Abstract
Pathogen-associated molecular patterns (PAMPs) are highly conserved motifs originating from microorganisms that act as ligands for pattern recognition receptors (PRRs), which are crucial for defense against pathogens. Thus, PAMP-mimicking vaccines may induce potent immune activation and provide broad-spectrum protection against microbes. Dextran encapsulation can regulate the surface characteristics of nanoparticles (NPs) and induces their surface modification. To determine whether dextran-encapsulated NPs can be used to develop antiviral vaccines by mimicking viral PAMPs, we synthesized NPs in a cyclohexane inverse miniemulsion (Basic-NPs) and further encapsulated them with dextran or tetramethylrhodamine isothiocyanate (TRITC)-dextran (Dex-NPs or TDex-NPs). We hypothesized that these dextran encapsulated NPs could activate innate immunity through cell surface or cytosolic PRRs. In vitro and in vivo experiments were performed using RAW 264.7 and C57BL/6 mice to test different concentrations and routes of administration. Only TDex-NPs rapidly increased retinoic acid-inducible gene I (RIG-I) at 8 h and directly bound to it, producing 120–300 pg/ml of IFN-α via the ERK/NF-κB signaling pathway in both in vitro and in vivo models. The effect of TDex-NPs in mice was observed exclusively with footpad injections. Our findings suggest that TRITC-dextran encapsulated NPs exhibit surface properties for RIG-I binding, offering potential development as a novel antiviral and anticancer RIG-I agonist.
Keywords: Pathogen-associated molecular pattern; Pathogen-associated molecular pattern molecules; Nanoparticles; Receptors, pattern recognition; Retinoic acid-inducible gene I; Interferon-alpha
INTRODUCTION
The application of nanotechnology in biomedical engineering has gained substantial interest in recent years (1,2). Nanoparticles (NPs) typically range in size from 1 to 100 nm, and various types of NPs, including lipid-based NPs (liposomes and micelles), organic/inorganic NPs, nanopolymers, emulsions, and virus-like particles, have been developed for novel pharmaceutical approaches such as drug delivery, cancer therapy, and bioimaging (3). Recently, NPs have also been employed in developing vaccines for various diseases. These vaccines primarily interact with the immune system by targeting innate immune responses and protecting against pathogens via molecular pattern recognition receptors (PRRs) (4). Engineered nanovaccines have emerged as a key field in vaccine development. These specialized NPs are constructed with Ags of interest or immunomodulatory materials through conjugation, encapsulation, adsorption, or simple mixing (5). NPs loaded with specific Ags may activate the humoral immune system to protect the host from corresponding pathogens (5). Furthermore, nanovaccines that capture pathogen-associated molecular patterns (PAMPs) elicit robust humoral immune responses and protect the host from a broad range of microbes, making them ideal candidates for novel antibacterial vaccines (6,7). Additionally, some NPs may stimulate innate immune responses by acting as ligands for PRRs. These PRR-engaging NPs are considered potential adjuvants or innovative vaccines, distinct from traditional vaccines (4,8).
The surface properties and patterns of NPs significantly aect their interactions with biomolecules, cells and microorganisms (9,10). Therefore, surface pattern-associated NPs exhibit distinct physical and chemical surface characteristics that mimic PAMPs or damage-associated molecular patterns (5,11). As a result, these NPs can activate various components of the innate immune system, including macrophages, dendritic cells, and other immune cells, through mechanisms similar to pathogen recognition (12,13). The interaction between NPs and the innate immune system is of particular interest due to its implications for the safety and efficacy of nanomedicines and nanomaterials (14). Understanding how different surface patterns influence immune activation can aid in designing NPs that are either more effective for therapeutic purposes or less likely to induce unwanted immune responses (4). Polysaccharides are used in NP surface modification due to their biocompatibility and versatility. They enhance NPs’ stability, circulation time, and cellular uptake, facilitating targeting of specific cells in immune responses (15). Different polysaccharides, such as polyethylene glycol (PEG) and dextran, are commonly introduced to enhance NP functionality. PEGylation, or coating NPs with PEG, prevents aggregation and reduces non-specific immune cell uptake. In contrast, dextran layers promote NP uptake by Ag-presenting cells, such as macrophages and dendritic cells, thus enhancing the immune response (15).
Retinoic acid-inducible gene I (RIG-I) is a critical molecule in the innate immune system that senses viral ss- or dsRNA (16) and activates downstream signaling cascades to induce the transcription of genes encoding type I IFNs, which confer an antiviral state by inhibiting viral replication and cell-to-cell transmission of the virus (17,18). RIG-I plays a central role not only in immunity against viral infections but also in anticancer immunity (17,18). Even in the absence of adaptive immunity, stimulation of RIG-I provides broad protection against viral infections in a type I IFN-dependent manner (19). Furthermore, the activation of RIG-I signaling in tumors induces cancer cell death and recruits immune cells to the tumor microenvironment, thereby enhancing the antitumor effects of cancer therapy (20). Thus, RIG-I is a potential target for immunomodulatory drugs. RIG-I agonists are mostly engineered as short nucleotides or RNA (18,21). RIG-I agonists are being explored for therapeutic applications in treating viral infections and cancer due to their ability to enhance immune responses (17,22,23).
RIG-I agonists face multiple barriers to therapeutic efficacy that are shared with many oligonucleotide therapies, including a short plasma half-life (i.e., minutes), high susceptibility to nuclease degradation, inefficient intracellular delivery, and, critically, degradation in lysosomes with minimal delivery to the cytosol where RIG-I is located (24,25). To overcome these barriers, research has highlighted the advantages of using NPs for the delivery of RIG-I agonists. NP-based delivery systems enhance the stability, efficacy, and targeting of RIG-I agonists (26,27,28). These NPs protect the agonists from degradation, facilitate cellular uptake, and allow for controlled release (29,30,31). Various materials, including lipids and polymers, can be used to optimize the NPs for specific therapeutic needs (32,33,34,35). The effectiveness of NP-delivered RIG-I agonists has recently been demonstrated in both antiviral applications and cancer treatments. In antiviral contexts, they enhance immune responses when used as adjuvants in vaccines against viral pathogens such as severe acute respiratory syndrome coronavirus 2 (36). In cancer treatment, NP formulations with RIG-I agonists have led to significant tumor growth inhibition and enhanced immune response against various cancers (37,38,39). Therefore, the integration of RIG-I agonists with NP delivery systems presents a promising and innovative approach for enhancing both antiviral vaccine development and cancer immunotherapy.
This study aimed to evaluate novel dextran-encapsulated NPs that mimic viral PAMPs for activating the RIG-I pathway and IFN production. We specifically found the ability of tetramethylrhodamine isothiocyanate (TRITC)-dextran encapsulated NPs (TDex-NPs) to stimulate RIG-I and IFN production both in vitro and in vivo. Our findings suggest that TDex-NPs are promising candidates for the development of innovative immunotherapeutic strategies with potential broad applications in antiviral therapy and cancer immunotherapy.
MATERIALS AND METHODS
Construction and preparation of NPs
A water soluble macroinitiator, poly (ethylene glycol) functionalized bromide (PEO5000-Br) was synthesized using a reported procedure (20). Using a similar procedure, 2-bromoisobutyryl bromide (0.46 g, 2.0 mmol, #252271; Sigma-Aldrich, St. Louis, MO, USA) was added dropwise to a solution of poly (ethylene glycol) monomethyl ether (Mn = 5,000 g/mol, 5.0 g, 1.0 mmol, #81323; Sigma-Aldrich) and triethylamine (0.2 g, 2.0 mmol, #471283; Sigma-Aldrich) in dichloromethane (75.0 ml, #3030-4404; Daejung, Siheung, Korea) at 0°C. The resulting solution was stirred for 16 h at 25°C. The product was isolated in hexane (#139386; Sigma-Aldrich) and dried in a vacuum dry oven for 24 h at 25°C (Basic-NPs). NPs encapsulated in dextran (average mol wt up to 11,600 Da, #00270; Sigma-Aldrich) (Dex-NPs) or TRITC-dextran (average mol wt up to 10,000, #R8881; Sigma-Aldrich) (TDex-NPs) were synthesized via ‘Activators Generated by Electron Transfer for Atom Transfer Radical Polymerization’ (21). PEO5000-Br, oligo monomethyl ether methacrylates (OEOMA 300), and poly (ethylene oxide) dimethacrylate (PEODMA) were used as a macroinitiator, biocompatible monomer, and crosslinker, respectively. Tris ([2-pyridyl] methyl) amine (TPMA), copper (II) bromide (CuBr2), and Span 80 were used as a ligand, catalyst, and surfactant, respectively. An aqueous solution of PEO5000-Br (74.4 mg, 0.014 mmol), OEOMA 300 (1.3 g, 4.3 mmol), PEODMA (97.5 mg, 0.13 mmol), TPMA (2.1 mg, 0.007 mmol), CuBr2 (1.6 mg, 0.007 mmol), encapsulation materials (Dextran or TRITC-dextran) (6.5 mg), and 3rd deionized water (1.4 ml) were mixed in a 50.0 ml Schlenk flask. To the aqueous solution, we added an organic solution of Span 80 (1.0 g) in cyclohexane (20.0 g). The mixture was sonicated for 20 min in an ice bath at 0°C to form a stable inverse miniemulsion. Oxygen was removed from the milky emulsion via nitrogen bubbling for 30 min. The reaction was initiated by adding an aqueous solution of ascorbic acid (0.028 mmol/ml, 0.004 mmol, 155 μl). Subsequently, the reaction was terminated by exposing to air after 24 h. The resulting NPs were separated via centrifugation (21,206 ×g, 20 min, 4°C), and decantation of the supernatant was repeated. The same procedure was performed twice adding tetrahydrofuran (#34865; Sigma-Aldrich) to remove residue reactants. For footpad injection, NPs were concentrated via centrifuging for 60 min at 21,206 ×g and 4°C. The supernatant was removed and 200 μl of PBS was added to achieve a final concentration of 7.5 mg/ml.
Dynamic light scattering (DLS) measurement
DLS measurements were conducted to determine the size distribution and hydrodynamic radius of the NPs. The samples were prepared by diluting the NP suspension in appropriate solvent to an optimum concentration, typically around 0.1 mg/ml, to ensure reliable scattering intensity. DLS measurements were performed using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK) equipped with a 633 nm laser at a scattering angle of 173°. Data were acquired at a fixed temperature of 25°C. Each measurement was repeated 3 times to ensure accuracy, with data being analyzed using the provided software to obtain the size distribution profile through cumulant analysis. Before the measurements, the instrument was calibrated using NIST-traceable standard sizes to ensure accuracy and reliability. Results are reported as the z-average diameter and polydispersity index, which provides insights into the uniformity of the particle size distribution.
Cell culture
RAW 264.7, a murine macrophage cell line, was obtained from the Korean Cell Line Bank and cultured in DMEM (LM001-01; WelGene, Gyeongsan, Korea) supplemented with 10% (v/v) FBS (26-140-079; Gibco, Waltham, MA, USA) and 1% penicillin/streptomycin (LS202-02; WelGene) at 37°C and 5% CO2. Cells (0.5×106 or 0.05×106) were cultured in a 35- or 24-well culture dish, respectively, and treated with polyinosinic-polycytidylic acid (poly[I:C]) (0.2 µg/ml, P1530; Sigma-Aldrich) or NPs at various doses or time points for further analysis. To examine their stability, TDex-NPs were kept at 4°C for 1 or 3 months, and cells were treated with fresh, 1-month-old, or 3-month-old TDex-NPs for 18 h at various doses, followed by further analysis. As the American Type Culture Collection recommends (40), RAW 264.7 cells were used up to a passage number of 18 for optimal performance related to their differentiation and stability.
Animal studies
Female C57BL/6 mice were obtained from Orient Bio, Inc. (Seongnam, Korea) and maintained under specific pathogen-free conditions with food and water available ad libitum. This study was approved by the Institutional Animal Care and Use Committee of the Konkuk University (KU IACUC Protocol # 17030). Separate groups of mice received intravenous (IV) or intraperitoneal (IP) injection of PBS or 200 μg of NPs, respectively. After 72 h, the mice were sacrificed, and the spleen, liver, lungs, lymph nodes (LNs), and thymus were harvested and prepared for immunoblotting or immunofluorescence assays. TDex-NPs (200 μg/100 µl) were intravenously or intraperitoneally injected into mice, and their sera collected at 0, 1, 3, 5, and 7 days were analyzed for various cytokines using ELISA. For footpad injection of NPs, 150 μg/20 μl of NPs was injected into the left footpad or both footpads of mice.
MTT assay
RAW 264.7 cells were cultured in 96-well plates and treated with 1, 10, 100, or 200 µg/ml of Basic-NPs, Dex-NPs, and TDex-NPs. After 18 h of treatment, 10 µg of MTT (Sigma-Aldrich) dissolved in 100 µl PBS was added to each well, and the plates were incubated for 2 h at 37°C. Subsequently, MTT was removed, and 100 µl of DMSO was added to each well. Absorbance was measured at 560 nm using a SpectraMax microplate reader (Molecular Devices, San Jose, CA, USA).
Immunofluorescence assay
RAW 264.7 cells were cultured on coverslips, washed with PBS, and fixed with 100% acetone (#179124; Sigma-Aldrich) at 25°C. After washing with PBS, cells were incubated overnight at 4°C with primary Abs against RIG-I (sc-376845, 1:100; Santa Cruz, Dallas, TX, USA). After washing with PBS, the cells were incubated with secondary Abs conjugated to the respective fluorescent dyes (e.g., Alexa Fluor 488 or 594, 1:4,000–5,000) and DAPI (D1306; Invitrogen, Waltham, MA, USA), followed by extensive washing with PBS. All primary and secondary Abs were diluted in PBS containing 3% CAS-BLOCK histochemical reagent (#008120; Invitrogen). The cells were mounted using Gel Mount Aqueous Mounting Medium (g0918; Sigma-Aldrich). For immunofluorescent staining of mouse tissues including LNs, spleen, liver, lung, and thymus, isolated tissues from C57BL/6 mice were embedded and frozen in Tissue-Tek® optimal cutting temperature compound (Sakura Finetechnical Co., Tokyo, Japan). The frozen tissues were then sectioned at a thickness of 10 µm using a cryostat (Leica CM3050 S or equivalent) and mounted onto Superfrost Plus microscope slides (Thermo Fisher Scientific, Waltham, MA, USA). Slides were air-dried for 30 min at 15°C–25°C to improve tissue adherence. For immunofluorescence staining, sections were initially rehydrated and washed three times in PBS for 5 min each. Blocking of nonspecific binding sites was performed by incubating the sections in a blocking buffer containing 3% CAS-BLOCK histochemical reagent in PBS for 1 h at room temperature. Slides were incubated with primary Abs (1:100) overnight at 4°C in a humidified chamber to ensure specific binding. After incubation, sections were washed three times with PBS for 5 min each to remove excess primary Abs. For detection, fluorophore-conjugated secondary Abs were diluted in blocking buffer and applied to the sections, which were then incubated in the dark for 1 h at room temperature. Following secondary Ab incubation, sections were counterstained with DAPI at a dilution of 1:5,000 in PBS for 5 min to visualize nuclei. Fluorescent images of the cells or tissue sections were captured using a deconvolution microscope (Olympus Korea Co. Ltd., Seoul, Korea) and processed with image analysis software such as ImageJ or MetaMorph software (MetaMorph Inc., San Jose, CA, USA).
Western blot analysis
Cells or tissues obtained from mice were lysed in radioimmunoprecipitation assay buffer supplemented with a protease inhibitor cocktail and phenylmethylsulfonyl fluoride (#52332; Sigma-Aldrich). The lysates were centrifuged for 5 min at 13,500 rpm (rcf), and supernatants were further quantified via bicinchoninic acid assay using a WelProt Protein Assay Kit (WelGene). Lysates were heated at 95°C for 10 min with 5× SDS sample loading buffer containing β-mercaptoethanol. Protein lysates were separated via 10%–12% SDS-PAGE electrophoresis and transferred to polyvinylidene fluoride membranes (IPVH08100; Millipore, Burlington, MA, USA) using standard wet blot procedures. Membranes were blocked for 1 h at room temperature with 5% skim milk dissolved in TBST solution and incubated overnight at 4°C with following primary Abs (1:500–1:2,000): RIG-I, GAPDH, TLR2, TLR3, TLR4, GAPDH, β-actin (Santa Cruz), ERK, pERK, AKT, pAKT, NF-κB, and pNF-κB (Cell Signaling Technology, Danvers, MA, USA). After incubation with the respective HRP-conjugated secondary Abs (1:10,000–20,000) (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) for 1 h at room temperature, HRP substrate (Millipore) was added, and the proteins were visualized via a chemiluminescent assay using the LAS-4000 imaging system (Fujifilm, Tokyo, Japan). The intensity of the bands in western blotting was accurately measured using ImageJ (http://rsb.info.nih.gov/ij/).
Immunoprecipitation
RAW 264.7 cells were lysed as described previously (41). and incubated with an Ab against RIG-I in a shaker at 4°C for 30 min. Lysates were centrifuged for 5 min at 4°C and 15,928 ×g, and the supernatant was collected. Either trypsin-EDTA (Gibco) or PBS was added to the supernatant, and the mixture was centrifuged for 5 min at 4°C, 13,000 rpm (rcf). Supernatants were collected and immunoblot analysis for RIG-I was performed as previously described (42).
ELISA
Cell culture supernatants from RAW 264.7 (1:2 diluted) and blood samples from venous sinuses of the mice were collected, and the levels of mouse IL-10 (BD Biosciences, Franklin Lakes, NJ, USA), IFN-α (PBL Assay Science, Piscataway, NJ, USA), and TNF-α (BioLegend, San Diego, CA, USA) were measured in duplicate using corresponding ELISA kits per the manufacturer’s instructions. The absorbance of the plates was measured at 450 nm using a SpectraMax microplate reader (Molecular Devices).
Statistical analysis
Data are presented as the mean ± SD. The statistical significance of differences among experimental groups was determined using one-way ANOVA or an unpaired t-test (p-value <0.05). All statistical analyses were performed using GraphPad Prism, version 9 (GraphPad Software, San Diego, CA, USA).
RESULTS
Construction and characterization of NPs
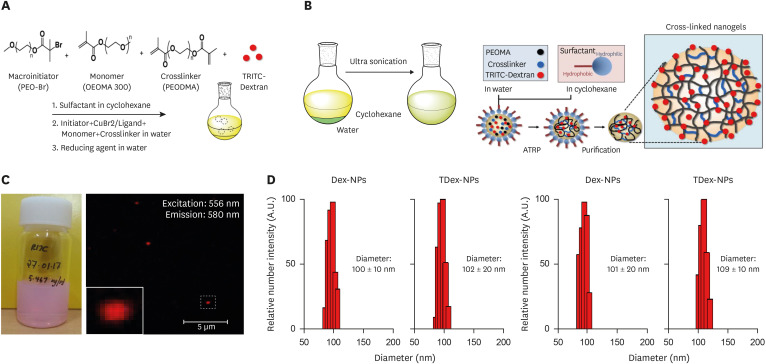
Basic-NPs, Dex-NPs, and TDex-NPs were synthesized via AGET-ATRP inverse miniemulsion (Fig. 1A and B). These NPs were not soluble in any solvent, including cyclohexane, tetrahydrofuran or water, indicating that crosslinking had occurred in the presence of PEODMA during polymerization. The purified TDex-NPs were present as a stable pink dispersion (Fig. 1C, left). In addition, a typical red optical fluorescence signal was detected at 556 nm excitation and 580 nm emission under a fluorescence microscope (Fig. 1C, right). These results indicated that TRITC-dextran was encapsulated by TDex-NPs. The particle size and size distribution of NPs were measured using DLS, and both Dex-NPs and TDex-NPs showed a narrow size distribution of about 100±10 nm and 102±20 nm in diameter, respectively (Fig. 1D). For the stability test of dextran encapsulated NPs, their size was measured after 90 days, which showed diameters (approximately 101±20 nm and 109±10 nm, respectively) similar to those obtained at the initial stage (Fig. 1E).
Figure 1. Characterization and stability of NPs loaded with dextran and TRITC-dextran.
(A) Schematic diagram depicting the synthesis of NPs via AGET-ATRP inverse miniemulsion. SPANPs were prepared without any additives (Basic-NPs), or with dextran (Dex-NPs) or TRITC-dextran (TDex-NPs). (B) The solubility of the synthesized NPs was tested in cyclohexane, THF, and water. (C) Image of purified TDex-NPs showing a stable pink dispersion (left). Fluorescence microscopy of TDex-NPs demonstrates a characteristic red optical fluorescence with excitation at 556 nm and emission at 580 nm (right). (D) Particle size and size distribution of Dex-NPs and TDex-NPs measured by DLS. (E) Stability test results showing the size of NPs after 30 days.
TDex-NPs activate RIG-I in RAW 264.7 cells with minimal toxicity
To examine whether NPs were toxic to cells, cells were treated with Basic-NPs, Dex-NPs, and TDex-NPs for 18 h at varying doses (0, 1, 10, 100, or 200 µg/ml), and cell viability was analyzed using the MTT assay. Although cells showed <80% viability at both 100 and 200 µg/ml of all NPs, TDex-NPs were less toxic than Basic-NPs and Dex-NPs at all doses examined (Supplementary Fig. 1A).
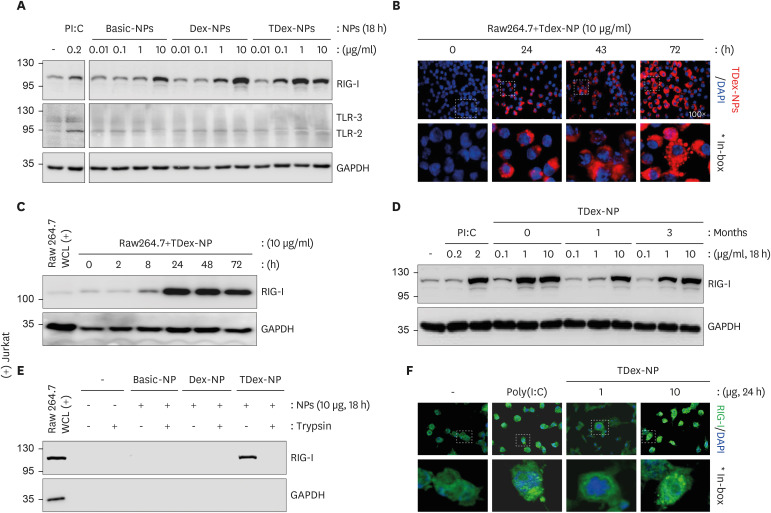
We examined whether Dex-NPs or TDex-NPs stimulated cytosolic PRRs. When RAW 264.7 cells were treated with Basic-NPs, Dex-NPs, and TDex-NPs for 18 h and immunoblotting experiments were performed for RIG-I, TLR-2, and TLR-3, TDex-NPs elicited the greatest increase in RIG-I levels at 1 µg/ml (over 3.5-fold higher than baseline), whereas both Basic-NPs and Dex-NPs showed a marked increase only at 10 µg/ml (Fig. 2A, top row). However, none of NPs increased both TLR-2 and TLR-3 (Fig. 2A, middle row). Given that TDex-NPs were loaded with TRITC-dextran, their uptake was analyzed in vitro using an immunofluorescence microscope (red). RAW 264.7 cells were treated with TDex-NPs for various durations. Its uptake was clearly evident at 24 h, and it was found to be further accumulated at 72 h (Fig. 2B).
Figure 2. Cellular toxicity, PRR stimulation, and RIG-I binding of TDex-NPs.
(A) RAW 264.7 cells were treated with 0.01, 0.1, 1, or 10 µg/ml of TDex-NPs for 18 h and their cell lysates were immunoblotted for TLR-2, TLR-3 or RIG-I. GAPDH was used as a loading control for protein quantity normalization. (B) RAW 264.7 cells were treated with TDex-NPs for various time points (0, 4, 8, 24, 48, and 72 h) and analyzed by immunofluorescence microscopy. The red fluorescence of TDex-NPs indicated its uptake. (C) As in (A), but RAW 264.7 cells were treated with 10 µg/ml of TDex-NPs for different time points (0, 2, 8, 24, 48, and 72 h) and their cell lysates were immunoblotted for RIG-I. (D) As in (C), but cells were treated with fresh, 1-, or 3-month-old TDex-NPs at doses of 0.1, 1, and 10 µg/ml for 18 h. (E) After treating RAW 264.7 cells with 10 µg/ml of Basic-NPs, Dex-NPs, or TDex-NPs for 18 h, the NPs were precipitated from cell lysates by centrifugation and immunoblotted for RIG-I with or without trypsinization. (F) RAW 264.7 cells were treated with 1 or 10 µg/ml of TDex-NPs for 24 h and subsequently immunostained for RIG-I (green). Distinct RIG-I spots were visible in the cytoplasm at the higher dose of 10 µg/ml.
The efficiency of TDex-NPs in increasing RIG-I expression was examined by treating cells with 10 µg/ml of TDex-NPs for various durations (0, 2, 8, 24, 48, and 72 h). The 10 μg contains approximately 6.02×1014 TRITC-dextran NPs. A rapid increase in RIG-I expression was noted at 8 h, with the maximum level achieved at 24 h (over 4- and 17-fold higher than baseline, respectively) (Fig. 2C). The stability of TDex-NPs over time was examined by treating cells with fresh, 1-, or 3-month-old TDex-NPs for 18 h at various doses, which were kept at 4°C (0.1, 1, and 10 µg/ml) and measuring RIG-I expression. Regardless of the storage period, all TDex-NPs increased RIG-I levels in a dose-dependent manner (Fig. 2D).
Viral dsRNA binding to RIG-I stimulates its increase (43). Therefore, we examined whether TDex-NPs taken up by macrophages directly bind to RIG-I. After the cells were treated with 10 µg/ml of Basic-NPs, Dex-NPs, or TDex-NPs for 18 h, NPs were precipitated from the respective cell lysates via centrifugation, and their lysates were immunoblotted for RIG-I with or without trypsinization. Notably, RIG-I binding was only evident in untrypsinized TDex-NPs, indicating that RIG-I is directly bound to the surface of TDex-NPs (Fig. 2E). To confirm the direct binding of RIG-I to TDex-NPs, the cells were treated with 1 or 10 µg/ml of TDex-NPs for 24 h and immunostained for RIG-I. Clear RIG-I spots (green) were observed in the cytoplasm with 10 µg/ml of TDex-NPs (Fig. 2F). Thus, TDex-NPs could directly bind RIG-I.
TDex-NPs induce INF-α secretion through the ERK/NF-κB signaling pathway in RAW 264.7 cells
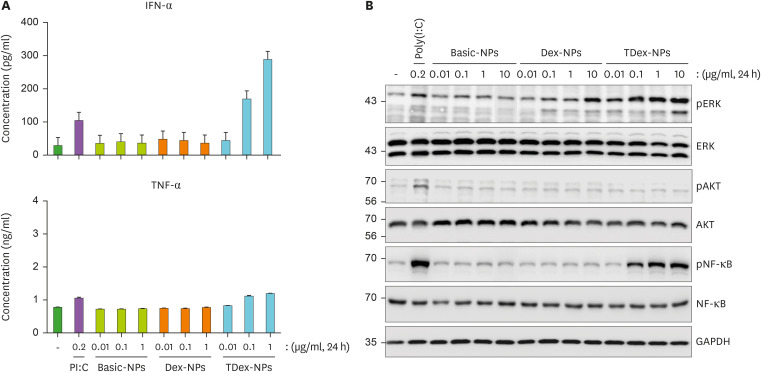
Upon ligand binding, RIG-I induces the expression of type I IFNs and proinflammatory cytokines, such as TNF-α and IL-6 (20,44). Therefore, we examined whether TDex-NP-mediated RIG-I activation induced IFN-α and TNF-α. RAW 264.7 cells were treated with Basic-NPs, Dex-NPs, or TDex-NPs at varying doses (0.01, 0.1, and 1 µg/ml) for 24 h, and their supernatants were analyzed for IFN-α and TNF-α using ELISA. IFN-α secretion was significantly increased only with 0.1 and 1 µg/ml of TDex-NPs in a dose-dependent manner (up to 180 and up to 300 pg/ml, respectively) (Fig. 3A), whereas no such effect was observed for TNF-α. As a negative control, IL-10 was analyzed. Although 0.1 and 1 µg/ml of TDex-NPs resulted in an increase in IL-10 levels, the difference was not significant (Supplementary Fig. 2A). IFN/RIG-I-mediated innate responses are positively associated with the ERK/NF-κB signaling pathway (45). Therefore, we examined whether RIG-I-mediated IFN-α secretion was positively associated with the ERK/NF-κB signaling pathway. RAW 264.7 cells were treated with Basic-NPs, Dex-NPs, or TDex-NPs at various doses (0.01, 0.1, 1, and 10 µg/ml) for 24 h, and their whole-cell lysates were immunoblotted to analyze the presence of ERK, AKT, NF-κB, and their phosphorylated forms. Only TDex-NPs activated ERK and NF-κB at doses of 0.1 µg/ml and above, and the increase was in a dose-dependent manner (Fig. 3B). This trend matched the increase in RIG-I activation when cells were treated with 0.1 µg or higher amounts of TDex-NPs (Fig. 2B).
Figure 3. TDex-NPs induces INF-α secretion through ERK/NF-κB signaling pathway in RAW 264.7 cells.
(A) RAW 264.7 cells were treated with Basic-NPs, Dex-NPs, and TDex-NPs at concentrations of 0.01, 0.1, and 1 µg/ml for 24 h. The supernatants were analyzed for IFN-α and TNF-α levels using ELISA (n=3). Data shown represent mean ± SEM. Statistical significance was determined using ANOVA followed by Tukey’s post hoc test. p<0.05 was considered statistically significant. (B) RAW 264.7 cells were treated with Basic-NPs, Dex-NPs, and TDex-NPs at concentrations of 0.01, 0.1, 1, and 10 µg/ml for 24 h. Whole cell lysates were analyzed by immunoblotting for ERK, AKT, NF-κB, and their phosphorylated forms. GAPDH was used as a loading control for protein quantity normalization.
Footpad injection of TDex-NPs induces IFN-α secretion through RIG-I activation
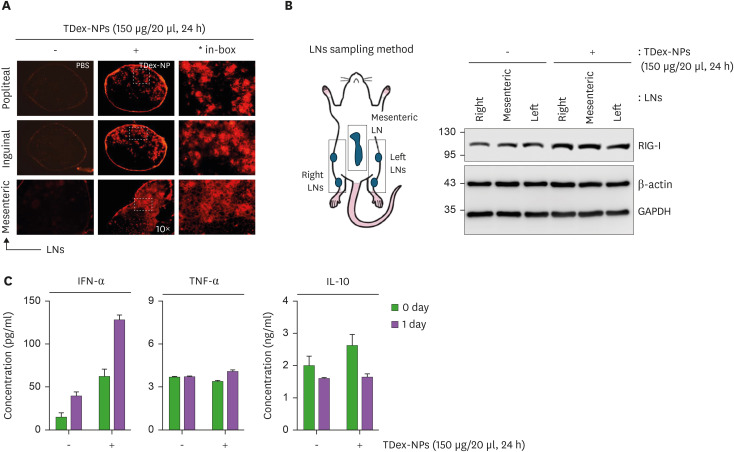
We examined whether the injection of TDex-NPs activates RIG-I leading to IFN-α secretion in vivo. First, 200 µg/ml of TDex-NPs were injected into mice intravenously or intraperitoneally. Mice were euthanized after 72 h, and the uptake of TDex-NPs was examined in cryosections of various tissues (spleen, liver, lung, mesenteric LNs, and thymus) using immunofluorescent microscopy. The TDex-NP signal was not detected in any tissues (Supplementary Fig. 3A). When immunoblotting was performed with the same tissues to analyze RIG-I, no changes were observed in the expression levels of RIG-I (Supplementary Fig. 3B). Mice were injected with 200 µg/ml of TDex-NPs, and their sera collected at 0, 1, 3, 5 and 7 days were analyzed for IFN-α, TNF-α, and IL-10 using ELISA. Consistent with the results presented in Supplementary Fig. 3A and B, there were no changes in the levels of IFN-α, TNF-α, and IL-10 (Supplementary Fig. 3C). Next, 150 µg of TDex-NPs was injected into both footpads of the mice for 24 h, and its uptake was examined in cryosections of popliteal, inguinal, and mesenteric LNs using immunofluorescence microscopy. TDex-NP uptake was evident in the medullary areas of all the LNs (Fig. 4A). Two popliteal, two inguinal, and a mesenteric LN were separately immunoblotted for RIG-I, and the increase was evident in all LNs (over 1.5-fold higher than baseline) (Fig. 4B). As footpad injection of TDex-NPs showed their uptake and an increase in RIG-I levels in LNs, it was hypothesized that TDex-NPs could induce IFN-α secretion, similar to the results observed in in vitro experiments (Fig. 3A). After the injection of 150 µg/20 µl of TDex-NPs into both footpads of mice, sera were harvested at 0 and 24 h and analyzed for IFN-α, TNF-α, and IL-10. A significant increase was noted only for IFN-α at 24 h (up to 130 pg/ml) (Fig. 4C).
Figure 4. In vivo evaluation of TDex-NP uptake, RIG-I activation, and IFN-α secretion.
(A) Mice were injected with PBS or 150 µg/ml of TDex-NPs into both footpads of mice. After 24 h, cryosections of popliteal, inguinal, and mesenteric LNs were analyzed using immunofluorescent microscopy. Representative images are shown (n=3). (B) As in (A), but popliteal, inguinal, and mesenteric LNs from mice were analyzed by immunoblotting for RIG-I. Actin and GAPDH were used as a loading control for protein quantity normalization. (C) Mice were injected with 150 µg/ml of TDex-NPs into both footpads, and sera were collected at 0 and 24 h post-injection. Sera were analyzed for IFN-α, TNF-α and IL-10 levels using ELISA (n=3, p<0.05 compared to control). Data shown represent mean ± SEM. Statistical significance was determined using ANOVA followed by Tukey’s post hoc test. p<0.05 was considered statistically significant.
DISCUSSION
As RIG-I is a cytosolic pathogen recognition receptor that binds to PAMP RNA (19), most RIG-I agonists are engineered into short nucleotides or RNA (18). To activate cytosolic RIG-I, RNA molecules must be first delivered to target tissue and then internalized by immune cells (18). However, due to the labile nature of RNA molecules, inhalation or direct IV injection of RNA results in poor therapeutic efficacy in vivo (46). The inherent negative charge and hydrophilicity of RNA molecules also limit their cellular uptake (47). Therefore, despite these RIG-I agonists being promising antiviral agents, vaccine adjuvants, and cancer immunotherapeutics (48), some oligoribonucleotides have shown limited clinical efficacy (49). There are several challenges associated with efficient delivery of ribonucleotides to achieve good therapeutic efficacy (50). Recently, integrating RIG-I agonists into NP-based vaccines has shown potential for creating broad-spectrum antiviral vaccines and cancer immunotherapy with enhanced safety and efficacy (24,51,52). Therefore, we constructed novel NPs with or without dextran (Dex-NPs) or TRITC-dextran (TDex-NPs) to mimic viral PAMPs (Fig. 1). TDex-NPs only activated RIG-I, followed by NF-κB/ERK activation and IFN-α production in vitro (Figs 2 and 3). Furthermore, foot-pad injection of TDex-NPs into mice induced RIG-I activation and IFN-α production in vivo (Fig. 4), despite not being short nucleotides or RNA. It was reported that RIG-I stimulation by recognition of PAMP dsRNA activated the NF-κB pathway, which resulted in the production of IFNs (17,53,54). Furthermore, the treatment with dextran-coated NPs markedly enhanced the phosphorylation level of ERK on human primary monocyte cells (55,56). It was reported that ERK activation is required for double-stranded RNA- and virus-induced interleukin-1 expression by macrophages, supporting a novel role for ERK in the regulation of the antiviral response of IL-1 expression and release by macrophages (57). Therefore, it is speculated that the direct binding of TDex-NPs to RIG-I activate the NF-κB signaling pathway followed by the production of IFNs, and parallelly, activate ERK in the regulation of the antiviral response of IL-1 expression and release by macrophages. Thus, our findings suggest that TDex-NPs are novel RIG-I agonists that can overcome the challenges associated with oligoribonucleotides and may be useful for developing antiviral or anticancer therapeutics.
Dextran is a complex branched polysaccharide–based neutral polymer chain unit varying in length from 1,000 to 2,000,000 Da. Dextran is a natural, biodegradable, and biocompatible nonionic polysaccharide and water-soluble material (58,59) that has wide applications in pharmaceutical, biomedical, and industrial fields (60,61,62,63). Surface functionalization with dextran is a useful and well-documented strategy that provides multiple advantages to nanocarriers (64), including stabilization to avoid self-agglomeration of neighboring NPs (65) and toxicity of magnetic particles (66,67), and antifouling properties against interference proteins (68). In particular, dextran-coated NPs exhibit bioadhesive properties (69) and enhanced uptake and intracellular delivery of small interfering RNAs in cultured cells (70). Therefore, it was expected that both Dex-NPs and TDex-NPs would be taken up by cells and activate the PRRs.
First, we examined whether dextran encapsulated NPs, such as Dex-NPs or TDex-NPs, could stimulate PRRs, such as RIG-I, TLR-2, and TLR-3. Notably, only TDex-NPs directly bound (Fig. 2E) and rapidly increased RIG-I from 8 h under in vitro conditions (over 4- and 17-fold higher than baseline) (Fig. 2A and C), and sequentially induced IFN-α secretion up to 300 pg/ml through the ERK/NF-κB signaling pathway (Fig. 3). Furthermore, these results were repeated in the in vivo experiment using footpad injection, with RIG-I increase (over 1.5-fold higher than baseline) (Fig. 4B) and IFN-α secretion (up to 130 pg/ml) (Fig. 4C). Although the antiviral therapeutic concentration of IFN-α may vary depending on the subtype of IFN-α, the virus to be treated, experimental conditions, and the pathophysiology of the specific disease, the IC50 values for IFN-α2a and IFN-α17 have been reported to be 14.6 pg/ml and 4.8 pg/ml against HCV-JFH1 in Huh7 cells, respectively (71). Therefore, it could be expected that the IFN-α concentration induced by the increase in RIG-I with TDex-NPs may exert a significant antiviral effect under in vitro or in vivo conditions, strongly suggesting that TDex-NPs could be developed as a novel RIG-I agonist.
RAW 264.7 murine macrophages express many PRRs on their cellular surface, such as TLR-1, TLR-2, and TLR-4 (72), and C-type lectin receptors such as Dectin-1, Dectin-2, and Mincle (52,73). However, these TLRs and CLRs are not dextran receptors. Furthermore, it was recently reported that non-targeted spherical NPs at a size of up to 100 nm were mainly internalized by clathrin-dependent endocytosis (74,75). Other reports have shown that dextran-coated NPs are mainly internalized by fluid phase endocytosis pathways without the mediation of a receptor (76,77). These results strongly suggest that TDex-NPs or Dex-NPs might be primarily taken up by phagocytosis of RAW 264.7 macrophages. Following which, only TDex-NPs can bind to RIG-I and activate its innate immune system. Besides the RIG-I pathway, there are other cytosolic PRRs that include nucleotide binding oligomerization domain-like receptors (NOD-like receptors) (78), AIM2-like receptors (79), cyclic GMP-AMP synthase (80), and nucleases and other DExD/H-box family helicases (81). Given that TDex-NPs activated the RIG-I pathway despite not being short nucleotides or RNA, TDex-NP could directly increase other cytosolic PRRs. Collectively, these results indicate that constructing NPs with polysaccharide-repeated surface patterns to mimic PAMPs could be a novel method for developing RIG-I agonists against a wide class of pathogens, in addition to existing RNA-based RIG-I agonists.
Varying the content of dextran can fine-tune or control the surface characteristics of NPs (82), and dextran encapsulation into NPs induces surface modification and thermal stability, implying utility for therapeutic purposes, including drug delivery (83). In addition, TRITC conjugation on dextran NPs increases the volume and compaction of associated complexes and induces the inverse proportion to molecular weight in intra-axonal mobility, probably due to an increase in intramolecular hydrophobic interactions (84). Also, it was recently found that KIN1148, a small molecule RIG-I agonist, directly binds to RIG-1 and thereby and serves to adjuvant broad multifaceted influenza virus vaccine immunity (22). These reports suggest that NPs encapsulated with TRITC-dextran might enhance the surface patterning and functionalization of NPs by the newly formed structural specificity on the surface of TDex-NPs induces direct binding to RIG-I. This leads to an earlier increase in RIG-I at 8 h of TDex-NP treatment (Fig. 2C) than at 12–16 h of treatment with other RIG-I agonists (85,86,87) and the activation of the RIG-I-mediated innate immune system (Fig. 2). Therefore, both previous reports and our results consistently demonstrated that TRITC conjugation on dextran NPs could affect the biological and physiological activity of TDex-NPs promoting the RIG-I-mediated innate immune system. These results suggest that fluorescent dyes could be utilized to improve the biological and physiological ability of various biomolecule carrier systems and to develop diagnostic and therapeutic tools other than as a fluorescent probe to study cell processes. Furthermore, conjugation with TRITC, which is an amine-reactive derivative of the hydrophobic uorescent dye tetramethylrhodamine isothiocyanate (84,88), facilitates the live monitoring of the location of NPs and their movement using fluorescence microscopy (Fig. 1C). Using these properties of TDex-NPs, we showed that they are taken up by cells to activate RIG-I-mediated innate immune systems both in vitro and in vivo (Figs. 2-4). Furthermore, as expected, and in agreement with previous results (89), TDex-NPs showed improved stability, with their RIG-I-activation properties being retained for up to 3 months (Figs. 1D, E, and 2D), which are beneficial features of this strategy for the development of RIG-I agonists.
Importantly, footpad injection led to the uptake of TDex-NPs in LNs (Fig. 4A) and induced RIG-I activation and IFN-α secretion in vivo (Fig. 4B and C). However, neither IV nor IP injections of TDex-NPs elicited any response in the examined tissues and sera (Supplementary Fig. 3). Optimizing the administration route of a therapeutic agent can improve biodistribution and alter its fate and efficacy in vivo (90). The immune activation resulting from different injection routes of NPs, such as IV, IP, and footpad injections, varies significantly based on how each route influences NP distribution, immune cell interactions, and inflammatory responses (91,92). Compared with IV and IP injections, footpad injection is highly localized, especially to regional LNs. Owing to the high density of Ag-presenting cells such as dendritic cells and macrophages in the footpad area, this route can induce a robust localized immune response with minimal and sustained systemic activation (93,94). Additionally, the lymphatic system plays a key role in the maintenance of an effective immune system and provides a unidirectional pathway from the peripheral tissues to the systemic circulation, which could be beneficial in certain immunotherapeutic applications (95). Therefore, the properties of TDex-NPs seem to require their localization to areas of high density for Ag-presenting cells in regional LNs to induce the RIG-I-mediated innate immune system. This means that administration via the lymphatic circulation is the optimal route for inducing the immunostimulatory properties of TDex-NPs in vivo rather than IV or IP injection, which lead to rapid systemic distribution.
In conclusion, we present a novel approach for developing efficient RIG-I agonists using TDex-NPs independent of RNA. The TDex-NPs construction method demonstrated here provides a versatile platform for developing RIG-I agonists with potential applications as antiviral or anticancer agents and vaccine adjuvants. The unique ability of TDex-NPs to bind directly to RIG-I and activate its innate immune response without relying on short nucleotides or RNA may help overcome challenges associated with existing RIG-I agonists, such as the manufacturing complexity and controlled release requirements of RNA. Further research is needed to enhance TDex-NP applications, including detailed mechanistic studies of their biological interactions, assessment of potential off-target effects, and antiviral efficacy testing through viral challenge experiments.
ACKNOWLEDGEMENTS
We would like to thank Editage (www.editage.co.kr) for English language editing (JOB CODE: KOUNI_5754). This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korea government (Ministry of Science and ICT, MSIT) (No. 2022R1A2C1009466 [Y.S.K], No. RS-2023-00280626 [H.S.H], and No. RS-2023-00213334 [S.W.Y]).
Abbreviations
- CuBr2
copper (II) bromide
- Dex-NP
nanoparticle constructed with dextran
- DLS
dynamic light scattering
- IP
intraperitoneal
- IV
intravenous
- LN
lymph node
- NP
nanoparticle
- OEOMA 300
oligo monomethyl ether methacrylates
- PAMP
pathogen-associated molecular pattern
- PEG
polyethylene glycol
- PEODMA
poly (ethylene oxide) dimethacrylate
- poly(I:C)
polyinosinic-polycytidylic acid
- PRR
pattern recognition receptor
- RIG-I
retinoic acid-inducible gene I
- TDex-NP
nanoparticle constructed with tetramethylrhodamine isothiocyanate-dextran
- TPMA
Tris ([2-pyridyl] methyl) amine
- TRITC
tetramethylrhodamine isothiocyanate
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Kang YS.
- Funding acquisition: Kang YS.
- Investigation: Kang YS.
- Methodology: Baek H, Yang SW, Kim MK, Kim D, Lee C, Kim S, Lee Y, Park M, Paik HJ.
- Project administration: Kang YS.
- Software: Yang SW.
- Supervision: Kang YS.
- Visualization: Baek H, Yang SW, Kim MK.
- Writing - original draft: Baek H, Yang SW.
- Writing - review & editing: Yang SW, Hwang HS, Kang YS.
SUPPLEMENTARY MATERIALS
Toxicity of Basic-NPs, Dex-NPs, and TDex-NPs in RAW 264.7 cells.
IL-10 production by Basic-NPs, Dex-NPs, and TDex-NPs in RAW 264.7 cells.
In vivo immune response after intravenous or intrapertoneal injection of TDex-NP into C57BL/6 mice.
References
- 1.Khan I, Saeed K, Khan I. Nanoparticles: properties, applications and toxicities. Arab J Chem. 2019;12:908–931. [Google Scholar]
- 2.Khan S, Mansoor S, Rafi Z, Kumari B, Shoaib A, Saeed M, Alshehri S, Ghoneim MM, Rahamathulla M, Hani U, et al. A review on nanotechnology: properties, applications, and mechanistic insights of cellular uptake mechanisms. J Mol Liq. 2022;348:118008 [Google Scholar]
- 3.Yao Y, Zhou Y, Liu L, Xu Y, Chen Q, Wang Y, Wu S, Deng Y, Zhang J, Shao A. Nanoparticle-based drug delivery in cancer therapy and its role in overcoming drug resistance. Front Mol Biosci. 2020;7:193. doi: 10.3389/fmolb.2020.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Hardie J, Zhang X, Rotello VM. Effects of engineered nanoparticles on the innate immune system. Semin Immunol. 2017;34:25–32. doi: 10.1016/j.smim.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aljabali AA, Obeid MA, Bashatwah RM, Serrano-Aroca Á, Mishra V, Mishra Y, El-Tanani M, Hromić-Jahjefendić A, Kapoor DN, Goyal R, et al. Nanomaterials and their impact on the immune system. Int J Mol Sci. 2023;24:2008. doi: 10.3390/ijms24032008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao T, Cai Y, Jiang Y, He X, Wei Y, Yu Y, Tian X. Vaccine adjuvants: mechanisms and platforms. Signal Transduct Target Ther. 2023;8:283. doi: 10.1038/s41392-023-01557-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou J, Kroll AV, Holay M, Fang RH, Zhang L. Biomimetic nanotechnology toward personalized vaccines. Adv Mater. 2020;32:e1901255. doi: 10.1002/adma.201901255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peek LJ, Middaugh CR, Berkland C. Nanotechnology in vaccine delivery. Adv Drug Deliv Rev. 2008;60:915–928. doi: 10.1016/j.addr.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi K, Wei JJ, Iida R, Ijiro K, Niikura K. Surface engineering of nanoparticles for therapeutic applications. Polym J. 2014;46:460–468. [Google Scholar]
- 10.El-Sherbiny IM, Abbas Y. Janus nano- and microparticles as smart drug delivery systems. Curr Pharm Biotechnol. 2016;17:673–682. doi: 10.2174/1389201017666160401145438. [DOI] [PubMed] [Google Scholar]
- 11.Li H, Chen X, Shen D, Wu F, Pleixats R, Pan J. Functionalized silica nanoparticles: classification, synthetic approaches and recent advances in adsorption applications. Nanoscale. 2021;13:15998–16016. doi: 10.1039/d1nr04048k. [DOI] [PubMed] [Google Scholar]
- 12.Boraschi D, Italiani P, Palomba R, Decuzzi P, Duschl A, Fadeel B, Moghimi SM. Nanoparticles and innate immunity: new perspectives on host defence. Semin Immunol. 2017;34:33–51. doi: 10.1016/j.smim.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Dobrovolskaia MA, Aggarwal P, Hall JB, McNeil SE. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol Pharm. 2008;5:487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Liu Z, Pang Y, Zhou H. The interaction between nanoparticles and immune system: application in the treatment of inflammatory diseases. J Nanobiotechnology. 2022;20:127. doi: 10.1186/s12951-022-01343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bamberger D, Hobernik D, Konhäuser M, Bros M, Wich PR. Surface modification of polysaccharide-based nanoparticles with PEG and dextran and the effects on immune cell binding and stimulatory characteristics. Mol Pharm. 2017;14:4403–4416. doi: 10.1021/acs.molpharmaceut.7b00507. [DOI] [PubMed] [Google Scholar]
- 16.Saito T, Gale M., Jr Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity. J Exp Med. 2008;205:1523–1527. doi: 10.1084/jem.20081210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20:537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thoresen D, Wang W, Galls D, Guo R, Xu L, Pyle AM. The molecular mechanism of RIG-I activation and signaling. Immunol Rev. 2021;304:154–168. doi: 10.1111/imr.13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kell AM, Gale M., Jr RIG-I in RNA virus recognition. Virology. 2015;479-480:110–121. doi: 10.1016/j.virol.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iurescia S, Fioretti D, Rinaldi M. The innate immune signalling pathways: turning RIG-I sensor activation against cancer. Cancers (Basel) 2020;12:3158. doi: 10.3390/cancers12113158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yong HY, Luo D. RIG-I-like receptors as novel targets for pan-antivirals and vaccine adjuvants against emerging and re-emerging viral infections. Front Immunol. 2018;9:1379. doi: 10.3389/fimmu.2018.01379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemann EA, Knoll ML, Wilkins CR, Subra C, Green R, García-Sastre A, Thomas PG, Trautmann L, Ireton RC, Loo YM, et al. A small molecule RIG-I agonist serves as an adjuvant to induce broad multifaceted influenza virus vaccine immunity. J Immunol. 2023;210:1247–1256. doi: 10.4049/jimmunol.2300026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Zhang H, Wang J, Chen J, Guo Z, Liu Y, Hua H. Exploiting RIG-I-like receptor pathway for cancer immunotherapy. J Hematol Oncol. 2023;16:8. doi: 10.1186/s13045-023-01405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobson ME, Becker KW, Palmer CR, Pastora LE, Fletcher RB, Collins KA, Fedorova O, Duvall CL, Pyle AM, Wilson JT. Structural optimization of polymeric carriers to enhance the immunostimulatory activity of molecularly defined RIG-I agonists. ACS Cent Sci. 2020;6:2008–2022. doi: 10.1021/acscentsci.0c00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanpouille-Box C, Hoffmann JA, Galluzzi L. Pharmacological modulation of nucleic acid sensors - therapeutic potential and persisting obstacles. Nat Rev Drug Discov. 2019;18:845–867. doi: 10.1038/s41573-019-0043-2. [DOI] [PubMed] [Google Scholar]
- 26.Zillinger T, Hartmann G. Targeted nanoparticle delivery of bifunctional RIG-I agonists to pancreatic cancer. Mol Ther. 2019;27:491–492. doi: 10.1016/j.ymthe.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright JF. AAV vector manufacturing process design and scalability - Bending the trajectory to address vector-associated immunotoxicities. Mol Ther. 2022;30:2119–2121. doi: 10.1016/j.ymthe.2022.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoo YJ, Lee CH, Park SH, Lim YT. Nanoparticle-based delivery strategies of multifaceted immunomodulatory RNA for cancer immunotherapy. J Control Release. 2022;343:564–583. doi: 10.1016/j.jconrel.2022.01.047. [DOI] [PubMed] [Google Scholar]
- 29.Angehrn Z, Haldna L, Zandvliet AS, Gil Berglund E, Zeeuw J, Amzal B, Cheung SYA, Polasek TM, Pfister M, Kerbusch T, et al. Artificial intelligence and machine learning applied at the point of care. Front Pharmacol. 2020;11:759. doi: 10.3389/fphar.2020.00759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carmona-Ribeiro AM. Supramolecular nanostructures for vaccines. Biomimetics (Basel) 2021;7:6. doi: 10.3390/biomimetics7010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamaly N, Yameen B, Wu J, Farokhzad OC. Degradable controlled-release polymers and polymeric nanoparticles: mechanisms of controlling drug release. Chem Rev. 2016;116:2602–2663. doi: 10.1021/acs.chemrev.5b00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou X, Zaks T, Langer R, Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehta M, Bui TA, Yang X, Aksoy Y, Goldys EM, Deng W. Lipid-based nanoparticles for drug/gene delivery: an overview of the production techniques and difficulties encountered in their industrial development. ACS Mater Au. 2023;3:600–619. doi: 10.1021/acsmaterialsau.3c00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dilliard SA, Siegwart DJ. Passive, active and endogenous organ-targeted lipid and polymer nanoparticles for delivery of genetic drugs. Nat Rev Mater. 2023;8:282–300. doi: 10.1038/s41578-022-00529-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lokugamage MP, Vanover D, Beyersdorf J, Hatit MZC, Rotolo L, Echeverri ES, Peck HE, Ni H, Yoon JK, Kim Y, et al. Optimization of lipid nanoparticles for the delivery of nebulized therapeutic mRNA to the lungs. Nat Biomed Eng. 2021;5:1059–1068. doi: 10.1038/s41551-021-00786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marx S, Kümmerer BM, Grützner C, Kato H, Schlee M, Renn M, Bartok E, Hartmann G. RIG-I-induced innate antiviral immunity protects mice from lethal SARS-CoV-2 infection. Mol Ther Nucleic Acids. 2022;27:1225–1234. doi: 10.1016/j.omtn.2022.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang-Bishop L, Wehbe M, Pastora LE, Yang J, Kimmel BR, Garland KM, Becker KW, Carson CS, Roth EW, Gibson-Corley KN, et al. Nanoparticle retinoic acid-inducible gene I agonist for cancer immunotherapy. ACS Nano. 2024;18:11631–11643. doi: 10.1021/acsnano.3c06225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cristóbal I, Santos A, Rojo F, García-Foncillas J. A complex microRNA regulatory network may control the HCP5/UTP3/c-Myc/VAMP3 signaling axis. Mol Ther. 2023;31:992–993. doi: 10.1016/j.ymthe.2023.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. 2020;17:395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucy TT, Mamun-Or-Rashid ANM, Yagi M, Yonei Y. Serial passaging of RAW 264.7 cells modulates intracellular AGE formation and downregulates RANKL-induced in vitro osteoclastogenesis. Int J Mol Sci. 2022;23:2371. doi: 10.3390/ijms23042371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim KM, Kim YS, Lim JY, Min SJ, Ko HC, Kim SJ, Kim Y. Intestinal anti-inflammatory activity of Sasa quelpaertensis leaf extract by suppressing lipopolysaccharide-stimulated inflammatory mediators in intestinal epithelial Caco-2 cells co-cultured with RAW 264.7 macrophage cells. Nutr Res Pract. 2015;9:3–10. doi: 10.4162/nrp.2015.9.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xian H, Xie W, Yang S, Liu Q, Xia X, Jin S, Sun T, Cui J. Stratified ubiquitination of RIG-I creates robust immune response and induces selective gene expression. Sci Adv. 2017;3:e1701764. doi: 10.1126/sciadv.1701764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Z, Xu X, Li J, Gu L, Yue Y, Sun F, Zhang X, Zhang T, Liu Y. RIG-I contributes to dsDNA-induced innate immune activation in human brain microvascular endothelial cells. Mol Immunol. 2022;152:78–85. doi: 10.1016/j.molimm.2022.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Plociennikowska A, Frankish J, Moraes T, Del Prete D, Kahnt F, Acuna C, Slezak M, Binder M, Bartenschlager R. TLR3 activation by Zika virus stimulates inflammatory cytokine production which dampens the antiviral response induced by RIG-I-like receptors. J Virol. 2021;95:e01050-20. doi: 10.1128/JVI.01050-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohamed A, Konda P, Eaton HE, Gujar S, Smiley JR, Shmulevitz M. Closely related reovirus lab strains induce opposite expression of RIG-I/IFN-dependent versus -independent host genes, via mechanisms of slow replication versus polymorphisms in dsRNA binding σ3 respectively. PLoS Pathog. 2020;16:e1008803. doi: 10.1371/journal.ppat.1008803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minchin S, Lodge J. Understanding biochemistry: structure and function of nucleic acids. Essays Biochem. 2019;63:433–456. doi: 10.1042/EBC20180038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Berg AIS, Yun CO, Schiffelers RM, Hennink WE. Polymeric delivery systems for nucleic acid therapeutics: approaching the clinic. J Control Release. 2021;331:121–141. doi: 10.1016/j.jconrel.2021.01.014. [DOI] [PubMed] [Google Scholar]
- 48.Palmer CR, Jacobson ME, Fedorova O, Pyle AM, Wilson JT. Environmentally triggerable retinoic acid-inducible gene I agonists using synthetic polymer overhangs. Bioconjug Chem. 2018;29:742–747. doi: 10.1021/acs.bioconjchem.7b00697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sardone V, Zhou H, Muntoni F, Ferlini A, Falzarano MS. Antisense oligonucleotide-based therapy for neuromuscular disease. Molecules. 2017;22:563. doi: 10.3390/molecules22040563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paunovska K, Loughrey D, Dahlman JE. Drug delivery systems for RNA therapeutics. Nat Rev Genet. 2022;23:265–280. doi: 10.1038/s41576-021-00439-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das M, Shen L, Liu Q, Goodwin TJ, Huang L. Nanoparticle delivery of RIG-I agonist enables effective and safe adjuvant therapy in pancreatic cancer. Mol Ther. 2019;27:507–517. doi: 10.1016/j.ymthe.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng B, Nguyen TM, Jayasinghe MK, Gao C, Pham TT, Vu LT, Yeo EYM, Yap G, Wang L, Goh BC, et al. Robust delivery of RIG-I agonists using extracellular vesicles for anti-cancer immunotherapy. J Extracell Vesicles. 2022;11:e12187. doi: 10.1002/jev2.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brisse M, Ly H. Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front Immunol. 2019;10:1586. doi: 10.3389/fimmu.2019.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Onomoto K, Onoguchi K, Yoneyama M. Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors. Cell Mol Immunol. 2021;18:539–555. doi: 10.1038/s41423-020-00602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Q, Miao T, Feng T, Yang C, Guo Y, Li H. Dextran-coated superparamagnetic iron oxide nanoparticles activate the MAPK pathway in human primary monocyte cells. Mol Med Rep. 2018;18:564–570. doi: 10.3892/mmr.2018.8972. [DOI] [PubMed] [Google Scholar]
- 56.Bharde AA, Palankar R, Fritsch C, Klaver A, Kanger JS, Jovin TM, Arndt-Jovin DJ. Magnetic nanoparticles as mediators of ligand-free activation of EGFR signaling. PLoS One. 2013;8:e68879. doi: 10.1371/journal.pone.0068879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maggi LB, Jr, Moran JM, Buller RM, Corbett JA. ERK activation is required for double-stranded RNA- and virus-induced interleukin-1 expression by macrophages. J Biol Chem. 2003;278:16683–16689. doi: 10.1074/jbc.M211744200. [DOI] [PubMed] [Google Scholar]
- 58.Dou H, Xu B, Tao K, Tang M, Sun K. The one-pot synthesis of dextran-based nanoparticles and their application in in-situ fabrication of dextran-magnetite nanocomposites. J Mater Sci Mater Med. 2008;19:2575–2580. doi: 10.1007/s10856-007-3202-7. [DOI] [PubMed] [Google Scholar]
- 59.Kawaguchi T, Hanaichi T, Hasegawa M, Maruno S. Dextran-magnetite complex: conformation of dextran chains and stability of solution. J Mater Sci Mater Med. 2001;12:121–127. doi: 10.1023/a:1008961709559. [DOI] [PubMed] [Google Scholar]
- 60.Priya S, Batra U, R N S, Sharma S, Chaurasiya A, Singhvi G. Polysaccharide-based nanofibers for pharmaceutical and biomedical applications: a review. Int J Biol Macromol. 2022;218:209–224. doi: 10.1016/j.ijbiomac.2022.07.118. [DOI] [PubMed] [Google Scholar]
- 61.Luanda A, Badalamoole V. Past, present and future of biomedical applications of dextran-based hydrogels: a review. Int J Biol Macromol. 2023;228:794–807. doi: 10.1016/j.ijbiomac.2022.12.129. [DOI] [PubMed] [Google Scholar]
- 62.Hu Q, Lu Y, Luo Y. Recent advances in dextran-based drug delivery systems: From fabrication strategies to applications. Carbohydr Polym. 2021;264:117999. doi: 10.1016/j.carbpol.2021.117999. [DOI] [PubMed] [Google Scholar]
- 63.Wang S, Fontana F, Shahbazi MA, Santos HA. Acetalated dextran based nano- and microparticles: synthesis, fabrication, and therapeutic applications. Chem Commun (Camb) 2021;57:4212–4229. doi: 10.1039/d1cc00811k. [DOI] [PubMed] [Google Scholar]
- 64.Tassa C, Shaw SY, Weissleder R. Dextran-coated iron oxide nanoparticles: a versatile platform for targeted molecular imaging, molecular diagnostics, and therapy. Acc Chem Res. 2011;44:842–852. doi: 10.1021/ar200084x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kunc F, Moore CJ, Sully RE, Hall AJ, Gubala V. Polycarboxylated dextran as a multivalent linker: synthesis and target recognition of the antibody-nanoparticle bioconjugates in PBS and serum. Langmuir. 2019;35:4909–4917. doi: 10.1021/acs.langmuir.8b03833. [DOI] [PubMed] [Google Scholar]
- 66.Ahmad T, Rhee I, Hong S, Chang Y, Lee J. Ni-Fe2O4 nanoparticles as contrast agents for magnetic resonance imaging. J Nanosci Nanotechnol. 2011;11:5645–5650. doi: 10.1166/jnn.2011.4502. [DOI] [PubMed] [Google Scholar]
- 67.Lee H, Yu MK, Park S, Moon S, Min JJ, Jeong YY, Kang HW, Jon S. Thermally cross-linked superparamagnetic iron oxide nanoparticles: synthesis and application as a dual imaging probe for cancer in vivo. J Am Chem Soc. 2007;129:12739–12745. doi: 10.1021/ja072210i. [DOI] [PubMed] [Google Scholar]
- 68.Moore CJ, Montón H, O’Kennedy R, Williams DE, Nogués C, Crean Née Lynam C, Gubala V. Controlling colloidal stability of silica nanoparticles during bioconjugation reactions with proteins and improving their longer-term stability, handling and storage. J Mater Chem B Mater Biol Med. 2015;3:2043–2055. doi: 10.1039/c4tb01915f. [DOI] [PubMed] [Google Scholar]
- 69.Khan MS, Gowda BHJ, Nasir N, Wahab S, Pichika MR, Sahebkar A, Kesharwani P. Advancements in dextran-based nanocarriers for treatment and imaging of breast cancer. Int J Pharm. 2023;643:123276. doi: 10.1016/j.ijpharm.2023.123276. [DOI] [PubMed] [Google Scholar]
- 70.Vocelle D, Chesniak OM, Malefyt AP, Comiskey G, Adu-Berchie K, Smith MR, 3rd, Chan C, Walton SP. Dextran functionalization enhances nanoparticle-mediated siRNA delivery and silencing. Technology (Singap) 2016;4:42–54. doi: 10.1142/S2339547816400100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dubois A, François C, Descamps V, Fournier C, Wychowski C, Dubuisson J, Castelain S, Duverlie G. Enhanced anti-HCV activity of interferon alpha 17 subtype. Virol J. 2009;6:70. doi: 10.1186/1743-422X-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kawauchi Y, Kuroda Y, Kojima N. Comparison of the carbohydrate preference of SIGNR1 as a phagocytic receptor with the preference as an adhesion molecule. Int Immunopharmacol. 2014;19:27–36. doi: 10.1016/j.intimp.2013.12.030. [DOI] [PubMed] [Google Scholar]
- 74.Rennick JJ, Johnston APR, Parton RG. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat Nanotechnol. 2021;16:266–276. doi: 10.1038/s41565-021-00858-8. [DOI] [PubMed] [Google Scholar]
- 75.Antonio LC, Ribovski L, Pincela Lins PM, Zucolotto V. The amount of dextran in PLGA nanocarriers modulates protein corona and promotes cell membrane damage. J Mater Chem B Mater Biol Med. 2022;10:8282–8294. doi: 10.1039/d2tb01296k. [DOI] [PubMed] [Google Scholar]
- 76.Berry CC, Wells S, Charles S, Aitchison G, Curtis AS. Cell response to dextran-derivatised iron oxide nanoparticles post internalisation. Biomaterials. 2004;25:5405–5413. doi: 10.1016/j.biomaterials.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 77.Wilhelm C, Billotey C, Roger J, Pons JN, Bacri JC, Gazeau F. Intracellular uptake of anionic superparamagnetic nanoparticles as a function of their surface coating. Biomaterials. 2003;24:1001–1011. doi: 10.1016/s0142-9612(02)00440-4. [DOI] [PubMed] [Google Scholar]
- 78.Motta V, Soares F, Sun T, Philpott DJ. NOD-like receptors: versatile cytosolic sentinels. Physiol Rev. 2015;95:149–178. doi: 10.1152/physrev.00009.2014. [DOI] [PubMed] [Google Scholar]
- 79.Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunol Rev. 2015;265:6–21. doi: 10.1111/imr.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li S, Cao L, Zhang Z, Kuang M, Chen L, Zhao Y, Luo Y, Yin Z, You F. Cytosolic and nuclear recognition of virus and viral evasion. Mol Biomed. 2021;2:30. doi: 10.1186/s43556-021-00046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramasundaram S, Saravanakumar G, Sobha S, Oh TH. Dextran sulfate nanocarriers: design, strategies and biomedical applications. Int J Mol Sci. 2022;24:355. doi: 10.3390/ijms24010355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pansailom N, Rattanamai S, Leepheng P, Rattanawarinchai P, Chattrairat K, Phromyothin D. Surface modification: PEG/dextran encapsulation of SPIONs. Mater Today Proc. 2017;4:6306–6310. [Google Scholar]
- 84.Zeini D, Glover JC, Knudsen KD, Nyström B. Influence of lysine and TRITC conjugation on the size and structure of dextran nanoconjugates with potential for biomolecule delivery to neurons. ACS Appl Bio Mater. 2021;4:6832–6842. doi: 10.1021/acsabm.1c00544. [DOI] [PubMed] [Google Scholar]
- 85.Thoresen DT, Galls D, Götte B, Wang W, Pyle AM. A rapid RIG-I signaling relay mediates efficient antiviral response. Mol Cell. 2023;83:90–104.e4. doi: 10.1016/j.molcel.2022.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burkart SS, Schweinoch D, Frankish J, Sparn C, Wüst S, Urban C, Merlo M, Magalhães VG, Piras A, Pichlmair A, et al. High-resolution kinetic characterization of the RIG-I-signaling pathway and the antiviral response. Life Sci Alliance. 2023;6:e202302059. doi: 10.26508/lsa.202302059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang M, Zhang S, Yang Z, Lin H, Zhu J, Liu L, Wang W, Liu S, Liu W, Ma Y, et al. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell. 2018;173:906–919.e13. doi: 10.1016/j.cell.2018.03.064. [DOI] [PubMed] [Google Scholar]
- 88.Murakami T, Okamoto H, Kim H. Structural and functional changes in high-density lipoprotein induced by chemical modification. Biomater Sci. 2015;3:712–715. doi: 10.1039/c4bm00402g. [DOI] [PubMed] [Google Scholar]
- 89.Gao S, Torrente-Rodríguez RM, Pedrero M, Pingarrón JM, Campuzano S, Rocha-Martin J, Guisán JM. Dextran-coated nanoparticles as immunosensing platforms: Consideration of polyaldehyde density, nanoparticle size and functionality. Talanta. 2022;247:123549. doi: 10.1016/j.talanta.2022.123549. [DOI] [PubMed] [Google Scholar]
- 90.Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R. Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov. 2021;20:101–124. doi: 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chenthamara D, Subramaniam S, Ramakrishnan SG, Krishnaswamy S, Essa MM, Lin FH, Qoronfleh MW. Therapeutic efficacy of nanoparticles and routes of administration. Biomater Res. 2019;23:20. doi: 10.1186/s40824-019-0166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Manolova V, Flace A, Bauer M, Schwarz K, Saudan P, Bachmann MF. Nanoparticles target distinct dendritic cell populations according to their size. Eur J Immunol. 2008;38:1404–1413. doi: 10.1002/eji.200737984. [DOI] [PubMed] [Google Scholar]
- 93.Taback B, Hashimoto K, Kuo CT, Chan A, Giuliano AE, Hoon DS. Molecular lymphatic mapping of the sentinel lymph node. Am J Pathol. 2002;161:1153–1161. doi: 10.1016/S0002-9440(10)64392-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chaney EJ, Tang L, Tong R, Cheng J, Boppart SA. Lymphatic biodistribution of polylactide nanoparticles. Mol Imaging. 2010;9:153–162. [PMC free article] [PubMed] [Google Scholar]
- 95.McLennan DN, Porter CJ, Charman SA. Subcutaneous drug delivery and the role of the lymphatics. Drug Discov Today Technol. 2005;2:89–96. doi: 10.1016/j.ddtec.2005.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Toxicity of Basic-NPs, Dex-NPs, and TDex-NPs in RAW 264.7 cells.
IL-10 production by Basic-NPs, Dex-NPs, and TDex-NPs in RAW 264.7 cells.
In vivo immune response after intravenous or intrapertoneal injection of TDex-NP into C57BL/6 mice.