and Extended Data Fig. 8c,d). Notable TM1 movement was measured in the NINJ1 A1 subunit, probably because TM1 of A1 interacts with A2 in the active state2–4. Without a neighbouring A3 subunit, TM1 of A2 showed modest movement in only one of three simulations (Extended Data Fig. 8d). Degen et al.2 reported TM1 displacement in simulations starting from an AlphaFold2-predicted model of a hypothetical wild-type NINJ1 side-to-side pentamer without face-to face dimerization. Similarly, we measured increased TM1 movement in a side-to-side pentamer of the cryo-EM-resolved NINJ1(K45Q) without face-to-face dimerization (Extended Data Fig. 8e,f). By contrast, five side-to-side copies of the inactive NINJ1(K45Q) face-to-face dimer showed minimal TM1 displacement (Extended Data Fig. 8g,h). These data, combined with our mutagenesis studies, suggest that dissociating NINJ1 face-to-face dimers might spontaneously lead to TM1 kink formation and subsequent NINJ1 activation.
Fig. 4. Proposed model for NINJ1 activation.
a, All-atom molecular dynamics simulations of NINJ1(K45Q) with face-to-face dimerization (see also Extended Data Figs. 5 and 8). Ribbon diagram of NINJ1 face-to-face dimers (A1, A2 in dark blue; B1, B2 in light blue), shown at the start of simulations. The last snapshot from each of the 3 independent 1-µs-long simulations is shown in grey. TM2 and TM3 are shown as transparent ribbons to highlight TM1. b, All-atom molecular dynamics simulations of NINJ1(K45Q) in the absence of autoinhibitory face-to-face dimerization (see also Extended Data Fig. 8). Ribbon diagram showing the first snapshot (in green) and the last snapshots (in grey) from each of the three independent 1-µs-long simulations. Transparency settings as in a. Arrow indicates TM1 movement. c, Extracellular view of the proposed model for NINJ1 activation. In the basal-state, NINJ1 face-to-face dimer trans-autoinhibits by sequestering the hydrophilic face. TM1 is unkinked and occludes the binding site for kinked TM1 observed in the active state. Upon stimulus, autoinhibition is released by dissociation of the face-to-face dimer and TM1 kink formation (curved arrows) to expose the PMR-inducing hydrophilic face.
Extended Data Fig. 8. MD simulations show TM1 movement upon dissociation of face-to-face dimers.
a, All-atom MD simulations of two side-to-side NINJ1 K45Q molecules with face-to-face dimerization, resolved by cryoEM (total of 4 NINJ1 molecules). One (out of two) NINJ1 face-to-face dimer is shown in blue at the start of simulations, and in grey at the end of a representative simulation run. The other copy of the NINJ1 dimer is not shown for clarity. Residues V38 on TM1 and H112 on TM3, used to track TM1 movement, are shown as spheres. Black dotted line indicates the distance between V38 and H112. b, Distance between V38 on TM1 and H112 on TM3 over the course of three 1 µs-long independent simulations measured for all 4 molecules in the dimer of dimers. Black dotted lines indicate a distance of 10 Å. c-d, Same as in a-b, but without face-to-face dimerization (total of 2 NINJ1 molecules). For simulation snapshots of a-d, see Fig 4a,b. e-h, All atom MD simulations of a hypothetical multimer composed of five side-to-side NINJ1 K45Q molecules with face-to-face dimerization (total of 10 NINJ1 molecules) (e,f) or without face-to-face dimerization (total of 5 NINJ1 molecules) (g,h). e, Ribbon diagram of NINJ1 multimer with face-to-face dimerization showing the first snapshot (in blue) and the last snapshots (in grey) from each of the three independent 1 µs-long simulations. TM2 and TM3 are shown as transparent ribbons to highlight TM1. f, Distance plots of TM1 movement in e, measured for 5 molecules. g, Ribbon diagram of NINJ1 multimer without face-to-face dimerization showing the first snapshot (in green) and the last snapshots (in grey) from each of the three independent 1 µs-long simulations. Arrows indicate TM1 movement. h, Distance plots of TM1 movement in g, measured for 5 molecules.
Our structural, functional and simulation data support the following model for NINJ1 activation (Fig. 4c). In the basal state, NINJ1 forms a face-to face dimer stabilized by unkinked TM1 helices. The dimer subunits autoinhibit in trans by mutually burying their membrane-rupturing hydrophilic faces. Autoinhibited NINJ1 face-to-face dimers occlude the binding site for a kinked TM1 from a neighbouring activated NINJ1 molecule2–4. In response to cell death stimuli, NINJ1 autoinhibition is released by dissociation of face-to-face dimers. This exposes the binding site and favours TM1 kinking to form active-state filaments. Our studies do not address the kinetics of NINJ1 conformational changes and thus also support a model in which NINJ1 TM1 kink formation precedes dimer dissociation. Notably, without face-to-face dimerization, the membrane-exposed hydrophilic face of NINJ1 is unleashed to execute PMR.
Discussion
NINJ1 is the effector of PMR in response to various cell death pathways1,7,13,14. The confluence of our structure–function findings identifies NINJ1 autoinhibition as the mechanism that prevents unprovoked PMR. The cytotoxic activity of the pore-forming proteins GSDMD and MLKL—responsible for pyroptosis and necroptosis, respectively—is also restrained by autoinhibition15–19. Our identification of autoinhibition by NINJ1 highlights a common autoinhibitory mechanism of restraining potent cell death executioners. Autoinhibition of GSDMD is released by caspase-mediated GSDMD cleavage15,16 and autoinhibition of MLKL is released by RIPK3-mediated MLKL phosphorylation20, whereas the mechanistic trigger that dissociates the autoinhibited NINJ1 dimer remains unknown. NINJ1 face-to-face dimer dissociation might be induced by structural changes in the plasma membrane that accompany cell death, such as membrane curvature or lipid packing defects21. Our structure reveals a polar pocket at the core of the NINJ1 face-to-face dimer that could be targeted by charged ions or a phospholipid head group. Collectively, our work provides a structural framework to delineate the molecular identity of the mechanistic trigger for NINJ1 activation.
Our crosslinking gel analysis showing dimeric endogenous NINJ1 does not exclude a potential tetramer or other lower-order oligomeric species as a minor population. Previous native PAGE1,8, super-resolution cellular imaging2,8 and MINFLUX nanoscopy3 approaches have not resolved large higher-order multimers of NINJ1 in the basal state. Nevertheless, TM2 and TM3 of the inactive-state side-to-side A1:A2 dimer structurally overlay with TM2 and TM3 of active-state filaments4 (Extended Data Fig. 9), suggesting that NINJ1 could form lower-order side-to-side multimers in cells and remain poised for activation.
Extended Data Fig. 9. Side-to-side dimers of active or inactive NINJ1 are structurally similar.
a, Overlay of inactive NINJ1 K45Q A1:A2 side-to-side dimers (blue) and active WT NINJ1 dimers (PDB ID: 8SZA, grey) shown as ribbons. TM helices are numbered. b, Close-up view of interacting residues, labelled and shown as sticks. Side-to-side dimerization in both structures is driven by hydrophobic packing between TM3 of A2 and TM1, TM2 of A1.
Our structure, supported by live-cell imaging (Fig. 2f–h), demonstrates that a population of NINJ1 exists in an N-in topology. However, our data do not preclude a population of NINJ1 that exists in an N-out topology1,22. The exact role of a potential N-out conformation will require further studies. One possibility is a two-step activation model for NINJ1: In step one, inactive-state N-out NINJ1 inserts its TM1 in the membrane to form intermediate-state N-in face-to-face dimers; in step 2, N-in face-to-face dimers dissociate and TM1 kinks to stabilize active-state NINJ1 multimers.
Two conflicting models of how NINJ1 executes PMR have been proposed2,3. In one report2, activated NINJ1 is proposed to remain localized to the plasma membrane. In this model, NINJ1 may assemble into large GSDMD-like pores with a hydrophilic conduit or long filaments that cap membrane edges to allow leakage of intracellular contents. In a second report3, active NINJ1 is proposed to be released into the extracellular space. In this model, NINJ1 forms a nanodisc-like assembly with a hydrophobic interior to excise chunks of the membrane bilayer resulting in the expulsion of cytoplasmic contents. Despite differences, both reports show that NINJ1 ruptures membranes via its hydrophilic face2,3. Our trans-autoinhibition model remains agnostic to the above models by illustrating that dimeric NINJ1 sequesters the hydrophilic face and that face-to-face dimer dissociation is necessary for NINJ1 activation.
Methods
Mice
Ninj1−/− mice with a C57BL/6 N background were previously described1. Wild-type mice were purchased from Charles River Laboratory. Mice were housed in individually ventilated cages within animal rooms maintained on a 14:10 h, light:dark cycle with ad libitum access to food and water. Animal rooms were temperature and humidity-controlled, between 20–26 °C and 30–70% respectively, with 10 to 15 room air exchanges per hour. BMDMs were collected from both male and female 6–10 week old mice. Three wild-type and three Ninj1−/− mice were used to account for biological variance, without randomization or blinding. All animal procedures were conducted under protocols approved by the Genentech Institutional Animal Care and Use Committee in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited facility in accordance with the Guide for the Care and Use of Laboratory Animals and applicable laws and regulations.
Mammalian cell culture and generation of stable cell lines
Wild-type and Ninj1−/− mouse BMDMs were collected and differentiated into macrophages in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) low-endotoxin foetal bovine serum (Omega Scientific) and 20% (v/v) L929-conditioned medium. On day 5 of differentiation, cells were replated and allowed to adhere overnight in a 24-well plate containing 0.5 × 106 cells per well. These cells were then primed with 1 µg ml−1 Pam3CSK4 for 5 h before lysis for native PAGE or SDS–PAGE. For expression and purification of all mouse NINJ1 constructs, Expi293F cells (Thermo Fisher Scientific) were cultured in Expi293 expression medium at 37 °C, 8% CO2. Expi293F cells were not authenticated and were not tested for Mycoplasma contamination. For imaging and cytotoxicity experiments, BALB/3T3 clone A31 (ATCC) and HEK293T (ATCC) cells, respectively, were maintained in complete DMEM containing 10% foetal bovine serum and penicillin/streptomycin. Ninj1-deficient BALB/3T3 cells were generated using CRISPR–Cas9 using the following pre-designed guides from IDT: Mm.Cas9.NINJ1.1.AC (5′-ACTGAGGAGTATGAGCTCAA-3′) and Mm.Cas9.NINJ1.1.AD (5′-CTCGTGCTGCAGATAGGAGT-3′). BALB/3T3 and HEK293T cells were authenticated by short tandom repeat profiling and regular single nucleotide polymorphism fingerprinting. BALB/3T3 and HEK293T cells tested negative for Mycoplasma.
Expression and purification of NINJ1
Full-length mouse NINJ1(K45Q) (Uniprot ID: O70131) followed by a tobacco etch virus (TEV) protease recognition sequence and a Flag epitope tag at the C terminus was cloned into a mammalian expression vector. This full-length construct was used for nanobody discovery, FSEC, SPR and cryo-EM. For epitope-specific nanobody selections, mouse NINJ1(K45Q) residues G30 to Q152, termed NINJ1(K45Q ΔN), followed by a TEV protease recognition sequence and a Flag epitope tag at the C terminus was cloned into a mammalian expression vector. Cells were seeded at 3 × 106 cells per ml and transfected per manufacturer’s protocol using the Expifectamine 293 transfection reagent (Thermo Fisher Scientific). Cells were treated with enhancer solutions 18 h after transfection and collected 24 h after enhancer addition. Cell pellets were stored at −80 °C until further use. Cells were thawed and washed with a hypotonic buffer (20 mM HEPES pH 7.5, 1 mM EDTA, 0.02 mg ml−1 leupeptin and 160 μg ml−1 benzamidine) before solubilising with 50 mM HEPES pH 7.5, 300 mM NaCl, 1% (w/v) LMNG (Anatrace), 0.1% (w/v) CHS Tris salt (Anatrace), 5 mM MgCl2, 2 mM ATP, 0.02 mg ml−1 leupeptin and 160 μg ml−1 benzamidine for 1.5 h at 4 °C. After ultracentrifugation, the supernatant was subjected to affinity purification using anti-Flag M2 resin (Sigma-Aldrich; A2220) pre-equilibrated with wash buffer (50 mM HEPES pH 7.5, 300 mM NaCl, 0.1% LMNG, 0.01% CHS). After extensive washing with wash buffer, protein was eluted with 50 mM HEPES pH 7.5, 150 mM NaCl, 0.005% LMNG, 0.0005% CHS and 0.2 mg ml−1 Flag peptide (Sigma-Aldrich). Purified NINJ1 was concentrated using a 50-kDa MWCO concentrator and loaded onto a Superdex 200 Increase 10/300 GL column (GE Healthcare) equilibrated with 20 mM HEPES pH 7.5, 150 mM NaCl and 0.001% LMNG, 0.0001% CHS. Peak fractions were concentrated using a 100-kDa MWCO concentrator. Samples were either used immediately for cryo-EM or were flash-frozen in liquid nitrogen and stored at −80 °C for nanobody discovery, FSEC or SPR. For nanodisc reconstitution, full-length mouse NINJ1(K45Q) was reconstituted into nanodiscs via an on-bead method. Cell pellets were thawed, washed with hypotonic buffer, and subsequently solubilised with 50 mM HEPES pH 7.5, 300 mM NaCl, 1% (w/v) dodecyl maltoside (DDM, Anatrace), 0.1% (w/v) CHS, 5 mM MgCl2, 2 mM ATP, 0.02 mg ml−1 leupeptin and 160 µg ml−1 benzamidine for 1.5 h at 4 °C. Post-solubilization, cell lysates were clarified using ultracentrifugation and the supernatant was applied to anti-Flag M2 resin. The resin was then washed with a buffer containing 50 mM HEPES pH 7.5, 300 mM NaCl, 0.1% DDM, 0.01% CHS before incubation with 1.2 mM of Soybean Polar Lipid Extract (Avanti) for 30 min. This was followed by the addition of Membrane Scaffold Protein MSP 1D1 (Sigma-Aldrich) at a final concentration of 0.6 mg ml−1. An hour later, 60 mg ml−1 of methanol-activated SM2 Bio-Beads (Bio-Rad) were added to the resin, and the mixture was rotated overnight at 4 °C. The resin was washed with 50 mM HEPES pH 7.5, 150 mM NaCl, and NINJ1 K45Q reconstituted into nanodiscs was eluted with 0.2 mg ml−1 Flag peptide. The eluted sample was concentrated and treated much like detergent-solubilized NINJ1, except that 20 mM HEPES pH 7.5, 150 mM NaCl was used as the running buffer for the Superdex 200 Increase 10/300 GL column.
Liposomal cargo-release assay
Stocks of 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC, Avanti Polar Lipids) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (sodium salt) (POPS, Avanti Polar Lipids) were prepared in chloroform from dry powder. A lipid mixture of 80% POPC and 20% POPS was generated, freeze dried, and hydrated with a solution of 10 mM Tris pH 8.0, 150 mM NaCl containing the cargo, LANCE Eu-W1024 Biotin (PerkinElmer). The suspension was sonicated in a water bath, freeze-thawed, and extruded using a Mini Extruder (Avanti Polar Lipids) fitted with a Nucleopore 0.4-μm membrane (Whatman) to yield large unilamellar vesicles. The liposomes were purified by eluting through a column packed with Pierce Streptavidin Agarose resin (Thermo Fisher Scientific). Liposomes were destabilized by addition of 0.0005% LMNG/0.00005% CHS (Anatrace). Cargo-release assay was set up by mixing destabilized liposomes (20 μM in lipid concentration, diluted from 6.4 mM stock) with 5 μM NINJ1 purified according to the protocol above or 0.5 μM Melittin peptide (Anaspec). Streptavidin-Alexa Fluor 647 (Thermo Fisher Scientific) was added to each well at a final concentration of 0.1 μM. Liposomes mixed with 0.0005% LMNG in ddH2O were used as a control. All samples were loaded into wells of a ProxiPlate (PerkinElmer). Time-resolved readout was recorded on an EnVision 2105 multimode plate reader (PerkinElmer) using EnVision Manager 1.14.3049.1193 (PerkinElmer). A pre-read of each plate was taken immediately after loading the wells, and each plate was subsequently incubated overnight at room temperature. After ~15 h of incubation, another reading was taken, followed by full digestion of the liposomes by addition of 1% CHAPSO (Anatrace) to each well. A final read was recorded representing 100% cargo release. Results were converted to percentage cargo released per well and background control subtracted.
Crosslinking assay
Wild-type and Ninj1−/− mouse BMDMs were seeded in 24-well plates (5.0 × 105 cells per well) and primed following the procedure described above. After media removal and a gentle wash with PBS solution, cells were incubated with 50 mM HEPES, 50 mM NaCl supplemented with the indicated concentration of glutaraldehyde (Polysciences) for 10 min at room temperature. 100 mM Tris 8.0 was added for 10 min to quench the crosslinking reaction before removal of the resulting solution. Cells were lysed by incubation with 50 mM HEPES 7.5, 150 mM NaCl, 0.25% LMNG, 0.025% CHS and 1× Complete protease inhibitor (Roche Applied Science) for 1 h at 4 °C. After ultracentrifugation in an Optima Max-XP Tabletop Ultracentrifuge (Beckman Coulter), the clarified lysate was diluted in 50 mM HEPES pH 7.5, 50 mM NaCl to 0.2 mg ml−1 and flash-frozen for subsequent immunoblotting.
Native PAGE, SDS–PAGE and immunoblotting
Purified SEC fractions of NINJ1(K45Q) were diluted in 20 mM HEPES pH 7.5, 50 mM NaCl, 0.001% LMNG, and 0.0001% CHS, mixed with 4× NativePAGE sample buffer (Thermo Fisher) and loaded onto a NativePAGE 4–16% gel (Thermo Fisher Scientific) for resolution via blue native PAGE using Coomassie G-250 (Thermo Fisher Scientific). Three-hundred picograms of protein was loaded per well. The samples were transferred to a polyvinylidene fluoride (PVDF) membrane for immunoblotting. Equivalent amounts of each peak were also loaded onto SDS–PAGE and similarly immunoblotted. 2 μg of lysate were loaded per well.
For the glutaraldehyde crosslinking assay, previously frozen lysates were resolved via blue native PAGE and immunoblotted following the procedure described for the SEC fractions. Equivalent amounts of lysate were loaded onto SDS–PAGE and immunoblotted for NINJ1 (clone 25, Genentech, 0.1 µg ml−1)1 and α-tubulin (Proteintech, 66031-1-Ig, lot 10020240, 1:10,000 dilution).
For NINJ1 splitGFP immunoblots, cell pellets were snap frozen and lysed in 1.5% GDN lysis buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 1.5% GDN) supplemented with Halt Protease Inhibitor (Thermo Fisher Scientific, 78438) and Halt Phosphatase Inhibitor (Thermo Fisher Scientific, 78427) for 2 h at 4 °C. Samples were denatured in NuPAGE LDS Sample Buffer for 30 min at room temperature (Thermo Fisher Scientific). For mutant NINJ1 immunoblots, cells were lysed in 10 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 2.5 mM MgCl2, 0.5 mM CaCl2, 5 μg ml−1 DNase (Qiagen), and 1× Complete Protease Inhibitor (Roche Applied Science) at 4 °C for 30 min. Antibodies recognized either anti-rabbit or anti-mouse NINJ1 (clone 25, 0.1 µg ml−1)1, GFP (Rockland, 600-101-215, lot 46825, 1:1,000 dilution), α-tubulin (Proteintech, 66031-1-Ig, lot 10020240, 1:10,000 dilution), mCherry (Abcam, AB167453, 1:1,000 dilution) and β-actin-HRP (Cell Signalling, 5125S, lot 7, 1:1,000 dilution). HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch and used at 1:10,000 dilution: goat anti-mouse (115-035-062, lot 162808), goat anti-rabbit (111-035-144, lot 160812) and donkey anti-goat (705-035-147, lot 161343).
Phage-displayed nanobody library design and construction
Phage-displayed nanobody library was constructed using Kunkel mutagenesis. The stop template for Kunkel mutagenesis was phagemid pS2202 harbouring the VH-4D5 domain followed by M13 minor coat protein p3, with stop codons on CDR-H1, CDR-H2 and CDR-H3, and mutations at H35G, Q39R, L45E and W47L as outlined before23. Mutagenic oligonucleotides were designed to simultaneously repair the stop codons and introduce mutations of CDR-H1, H2 and H3 (Supplementary Table 1).
NINJ1-specific nanobody discovery
Phage-displayed nanobody library was cycled through 5 rounds of binding selections with decreasing concentration of Flag-tagged NINJ1(K45Q) purified in LMNG/CHS at 850, 400, 120, 60 and 30 nM in selection buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 0.01% LMNG, 0.001% CHS, 0.5% BSA) according to a previously described protocol24. Pierce anti-DYKDDDDK magnetic agarose beads (Thermo Fisher Scientific) were used in the first two rounds. From the third round on, anti-Flag M2 antibody (Sigma-Aldrich) immobilized NUNC 96-well Maxisorp immunoplates were used. The phage pools from rounds 2–5 were pre-cleared with 2× 20 µl of anti-Flag magnetic agarose to remove non-specific binders. Phage particles were eluted by incubating 1.5 mg ml−1 of 3× Flag peptide for 10 min at room temperature. After five rounds of binding selection, individual phage clones were analysed in a high throughput spot phage ELISA using plate-immobilized anti-Flag M2 antibody and captured NINJ1(K45Q)24. Non-specific binding was defined as phage particle binding to either anti-Flag M2 antibody, or BSA or a control membrane protein. Phage clones binding to NINJ1(K45Q) with a signal >0.5 and signal/noise of >4 were selected for further ELISA screening. Clones that bind to Flag-tagged NINJ1(K45Q ΔN) purified in LMNG/CHS and to Flag-tagged NINJ1(K45Q) reconstituted in nanodiscs were considered as positive clones for FSEC and cryo-EM analyses.
Nanobody purification
The sequence for Nb538 was amplified from the phage display vector and cloned into a bacterial expression vector with a C-terminal hexahistidine tag after a DKTHTGGSSGGS linker sequence. The construct was transformed into BL21(DE3) Escherichia coli, grown in terrific broth medium supplemented with 2 mM MgCl2, 0.1% w/v glucose and 50 µg ml−1 Carbenicillin at 37 °C. Culture was induced at an optical density (OD) at 600 nm of 0.4 with 400 μM IPTG. After induction, cells were grown for 4 h at 30 °C. The bacterial cell pellet was freeze-thawed and resuspended in 5 ml of resuspension buffer (200 mM Tris pH 8.0, 500 mM sucrose, 0.5 mM EDTA, 0.1 mg of lysozyme, 1 µl Benzonase Nuclease (Sigma-Aldrich)) per gram of bacterial pellet. After stirring for 30 min at room temperature, two volumes of MilliQ water were added to induce hypotonic lysis of the periplasm and the solution was further stirred at room temperature for 45 min. The resulting lysate was supplemented with 150 mM NaCl, before centrifugation at 30,000g for 25 min at 4 °C. The supernatant was loaded over Protein A resin (Genscript), washed extensively with 20 mM Tris pH 8.0, 150 mM NaCl, and eluted with 100 mM Glycine pH 3.0. The eluate was neutralized with 400 mM Tris pH 8.0, 250 mM NaCl. Purified Nb538 was flash-frozen in small aliquots in liquid nitrogen and stored at −80 °C until further use.
Fluorescence size-exclusion chromatography
FSEC experiments were performed in 20 mM HEPES pH 7.5, 150 mM NaCl, 0.001% LMNG/CHS. Purified His-tagged nanobodies (0.5 µM) were incubated with NINJ1(K45Q) purified in LMNG/CHS (SEC peak 2 fractions, 1 µM) and His-Glow reagent25 (1.1 µM) at room temperature for 20 min. After 20 min, 100 µl reaction mixture was injected onto a BioSep SEC-s3000 LC column (Phenomenex, 00H-2146-K0) at 4 °C. His-Glow fluorescence was measured with an in-line Jasco FP2020 at an excitation wavelength of 490 nm and an emission wavelength of 520 nm. SEC data were collected using Cytiva Unicorn v7.0.
Surface plasmon resonance
SPR experiments were performed using a Biacore S200 instrument at 25 °C in 20 mM HEPES pH 7.5, 150 mM NaCl, 0.01% LMNG/CHS. All experiments were performed on a Sensor Chip CM5 (Cytiva, 29104988) covalently immobilized with an anti-His antibody using the His Capture Kit (Cytiva, 28995056) on all four flow cells. Binding was performed in a multi-cycle kinetic experiment. In each cycle, flow cell 1 of the chip was used as a no ligand control. His-tagged Nb538 was immobilized on flow cell 2, and His-tagged NbAlfa, a control for non-specific binding, was immobilized on flow cell 3 to get a binding response of about 200–250 response units after 100 s of stabilization. In each cycle, analyte binding was performed by flowing a two-fold dilution series of 190 nM NINJ1(K45Q) or 386 nM wild-type NINJ1 purified in LMNG/CHS, over all four cells, with a 30 µl min−1 flow rate, 60 s contact time, followed by 360 s of dissociation. The CM5 chip immobilized with anti-His antibody was regenerated by injecting 10 mM glycine-HCl at pH 1.5 (Cytiva, BR100354) between cycles. Binding experiments were performed in triplicate. All data were analysed on the Biacore S200 evaluation software, and the signal for Nb538 binding to NINJ1 was double referenced with signal from buffer injection, and signal from analyte over the sensor without any ligand (flow cell 1). Kinetic parameters were determined using a 1:1 fitting model.
Cryo-EM sample preparation and data collection
Purified NINJ1(K45Q) peak 2 (3 µM final) was mixed with purified Nb538 (15 µM final) for 1 h at 4 °C and loaded onto a Superdex 200 Increase 10/300 GL column (GE Healthcare). Fractions corresponding to the NINJ1(K45Q)–Nb538 complex were concentrated using a 100-kDa MWCO concentrator to 0.4 mg ml−1 and immediately used for grid preparation. Prior to sample application, a holey gold grid (UltrAuFoil 25 nm R 1.2/1.3; Quantifoil, Großlöbichau) was subjected to glow discharge using a Solarus plasma cleaner (Gatan). 4 µl of purified sample at 0.4 mg ml−1 was pipetted onto to the grid which was then blotted and plunge frozen in liquid ethane using a Vitrobot (Thermo Fisher Scientific), at a temperature of 4 °C, 100% relative humidity, a blot force of 5, and a 3 s blot time.
For both NINJ1(K45Q) and NINJ1(K45Q)–Nb538, electron microscopy data were collected with a Titan Krios G2 equipped with a Falcon4i direct electron detector, SelectrisX energy filter, and EPU automation software (Thermo Fisher Scientific). In total, 4,576 movies (NINJ1(K45Q)) and 11,802 movies (NINJ1(K45Q)–Nb538) were recorded at an operating voltage of 300 kV, 165,000× nominal magnification (calibrated pixel size 0.731 Å per pixel), energy filter slit width 20 eV, defocus range −0.8 to −2.6 μm, and a total electron fluence of ~40 e− Å2 (Extended Data Table 1 and Extended Data Fig. 2).
Cryo-EM data processing
CryoSPARC was used for all image processing26. For both the NINJ1(K45Q) and NINJ1(K45Q)–Nb538 datasets, CryoSPARC Live was used for movie motion correction, CTF estimation and initial ab initio blob particle picking (Extended Data Fig. 4b,c). For 3D reconstruction, we used 2D classes showing NINJ1 in a dimer of dimers configuration due to the availability of more diverse views. For the NINJ1(K45Q)–Nb538 dataset, after 2D classification and 3D ab initio and heterogenous refinement produced a low-resolution electron microscopy map of the complete complex, this low-resolution map was used for template-based picking. An additional sequence of 2D and 3D processing, including rebalancing 2D classes to account for a degree of preferred orientation, resulted in an improved 3D map that was used as a picking template and initial 3D model for the final image processing sequence. Using this template, 1,568,603 particles were picked, and 2D classification and 3D heterogenous refinement identified a final class of 102,877 particles. After reference-based motion correction, non-uniform refinement (including global tilt/trefoil and per-particle defocus CTF refinement) produced a map with a global resolution of ~3.0 Å, as measured by a Fourier shell correlation cutoff of 0.143 between independently processed half-maps27. Local resolution varied from ~2.7 to 3.9 Å (Extended Data Fig. 5). Map interpretability was further improved by density modification using Phenix Resolve cryo-EM28. Final data processing statistics are reported in (Extended Data Table 1). For the NINJ1(K45Q) dataset without Nb538, a similar data processing scheme was used(Extended Data Fig. 2). First, particles from the initial ab initio blob particle picking were sorted using 2D classification and 3D ab initio and heterogenous refinement to produce a low-resolution and anisotropic 3D map, which was then used for template-based particle picking, identifying 2,163,892 particles. Subsequent 2D classification and 3D ab initio and heterogenous refinement resulted in a final class of 319,331 particles, and map interpretability was improved using DeepEMhancer29. For apo NINJ1(K45Q) without Nb538, the final map was highly anisotropic and hence not used for model building.
Model building and refinement
Separate AlphaFold29 models of NINJ1 and Nb538 lacking complementarity determining regions (CDRs) were fitted into the final density-modified cryo-EM map using UCSF ChimeraX. A near-complete model was built and refined using ISOLDE30. The model was refined using real-space refinement in Phenix v1.2131. ChimeraX v.1.7.132 and PyMOL v.2.5.2 (Schrödinger) were used for structure visualization and figure preparation.
Immunofluorescence and live-cell imaging
Mouse GFP11–NINJ1, NbAlfa–GFP1–10 and Nb538–GFP1–10 cDNA were synthesized and subcloned into pcDNA3.1/Zeo(+) (Genscript). Site directed mutagenesis was performed on mouse GFP11–NINJ1 to generate A42W, K45R, S46A and N119Q mutations, respectively (Genscript). These constructs were transiently transfected into Ninj1-deficient BALB/3T3 cells, along with mCherry (Clontech, 632523) as a marker for transfected cells. In brief, Ninj1-deficient BALB/3T3 cells (9,000 per well) were plated on 8-well chamber slides (Ibidi, 80806). Cells were allowed to adhere overnight and then transiently transfected with 100 ng mCherry, 200 ng GFP11-Ninj1, 200 ng of either NbAlfa–GFP1–10 or Nb538–GFP1–10, 0.5 µl Lipofectamine LTX, and 0.25 µl Plus Reagent (Thermo Fisher Scientific). For splitGFP NINJ1 dimer-destabilizing mutants, cells were transiently transfected with 100 ng mCherry, 200 ng GFP11–NINJ1 or indicated GFP11–Ninj1 mutants, 200 ng of either NbAlfa–GFP1–10 or Nb538–GFP1–10, and 0.75 µl TransIT-X2 Reagent (Mirus Bio). 24 h after transfection, cells were imaged in Opti-MEM using a Leica SP8 laser scanning confocal microscope equipped with a 63×/1.40 HC PL APO CS2 oil immersion objective. Samples were maintained at 37 °C and 5% CO2 throughout imaging using an environmental control chamber and gas mixer (OkoLab). Images were processed in Fiji software. Mean average GFP fluorescence was quantified at the plasma membrane of transfected cells using Fiji. Fluorescence was normalized to the empty vector control. High expression of the NINJ1 dimer-dissociating mutants GFP11–NINJ1(A42W) and GFP11–NINJ1(K45R) caused cellular toxicity in Ninj1-deficient BALB/3T3s. Thus, for quantification of splitGFP NINJ1 dimer-destabilizing mutants, expression of mCherry (mean intensity between 30–60 arbitrary units using the Measure function in Fiji software) was used to threshold for cells with similar transfection levels.
cDNAs encoding non-tagged mouse NINJ1 and its mutants were synthesized and subcloned into pBCMV_Blasticidin (Genscript). For fixed-cell imaging of NINJ1 mutant localization, Ninj1-deficient BALB/3T3 cells were transfected with 100 ng mCherry, 100 ng of indicated NINJ1 plasmid, and 0.75 µl TransIT-X2 Reagent. Cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, permeabilized with 0.05% saponin, and incubated in blocking serum (PBS supplemented with 0.05% saponin and 10% normal donkey serum (Jackson ImmunoResearch)) for 1 h at room temperature. Cells were then incubated with anti-mouse NINJ1 clone 80 (rabbit IgG2b raised against the N-terminal extracellular domain; Genentech, 2 µg mL−1) diluted in blocking serum for 2 h at room temperature. NINJ1 antibody signal was detected using AF647 anti-rabbit secondary (Thermo Fisher Scientific, A31573, lot 2674379, 1:500) incubated for 1 h at room temperature. Nuclei were stained with DAPI (Thermo Fisher Scientific, D1306, lot 2462719, 1 µg ml−1). High-resolution images were acquired using a Leica SP8 laser scanning confocal microscope equipped with a 63×/1.40 HC PL APO CS2 oil immersion objective.
Cytotoxicity and YOYO-1 assay
We selected structure-based mutants as follows. To destabilize face-to-face dimers, we mutated amino acids to introduce bulky side chains to introduce a steric clash in the dimer interface, whereas to stabilize dimers we introduced complementary mutations without bulky side chains. We introduced mutations in amino acids which do not contribute to the active-state NINJ1 filament interface observed in PDB: 8SZA. For transient expression in HEK293T cells, 2 × 104 cells were transfected with 50 ng of plasmid encoding non-tagged mouse NINJ1 or indicated mutants with 0.16 μl Lipofectamine 2000 (Thermo Fisher Scientific) per well in Costar 96-well white flat bottom plates (Fisher Scientific). At 16 h after transfection, cytotoxicity was measured by CellTiter-Glo assay (Promega).
YOYO-1 dye (Thermo Fisher Scientific) was added at a final concentration of 200 nM at the time of transfection. IncuCyte S3 images were scanned in the green channel every hour for at least 20 h and at 10× magnification. Nuclear-ID Red DNA stain (Enzo Life Sciences) was added after the last time point and scanned in the red channel. IncuCyte S3 2019A software was used to determine the total number of YOYO-1+ cells and Nuclear-ID+ cells (total cells). The percentage of YOYO-1+ cells was calculated as the number of YOYO-1+ cells divided by the total number of Nuclear-ID+ cells. Each independent replicate was performed on a separate day. All data points in any given figure panel come from one plate to rule out plate-to-plate variation.
Molecular dynamics simulations setup
The cryo-EM structure of NINJ1(K45Q), with Nb538 removed, was prepared for simulation using the 2023-1 release of Maestro (Schrödinger). To measure TM1 movement, four models generated from the cryo-EM structure of NINJ1(K45Q) were used. The first model included two side-to-side NINJ1(K45Q) molecules in the presence of face-to-face dimerization (total 4 NINJ1 molecules as a side-to-side dimer of face-to-face dimers). The second model was generated from the first model by deleting the face-to-face dimers (total of two NINJ1 molecules as a side-to-side dimer). For the third model, five hypothetical side-to-side NINJ1(K45Q) molecules were generated in the presence of face-to-face dimerization (total ten NINJ1 molecules as a side-to-side pentamer of face-to-face dimers). The fourth model was generated from the third model by deleting the face-to-face dimers (total of five NINJ1 molecules as side-to-side multimers). For simulations with wild-type NINJ1, all four Gln45 residues in the cryo-EM resolved NINJ1(K45Q) dimer of dimers model were mutated back to Lys45. For all models, N and C termini were capped with acetyl and N-methyl amide groups respectively using Protein Preparation Wizard module33. Protonation states for His, Glu, Asp and conformational flips of His, Asn, Gln side chains were optimized using PROPKA33 at pH 7.4. Restrained minimization was carried out using the OPLS4 force field34, with heavy atoms converging to a root-mean-square deviation (r.m.s.d.) of 0.3 Å. The prepared system was finally embedded into a pre-equilibrated POPC membrane with an initial orientation based on the OPM database, solvated with SPC waters and neutralized with 0.15 M sodium chloride ions.
Molecular dynamics simulations protocol and analysis
Molecular dynamics simulations were carried out using the Desmond suite (Schrödinger) with OPLS4 force field35. Three independent simulations were performed, each undergoing an equilibration stage followed by a 1 µs production run. The membrane relaxation protocol in the Schrödinger software was used to equilibrate the system.
Snapshots from each trajectory were saved every 1,000 ps during the production phase of the simulation. Simulation frames were analysed after aligning all frames to the transmembrane helices of the starting structure. The r.m.s.d. and distances were analysed using simulation event analysis and figures were generated with PyMOL (Schrödinger) and Seaborn Python package36. All molecular dynamics analysis was done with three independent runs starting from randomized initial velocity (total 3 × 1 μs = 3 μs). The r.m.s.d. plots were generated with the Seaborn Python package with the mean and standard deviation calculated from all three molecular dynamics replicates.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-024-08273-4.
Supplementary information
Supplementary Table 1
Source data
Acknowledgements
The authors thank P. Joshi, C. Tam and the Biomolecular Resources Department at Genentech for cloning and protein expression support; W. Phung and W. Sandoval for native mass spectrometry; and P. Dutka, J. Nicoludis and N. Skelton for helpful discussions.
Extended data figures and tables
Author contributions
V.M.D. and I.D. designed the study. S.P. expressed NINJ1. S.P., K.A. and I.D. purified NINJ1 constructs. L.Z. performed yeast display for nanobody selection with input from I.D. and Y.Z. L.Z. performed ELISA screening and cloned nanobody constructs. I.D. expressed and purified nanobodies and performed FSEC. S.G. performed SPR experiments with input from I.D. I.D. generated the NINJ1(K45Q)–Nb538 complex and prepared cryo-EM grids. M.C.J. screened cryo-EM grids, followed by cryo-EM data collection. M.C.J., I.D. and S.P. performed extensive image processing with input from A.R. I.D. and S.P. built and refined models of NINJ1(K45Q) in complex with Nb538. M.C.J., S.P. and I.D. analysed cryo-EM data and models, and prepared figures and tables. M.E.T. and I.B.S. isolated and cultured primary macrophages. S.P. performed native PAGE experiments. M.E.T. performed and analysed live-cell and fixed-cell imaging experiments and associated immunoblots, and prepared figures. I.B.S. generated Ninj1−/− BALB/3T3 cells. Y.Y. set up, performed and analysed molecular dynamics simulations. Y.Y. prepared r.m.s.d. and distance plots. P.A.C., I.B.S. and N.K. performed cytotoxicity assays, analysed data and performed immunoblots. N.K. prepared figures for cytotoxicity assays. K.A. performed cargo-release assays. I.D. wrote the manuscript with input from all authors. The overall project was supervised by K.N., V.M.D. and I.D.
Peer review
Peer review information
Nature thanks Michelle Dunstone and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
Coordinates for NINJ1(K45Q) bound to Nb538 have been deposited in the RCSB Protein Data Bank (PDB) under accession code 9BIA. Corresponding electron microscopy density map has been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-44585. All other data that support the findings of this study are available from the corresponding authors upon request. This paper makes use of PDB accession code 8SZA. Source data are provided with this paper.
Competing interests
The following authors are employees of Genentech: S.P., M.E.T., M.C.J., Y.Y., L.Z., K.A., I.B.S., S.G., P.A.C., Y.Z., A.R., K.N., N.K., V.M.D. and I.D.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sergei Pourmal, Melissa E. Truong, Matthew C. Johnson, Ying Yang
Contributor Information
Vishva M. Dixit, Email: dixit@gene.com
Ishan Deshpande, Email: deshpani@gene.com.
Extended data
is available for this paper at 10.1038/s41586-024-08273-4.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-024-08273-4.
References
- 1.Kayagaki, N. et al. NINJ1 mediates plasma membrane rupture during lytic cell death. Nature591, 131–136 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Degen, M. et al. Structural basis of NINJ1-mediated plasma membrane rupture in cell death. Nature618, 1065–1071 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David, L. et al. NINJ1 mediates plasma membrane rupture by cutting and releasing membrane disks. Cell187, 2224–2235 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahoo, B., Mou, Z., Liu, W., Dubyak, G. & Dai, X. How NINJ1 mediates plasma membrane rupture and why NINJ2 cannot. Preprint at bioRxiv10.1101/2023.05.31.543175 (2023). [DOI] [PMC free article] [PubMed]
- 5.Kayagaki, N., Webster, J. D. & Newton, K. Control of cell death in health and disease. Annu. Rev. Pathol.19, 157–180 (2024). [DOI] [PubMed] [Google Scholar]
- 6.Kayagaki, N. et al. Inhibiting membrane rupture with NINJ1 antibodies limits tissue injury. Nature618, 1072–1077 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramos, S., Hartenian, E., Santos, J. C., Walch, P. & Broz, P. NINJ1 induces plasma membrane rupture and release of damage-associated molecular pattern molecules during ferroptosis. EMBO J.10.1038/s44318-024-00055-y (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borges, J. P. et al. Glycine inhibits NINJ1 membrane clustering to suppress plasma membrane rupture in cell death. eLife11, e78609 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at bioRxiv10.1101/2021.10.04.463034 (2022).
- 11.Cabantous, S., Terwilliger, T. C. & Waldo, G. S. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol.23, 102–107 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Götzke, H. et al. The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat. Commun.10, 4403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han, J.-H. et al. NINJ1 mediates inflammatory cell death, PANoptosis, and lethality during infection conditions and heat stress. Nat. Commun.15, 1739 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dondelinger, Y. et al. NINJ1 is activated by cell swelling to regulate plasma membrane permeabilization during regulated necrosis. Cell Death Dis.14, 755 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi, J. et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature526, 660–665 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Kayagaki, N. et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature526, 666–671 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Murphy, J. M. et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity39, 443–453 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Hildebrand, J. M. et al. Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc. Natl Acad. Sci. USA111, 15072–15077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su, L. et al. A plug release mechanism for membrane permeation by MLKL. Structure22, 1489–1500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun, L. et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell148, 213–227 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Nagata, S., Suzuki, J., Segawa, K. & Fujii, T. Exposure of phosphatidylserine on the cell surface. Cell Death Differ.23, 952–961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araki, T. & Milbrandt, J. Ninjurin, a novel adhesion molecule, is induced by nerve injury and promotes axonal growth. Neuron17, 353–361 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Barthelemy, P. A. et al. Comprehensive analysis of the factors contributing to the stability and solubility of autonomous human VH domains. J. Biol. Chem.283, 3639–3654 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Tonikian, R., Zhang, Y., Boone, C. & Sidhu, S. S. Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat. Protoc.2, 1368–1386 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Xu, H. et al. in High-Throughput Protein Production and Purification: Methods and Protocols (ed. Vincentelli, R.) 389–402 (Springer, 2019).
- 26.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger, T. C., Ludtke, S. J., Read, R. J., Adams, P. D. & Afonine, P. V. Improvement of cryo-EM maps by density modification. Nat. Methods17, 923–927 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol.4, 874 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meng, E. C. et al. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci.32, e4792 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R. & Sherman, W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des.27, 221–234 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Lu, C. et al. OPLS4: improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput.17, 4291–4300 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Bowers, K. J. et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proc. 2006 ACM/IEEE Conference on Supercomputing 84–es (ACM, 2006).
- 36.Waskom, M. seaborn: statistical data visualization. J. Open Source Softw.6, 3021 (2021). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Data Availability Statement
Coordinates for NINJ1(K45Q) bound to Nb538 have been deposited in the RCSB Protein Data Bank (PDB) under accession code 9BIA. Corresponding electron microscopy density map has been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-44585. All other data that support the findings of this study are available from the corresponding authors upon request. This paper makes use of PDB accession code 8SZA. Source data are provided with this paper.