Abstract
Major intrinsic proteins (MIPs) facilitate the passive transport of small polar molecules across membranes. MIPs constitute a very old family of proteins and different forms have been found in all kinds of living organisms, including bacteria, fungi, animals, and plants. In the genomic sequence of Arabidopsis, we have identified 35 different MIP-encoding genes. Based on sequence similarity, these 35 proteins are divided into four different subfamilies: plasma membrane intrinsic proteins, tonoplast intrinsic proteins, NOD26-like intrinsic proteins also called NOD26-like MIPs, and the recently discovered small basic intrinsic proteins. In Arabidopsis, there are 13 plasma membrane intrinsic proteins, 10 tonoplast intrinsic proteins, nine NOD26-like intrinsic proteins, and three small basic intrinsic proteins. The gene structure in general is conserved within each subfamily, although there is a tendency to lose introns. Based on phylogenetic comparisons of maize (Zea mays) and Arabidopsis MIPs (AtMIPs), it is argued that the general intron patterns in the subfamilies were formed before the split of monocotyledons and dicotyledons. Although the gene structure is unique for each subfamily, there is a common pattern in how transmembrane helices are encoded on the exons in three of the subfamilies. The nomenclature for plant MIPs varies widely between different species but also between subfamilies in the same species. Based on the phylogeny of all AtMIPs, a new and more consistent nomenclature is proposed. The complete set of AtMIPs, together with the new nomenclature, will facilitate the isolation, classification, and labeling of plant MIPs from other species.
The hydrophobic interior of a membrane constitutes a barrier for the rapid flow of small polar molecules. The large and evolutionary conserved family of major intrinsic proteins (MIPs) has evolved to facilitate the passive flow of small polar molecules like water and/or glycerol across membranes in all types of organisms ranging from bacteria to fungi, animals, and plants. MIPs that specifically transport water are named aquaporins (AQPs). The permeability of membranes is actively controlled by the regulation of the amount of different MIPs present but also in some cases by phosphorylation/dephosphorylation of the channels. Plant MIPs can be classified into different subfamilies based on their sequence similarity. Two of the subfamilies are named after their main location in the cell, plasma membrane intrinsic proteins (PIPs) and tonoplast intrinsic proteins (TIPs). The third subfamily has been named NOD26-like MIPs (NLMs; Weig et al., 1997) or NOD26-like intrinsic proteins (NIPs; Heymann and Engel, 1999). NOD26, the first identified member of this subfamily, is located in the peribacteroid membrane of nitrogen-fixating symbiosomes in root nodules in soybean (Glycine max; Fortin et al., 1987). The functions of these three subfamilies in plants have been extensively reviewed (Kjellbom et al., 1999; Johansson et al., 2000; Santoni et al., 2000; Maurel and Chrispeels, 2001). A novel fourth MIP subfamily recently was proposed in plants (U. Johanson, at the MIP 2000 meeting in Göteborg, Sweden, July 2000; Chaumont et al., 2001; U. Johanson and S. Gustavsson, unpublished data). This subfamily was named small basic intrinsic proteins (SIPs) because the proteins are relatively small, similar to TIPs, but different from TIPs because they are basic like the PIPs and many of the NLMs. Neither the substrate specificity nor the intracellular localization of SIPs is known.
The structures of a glycerol facilitator (GlpF) from Escherichia coli and a water channel (AQP1) from man were recently determined at 2.2 and 3.8 Å resolution, respectively (Fu et al., 2000; Murata et al., 2000). Despite the huge evolutionary distance and the difference in substrate specificity, the overall fold is very similar in the two MIPs (Unger, 2000). MIPs have an internal symmetry as a consequence of a direct repeat in the sequence. Each one-half of a MIP consists of two transmembrane helices followed by a conserved loop with the amino acid motif Asn-Pro-Ala (NPA) and ended with a third transmembrane helix. Both the N terminus and the C terminus as well as the first NPA box are located on the cytoplasmic face of the membrane. Due to the uneven number of transmembrane helices in the repeat, the orientation of the transmembrane helices and the NPA motif in the second half of the protein is reversed as compared with the first one-half. The sequences immediately after the NPA boxes in fact form two half-transmembrane helices that are inserted into the membrane from opposite sides and connect to each other via an interaction between the two NPA boxes at the N-terminal end of the two short helices. The transmembrane regions together form a pore that allows the substrate to circumvent the hydrophobic part of the membrane. Most of the polar interaction sites inside the pore of GlpF are formed by the highly conserved NPA regions, which together form a polar stripe that allow glycerol to pass through the pore. However, alignments of SIPs with GlpF and AQP1 show that the polar lining inside SIPs is different, which suggests that SIPs have a different substrate specificity compared with GlpF and AQP1 (U. Johanson and S. Gustavsson, unpublished data). MIP structures will become indispensable tools in the work of trying to understand functional consequences of differences in primary structure between different classes of MIPs.
The nomenclature for plant MIPs is confusing and varies between different species and also between the different MIP subfamilies. The individual names sometimes reflect how the MIP is induced, the size of the protein, similarity to other MIPs, or is totally uninformative. In most plant species where MIPs have been isolated, there are only a few MIPs known. On this patchy knowledge it is difficult to construct a consistent and systematic nomenclature. The Arabidopsis genomic sequence provides a unique opportunity to obtain the complete set of MIPs in a plant for the first time and this will provide a framework for classification of MIPs from other plants. However, the names of MIPs from Arabidopsis (AtMIPs) are not always consistent or informative about phylogenetic relations. This could be achieved with a new nomenclature of AtMIPs that is based on phylogenetic analyses and where the names in a systematic way reflect distinct clades that are evolutionarily stable. The new nomenclature should confine to the recommendations set up by the Commission on Plant Gene Nomenclature (CPGN) but it should also retain as much as possible of the old names to facilitate the translation of old names into new.
In this paper, the complete set of AtMIPs is described and analyzed with different phylogenetic methods. A new and more consistent AtMIP nomenclature is presented. In addition, the structures of all AtMIP genes are presented and compared with the sequence-based phylogenetic analysis. A short summary of what is known about the functional aspects of plant MIPs is also included. It is our hope that this will facilitate the classification of other plant MIPs and that the suggested nomenclature will be accepted as a standard and used in other plants.
RESULTS
Identification
The completed sequence of the Arabidopsis genome has made it possible to identify all MIP genes in a plant (Arabidopsis Genome Initiative, 2000). Thirty-five different genes encoding complete AtMIPs were found by BLAST searches in GenBank (Table I). In addition, five genes encoding partial MIP-like sequences were found. These genes are either partial or interupted by premature stop codons and were thus considered as nonfunctional pseudogenes because they lacked the complete set of transmembrane regions that are conserved in all MIPs. Searches with TFASTA resulted in two additional pseudogenes, both similar to parts of F5I10.2, but no additional full-length genes were found. To identify new AtMIP genes and to assure a correct annotation of the coding sequence in the genes, each gene was compared with previously identified genes and cloned cDNAs encoding AtMIPs. The annotation of the genomic sequence was correct for most AtMIP genes. However, for a few genes a different annotation of the coding sequence in the genomic sequence was favored either by cDNA sequences or due to homology reasons. These alternative assignations of exons, specified in Table I, are used in all translations and analyses in this paper.
Table I.
Proposed new names for all Arabidopsis MIPs
| New Namea | Johanson et al.b | Weig et al.c | AGI Gened | BAC Genee | Accession No.f | Expressed Sequence Tag (EST)g | Commentsh |
|---|---|---|---|---|---|---|---|
| PIP1;1 | PIP1a | PIP1a | At3g61430 | F2A19.30 | CAB71073 | Yes | |
| PIP1;2 | PIP1b | PIP1b | At2g45960 | F4I18.6 | AAC28529 | Yes | AthH2, TMP-A |
| PIP1;3 | PIP1c | PIP1c | At1g01620 | F22L4.16 | AAF81320 | Yes | TMP-B |
| PIP1;4 | PIP1e | TMP-C | At4g00430 | F5I10.2 | AAF02782 | Yes | Exon 2 ends at 86,121, not 86,097; compare with TMP-C. |
| PIP1;5 | PIP1d | pCR55 | At4g23400 | F16G20.100 | CAA20461 | Yes | |
| PIP2;1 | PIP2a | PIP2a | At3g53420 | F4P12_120 | CAB67649 | Yes | |
| PIP2;2 | PIP2b | PIP2b | At2g37170 | T2N18.7 | AAD18142 | Yes | |
| PIP2;3 | PIP2c | RD28 | At2g37180 | T2N18.6 | AAD18141 | Yes | |
| PIP2;4 | PIP2f | T04164 | At5g60660 | MUP24.8 | BAB09839 | Yes | |
| PIP2;5 | PIP2d | – | At3g54820 | F28P10.200 | CAB41102 | Yes | |
| PIP2;6 | PIP2e | T22419 | At2g39010 | T7F6.18 | AAC79629 | Yes | |
| PIP2;7 | PIP3a | PIP3 | At4g35100 | M4E13.150 | CAA17774 | Yes | Salt-stress-inducible MIP |
| PIP2;8 | PIP3b | – | At2g16850 | F12A24.3 | AAC64216 | Yes | |
| – | Pseudo PIP3 | – | At2g16830 | F12A24.1 | AAC64228 | ? | Pseudogene encoding 26 amino acids with second NPA. Similar to F12A24.3. |
| – | – | – | – | T8O11i | 77,410–77,455 | ? | Partial F5I10.2-like sequence encoding helix 4. |
| – | – | – | – | F5I10i | 77,564–77,656 | ? | Partial F5I10.2-like sequence encoding loop B and helix 3. |
| TIP1;1 | γTIP1 | γTIP | At2g36830 | T1J8.1 | AAD31569 | Yes | |
| TIP1;2 | γTIP2 | Z30833, T21060, T22237 | At3g26520 | MFE16.3 | BAB01832 | Yes | Salt-stress-inducible TIP, γTIP2 |
| TIP1;3 | γTIP3 | – | At4g01470 | F11O4.1 | AAC62778 | ? | No introns |
| TIP2;1 | δTIP1 | δTIP | At3g16240 | MYA6.5 | BAB01264 | Yes | |
| TIP2;2 | δTIP2 | Z18142 | At4g17340 | d14705w | CAB10515 | Yes | |
| TIP2;3 | δTIP3 | T20432, T76151 | At5g47450 | MNJ7.4 | BAB09071 | Yes | |
| TIP3;1 | αTIP | αTIP | At1g73190 | T18K17.14 | AAG52132 | Yes | |
| TIP3;2 | βTIP | βTIP | At1g17810 | F2H15.4 | AAF97261 | Yes | |
| TIP4;1 | ɛTIP | T21742 | At2g25810 | F17H15.16 | AAC42249 | Yes | Similar to NtTIPa |
| TIP5;1 | ζTIP | – | At3g47440 | T21L8.190 | CAB51216 | ? | |
| – | Pseudo-γTIP2 | – | – | F7P1i | 79,395–79,728 | ? | Pseudogene similar to MFE16.3, truncated, NPV instead of NPA, no exon 2. |
| – | Pseudo-δTIP | – | At1g52180 | F9I5.3 | AAF29403 | ? | Pseudogene, only exon 2, 124 amino acids. |
| NIP1;1 | NLM1 | NLM1 | At4g19030 | F13C5.200 | CAA16760 | Yes | Misannotated: delete amino acids 205 through 216; exon 4 starts at 86,879, not 86,915. |
| NIP1;2 | NLM2 | NLM2 | At4g18910 | F13C5.80 | CAA16748 | Yes | |
| NIP2;1 | NLM3 | NLM4 | At2g34390 | T31E10.27 | AAC26712 | Yesj | |
| NIP3;1 | NLM9 | – | At1g31880k | F5M6.28 | AAG50717 | ? | F5M6.28 is missing exon 1; exon 1, 38,681 through 38,550; exon 2 starts at 38,310, long C terminus. |
| NIP4;1 | NLM4 | – | At5g37810 | K22F20.9 | BAB10360 | ? | |
| NIP4;2 | NLM5 | NLM5 | At5g37820 | K22F20.10 | BAB10361 | Yesj | |
| NIP5;1 | NLM6 | NLM8 | At4g10380 | F24G24.180 | CAB39791 | Yes | Long N terminus. |
| NIP6;1 | NLM7 | NLM7 | At1g80760 | F23A5.11 | AAF14664 | Yes | Long N terminus. |
| NIP7;1 | NLM8 | NLM6 | At3g06100 | F28L1.3 | AAF30303 | Yes | Intron 3, 8,151 through 7,945, avoids TT acceptor splice site, restores conserved length of exon 4. |
| – | Pseudo-NLM3 | NLM3 | At2g29870 | T27A16.3 | AAC35214 | ? | Pseudogene encoding 139 amino acids, missing first NPA. |
| – | Pseudo-NLM9 | – | At2g21020 | F26H11.22 | AAD29814 | ? | Pseudogene, version 3 missing second NPA. Alternate annotation results in two NPAs, but missing helix 3 and 4, long C terminus. |
| SIP1;1 | SIP1a | – | At3g04090 | T6K12.29 | AAF26804 | Yes | |
| SIP1;2 | SIP1b | – | At5g18290 | MRG7.25 | BAB09487 | Yes | |
| SIP2;1 | SIP2 | – | At3g56950 | F24I3.30 | CAB72165 | Yes |
Proposed new names for Arabidopsis MIPs. When referring to the corresponding gene, the name is written in italics. Genes encoding partial MIP-like sequences without all transmembrane regions are regarded as nonfunctional pseudogenes. Pseudogenes are not included in the proposed nomenclature as indicated by “–”.
Nomenclature used in Johansson et al. (2000) and in U. Johanson and S. Gustavsson (unpublished data).
Nomenclature used in Weig et al. (1997) and Weig and Jakob (2000a, 2000b).
Arabidopsis Genome Initiative (AGI) gene nomenclature; At3g56950 indicates gene no. 5,695 (counting from top) on chromosome 3.
Gene name used in annotation of bacterial artificial chromosome (BAC) or P1 sequences.
Protein accession no. for a translation of the corresponding gene. When a gene annotation is missing, the location in the BAC is given in base pairs.
Matching EST found in databases. ?, Not found.
Alternative exon/introns positions proposed and used in this paper and odd features of the gene or the encoded protein. Some older names found in the literature (Kaldenhoff et al., 1993; Shagan and Bar-Zvi, 1993; Shagan et al., 1993; Pih et al., 1999) and in Genbank accession no. AF057137.
Not annotated.
cDNA cloned by reverse transcriptase-PCR (Weig and Jakob, 2000b).
At1g31880 has 285 amino acids added at the N terminus, previously annotated as a separate gene (F5M6.11). At1g31880 is missing 29 amino acids close to the C terminus.
In total, 15 new AtMIPs have been identified from genomic sequence generated in the Arabidopsis Genome Initiative. All of these except AtNIP3;1 have been described elsewhere (U. Johanson, at the MIP 2000 meeting in Göteborg, Sweden, July 2000; Johansson et al., 2000; Weig and Jakob, 2000a, 2000b; Chaumont et al., 2001). Based on the phylogeny of AtMIPs a new and more consistent nomenclature is proposed (Table I; see below).
Phylogeny and New Nomenclature
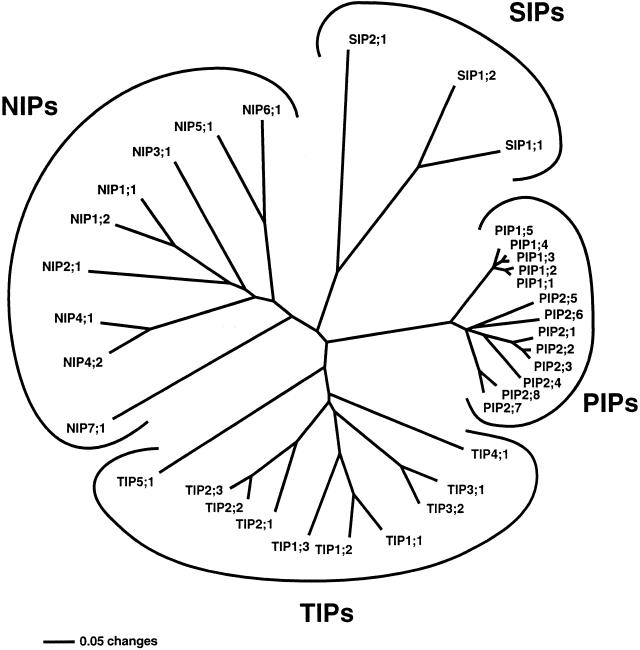
Using the full length of the alignment of all AtMIPs, the distance method resulted in one tree (Fig. 1). The AtMIPs are clearly divided into four distinct subfamilies, PIPs, TIPs, NIPs, and SIPs. The old names for these subfamilies are retained except for the former NLM subfamily, where NIP is preferred to make the nomenclature more uniform (Heymann and Engel, 1999). Each subfamily can be further subdivided into groups of related proteins. The proposed names of AtMIPs consist of the subfamily name followed by a number indicating the group to which the MIP belongs and a second number identifying the individual MIP in the group. To achieve approximately the same divergence in all groups, a maximum distance of 30% was accepted within each group. The distance was chosen arbitrarily to retain most of the groups that have been used in previous nomenclatures. This maximum divergence is low enough to resolve the AtPIPs into AtPIP1s and AtPIP2s, but on the other hand the former AtPIP3a and AtPIP3b are not divergent enough to form a group of their own and are instead named AtPIP2;7 and AtPIP2;8, respectively. In the TIP subfamily, AtαTIP and AtβTIP form the new AtTIP3 group because these proteins are very similar. All the other AtTIP groups remain the same, but AtγTIPs become AtTIP1s and AtδTIPs become AtTIP2s, whereas the very different εTIP and ζTIP form the separate groups AtTIP4;1 and AtTIP5;1, respectively. This numbering of the TIP groups follows the nomenclature used in maize (Zea mays) where the second largest set of MIPs has been identified (Chaumont et al., 2001). Within the AtNIPs, no groups have been reflected in the old nomenclature. Only two pairs of AtNIPs are similar enough to form groups according to the criterium of a maximum distance of 30%. For AtSIPs, the division into SIP1s and SIP2s is retained (U. Johanson, at the MIP 2000 meeting in Göteborg, Sweden, July 2000; Chaumont et al., 2001; U. Johanson and S. Gustavsson, unpublished data). In summary, there are only two AtPIP and AtSIP groups, respectively, but five and seven AtTIP and AtNIP groups, respectively. This is a consequence of the large variation in divergence in these subfamilies. The AtPIPs constitute a much more homogenous subfamily than any of the other subfamilies. If the maximum divergence were set lower to try to resolve AtPIP2;7 and AtPIP2;8 as a separate group, then AtTIP1;3 would also form a group of its own. The recently discovered AtSIPs also have substantial sequence variation within the subfamily and are at the same time very different to the other plant MIPs, as illustrated by the long branches in the tree (Fig. 1; Chaumont et al., 2001). A lower limit of divergence would result in three AtSIP groups.
Figure 1.
Phylogenetic comparison of the complete set of 35 different MIPs encoded in the Arabidopsis genome. Plant MIPs are divided into four distinct subfamilies: PIPs, TIPs, NIPs, and SIPs. Similar proteins within a subfamily, with a maximum of 30% divergence, are clustered in monophyletic groups. The first and the last digit in the protein name identify the group and the individual gene product, respectively. This tree was obtained using the whole alignment and the distance method. Omitting the none-conserved N- and C-terminal regions from the phylogenetic analysis does not break the defined groups; only the relative positions of PIP2;6 and TIP3s is changed. In this case PIP2;6 forms a separate branch between PIP2;4 and PIP2;5 and TIP3s branch between TIP2s andTIP4;1. The bar indicates the mean distance of 0.05 changes per amino acid residue.
To suggest a new nomenclature that can be useful in the classification of MIPs from other plants species, it is important that the groups that are recognized in Arabidopsis are stable and not dependent on any particular phylogenetic method. The distance tree (Fig. 1) was compared with the two shortest trees generated with the parsimony method (not shown). The topologies of all three trees are virtually identical with some minor variation in the phylogeny of AtPIP2s. Another small difference is that AtTIP2s are basal to AtTIP4;1 in parsimony trees. However, all the defined groups were stable and were not split by any of the methods.
The alignment of cytoplasmic N- and C-terminal regions of AtMIPs contains many gaps because these regions are highly variable even within the subfamilies. A meaningful alignment of homologous sites can only be done to the closest AtMIP homologs. It is possible that these regions are also less conserved when AtMIPs are compared with orthologues in other species. To investigate whether these highly variable regions are critical for the classification of AtMIPs, these regions were excluded from the analyses. Although there is some variation in how the groups are positioned in the trees, the individual groups remain stable regardless if the distance or the parsimony method is used.
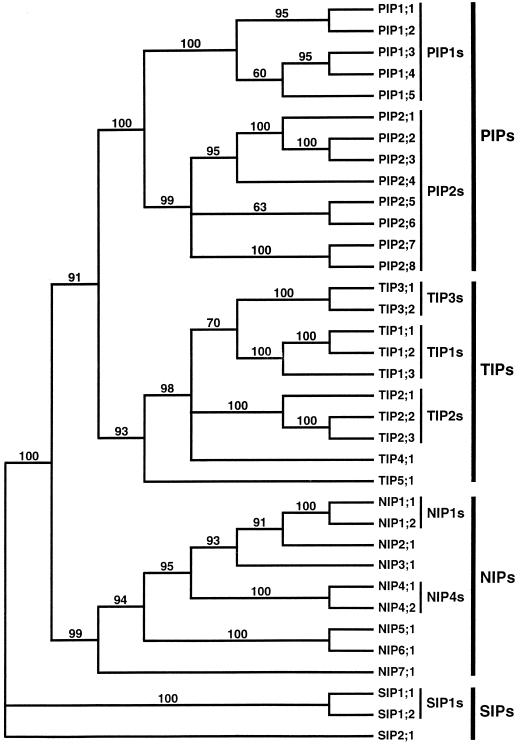
A method to measure the stability of a phylogeny is to calculate bootstrap values. Bootstrap tests were performed on the full-length alignment of AtMIPs. The result of 100 replicates with the distance method is shown in Figure 2. Using the distance method all the subfamilies are supported in at least 93% of the replicates. Bootstrap values generated with the parsimony method are in general lower, with the weakest support for the AtTIP clade, only 75%. However, the defined groups are very stable in both methods, with bootstrap values equal to or higher than 99% and 89% in the distance and parsimony methods, respectively. From the bootstrap values it is clear that the internal phylogeny of AtPIP2s and the relationship of the AtTIP groups are not very stable. These low bootstrap values are consistent with the observed variation between phylogenetic trees generated with different methods (see above).
Figure 2.
Bootstrap majority-rule consensus tree generated with the distance method. A bootstrap value of 100% indicates branches that were supported in all replicates of resampling of data. Branches with a bootstrap value of less than 50% are collapsed.
Gene Structure
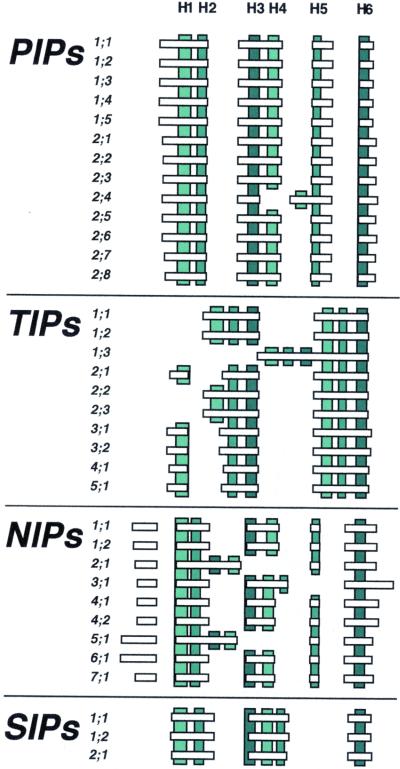
A comparison of the gene structures of all the AtMIP genes shows that the exact locations of the introns in the genes are unique and conserved within each subfamily (Fig. 3). Also, the number of introns is in general specific for each subfamily. All the AtPIPs have three introns, AtSIPs have two, and most AtNIPs have four introns. In AtTIPs, the pattern is more varied; one-half of the genes have two introns and the other one-half have just a single or no intron. Based on the phylogenetic tree in Figure 1, the simplest explanation for the observed gene structures among AtTIPs is that intron 1 has been lost independently in AtTIP1s and in two of the AtTIP2s. These were probably independent events because there is no sign of a higher similarity in this one-half of the genes that would suggest that the intron-less half was copied from one group to the other. In AtTIP1;3, the second intron also has been subsequently lost. In the same manner, the deviations from the conserved gene structure in AtNIPs suggest that the second intron has been lost independently in AtNIP2;1 and AtNIP5;1. In addition, the third intron has been lost in AtNIP3;1. Given the high similarity among the AtPIPs, it is not surprising that there is only one exception from the conserved gene structure in this subfamily. In AtPIP2;4, the position of intron 2 has changed as the result of an insertion of a new intron and a loss of the old intron 2.
Figure 3.
Schematic structure of MIP encoding genes in Arabidopsis. Horizontal bars and gaps depict exons and intron positions, respectively. Parts encoding transmembrane helices H1 to H6 according to an alignment with GlpF are indicated by vertical bars. The color on the vertical bars shows homologous transmembrane helices in the first and second halves of the MIPs. The exons and transmembrane helices are drawn to scale but the positions of helices are schematic. Helices encoded on two exons are only indicated on the exon where the major part is encoded. Small indels in the alignment of different genes, positioned between two helices on the same exon, are not shown.
Focusing on the initial gene structure in the subfamilies, before loss of introns, there are some common features in the organization of how the transmembrane helices are encoded on the exons. All subfamilies except AtTIPs have helix 1 and 2 encoded together on a separate exon, helix 3 and 4 on the next, and helix 6 alone on the last exon. This suggests that a common ancestor to the PIPs, NIPs, and SIPs had this type of gene structure. The encoding of helix 5 is more varied. In AtPIPs and AtNIPs, helix 5 is coded by a separate exon but in AtTIPs and AtSIPs helix 5 is encoded together with helix 4 and 6, and with 3 and 4, respectively.
DISCUSSION
Identification, Phylogeny, and New Nomenclature
For a newly identified protein or a gene, the sequence is often the first and sometimes the only thing that is known. In most cases a BLAST search is done to try to classify the gene or protein to get an idea about the function of the protein from homologs that are already characterized. To facilitate the identification of the most similar AtMIPs to isolated MIPs or MIP genes from other plant species, a comprehensive list of all AtMIPs was compiled (Table I). This list also provides important information on which MIPs can also be expected in other plants and can be used as a starting point for searches of MIPs in other species.
Thirty-five different complete AtMIPs were identified from genomic sequences. Thirty-three of these AtMIPs have also been identified in searches with the MIP pFAM motif, PF00230, by the Munich Information center for Protein Sequences (http://mips.gsf.de/proj/thal/db/tables/tables_func_main.html#pfam). All 35 genes and three of the pseudogenes are listed as MIPs at Ian Paulsen's (University of California, San Diego) Web site (http://www.biology.ucsd.edu/∼ipaulsen/transport/) and topological analyses with hydropathy plots are available at http://www.cbs.umn.edu/Arabidopsis/atprotdb2/famMIP.htm. Plant MIPs are subdivided into the four subfamilies of PIPs, TIPs, NIPs, and SIPs (U. Johanson, at the MIP 2000 meeting in Göteborg, Sweden, July 2000; Chaumont et al., 2001; U. Johanson and S. Gustavsson, unpublished data). In Arabidopsis, there are 13 PIPs, 10 TIPs, nine NIPs, and three SIPs. Of these 35 AtMIPs, only AtNIP3;1 has not been reported before. In total, 15 of the 35 AtMIPs were first identified from the genomic sequence of Arabidopsis due to their low expression levels (Johansson et al., 2000; Weig and Jakob, 2000b; Chaumont et al., 2001; U. Johanson and S. Gustavsson, unpublished data). It is interesting to note that all three AtSIPs and eight out of nine AtNIPs are included in these 15 lowly expressed AtMIPs. In contrast, only two AtPIPs and two AtTIPs of 13 and 10 proteins, respectively, were not first identified as cDNA clones but revealed by analyses of genomic sequence. Expression has still not been verified for two of the AtTIPs and for two of the AtNIPs. One of the AtMIP genes without an EST is AtTIP1;3. This gene is odd because there are no introns, which could indicate that it is a retropseudogene (see below). AtTIP5;1, the other AtTIP with a missing EST, has the most different sequence compared with other AtTIPs. However, all four AtMIPs genes without a matching EST encode all the transmembrane regions that are required for a functional MIP. This is in contrast to the partial MIP sequences that are encoded in the seven pseudogenes.
A large set of cDNAs encoding different MIPs in maize was identified recently and analyzed (Chaumont et al., 2001). In maize, there are at least 14 PIPs, 13 TIPs, five NIPs, and three SIPs if partial cDNA clones are taken into account. This can be compared with Arabidopsis where there are 13 PIPs, 10 TIPs, nine NIPs, and three SIPs. The numbers of MIPs in each subfamily are very similar in maize and Arabidopsis. The only real difference is that there are more NIPs known in Arabidopsis than in maize. This is not unexpected because all the identified NIPs are rare among ESTs both in maize and Arabidopsis (Chaumont et al., 2001), suggesting that there are more NIP genes to identify in maize.
Each subfamily is subdivided into groups of related proteins. A maximum divergence of 30% was used as a cutoff value to define the groups in a consistent way in all subfamilies in the new nomenclature. This limit of divergence was chosen because most groups in the older nomenclature were in this way retained in the new nomenclature. This is convenient because it makes it relatively easy to convert the old names into the new nomenclature when reading older literature. At the same time the format of the new names is different and this will exclude any doubt about what nomenclature is used. It is often impossible to identify the exact orthologue to an AtMIP in another species because there are too many very similar paralogues or because all MIPs are not known in that species. One idea with the groups is that it should be possible to sort new MIPs from other plants into the AtMIP groups. The new MIP can then be named according to subfamily and group. However, within a group new proteins are numbered in the order they are discovered without any ambition to reflect a higher similarity to a specific AtMIP within the group. This will result in informative names that reflect the phylogenetic groups in a stable way and do not need to be changed when more isoforms are found. To achieve a clear classification, it is important that all the groups are distinctly resolved from each other in phylogenetic analyses. The defined groups in Arabidopsis are stable with relatively high bootstrap values and do not change with different phylogenetic methods or exclusion of the most variable regions in the alignment. Most of the defined groups have already been shown to be evolutionarily conserved because they are found both in monocotyledons and dicotyledons (Karlsson et al., 2000; Chaumont et al., 2001). Thus, these groups will also be helpful in classification of MIPs from relatively distantly related plants.
The number of groups in each subfamily varies from only two in AtPIPs and AtSIPs to five and seven in AtTIPs and AtNIPs, respectively. The large variation is a reflection of the amount of sequence divergence and the number of proteins in each subfamily. The AtMIPs in the largest subfamily, the AtPIPs, are all very similar, suggesting that this subfamily has expanded relatively recently. Based on the comparison of maize and Arabidopsis PIPs, it has been suggested that the PIP genes were multiplied independently after the split of monocotyledons and dicotyledons (Chaumont et al., 2001). This would imply that there has been a recent selection for an amplification of PIP genes. It is not clear what event or adaptation required more PIP genes independently in maize and Arabidopsis. An alternative explanation would be that the expansion of PIPs happened before the split of monocots and dicots and that there has been a concerted evolution of PIPs in both monocots and dicots, resulting in the large number of internally very similar PIPs in both species.
Gene Structure
Comparisons of gene structures may result in additional information on phylogenetic relations and the evolution of a gene family. Studies of the genes encoding NOD26, AtTIP3;1, AtTIP1;1, and TobRB7, an AtTIP2;2 homolog from tobacco (Nicotiana tabacum), showed that all the three TIP genes have a very similar organization, whereas the gene structure of NOD26 is different (Miao and Verma, 1993). The authors observed that the introns were preferentially located in regions encoding loops connecting the transmembrane helices and suggested that the transmembrane regions encoded by a single exon act as a functional domain.
Analyses of all AtMIP gene structures confirm that both the position as well as the number of introns are remarkably well conserved within each subfamily. In most cases it is possible to classify a MIP gene to a subfamily just based on a single intron position. However, there are a few genes were the intron pattern deviates from the characteristic pattern of the subfamily. Based on the phylogenetic tree, it is inferred that introns have been lost independently in total seven times: three times in AtTIPs, three times in AtNIPs, and only once in AtPIPs. In contrast, there is only one example of an insertion of an intron in a AtMIP gene. It is interesting that this gene, AtPIP2;4, is the same PIP gene that has also lost an intron, resulting in a gene of similar length as other AtPIPs. The mechanism of intron loss is likely to include reverse transcription of mRNA followed by homologous recombination or recombination into a new locus. In this context, it is interesting to note that there is a retrotransposon-like element (accession no. AAC62785) only 5 kb upstream of AtTIP1;3, the only AtMIP gene totally lacking introns. In the AtMIP gene family, there is a trend to lose introns. This is contrary to the idea that plant genomes in general have an intrinsic predisposition for “genomic obesity” (Bennetzen and Kellogg, 1997). However, it has been observed that angiosperm weeds have smaller genomes than other plants. On this basis, it has been suggested that there is a strong selection for small genomes in rapidly cycling weeds like Arabidopsis (Bennett et al., 1998). In tobacco, which has an approximately 30 times larger genome than Arabidopsis (Arumuganathan and Earle, 1991), the gene encoding TobRB7, an AtTIP2;2 homolog (Karlsson et al., 2000), has not lost the first intron (Miao and Verma, 1993). Thus, the observed tendency to lose introns might not be relevant to other plant species with no apparent selection for reduction of their genome size.
It has been demonstrated recently that many of the groups defined in each AtMIP subfamily are present also in monocotyledons, i.e. they were formed before the split of the monocotyledons and the dicotyledons (Chaumont et al., 2001). This would argue that the gene structure that is common to the groups of an AtMIP subfamily would also have been present in a common ancestor to monocotyledons and dicotyledons. Hence, many monocotyledons and dicotyledons MIP genes are likely to have remnants of the gene structure characteristic for each Arabidopsis subfamily.
The similar organization of transmembrane encoding regions on exons among non-TIP-AtMIPs raises several interesting questions. Are PIPs, NIPs, and SIPs more closely related to each other than to TIPs? Comparisons of protein sequence do not support this, but rather favor that SIPs are the most different subfamily. If the SIPs are used to root the tree in Figure 2, PIPs and TIPs are more closely related than PIPs and NIPs. It is possible that a common ancestor to all plant MIP genes had a similar organization to the one found in AtPIPs and AtNIPs and that this was later lost partially in SIPs and totally in TIPs. In an alternate manner, the SIP gene structure was present in the ancestor of all plant MIPs and subsequently the ancestor of PIPs, TIPs, and NIPs gained an extra intron resulting in the present gene structure of NIPs and PIPs. Later, this gene structure was lost in TIPs. However, the exact location of introns is not the same in subfamilies with a similar organization. This could suggest that the similar organization of transmembrane-encoding regions on exons instead is the result of homoplasy and not of a common origin. It has been recognized that exons in a gene sometimes correspond to functional or structural domains of the encoded protein (Go, 1981). Whether the putative domains found in AtMIPs reflect the insertion and folding of the MIPs in the membrane or represent genetic building blocks used to assemble different MIP subfamilies can only be speculated. However, it is interesting to note that the position of intron 2 in AtTIPs marks the internal symmetry caused by the sequence similarity between the first and second halves in all MIPs.
Subcellular Localization of MIPs
The identification of four major subfamilies within the MIP gene family almost implies different physiological roles of the encoded proteins. These proteins are found in at least two subcellular compartments, the tonoplast (TIPs) and the plasma membrane (PIPs). Whether the prediction of the subcellular localization based on sequence similarity always holds true has been questioned (Barkla et al., 1999) and should be verified for each protein. Although the AQP activities of TIPs and PIPs are comparable in heterologous expression systems, these two subfamilies of membrane proteins could have different physiological roles because no hydrostatic pressure gradient exists across the tonoplast, whereas an enormous pressure gradient of several bars at the plasma membrane maintains the turgor of individual cells and presumably the shape of cell collectives and plant tissues. Separating the cytoplasm from the apoplast is certainly a different task compared with the compartmentation of cytosol and vacuole.
However, under certain circumstances the physiological roles of TIPs and PIPs can overlap. An example is expansion growth of plant cells where the majority of water has to be transferred from the apoplast via the cytosol into the vacuole. Plasmalemmasomes, invaginations of the plasma membrane, have been proposed to function in situations of high fluxes and PIP1 AQPs have been identified in these structures (Robinson et al., 1996), but to date corresponding TIPs in the tonoplast adjacent to plasmalemmasomes are not known.
Although the NIP prototype, NOD26 from soybean, is inserted into the peribacteroid membrane of root nodules (Fortin et al., 1987), the physiological roles of the Arabidopsis homologues, AtNIPs, are unknown.
Transport Activities of MIPs
The transport activities of MIPs, water channels or mixed-functional water channels/solute transporters, correlate with the subfamilies of MIPs, although some exceptions have been reported (see below). Members of the PIP2 group are described as “good” AQPs in the Xenopus laevis expression system, whereas PIP1 proteins often cause lower osmotic water permeability (Pf) values in this expression system (Johansson et al., 1998; Chaumont et al., 2000). It has been speculated that PIP1 AQPs could be responsible for the transport of yet unidentified solutes across the plasma membrane. It is interesting that NtAQP1 from tobacco, which is very similar to AtPIP1:3, has been reported to transport glycerol in addition to water in X. laevis oocytes (Biela et al., 1999). In addition, NtTIP1 (tobacco) and LIMP1 (Lotus japonicus) have been shown to transport water, glycerol, and urea (only NtTIP1) when expressed in X. laevis oocytes (Gerbeau et al., 1999; Guenther and Roberts, 2000). The closest Arabidopsis relative of NtTIP1, AtTIP4;1, has not been functionally tested. The closest relative of LIMP1, AtTIP1;1, shows no glycerol transport activity (Maurel et al., 1993). Mixed transport activities have been shown for NIPs from different organisms. NOD26 from soybean forms a functional water channel and glycerol permease in X. laevis oocytes (Rivers et al., 1997; Dean et al., 1999). The Arabidopsis homologue, AtNIP1;1 is an AQP when expressed in X. laevis oocytes (Weig et al., 1997). Moreover, AtNIP1;1 and AtNIP1;2 form functional glycerol permeases when expressed in baker's yeast (Saccharomyces cerevisiae; Weig and Jakob, 2000a). LIMP2, a NIP from L. japonicus with high similarity to AtNIP1;2, expresses water and glycerol transport activities in X. laevis oocytes (Guenther and Roberts, 2000).
Expression Analysis
With few exceptions, a strict organ-specific expression has not been found for Arabidopsis MIP genes. However, preferential expression in seeds/embryos, roots, and shoots has been found for some TIP genes. AtTIP3;1 is a seed- and embryo-specific AQP not only in Arabidopsis, but also in other plants such as Phaseolus vulgaris and Ricinus communis (Johnson et al., 1989; van de Loo et al., 1995). AtTIP1;1 is expressed mainly in the elongation zone of roots and to lower levels in various shoot organs (Höfte et al., 1992; Ludevid et al., 1992). It is interesting that this elongation-associated AQP can be induced by gibberellic acid, which is known to promote cell growth in Arabidopsis dwarf mutants (Phillips and Huttly, 1994). In contrast, AtTIP2;1 is mainly expressed in shoots and to a lower extent in roots (Daniels et al., 1996).
Members of the PIP subfamily do not show any preferential expression in certain organs. All abundant PIPs described by Kammerloher et al. (1994) are more or less equally expressed in shoots and roots of Arabidopsis plants. However, AtPIP2;7 was detected as the most abundant plasmalemma AQP in a photoautotrophic Arabidopsis cell culture (Weig et al., 1997). This could indicate that this cell culture represents a specific cell line of Arabidopsis leaves where normally AtPIP2;7 is mainly expressed or it may reflect that gene expression is deregulated in cells growing in cell suspension cultures.
The situation is quite different for some NIPs: AtNIP1;1 and AtNIP4;1 seem to be exclusively expressed in Arabidopsis roots (Weig et al., 1997; Weig and Jakob, 2000a, 2000b). AtNIP1;2 and AtNIP4;2 transcripts were also found in roots, but also in leaves, stems, and flowers of an adult Arabidopsis plant.
Whether the same expression patterns that have been identified for AtMIPs can be expected to be found for ortholgues in other plants remains to be seen. Results for SoδTIP, an AtTIP2;1 homolog from spinach (Spinacia oleracea), suggest that is not the case. The expression pattern of SoδTIP was completely different compared with the expression of putative orthologues in other plants. Instead, it was more similar to AtTIP1;1 (Karlsson et al., 2000). However, a major obstacle in these comparisons is that it is hard to establish that any two sequences are true orthologues. It is always possible that the wrong homologous protein is used in comparisons and that there are very similar paralogues with a consistent expression pattern.
Regulation of Transport Activity
Regulation of the transport activity has been shown for a few examples in three out of the four subfamilies so far.
NOD26 (a NIP from soybean) can be phosphorylated by a calcium-dependent protein kinase (Weaver et al., 1991). This protein has later been proposed to from an ion channel whose activity is regulated by phosphorylation (Weaver et al., 1994; Lee et al., 1995). Whether the AQP and glycerol permease activity of this protein (see above) is affected by its phophorylation state is not known. The phosphorylated Ser residue, Ser262, of NOD26 is conserved in most but not all NIPs from Arabidopsis (Johansson et al., 2000).
Also, αTIP from P. vulgaris has been shown to be phosphorylated by a membrane-bound calcium-dependent protein kinase (Johnson and Chrispeels, 1992). Ser7 in the N-terminal domain has later been identified as the in vivo target of the protein kinase, although mutation of two other Ser residues (Ser23 and Ser99) also affects the AQP activity of αTIP in X. laevis oocytes (Maurel et al., 1995). The closest relatives of the P. vulgaris AQP in Arabidopsis, AtTIP3;1 and AtTIP3;2, contain Ser residues close to the N terminus, but in a reversed context (SARR in AtTIP3;1 and AtTIP3;2 versus RRYS in P. vulgaris αTIP; Johnson and Chrispeels, 1992). AtTIP3;1 has been reported not to be phosphorylated (Maurel et al., 1997), suggesting that protein phosphorylation at this site is not involved in the regulation of the Arabidopsis homologues.
In spinach, the PIP2 member PM28A is phosphorylated by a calcium-dependent and membrane-associated protein kinase that leads to activation of the AQP activity as determined in X. laevis oocytes (Johansson et al., 1998). In vivo, a low apoplasmic water potential causes a dephosphorylation of Ser274 of PM28A. A mutant form of PM28A, where Ser274 was mutated to Ala, displayed a reduced membrane water permeability compared with wild type, when expressed in the X. laevis oocyte system in the presence of phosphatase inhibitors (Johansson et al., 1998) Thus, a decrease in plasma membrane water permeability, due to the dephosphorylation of plasma membrane AQPs, could slow down water loss from the cells and therefore allow the plant to cope with a lowered apoplastic water potential. The phosphorylation site (Ser274) of PM28A is strongly conserved in all Arabidopsis PIP2 AQPs. No comparable phosphorylation studies have been performed with the Arabidopsis proteins.
Working Group at the CPGN
To further extend the nomenclature for MIP genes to other plant species, a working group has been established with the CPGN, a section of the International Society for Plant Molecular Biology, and a Web site has been set up (http://mbclserver.rutgers.edu/CPGN/AquaporinWeb/Aquaporin.group.html). In addition to this article, the aim of the working group is to provide up-to-date information on the MIP gene family not only from Arabidopsis but also from other plant species. Others are invited to join the working group or send comments related to the MIP nomenclature for further public discussion. Moreover, the Web site will provide multiple sequence alignments of MIP sequences as well as dendrograms and lists of members of the gene family. Therefore, the complete AtMIP family could serve as a starting point for anyone who wants to identify the relation of new MIP sequences within the gene family. This information is also available at: http://www.plantae.lu.se/AtMIPs/index. The “Aquaporin Web site” at CPGN will also provide a download of the complete set of Arabidopsis DNA and protein sequences in formats (Fasta, GCG, and GenBank) that can be used with sequence analysis software packages. This service should help individual researchers to quickly compare new MIP sequences with the complete set of the Arabidopsis family.
MATERIALS AND METHODS
MIP genes and ESTs were identified and analyzed by BLAST or TBLASTN searches at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov:80/blast/blast.cgi) or at The Arabidopsis Information Resource (TAIR; www.Arabidopsis.org/blast/). The two most divergent MIPs from each subfamily were used in additional searches for more distant MIP homologs with TFASTA at The Arabidopsis Information Resource (http://www.Arabidopsis.org/cgi-bin/fasta/TAIRfasta.pl). All protein sequences analyzed are based on translations of genomic sequences from Arabidopsis ecotype Columbia. Use of alternative exon/intron borders compared with the annotation in GenBank are specified in Table I. These alternative borders were supported either by cDNA clones, sequence homology, or conserved exon size and a canonical acceptor site.
MacVector 7.0 (Oxford Molecular Ltd, Oxford) was used to translate sequences and ClustalW version 1.4 (Thompson et al., 1994) included in MacVector 7.0 was used to generate multiple alignments of translated sequences using the blosum30 matrix and slow mode. Open gap penalty and extend gap penalty were set to 10.0 and 0.05, respectively. The resulting ClustalW alignments were identical, regarding positions of gaps and alignments of residues, independent of the submission order of sequences. Alignments were manually inspected and adjusted to fit to conserved residues (Heymann and Engel, 2000). Gaps were preferentially located to loop regions connecting transmembrane helices as defined in an alignment with GlpF (accession no. 11514194). The alignment that forms the basis for all the phylogenetic analyses here is available at http://mbclserver.rutgers.edu/CPGN/AquaporinWeb/Aquaporin.group.html.
PAUP*4.0b4a (Swofford, 2000) was used in phylogenetic analyses of the 399-character-long alignment of 35 different AtMIPs. Cytoplasmic N- and C-terminal regions were defined as character 1 to 86 and 329 to 399, respectively, based on the alignment with GlpF. Both the maximum parsimony and a distance (minimum evolution) method were employed in heuristic searches for the shortest unrooted trees. The starting trees in the distance method were obtained by neighbor joining. Gaps were treated as missing characters and the tree-bisection-reconnection option was used as branch-swapping algorithm. Pair-wise distances expressed as mean character differences for the complete alignment, adjusted for missing data, were compared to obtain a similar maximum divergence in the different groups of proteins. One hundred replicates with full heuristic searches were performed in bootstrap tests of phylogenetic trees.
ACKNOWLEDGMENTS
We are grateful to many of the researchers working with plant MIPs for valuable discussions and comments on the new MIP nomenclature in plants. Francois Chaumont (Université Catholique de Louvain, Louvain-La-Neuve, Belgium) is also acknowledged for sharing unpublished data. Carl Price (CPGN, Waksman Institute, Piscataway, NJ) has provided helpful comments on gene nomenclature in general and also assisted in the setup of the “Aquaporin Web site” at CPGN.
Footnotes
This work was supported by the Swedish Council for Forestry and Agricultural Research, by the Swedish Natural Science Research Council, by the European Union-Biotech Program (grant no. BIO4–CT98–0024), by the Swedish Strategic Network for Plant Biotechnology, and by the Deutsche Forschungsgemeinschaft.
LITERATURE CITED
- Arabidopsis Genome Initiative. Analysis of the genome of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- Barkla BJ, Vera-Estrelle R, Pantoja O, Kirch H-H, Bohnert HJ. Aquaporins localization: how valid are the TIP and PIP labels? Trends Plant Sci. 1999;4:86–88. doi: 10.1016/s1360-1385(99)01388-6. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ, Hanson L. DNA amounts in two samples of angiosperm weeds. Ann Bot. 1998;82:121–134. [Google Scholar]
- Bennetzen JL, Kellogg EA. Do plants have a one-way ticket to genomic obesity? Plant Cell. 1997;9:1509–1514. doi: 10.1105/tpc.9.9.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biela A, Grote K, Otto B, Hoth S, Hedrich R, Kaldenhoff R. The Nicotiana tabacumplasma membrane aquaporin NtAQP1 is mercury-insensitive and permeable for glycerol. Plant J. 1999;18:565–570. doi: 10.1046/j.1365-313x.1999.00474.x. [DOI] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Jung R, Chrispeels MJ. Plasma membrane intrinsic proteins from maize cluster in two sequence subgroups with differential aquaporin activity. Plant Physiol. 2000;122:1025–1034. doi: 10.1104/pp.122.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Wojcik E, Chrispeels MJ, Jung R. Aquaporins constitutes a large and highly divergent protein family in maize. Plant Physiol. 2001;125:1206–1215. doi: 10.1104/pp.125.3.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Chaumont F, Mirkov TE, Chrispeels MJ. Characterization of a new vacuolar membrane aquaporin sensitive to mercury at a unique site. Plant Cell. 1996;8:587–599. doi: 10.1105/tpc.8.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean RM, Rivers RL, Zeidel ML, Roberts DM. Purification and functional reconstitution of soybean nodulin 26: an aquaporin with water and glycerol transport properties. Biochemistry. 1999;38:347–353. doi: 10.1021/bi982110c. [DOI] [PubMed] [Google Scholar]
- Fortin MG, Morrison NA, Verma DPS. Nodulin-26, a peribacteroid membrane nodulin, is expressed independently of the development of the peribacteroid compartment. Nucleic Acids Res. 1987;15:813–824. doi: 10.1093/nar/15.2.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Libson A, Miercke LJ, Weitzman C, Nollert P, Krucinski J, Stroud RM. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- Gerbeau P, Guclu J, Ripoche P, Maurel C. Aquaporin Nt-TIPa can account for the high permeability of tobacco cell vacuolar membrane to small neutral solutes. Plant J. 1999;18:577–587. doi: 10.1046/j.1365-313x.1999.00481.x. [DOI] [PubMed] [Google Scholar]
- Go M. Correlation of DNA exonic regions with protein structural units in hemoglobin. Nature. 1981;291:90–92. doi: 10.1038/291090a0. [DOI] [PubMed] [Google Scholar]
- Guenther JF, Roberts DM. Water-selective and multifunctional aquaporins from Lotus japonicusnodules. Planta. 2000;210:741–748. doi: 10.1007/s004250050675. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Engel A. Aquaporins: phylogeny, structure, and physiology of water channels. News Physiol Sci. 1999;14:187–193. doi: 10.1152/physiologyonline.1999.14.5.187. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Engel A. Structural clues in the sequences of the aquaporins. J Mol Biol. 2000;295:1039–1053. doi: 10.1006/jmbi.1999.3413. [DOI] [PubMed] [Google Scholar]
- Höfte H, Hubbard L, Reizer J, Ludevid D, Herman EM, Chrispeels MJ. Vegetative and seed-specific forms of tonoplast intrinsic protein in the vacuolar membrane of Arabidopsis thaliana. Plant Physiol. 1992;99:561–570. doi: 10.1104/pp.99.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson I, Karlsson M, Johanson U, Larsson C, Kjellbom P. The role of aquaporins in cellular and whole plant water balance. Biochim Biophys Acta. 2000;1465:324–342. doi: 10.1016/s0005-2736(00)00147-4. [DOI] [PubMed] [Google Scholar]
- Johansson I, Karlsson M, Shukla VK, Chrispeels MJ, Larsson C, Kjellbom P. Water transport activity of the plasma membrane aquaporin PM28A is regulated by phosphorylation. Plant Cell. 1998;10:451–459. doi: 10.1105/tpc.10.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Chrispeels MJ. Tonoplast-bound protein kinase phosphorylates tonoplast intrinsic protein. Plant Physiol. 1992;100:1787–1795. doi: 10.1104/pp.100.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Herman EM, Chrispeels MJ. An abundant, highly conserved tonoplast protein in seeds. Plant Physiol. 1989;91:1006–1013. doi: 10.1104/pp.91.3.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldenhoff R, Kolling A, Richter G. A novel blue light- and abscisic acid-inducible gene of Arabidopsis thalianaencoding an intrinsic membrane protein. Plant Mol Biol. 1993;23:1187–1198. doi: 10.1007/BF00042352. [DOI] [PubMed] [Google Scholar]
- Kammerloher W, Fischer U, Piechottka GP, Schäffner AR. Water channels in the plant plasma membrane cloned by immunoselection from a mammalian expression system. Plant J. 1994;6:187–199. doi: 10.1046/j.1365-313x.1994.6020187.x. [DOI] [PubMed] [Google Scholar]
- Karlsson M, Johansson I, Bush M, McCann MC, Maurel C, Larsson C, Kjellbom P. An abundant TIP expressed in mature highly vacuolated cells. Plant J. 2000;21:83–90. doi: 10.1046/j.1365-313x.2000.00648.x. [DOI] [PubMed] [Google Scholar]
- Kjellbom P, Larsson C, Johansson I, Karlsson M, Johanson U. Aquaporins and water homeostasis in plants. Trends Plant Sci. 1999;4:308–314. doi: 10.1016/s1360-1385(99)01438-7. [DOI] [PubMed] [Google Scholar]
- Lee JW, Zhang YX, Weaver CD, Shomer NH, Louis CF, Roberts DM. Phosphorylation of nodulin 26 on serine 262 affects its voltage-sensitive channel activity in planar lipid bilayers. J Biol Chem. 1995;270:27051–27057. doi: 10.1074/jbc.270.45.27051. [DOI] [PubMed] [Google Scholar]
- Ludevid D, Höfte H, Himelblau E, Chrispeels MJ. The expression pattern of the tonoplast intrinsic protein γ -TIP in Arabidopsis thalianais correlated with cell enlargement. Plant Physiol. 1992;100:1633–1639. doi: 10.1104/pp.100.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Chrispeels M, Lurin C, Tacnet F, Geelen D, Ripoche P, Guern J. Function and regulation of seed aquaporins. J Exp Bot. 1997;48:421–430. doi: 10.1093/jxb/48.Special_Issue.421. [DOI] [PubMed] [Google Scholar]
- Maurel C, Chrispeels MJ. Aquaporins: a molecular entry into plant water relations. Plant Physiol. 2001;1251:135–138. doi: 10.1104/pp.125.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Kado RT, Guern J, Chrispeels MJ. Phosphorylation regulates the water channel activity of the seed-specific aquaporin α TIP. EMBO J. 1995;14:3028–3035. doi: 10.1002/j.1460-2075.1995.tb07305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Reizer J, Schroeder JI, Chrispeels MJ. The vacuolar membrane protein γ -TIP creates water specific channels in Xenopusoocytes. EMBO J. 1993;12:2241–2247. doi: 10.1002/j.1460-2075.1993.tb05877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao G-H, Verma DPS. Soybean nodulin-26 gene encoding a channel protein is expressed only in the infected cells of nodules and is regulated differently in roots of homologous and hetrologous plants. Plant Cell. 1993;5:781–794. doi: 10.1105/tpc.5.7.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- Phillips AL, Huttly AK. Cloning of two gibberellin-regulated cDNAs from Arabidopsis thaliana by subtractive hybridization: expression of the tonoplast water channel, γ TIP, is increased by GA3. Plant Mol Biol. 1994;24:603–615. doi: 10.1007/BF00023557. [DOI] [PubMed] [Google Scholar]
- Pih KT, Kabilan V, Lim JH, Kang SG, Piao HL, Jin JB, Hwang I. Characterization of two new channel protein genes in Arabidopsis. Mol Cells. 1999;9:84–90. [PubMed] [Google Scholar]
- Rivers RL, Dean RM, Chandy G, Hall JE, Roberts DM, Zeidel ML. Functional analysis of nodulin 26, an aquaporin in soybean root nodule symbiosomes. J Biol Chem. 1997;272:16256–16261. doi: 10.1074/jbc.272.26.16256. [DOI] [PubMed] [Google Scholar]
- Robinson DC, Sieber H, Kammerloher W, Schäffner AR. PIP1 aquaporins are concentrated in plasmalemmasomes of Arabidopsis thalianamesophyll. Plant Physiol. 1996;111:645–649. doi: 10.1104/pp.111.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni V, Gerbeau P, Javot H, Maurel C. The high diversity of aquaporins reveals novel facets of plant membrane functions. Curr Opin Plant Biol. 2000;3:476–481. doi: 10.1016/s1369-5266(00)00116-3. [DOI] [PubMed] [Google Scholar]
- Shagan T, Bar-Zvi D. Nucleotide sequence of an Arabidopsis thalianaturgor-responsive cDNA clone encoding TMP-A, a transmembrane protein containing the major intrinsic protein motif. Plant Physiol. 1993;101:1397–1398. doi: 10.1104/pp.101.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shagan T, Meraro D, Bar-Zvi D. Nucleotide sequence of an Arabidopsis thalianaturgor-responsive TMP-B cDNA clone encoding transmembrane protein with a major intrinsic protein motif. Plant Physiol. 1993;102:689–690. doi: 10.1104/pp.102.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- Thompson JD, Higgins DG, Gibson Clustal W: improving the sensitivity of the progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger VM. Fraternal twins: AQP1 and GlpF. Nature Struct Biol. 2000;7:1082–1084. doi: 10.1038/81914. [DOI] [PubMed] [Google Scholar]
- van de Loo FJ, Turner S, Somerville C. Expressed sequence tags from developing castor seeds. Plant Physiol. 1995;108:1141–1150. doi: 10.1104/pp.108.3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CD, Crombie B, Stacey G, Roberts DM. Calcium-dependent phosphorylation of symbiosome membrane proteins from nitrogen-fixing soybean nodules: evidence for phosphorylation of nodulin-26. Plant Physiol. 1991;95:222–227. doi: 10.1104/pp.95.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CD, Shomer NH, Louis CF, Roberts DM. Nodulin 26, a nodule-specific symbiosome membrane protein from soybean, is an ion channel. J Biol Chem. 1994;269:17858–17862. [PubMed] [Google Scholar]
- Weig A, Deswarte C, Chrispeels MJ. The major intrinsic protein family of Arabidopsishas 23 members that form three distinct groups with functional aquaporins in each group. Plant Physiol. 1997;114:1347–1357. doi: 10.1104/pp.114.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weig AR, Jakob C. Functional identification of the glycerol permease activity of Arabidopsis thaliana NLM1 and NLM2 proteins by heterologous expression in Saccharomyces cerevisiae. FEBS Lett. 2000a;481:293–298. doi: 10.1016/s0014-5793(00)02027-5. [DOI] [PubMed] [Google Scholar]
- Weig AR, Jakob C. Functional characterization of Arabidopsis thalianaaquaglyceroporins. In: Hohmann S, Nielsen S, editors. Molecular Biology and Physiology of Water and Solute Transport. Dordrecht, The Netherlands: Kluwer Academic/Plenum Publishers; 2000b. pp. 365–372. [Google Scholar]