Abstract
Background
The serotonin type 3 receptor (5-HT3R) is a member of a superfamily of ligand gated ion channels. All members of this family share a large degree of sequence homology and presumably significant structural similarity. A large number of studies have explored the structure-function relationships of members of this family, particularly the nicotinic and GABA receptors. This information can be utilized to gain additional insights into specific structural and functional features of other receptors in this family.
Results
Thirteen amino acids in the mouse 5-HT3ASR that correspond to the putative E binding loop of the nicotinic α7 receptor were chosen for mutagenesis. Due to the presence of a highly conserved glycine in this region, it has been suggested that this binding loop is comprised of a hairpin turn and may form a portion of the ligand-binding site in this ion channel family. Mutation of the conserved glycine (G147) to alanine eliminated binding of the 5-HT3R antagonist [3H]granisetron. Three tyrosine residues (Y140, Y142 and Y152) also significantly altered the binding of 5-HT3R ligands. Mutations in neighboring residues had little or no effect on binding of these ligands to the 5-HT3ASR.
Conclusion
Our data supports a role for the putative E-loop region of the 5-HT3R in the binding of 5-HT, mCPBG, d-tc and lerisetron. 5-HT and mCPBG interact with Y142, d-tc with Y140 and lerisetron with both Y142 and Y152. Our data also provides support for the hypothesis that this region of the receptor is present in a loop structure.
Background
Sequence homology between the serotonin type 3 receptor (5-HT3R), the nicotinic acetylcholine receptor (nAChR), the GABAA receptor and the glycine receptor suggests a large amount of structural similarity within this superfamily of ligand gated ion channels[1-3]. This presumed structural homology can be used to guide site directed mutagenesis studies of particular receptor subtypes. On a gross level, all members of this superfamily assemble as pentameric receptors [4]. In some cases, receptors can be assembled from a single subunit (5-HT3AR and nicotinic α7 receptors) [1,5-8]. In other cases, at least two different subunits are required [2,4,9]. The ligand binding site is thought to be formed at the junction between two subunits [2,4].
Based on data obtained from affinity labeling and site directed mutagenesis studies, several regions of the nicotinic acetylcholine receptor have been implicated in ligand binding [10]. Six essential loops appear to contribute to the binding site with the position of each loop differing slightly depending on whether the receptor is heteromeric or homomeric[4]. For homomeric receptors such as the α7 nAChR, the loops are identified as A, B, C, D, E and F [4,11]. The amino acids that interact with ligands presumably extend into the binding site from these loops. Subunits are composed of 2 faces (+ and -) with the + face of one subunit forming one side of the binding site and the – face of another subunit forming the complimentary side. Some binding loops (A, B and C) are present on the – face while the remaining loops (D and E) are present on the + face. [4]. The individual amino acids that form binding interactions with the functional groups present on a ligand are likely to differ for each receptor subtype, reflecting the specificity of a particular binding site, however, the overall structure of the binding domain may be similar even for binding sites with different ligand specificities. Identification of ligand specificity requires identification of both the location or structure of individual binding site loops and the amino acids present in a particular receptor subtype.
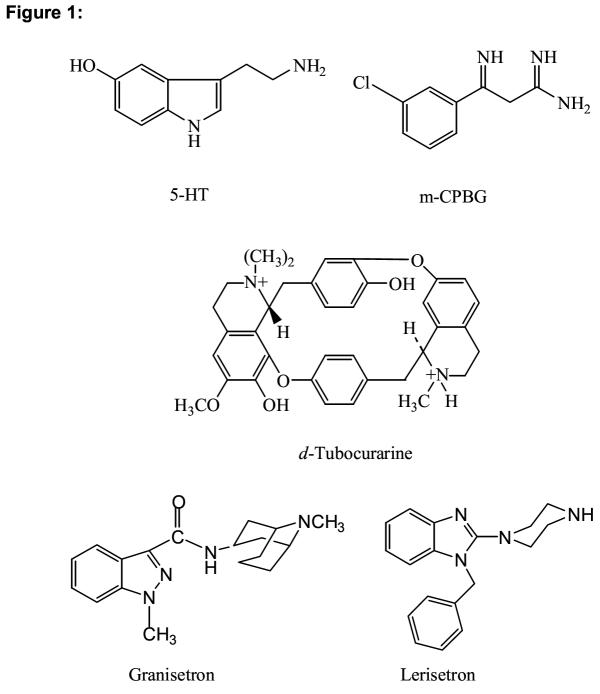
The purpose of this study is to extend the information available from other members of this receptor family to the 5-HT3R. Sequence homology and a presumed structural similarity to other ligand gated ion channels suggests that the E loop region of the 5-HT3AR forms part of the ligand binding domain for 5-HT3R ligands. This region extends from Y140 to K153 and is shown in Table 1. The homologous sequences of other representative members of this family are also shown. In the center of this region is a critical glycine residue that is thought to play a role in establishing a hairpin loop [12]. Recent x-ray crystallographic data obtained from an ACh binding protein (AChBP) shows a loop structure in this region resulting from a 3 residue turn containing a glycine homologous to G147 of the 5-HT3R. On either side of this putative turn region are residues that have been identified as important to receptor binding [12-18]. The formation of this loop structure brings amino acids on either side of glycine into close proximity and may form a binding pocket that will accommodate one or more functional groups. In order to identify the interaction of amino acids in this binding loop with 5-HT3R ligands, we have constructed alanine mutations of residues throughout this region and evaluated the alteration in binding affinity of 5 different classes of 5-HT3R ligands (Figure 1). Our data identifies 3 tyrosine residues that appear to interact selectively with each structural class and supports the existence of a loop structure in this region of the receptor.
Table 1.
Sequence Comparison of Putative Binding Loop Region.
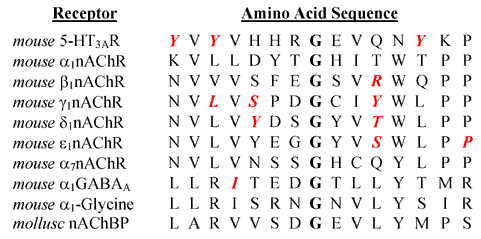 |
Primary amino acid sequence of representative members of the LGIC superfamily showing amino acid residues on either side of the conserved glycine residue. The amino acids shown span the region from Y140 – P154 of the 5-HT3R. The conserved glycine is shown in bold. Amino acids identified in the binding site of other LGIC subunits and the tyrosines identified in this study are shown in red, bold italics [11-17].
Figure 1.
5-HT3R ligands.
Results
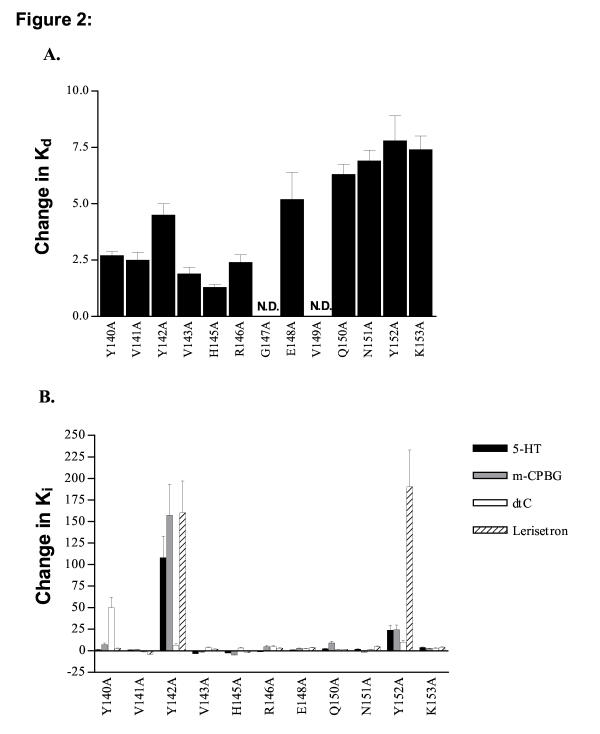
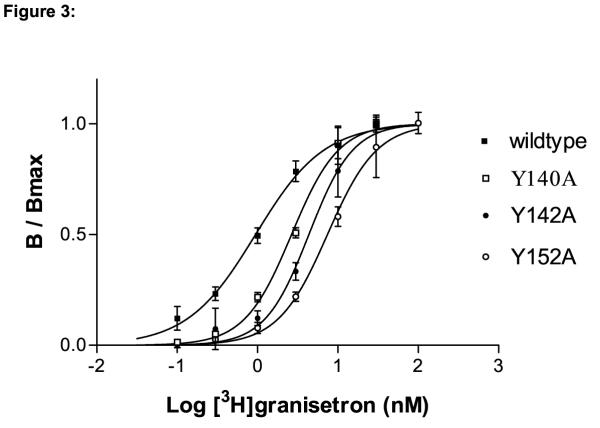
All mutant receptors were tested for their ability to bind the 5-HT3R antagonist [3H]granisetron. Table 2 shows the Kd values for wildtype mouse 5-HT3ASRs and the 13 alanine mutations we evaluated. [3H]granisetron is a potent antagonist of the wt 5-HT3R (Kd = 0.98 ± 0.12 nM). This value agrees with published data for this compound [19,20]. Bmax values range from the 5.5 pmoles/mg protein observed for E148A to 0.30 pmoles/mg protein for the K153A mutation, indicating some variability in expression of the different receptors. In general, however, receptor expression was similar to that reported by other laboratories [1,20-22]. No detectable binding was observed for G147A and V149A mutant receptors. For all other mutants, decreases in binding affinity (increased Kd) were observed although the magnitude of the change was less than 10 fold in all cases. A bar graph showing the change in Kd value resulting from each alanine mutation is shown in Figure 2A. The largest decreases in binding affinity were observed for Y142A (4.6 fold, Figure 3), E148A (5.3 fold) and Q150A – K153A (6 – 8 fold).
Table 2.
[3H]Granisetron binding to wildtype and mutant 5-HT3ASRs.
| Receptor | Kd (nM) [3H]granisetron | Bmax (pmol/mg protein) | n |
| Wildtype | 0.98 ± 0.12 | 3.2 ± .13 | 1 ± 0.12 |
| Y140A | 2.7 ± 0.19 | 0.44 ± .08 | 1.5 ± 0.14 |
| V141A | 2.5 ± 0.35 | 0.32 ± .04 | 1.3 ± 0.22 |
| Y142A | 4.5 ± 0.5 | 0.97 ± .14 | 1.6 ± 0.27 |
| V143A | 1.9 ± 0.28 | 1.1 ± .04 | 0.97 ± 0.13 |
| H145A | 1.3 ± 0.13 | 0.81 ± .07 | 1.38 ± 0.17 |
| R146A | 2.4 ± 0.34 | 1.0 ± .16 | 1.26 ± 0 .21 |
| G147A | - | - | - |
| E148A | 5.2 ± 1.2 | 5.5 ± 1.2 | 0.93 ± 0.19 |
| V149A | - | - | - |
| Q150A | 6.3 ± 0.45 | 0.56 ± .09 | 1.19 ± 0 .1 |
| N151A | 6.9 ± 0.48 | 4.9 ± 1.1 | 1.66 ± 0.16 |
| Y152A | 7.8 ± 1.1 | 0.59 ± .10 | 1.4 ± 0.24 |
| K153A | 7.4 ± 0.61 | 0.30 ± .08 | 1.51 ± 0.17 |
The Kd, Bmax and Hill coefficient (n) were determined using non-linear curve fitting of binding data as described in the methods section. Each value represents the combined data from at least four experiments and is represented as ± SE. The Kd values were used to calculate the ratios plotted in Figure 2, indicating the change in binding affinity as a result of each individual mutation. A plot of the data obtained from Y140A, Y142A and Y152A is shown in Figure 3.
Figure 2.
Changes in Ki or Kd (for binding of [3H]granisetron) as a result of alanine mutations. The values shown are the ratio of the Kd or Ki for wildtype and mutant receptors. The length of each bar illustrates the magnitude of change in either the Kd or Ki relative to wildtype while the direction indicates an increase (up) or decrease (down) in the Kd or Ki. A. Most alanine mutations produced little change in Kd. No binding was detected for the G147A and V149A receptors therefore no Kd could be determined. B. Changes in Ki were small for most receptors although large changes were observed for select ligands at Y140A, Y142A and Y152A receptors. The relative increases in Ki for these amino acids are as follows: 5-HT: Y140A 1.3 ± 0.30 fold, Y142A 110 ± 25 fold, Y152A 24 ± 5.6 fold mCPBG: Y140A 7.3 ± 1.7 fold, Y142A 160 ± 36 fold, Y152A 24 ± 5.6 fold d-tc: Y140A 50 ± 12 fold, Y142A 6.5 ± 1.5, Y152A 10.0 ± 2.3 fold lerisetron: Y140A 4.6 ± 1.1 fold, Y142A 160 ± 37 fold, Y152A 190 ± 43 fold
Figure 3.
Saturation binding isotherms for binding of [3H]granisetron to wildtype, Y140A, Y142A and Y152A receptors. The data shown represent the specific binding of [3H]granisetron as determined from at least four identical experiments. Specific binding was determined as the fraction of total binding not inhibited by a saturating concentration of mCPBG or MDL-72222. Data were combined and fitted using non-linear curve fitting. Only small shifts in binding affinity to [3H]granisetron were observed on all three mutants. Kd values, Bmax and Hill coefficients are shown in Table 2.
Inhibition binding assays were also conducted. Four test compounds with structures representative of the major classes of 5-HT3R ligands were chosen: serotonin (5-HT, the endogenous agonist), m-chlorophenylbiguanide (mCPBG, agonist), d-tubocurarine (d-tc, antagonist) and lerisetron (antagonist). The Ki values for inhibition of [3H]granisetron binding by all four compounds are shown in Table 3. Little change in Ki value was observed for the majority of mutations. The values highlighted in bold in Table 3 represent the Ki values for inhibition of [3H]granisetron binding on mutant receptors that increased over 10 fold compared to the Ki obtained for wildtype receptors. The bar chart in Figure 2B illustrates the changes in Ki resulting from each mutation on the test compounds. Ratios of Ki are shown as positive for increases in Ki on mutant receptors versus wildtype and negative for decreases. A positive change thus corresponds to a decrease in binding affinity for the compound as a result of the mutation. Large decreases in binding affinity were observed for select compounds only on the Y140A, Y142A and Y152A mutations.
Table 3.
Ki values for Inhibition of [3 H]granisetron binding to wildtype and mutant 5-HT3ASRs.
| Ki (Inhibition of [3H]granisetron binding) nM | ||||
| Receptor | 5-HT (nM) | MCPBG (nM) | d-tc (nM) | Lerisetron |
| Wildtype | 74 ± 17 | 3.0 ± 0.55 | 6 ± 2.1 | 0.8 ± 0.19 |
| Y140A | 97 ± 20 | 22 ± 6.1 | 300 ± 120 | 2.1 ± 0.33 |
| V141A | 82 ± 9.2 | 4.1 ± 0.50 | 4.7 ± 0.87 | 0.20 ± 0.02 |
| Y142A | (8 ± 2.0) × 103 | 470 ± 90 | 39 ± 19 | 130 ± 28 |
| V143A | 22 ± 2.2 | 2.0 ± 0.44 | 22 ± 2.6 | 1.5 ± 0.41 |
| H145A | 29 ± 6.5 | 0.60 ± 0.10 | 19 ± 2.6 | 0.42 ± 0.04 |
| R146A | 61 ± 14 | 15 ± 4.9 | 29 ± 4.9 | 2.3 ± 0.31 |
| E148A | 73 ± 11 | 8.2 ± 1.5 | 14 ± 1.7 | 2.8 ± 0.53 |
| Q150A | 170 ± 29 | 27 ± 3.9 | 9.3 ± 1.3 | 1.1 ± 0.21 |
| N151A | 140 ± 20 | 1.8 ± 0.18 | 9.3 ± 1.4 | 3.7 ± 0.62 |
| Y152A | (1.8 ± 0.4) × 103 | 73 ± 26 | 60 ± 19 | 150 ± 36 |
| K153A | 270 ± 34 | 7.7 ± 2.0 | 17 ± 3.0 | 3.0 ± 0.66 |
IC50 values were determined from competition binding data using [3H]granisetron as described in the methods section. Ki values were calculated from the IC50 using the Cheng-Prusoff equation, the Kd value, and the concentration of [3H]granisetron (typically equal to the Kd). Each value represents the combined data from at least four experiments and is represented as Ki ± SE. Values in bold type indicate changes in Ki greater than or equal to 10 fold on mutant vs. wt receptors.
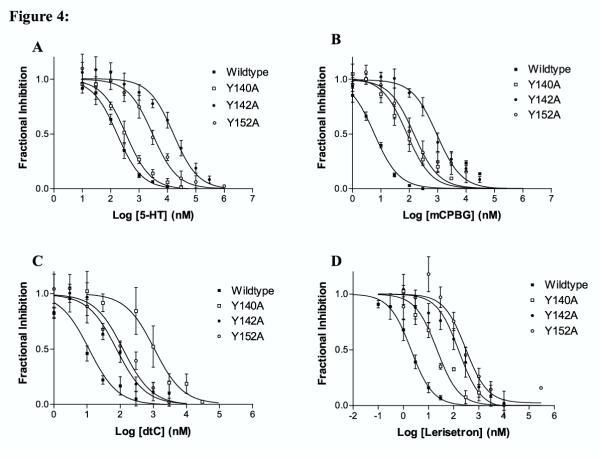
A more detailed analysis of the competition binding data obtained for the Y140A, Y142A and Y152A mutations is shown in Figure 4. For 5-HT, the Y142A mutation produced a 110 fold increase in Ki and Y152A produced a 24 fold increase. No change in Ki was observed for the Y140A mutation (Figure 4A). The 5-HT3R agonistmCPBG showed a similar profile for the changes in Ki values resulting from mutations of the three tyrosines (Figure 4B). As was observed for 5-HT, the Y142A mutation produced a large increase in Ki (160 fold) while the Y14 0A and Y152A mutations produced only 7 and 24 fold changes respectively.
Figure 4.
Inhibition of [3H]granisetron binding by 5-HT3R ligands. The data represent the combined results of at least four experiments. Assays were conducted as described in Materials and Methods. Fractional response is the fraction of [3H]granisetron binding obtained in the presence and absence of inhibitor. An IC50 value was determined from the data using non-linear curve fitting as described in the methods and the Ki value calculated using the Cheng-Prusoff equation, the Kd as determined in Figure 3 and the concentration of ligand used to obtain the inhibition data. Kd values are reported in Table 3. A. 5-HT B. mCPBG C. dtC D. Lerisetron
The Ki value for d-tc inhibition of [3H]granisetron binding was altered only slightly by the Y142A or Y152A mutations (6.5 fold and 10 fold changes respectively). The Y140A mutation, however, produced a 50 fold increase in the Ki compared to wildtype receptors (Figure 4C).
The Ki value for lerisetron inhibition of [3H]granisetron binding was increased 160 fold by the Y142A mutation and 190 fold by the Y152A mutation. Only a 4.6 fold change in Ki resulted from the Y140A mutation. Lerisetron was the only compound for which a large increase in the Ki was observed on the Y152A mutation (Figure 4D). This mutation produced smaller changes in Ki for 5-HT and mCPBG (24 fold for both) and only a 10 fold change for d-tc.
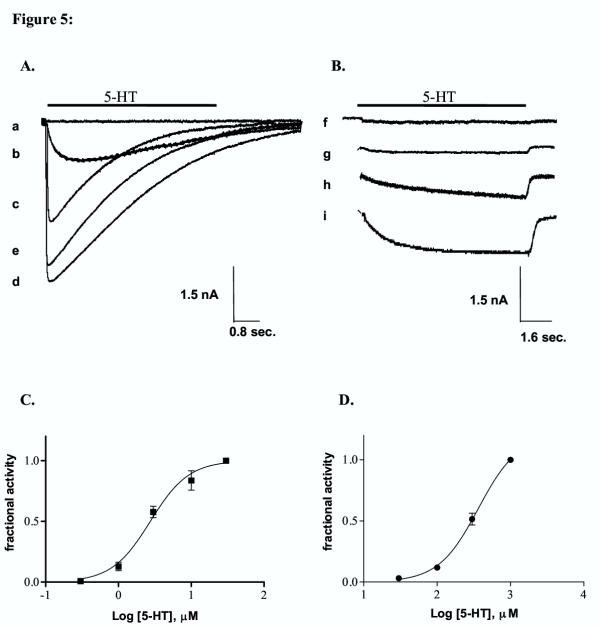
In whole cell patch clamp studies, 5-HT perfusion of cells transfected with Y140A and Y142A cDNA produced no responses at 5-HT concentrations of up to 1 mM although specific binding to these receptors was identified in receptor binding studies. Unlike Y140A and Y142A, Y152A receptors responded to application of 5-HT. Due to the low potency of 5-HT on these receptors, only a portion of the concentration response curve could be determined (up to 1 mM). The EC50 value was estimated as greater than 370 μM (Figure 5). This value shows a greater than 140 fold increase in EC50 compared to wildtype receptors; a larger change than was observed for the Ki (24 fold). The most dramatic change observed for whole cell currents was an alteration in the kinetics of the response elicited by application of 5-HT. Y152A mutant receptors displayed much slower rise times compared to wildtype receptors at all concentrations tested. Peak wt responses were typically obtained in less than 80 ms while Y152A responses required several seconds to plateau. Desensitization kinetics were also altered. While wt receptors desensitized rapidly, mutant receptors showed no desensitization during the 8 s perfusion time.
Figure 5.
Whole cell patch clamp recording from tsA 201 cells transfected with wildtype and Y152A cDNA. Data were obtained using whole cell patch clamp on tsA201 cells transfected with wt or Y152A mutant 5-HT3ASR cDNA. Cells were clamped at a holding potential of -60 mV and 5-HT applied with a rapid perfusion system. The application time for 5-HT is indicated by the bar above the curves. The responses indicated in A and B are representative examples of at least 4 sets of experiments. A. Wildtype 5-HT3ASR. 5-HT was applied as indicated for a 4 s duration. The concentrations of 5-HT used and the response amplitudes are: a. 0.3 μM (0.03 nA) b. 1.0 μM (0.96 nA) c. 3.0 μM (2.5 nA) d. 30.0 μM (4.4 nA), e. 100.0 μM (3.9 nA). B. Y152A mutant receptors. 5-HT was applied as indicated for an 8 s duration. The curves in B were offset vertically for clarity. The dashed line in each curve indicates the baseline level for that response. The concentrations of 5-HT used and the response amplitudes are: f. 3.0 μM (0.10 nA) g. 100 μM (0.14 nA) h. 300.0 μM (0.57 nA) i. 1.0 mM (1.0 nA). C. Concentration response curve for 5-HT activation of wt 5-HT3ASRs. Each data point represents the combined results of 4 experiments. Responses were normalized to the response obtained at 30 μM and the curves fit as described in the methods section. EC50 = 2.7 ± 0.25 μM, n = 1.6 ± 0.22. D. Concentration response curve for 5-HT activation of Y152A 5-HT3ASRs. Each data point represents the combined results of 4 experiments. The response did not plateau in the range of concentrations used and hence responses were normalized to the maximum concentration of 5-HT (1 mM). The EC50 was estimated by fitting the data to the following equation: I = Imax/(1 +(EC50/ [C])n), where is the normalized current at 5-HT concentration [C], EC50 is the concentration of 5-HT needed to obtain half maximal activation and n is the apparent Hill coefficient. Imax in this case represents the estimated maximum current relative to the response obtained at 1 mM. To obtain the curve shown in the figure, the Hill coefficient was fixed at the wt value of 1.6. The EC50 value obtained from this fit is 370 ± 27 μM.
Discussion
The putative E-loop region of the LGIC family of receptors is homologous to residues Y140 through K153 in the 5-HT3R [4,12-18]. Structure-function studies of this region have been conducted in several other members of this family of receptors including GABAA and nAChR subtypes. In each case, residues have been identified that alter either the binding of selective ligands or receptor function [4,11-18]. In order to determine if this loop also contains residues critical to the structure or function of the 5-HT3R, we have constructed alanine mutations of amino acids throughout the homologous region of the mouse 5-HT3ASR and investigated the affects on binding of 5 different structural classes of 5-HT3R ligands. We have identified three tyrosine residues that appear to play a role in binding of selective ligands to this receptor. In addition, our data support the existence of a loop structure in this region as has been hypothesized for the nAChR and identified in a homologous AChBP [12,23,24].
Representative members of 5 major structural classes of 5-HT3R ligands were tested on all mutants (Figure 1). These ligands include the antagonists [3H]granisetron, d-tc and lerisetron, and the agonists 5-HT and mCPBG. Most of the mutations tested produced only minor changes in binding affinity for these ligands. Large changes in binding are only apparent on Y140A, Y142A and Y152A receptors for select ligands. The resulting effects of individual mutations are specific to particular structural classes of ligands. The Y140A mutation altered the Ki for d-tc inhibition, but did not alter the Ki obtained for any other compound tested, while Y142A altered the Ki for mCPBG, 5-HT and lerisetron but had little if any effect on inhibition by d-tc. These data indicate the highly specific nature of the effects introduced by the alanine mutations and appear to reflect specific changes in ligand/receptor interaction.
Binding of [3H]granisetron is altered only slightly by the alanine mutations introduced in this study (<10 fold). This result indicates the lack of involvement of amino acids in this binding loop in the binding of [3H]granisetron. It is apparent, however, that [3H]granisetron does occupy the same binding cleft as other 5-HT3R ligands as evidenced by the ability of 5-HT, mCPBG, d-tc and lerisetron to displace it from the binding site. The lack of any large change in binding of granisetron supports our contention that there is little global structural perturbation of the binding site resulting from the introduction of each individual alanine mutation.
The Ki for inhibition of [3H]granisetron binding by 5-HT was increased 110 fold by the Y142A mutation. A similar increase was also observed for mCPBG (160 fold). In contrast, the Y140A and Y152A mutations produced relatively small changes in the Ki. Since both compounds are 5-HT3R agonists, they are likely to share the same binding interactions. One of these interactions appears to be with Y142. Other studies have also identified binding site interactions for agonists. R91, E106, F107, W183 and several residues adjacent to the M1 region have all been demonstrated to alter the action of 5-HT and/or mCPBG [20,21,25,26]. Within the crystal structure of the AChBP, homologous residues are located in the apparent binding site [24]. The effects of agonist on the 5-HT3R are mediated by their interaction with these binding site residues, resulting in stable receptor conformations, including the channel open state. Identification of interacting amino acids and their location in the tertiary structure of the receptor may provide clues to the mechanism of channel opening. For example, amino acids homologous to Y142 and W183 (R104 and W143 respectively) are in close proximity in the AChBP, although on complementary faces of the receptor subunits [24]. Spier et. al. have suggested that W183 may be involved in a cation-π interaction with the amino group of 5-HT and mCPBG [26]. If W183 and Y142 are located near each other but on opposite faces of the binding site in the 5-HT3R, then they could potentially act in concert to help stabilize a conformation of the receptor leading to channel opening.
d-Tubocurarine inhibition was uniquely altered by the Y140A mutation. An increase in Ki of 50 fold was observed for d-tc on this mutant. Little if any change resulted for any other mutation. Thus, while d-tc may form an interaction with the receptor at this binding loop, it appears to interact with Y140 rather than Y142 or Y152. This difference in binding site interactions of the antagonist d-tc and the agonists 5-HT and mCPBG could be the result of a slightly different positioning of d-tc in the binding site. The antagonists d-tc and [3H]granisetron also interact differently with this region since [3H]granisetron binding was not altered by mutations at any of the amino acids tested. While the binding of d-tc appears to involve some interaction with this binding loop, the binding of [3H]granisetron does not. Previous studies have indicated a point of overlap between [3H]granisetron and d-tc at W89 of the 5-HT3ASR. [20]. These studies indicate that the binding regions for these antagonists are partially overlapping at W89 but not at Y140. In addition to Y140 and W89, d-tc has also been observed to interact with D206 and several other residues in loop C [27]. The d-tc binding site appears to involve interaction with at least three different binding loops (A, E and C) although additional interacting amino acids may be found. Data obtained for d-tc is particularly valuable due to the rigid nature of this molecule. Determination of interacting functional groups and their relationship to individual amino acids could enable d-tc to be used as a molecular ruler to determine relative positions of these amino acids.
Lerisetron inhibition of [3H]granisetron binding to mutant receptors was also investigated. Lerisetron is a potent 5-HT3R antagonist first synthesized by Orales et. al.[28]. As was observed for 5-HT3R agonists, lerisetron binding was altered by the Y142A mutation. This mutation produced an increase in the Ki of 160 fold compared to wildtype receptors. This increase in Ki indicates an important interaction of lerisetron with Y142 and a similarity between the binding location of lerisetron, 5-HT and mCPBG. Lerisetron is the only antagonist tested that shares a binding site interaction in this region with agonists. In contrast to 5-HT and mCPBG, however, lerisetron also interacts with Y152 as indicated by the 190 fold increase in Ki on Y152A mutant receptors. Since neither d-tc nor [3H]granisetron interacts with Y142 or Y152, there appears to be a difference between the interactions formed by these antagonists compared to lerisetron. The binding sites of the three antagonists tested differ with respect to this binding loop. Since the role of a competitive antagonist is simply to block the binding of agonists and prevent channel opening, the specific amino acids that interact with the ligand can vary for different antagonists. This is less likely to be the case with agonists since they must produce a conformational change in the protein to exert their effects.
Only two mutant receptors failed to bind [3H]granisetron; G147A and V149A. G147 is the conserved glycine in this putative binding loop. Chirara et. al. have suggested that the highly conserved nature of the glycine in this region may indicate the existence of a loop structure consisting of either γ or a loose three residue-turn in the nAChR [12]. Either of these turns would bring the two putative β-strands together such that γL109, γY111 and γS115 and γY117 are all on the same side of an antiparallel β-sheet. These residues have been identified by affinity labeling, site-directed mutagenesis or cysteine substitution to lie on the same surface. A classic 2-residue β-turn would place these residues on opposite surfaces [12]. Substitution of the conserved glycine by alanine may disrupt the structure of this region and prevent assembly or expression of the receptor. The recent determination of the crystal structure of an AChBP supports this hypothesis. The AChBP displays a large amount of homology to the amino terminal of LGIC receptors and thus may be similar in structure [20,24]. The crystal structure of this protein reveals a loose 3 residue turn incorporating the conserved glycine residue [24]. Homologous residues in other LGIC subunits have also been identified and are shown in Figure 1. The residues identified in this study as altering binding affinity of 5-HT3R ligands would also be present on the same surface if this structure is present in the 5-HT3R. While Y140 lies somewhat outside the region identified by Chiara in the nAChR (homologous to γN107), γL109 and γL119 are homologous to Y142 and Y152 of the 5-HT3ASR. The ability of lerisetron to interact with both Y142 and Y152 also supports the hypothesis that these two amino acids are present in a loop structure since the eight intervening residues would position Y142 and Y152 too far apart to permit them both to interact with a single ligand even if they were interacting with functional groups on opposite ends of the molecule. A loop structure would bring them into closer proximity and permit interaction with the small molecule lerisetron.
All three tyrosine mutations were investigated using a whole cell patch clamp assay to determine if functional changes could be observed. Whole cell responses could not be obtained for Y140A or Y142A, although specific binding of [3H]granisetron was observed. These data suggest that, while the receptors do assemble and are capable of binding [3H]granisetron, they are either not transported to the cell surface or are non-responsive to 5-HT at concentrations of 1 mM or less.
Y152A does produce functional channels however they display distinctly altered response kinetics when compared to wildtype receptors. Y152A responses do not show the rapid rise times observed in wt receptors. The extremely slow rise times observed for Y152A receptors may indicate a change in rate constants preceding channel opening. These changes in the rate constants for either agonist binding or channel opening also produce a 140 fold decrease in the observed EC50 for 5-HT activation. The slow rise is followed by a non-desensitizing phase of the response that is dramatically different from the fast desensitization observed for wt 5-HT3ASRs. Lack of desensitization could result from either a stabilization of the open state of the channel or a destabilization of the desensitized state. Mutations of homologous or nearby residues in both the nAChR and GABAA receptors have also been demonstrated to alter the agonist response. Mutation of the homologous residue in the GABAA receptor γ-subunit (T142) to serine altered the efficacy of the agonist Flumazenil, converting it to a partial agonist [29]. In the nAChR, mutation of mouse and rat εP121 to leucine altered both the binding of acetylcholine and the stability of the open state of the channel. εP121 is homologous to P154 in the 5-HT3R and is only two residues away from Y152. The authors of this study concluded that this portion of the acetylcholine binding site was closely linked to the channel opening region of the receptor [30]. It is reasonable to conclude that the homologous region in the 5-HT3R may perform a similar function. The link between an agonist binding domain and a conformational change leading to channel opening is not unexpected since the two must obviously be linked. If binding to this region of the receptor is shown to be a critical step between the binding of agonists and the opening of the channel, further investigation of the amino acids in this loop may provide valuable clues to molecular basis of this process.
Conclusions
Our data indicate an important role for this putative binding site loop in the interaction of the 5-HT3R with different ligands and illustrate the difference in binding of different structural classes of ligands. Each structural class shows different patterns of interaction with amino acids in this region of the receptor. [3H]granisetron does not appear to interact with any of the amino acids tested while d-tc interacts with only Y140, mCPBG and 5-HT with Y142 and lerisetron with both Y142 and Y152. Similar selective effects have been observed on other residues including W183, W89, F107 and E106 among others[20,21,25,26]. Our data in conjunction with those of other laboratories indicates the differences in orientation of different ligands within the same binding cleft. These differences in orientation result in different amino acid/functional group interactions. As the structural detail of these interactions emerges, these differences could potentially be exploited to produce more potent and specific ligands. For example construction of a "hybrid" ligand that combines the interactions of [3H]granisetron with those of d-tc, 5-HT or mCPBG could produce an antagonist with increased affinity due to the additional binding energy of these interactions and a greater specificity since it would utilize more structural features of the binding site.
The requirement of a glycine at position 147 and the ability of lerisetron to interact with both Y142 and Y152 also support the hypothesis that the secondary structure in this region of the receptor is formed by a loop structure. If the loose 3 residue turn proposed by Chiara et. al. and shown for the AChBP is present in the 5-HT3R, then Y140, Y142 and Y152 would be present on the same side of the sheet and all three would be capable of extending into the binding site. A similar observation has been made for the nAChR where γS111, γY117, γL119, δR113 and δT119 of the mouse nAChR receptor and γL109 and γY111 of the torpedo nAChR are all thought to be present in the binding site [12-18].
Materials and Methods
Materials
[3H]granisetron was purchased from New England Nuclear, 5-HT from Spectrum, and mCPBG and d-tc from Research Biochemical International. Lerisetron was provided by Dr. Karen Kirschbaum at The University of Louisiana at Monroe, Monroe, LA. All other reagents were obtained from commercial sources.
Site directed mutagenesis
Wild type 5-HT3AS mouse receptor cDNA was obtained from Dr. Michael White [20]. Mutant receptors were constructed using either the Quick Change Mutagenesis kit (Stratagene) or the Altered Sites Mutagenesis kit (Promega). All mutations were confirmed by commercial DNA sequencing.
Cell culture and transfection
tsA201 cells, a derivative of HEK293 cells, were seeded at a density of 5 × 106 cells/100 mm dish. Cells were grown in DMEM medium containing 10% FBS, 100 units/ml penicillin/streptomycin for nine hours in 5% CO2 and transfected with 10 μg mouse 5-HT3ASR cDNA per 100 mm dish using the calcium phosphate technique (New Life Technologies, NY). Media was changed 12–14 hrs after transfection. The cells were allowed to grow for another 24 hours and then harvested.
For whole cell patch clamp experiments, tsA201 cells were seeded to a density of 0.25 × 106 cells/60 mm dish. Cells were grown in DMEM culture medium containing 10% FBS and 100 units/ml penicillin/streptomycin for 12 hours prior to transfection. Transfections were performed using Superfect Transfection Reagent (Qiagen, CA). Ten μg of cDNA were mixed with DMEM medium containing no serum or antibiotics in a volume of 150 μl. Twenty μl of Superfect reagent were then added and the mixture incubated at room temperature for 15 min. The reaction was terminated by adding 1 ml of DMEM medium containing 10% FBS and 100 units/ml penicillin/streptomycin and the entire mixture added to cells in the 60 mm dish. Cells were exposed to Superfect Reagent for 3 hours. At that time, the reagent was replaced with DMEM medium containing 10% FBS and 100 units/ml penicillin/streptomycin and incubated for an additional 24 hours prior to use.
Binding assays
Transfected cells were scraped from the dishes, washed twice with Dulbecco's PBS (New Life Technologies, NY), then resuspended in 1.0 ml ice cold PBS/100 mm dish. Cells were either used fresh or frozen at this step until needed. Immediately prior to use, cells were homogenized on ice in PBS using a glass tissue homogenizer then centrifuged at 35 000 × g for 30 minutes in a Beckman JA20 rotor (4°C). Membranes were washed once more with PBS at 4°C then resuspended in 1 ml PBS/100 mm dish. Protein content was determined using a Lowry assay (Sigma. Diagnostics, St. Louis, MO). Membranes were initially prepared and Bmax and Kd values determined in the presence and absence of a cocktail of protease inhibitors (Complete Protease Inhibitor Cocktail, Roche Diagnostics, Mannheim Germany). No change in Bmax was observed as a result of omitting the protease inhibitor (Bmax (+ protease inhibitor)= 3.1 ± 0.11 pmoles/mg protein vs Bmax (- protease inhibitor) = 3.2 ± 0.13 pmoles/mg protein.) hence all assays were performed in the absence of the cocktail. Binding assays were performed in PBS.
For Kd determinations, 100 μl of homogenate was incubated at 37°C for 1 hour with varying concentrations of [3H]granisetron (NEN, MA). Specific binding of [3H]granisetron was determined as the bound [3H]granisetron not displaced by a saturating concentration of a competing ligand. Kd values were determined by fitting the binding data to the following equation using Graphpad PRISM (San Diego CA): B = Bmax [L]n / ([L]n + Kn), where B is bound ligand, Bmax is the maximum binding at equilibrium L is the free ligand concentration and n is the Hill coefficient.
Ki determinations, 100 μl of homogenate was incubated at 37°C for 2 hours with varying concentrations of inhibitor and [3H]granisetron (NEN, MA). Binding was terminated by rapid filtration onto a GF/B filters. The IC50 values were calculated by fitting the data to the following equation using Graphpad PRISM (San Diego CA): θ = 1/ (1+(L/IC50)), where θ is the fractional amount of [3H]granisetron bound in the presence of inhibitor at concentration L as compared to the amount of [3H]granisetron bound in the absence of inhibitor. IC50 is the concentration at which θ = 0.5. The Ki is calculated from the IC50 value using the Cheng-Prusoff equation.
Electrophysiological Recordings
Transfected cells were transferred to a recording chamber containing extracellular solution (140 mM NaCl, 1.7 mM MgCl2, 5 mM KCl, 1.8 mM CaCl2, 25 mM HEPES pH 7.4). Patch electrodes of resistance 2.5–3.0 MΩ were filled with filtered intracellular solution containing 145 mM KCl, 2 mM MgCl2, 1 mM EGTA, 25 mM HEPES (pH 7.4). Cells were clamped in whole cell configuration at a holding potential of -60 mV. A continuous extracellular solution flow (0.8 ml/min) was maintained throughout the recording procedure. 5-HT was dissolved in extracellular solution and delivered to cells using a rapid perfusion system (Warner Instruments, Hamden, CT) at a rate matching the extracellular solution flow rate. The drug perfusions lasted for a period varying from 4 to 8 seconds. Currents elicited by agonist application were measured using an Axopatch 200 B amplifier (Foster City, CA). The data were plotted and analyzed by non-linear curve fitting (Graphpad PRISM, San Diego CA) according to the following equation: I = 1/(1 +(EC50/ [C])n), where is the normalized current at 5-HT concentration [C], EC50 is the concentration of 5-HT needed to obtain half maximal activation and n is the apparent Hill coefficient.
Acknowledgments
Acknowledgements
This work was supported by The University of Louisiana at Monroe, College of Pharmacy and grants from the Southeast Affiliate of the American Heart Association. We would also like to gratefully acknowledge Dr. Karen Kirschbaum at The University of Louisiana at Monroe, Monroe, LA for providing us with the lerisetron used in these studies.
Contributor Information
Padmavati Venkataraman, Email: padma964@hotmail.com.
Srinivasan P Venkatachalan, Email: pv_srinivasan@yahoo.com.
Prasad R Joshi, Email: joship@tribe.ulm.edu.
Mani Muthalagi, Email: muthalagiss@yahoo.com.
Marvin K Schulte, Email: pyschulte@ulm.edu.
References
- Maricq AV, Peterson AS, Brake AJ, Myers RM, Julius D. Primary structure and functional expression of the 5-HT3R, serotonin gated ion channel. Science. 1991;397:359–363. doi: 10.1126/science.1718042. [DOI] [PubMed] [Google Scholar]
- Karlin A, Akabas MH. Towards a structural basis for the function of nicotinic acetylcholine recetors and their cousins. Neuron. 1995;15:1231–1244. doi: 10.1016/0896-6273(95)90004-7. [DOI] [PubMed] [Google Scholar]
- Schofield PR, Darlison MG, Fujita N, Burt DR, Stephenson FA, Rodriguez H, Rhee LM, Rmachandran J, Reale V, Glencorse TA, Seeburg PH, Barnhard EA. Sequence and functional expression of the GABAA receptor shows a ligand-gated receptor super-family. Nature. 1987;328:221–227. doi: 10.1038/328221a0. [DOI] [PubMed] [Google Scholar]
- Arias HR. Localization of agonist and competitive antagonist binding sites on nicotinic acetylcholine receptors. Neurochem Intl. 2000;36:595–645. doi: 10.1016/S0197-0186(99)00154-0. [DOI] [PubMed] [Google Scholar]
- Hope AG, Downie DL, Sutherland L, Lambert JJ, Peters JA, Burchell B. Cloning and functional expression of an apparent splice variant of the murine 5-HT3 receptor A subunit. E J Pharm. 1993;245:187–192. doi: 10.1016/0922-4106(93)90128-V. [DOI] [PubMed] [Google Scholar]
- Belelli D, Balcarek JM, Hope AG, Peters JA, Lambert JJ, Blackburn TP. Cloning and functional expression of a human 5-hydroxytryptamine type 3AS receptor subunit. Mol Pharm. 1995;48:1954–1062. [PubMed] [Google Scholar]
- Couturier S, Bertrand D, Matter JM, Hernandez MC, Bertrand S, Millar N, Valera S, Barkas T, Ballivet M. A neuronal nicotinic acetylcholine receptor subunit (alpha 7) is developmentally regulated and forms a homo-oligomeric channel blocked by alpha-BTX. Neuron. 1990;5:847–56. doi: 10.1016/0896-6273(90)90344-f. [DOI] [PubMed] [Google Scholar]
- Seguela P, Wadiche J, Dineley-Miller K, Dani JA, Patrick JW. Molecular cloning, functional properties, and distribution of rat brain alpha 7: a nicotinic cation channel highly permeable to calcium. J Neurosci. 1990;13:596–604. doi: 10.1523/JNEUROSCI.13-02-00596.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin AE, Huvar R, D'Andrea MR, Pyati J, Zhu JY, Joy KC, Wilson SJ, Galindo JE, Glass CA, Luo L, Jackson MR, Lovenberg TW, Erlander MG. The pharmacological and functional characteristics of the serotonin 5-HT3A receptor are specifically modified by a 5-HT3B receptor subunit. J Biol Chem. 1999;274:30799–30810. doi: 10.1074/jbc.274.43.30799. [DOI] [PubMed] [Google Scholar]
- A Devillers-Thiery, Galzi JL, Eisele JL, Bertrand S, Bertrand D, JP Changeux. Functional architecture of the nicotinic acetylcholine receptor: A prototype of ligand gated ion channels. J Membrane Biol. 1993;136:97–112. doi: 10.1007/BF02505755. [DOI] [PubMed] [Google Scholar]
- Corringer P, Le Nove're N, Changeux J. Nicotinic receptors at the amino acid level. Annu Rev Pharmacol Toxicol. 2000;40:431–58. doi: 10.1146/annurev.pharmtox.40.1.431. [DOI] [PubMed] [Google Scholar]
- Chiara DC, Xie Y, Cohen JB. Structure of the agonist-binding sites of the torpedo nicotinic acetylcholine receptor: affinity-labelling and mutational analysis identify gamma Tyr-111/delta Arg-113 as antagonist affinity determinants. Biochemistry. 1999;38:6689–6698. doi: 10.1021/bi9901735. [DOI] [PubMed] [Google Scholar]
- Sine SM. Molecular dissection of subunit interfaces in the acetylcholine receptor: identification of residues that determine curare selectivity. ProcNatlAcadSci. 1993;90:9436–9440. doi: 10.1073/pnas.90.20.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sine SM, Kreienkamp HJ, Bren N, maeda R, Taylor P. Molecular dissection of subunit interfaces in the acetylcholine receptor: identification of determinants of alpha-conotoxin M1 selectivity. Neuron. 1995;15:205–211. doi: 10.1016/0896-6273(95)90077-2. [DOI] [PubMed] [Google Scholar]
- Sine SM. Identification of equivalent residues in the, γ, δ δ and ε subunits of the nicotinic receptor that contribute to α-bungarotoxin binding. JBiolChem. 1997;272:23521–23527. doi: 10.1074/jbc.272.38.23521. [DOI] [PubMed] [Google Scholar]
- Gu Y, Camacho P, Gardner P, Hall ZW. Identification of two amino acid residues in the ε subunit that promote mammalian muscle acetylcholine receptor assembly in COS cells. Neuron. 1991;6:879–887. doi: 10.1016/0896-6273(91)90228-r. [DOI] [PubMed] [Google Scholar]
- Kreienkamp HJ, Maeda RK, Sine SM, Taylor P. Intersubunit contacts governing assembly of the mammalian nicotinic acetylcholine receptor. Neuron. 1995;14:635–644. doi: 10.1016/0896-6273(95)90320-8. [DOI] [PubMed] [Google Scholar]
- Westh-Hansen SE, Rasmussen PB, S Hastrup, J Nabekura, K Noguchi, N Akaike, Witt MR, Nielsen M. Decreased agonist sensitivity of human GABA(A) receptors by an amino acid variant, isoleucine to valine, in the alpha1 subunit. Eur J Pharmacol. 1997;329:253–257. doi: 10.1016/S0014-2999(97)10133-9. [DOI] [PubMed] [Google Scholar]
- Green T, Stauffer KA, Lummis SCR. Expression of recombinant homo-oligomeric 5-hydroxytryptamine3 receptors provides new insights into their maturation and structure. J Biol Chem. 1995;270:6056–6061. doi: 10.1074/jbc.270.11.6056. [DOI] [PubMed] [Google Scholar]
- Yan D, Schulte MK, Bloom KE, White MM. Structural features of the ligand-binding domain of the serotonin 5HT3R. J Biol Chem. 1999;274:5537–5541. doi: 10.1074/jbc.274.9.5537. [DOI] [PubMed] [Google Scholar]
- Boess FG, Steward LJ, Steele JA, Liu D, Reid J, Glencorse TA, Martin IL. Analysis of the ligand binding site of the 5-HT3R using site directed mutagenesis: Importance of glutamate 106. Neuropharm. 1997;36:637–647. doi: 10.1016/S0028-3908(97)00044-0. [DOI] [PubMed] [Google Scholar]
- Lummis SC, Sepulveda MI, Kilpatrick GJ, Baker J. Characterization of [3H]meta-chlorophenylbiguanide binding to 5-HT3 receptors in N1E-115 neuroblastoma cells. Eur J Pharmacol. 1993;243:7–11. doi: 10.1016/0014-2999(93)90160-J. [DOI] [PubMed] [Google Scholar]
- Smit AB, Syed NI, Schaap D, van Minnen J, Klumperman J, Kits KS, Lodder H, van der Schors RC, van Elk R, Sorgedrager B, Brejc K, Sixma TK, Geraerts WPM. A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature. 2001;411:261–268. doi: 10.1038/35077000. [DOI] [PubMed] [Google Scholar]
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van der Oost J, Smit AB, Sixma TK. Crystal structure of an Ach-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- Steward LJ, Boess FG, Steele JA, Liu D, Wong N, Martin IL. Importance of phenylalanine 107 in agonist recognition by the 5-hydroxytryptamine3A receptor. Mol Pharm. 2000;57:1249–1255. [PubMed] [Google Scholar]
- Spier AD, Lummis SCR. The role of tryptophan residues in the 5-hydroxytryptamine3 receptor ligand binding domain. J Biol Chem. 2000;275:5620–5625. doi: 10.1074/jbc.275.8.5620. [DOI] [PubMed] [Google Scholar]
- Hope AG, Belelli D, Mair ID, Lambert JJ, Peters JA. Molecular determinanats of (+)tubocurarine binding at recombinant 5-hydroxytryptamine3A receptor subunits. Mol Pharm. 1999;55:1037–1043. doi: 10.1124/mol.55.6.1037. [DOI] [PubMed] [Google Scholar]
- Orjales A, Mosquera R, Labeaga L, Rodes R. New 2-Piperazinylbenzimidazole derivatives as 5HT3 antagonists. Synthesis and pharmacological Evaluation. J Med Chem. 1997;40:586–593. doi: 10.1021/jm960442e. [DOI] [PubMed] [Google Scholar]
- Mihic JS, Whiting PJ, Klein RL, Wafford KA, Harris RA. A single amino acid of the human γ-aminobutyric acid type A receptor γ2 subunit determines benzodiazepine efficacy. J Biol Chem. 1994;269:32768–32773. [PubMed] [Google Scholar]
- K Ohno, Wang H, Milone M, Bren N, Brengmean JM, Nakano S, Quiram P, Pruitt JN, Nine SM, Engel AG. Congenital myasthenic syndrome caused by decreased agonist binding affinity due to a mutation in the acetylcholine receptor ε subunit. Neuron. 1996;17:157–170. doi: 10.1016/s0896-6273(00)80289-5. [DOI] [PubMed] [Google Scholar]