Abstract
Geranylgeranyltransferase-I (GGT-I) is a heterodimeric enzyme that shares a common α-subunit with farnesyltransferase (FTase) and has a distinct β-subunit. GGT-I preferentially modifies proteins, which terminate in a CaaL box sequence motif. Cloning of Arabidopsis GGT-I β-subunit (AtGGT-IB) was achieved by a yeast (Saccharomyces cerevisiae) two-hybrid screen, using the tomato (Lycopersicon esculentum) FTase α-subunit (FTA) as bait. Sequence and structure analysis revealed that the core active site of GGT-I and FTase are very similar. AtGGT-IA/FTA and AtGGT-IB were co-expressed in baculovirus-infected insect cells to obtain recombinant protein that was used for biochemical and molecular analysis. The recombinant AtGGT-I prenylated efficiently CaaL box fusion proteins in which the a2 position was occupied by an aliphatic residue, whereas charged or polar residues at the same position greatly reduced the efficiency of prenylation. A polybasic domain proximal to the CaaL box motif induced a 5-fold increase in the maximal reaction rate, and increased the affinity of the enzyme to the protein substrate by an order of magnitude. GGT-I retained high activity in a temperature range between 24°C and 42°C, and showed increased activity rate at relatively basic pH values of 7.9 and 8.5. Reverse transcriptase-polymerase chain reaction, protein immuno-blots, and transient expression assays of green fluorescent protein fusion proteins show that GGT-IB is ubiquitously expressed in a number of tissues, and that expression levels and protein activity were not changed in mutant plants lacking FTase β-subunit.
Protein prenylation involves the covalent attachment of the C-15 isoprene farnesyl or the C-20 isoprene geranylgeranyl groups to the C-terminal end of some proteins. Prenylation facilitates protein adherence to membranes and, for some proteins that have been studied in detail, is also required for their function (Schafer and Rine, 1992; Zhang and Casey, 1996; Rodríguez-Concepción et al., 1999a; Yalovsky et al., 1999; Sinensky, 2000).
The prenylation reaction is catalyzed by three protein prenyl transferases, a farnesyltransferase (FTase), geranylgeranyltransferase-I (GGT-I) and -II (GGT-II) (Rab geranylgeranyltransferase; Zhang and Casey, 1996; Rodríguez-Concepción et al., 1999a; Yalovsky et al., 1999). FTase and GGT-I are heterodimeric enzymes that share a common α-subunit but have distinct β-subunits that determine substrate specificity (Zhang and Casey, 1996). Both enzymes recognize a conserved C-terminal amino acid sequence motif known as CaaX box in which “C” is Cys, “a” represents an usually aliphatic amino acid, and “X” can be any amino acid in the case of FTase but is almost exclusively a Leu when prenylated by GGT-I (Schafer and Rine, 1992; Zhang and Casey, 1996; Rodríguez-Concepción et al., 1999a; Yalovsky et al., 1999). The presence of a polybasic domain rich in Arg and Lys proximal to the CaaX box greatly increases substrate affinity of GGT-I (James et al., 1995).
The activities of FTase and GGT-I are in part promiscuous and GGT-I can prenylate FTase substrates, albeit inefficiently (Ohya et al., 1991; Trueblood et al., 1993; Armstrong et al., 1995). GGT-I can use either geranylgeronyl diphosphate (GGPP) or farnesyl diphosphate (FPP) as prenyl group donors but the efficiency of substrate transfer depends on the substrate protein (Armstrong et al., 1995; Yokoyama et al., 1995, 1997). Three single mutations in RAM1 (yeast [Saccharomyces cerevisiae] FTase β-subunit [FTB] gene) convert the protein substrate specificity of FTase to that of GGT-I (Del Villar et al., 1997). Detailed sequence analysis will be required, however, to verify the conservation of these three amino acid residues between FTase and GGT-I.
Both FTase and GGT-I require Zn2+ ions for their catalytic activity (Reiss et al., 1992; Yokoyama et al., 1995; Zhang and Casey, 1996). In rat FTase, three amino acid residues in the β-subunit (Asp-297, Cys-299, and His-363) serve as ligands to the catalytic zinc (Park et al., 1997; Strickland et al., 1998). These residues are conserved in yeast and plant FTBs (Yalovsky et al., 1997). It is not yet known whether the same residues serve as zinc ligands in GGT-I β-subunits.
Mutations in GGT-I β-subunit gene are lethal in the budding yeast S. cerevisiae (Ohya et al., 1991; Trueblood et al., 1993) and Drosophila melanogaster (Therrien et al., 1995), and in the fission yeast Schizosacchromyces pombe when cells are grown at a restrictive temperature of 37°C (Díaz et al., 1993). In S. cerevisiae, GGT-I β is encoded by CDC43, which was also called CAL1 (Ohya et al., 1991). cdc43− (cal1−) mutant cells can be rescued by growing the cells in the presence of Ca2+ (Ohya et al., 1991) or by over expression of CDC42, a GGT-I substrate (Trueblood et al., 1993). S. pombe cwg2+ (GGT-I β) mutants are defected in β-d-glucan synthesis and can be rescued when grown at 37°C in high osmotic pressure growth media (Díaz et al., 1993). In contrast to the lethality caused by mutations in the GGT-I β-subunit gene, yeast and plant FTB gene deletion mutants are viable, although they show several growth, mating, and developmental defects (Goodman et al., 1988; Schafer et al., 1990; Cutler et al., 1996; Bonetta et al., 2000; Yalovsky et al., 2000a; Ziegelhoffer et al., 2000). These results indicate that GGT-I can partially suppress FTase β-mutations, but that in most cases where mutants have been identified, excluding S. pombe (Díaz et al., 1993), FTase cannot compensate for GGT-I loss of function.
Although GGT-I-like activity had been demonstrated in plants several years ago (Randall et al., 1993), only very little is known about plant GGT-I. A plant gene encoding GGT-IB has not been cloned and GGT-I activity has not been characterized in detail. Considering the number of geranylgeranylated plant proteins identified to date (Dykema et al., 1999; Rodríguez-Concepción et al., 1999a; Yalovsky et al., 1999), the role of GGT-I in various cellular processes is undoubtedly complex. Similar to their mammalian and yeast counterparts, most known members of the Rac-related Rop family of small GTPases in plants have conserved C-terminal CaaL box motifs and a polybasic sequence domain proximal to the prenyl acceptor-Cys. Direct geranylgeranylation has been demonstrated for two members of this family (Lin et al., 1996; Trainin et al., 1996), but it is likely that most Rops are substrates for GGTase-I. Rac GTPases have been implicated in the reorganization of the actin cytoskeleton through activation of phosphatidylinositol 4-phosphate 5-kinase. It is likely that Rop proteins have a related function in the regulation of polar growth in plant cells because, similar to the fission yeast homolog, overexpression of an Arabidopsis Rop protein induces isotropic growth in fission yeast, and the protein is found at the site of growth (Li et al., 1998). In plants, certain Rop proteins localize to the tip of the growing pollen tube, consistent with a role of the protein in polarized cell growth (Lin et al., 1996).
The effect of geranylgeranylation on membrane localization of GGT-I target proteins in plants is now best understood for CaM53, a novel type of calmodulin protein that is not found in yeast or mammalian cells. CaM53 has a typical calmodulin domain but contains a C-terminal extension of 34 amino acids rich in Lys and Arg and a typical GGTase-I CaaL motif (Rodríguez-Concepción et al., 1999b). Prenylated CaM53 localizes to the plasma membrane but the Cys acceptor mutant protein accumulates in the nucleus (Rodríguez-Concepción et al., 1999b; Yalovsky et al., 1999).
To provide further insight into plant GGT-I function, we cloned the gene encoding Arabidopsis GGT-I β-subunit (AtGGT-IB) and co-expressed it together with AtFTA/GGT-IA. The purified recombinant protein was used to study GGT-I substrate specificity and other kinetic parameters that determine the enzymatic activity. The expression pattern and activity of GGT-I both in wild-type (wt) and era1-2 mutant Arabidopsis plants in which the entire gene encoding Arabidopsis FTB (AtFTB) is deleted (Cutler et al., 1996) were studied using specific antibodies (Abs) that were raised against AtGGT-IB together with a green fluorescent protein (GFP)-CaM53 fusion protein.
RESULTS
Cloning of AtGGT-IB
The gene encoding AtGGT-IB was cloned in a yeast two-hybrid screen using the tomato (Lycopersicon esculentum) FTase α-subunit (LeFTA) as bait. A total of 111 His+ β-galactosidase+-positive clones were isolated. Fifty clones developed strong blue color indicating strong interactions between the LeFTA bait and the prey proteins. The sequences of 32 of these strong positive clones were determined. Three out of the 32 sequenced clones encoded AtGGT-IB. The sequences of all three AtGGT-IB clones were identical both in their open reading frame and 3′-untranslated regions. It is interesting that seven other clones that were sequenced encoded AtFTB and all contained identical sequences. The other 22 clones, which were sequenced, encoded different proteins.
The AtGGT-IB cDNA encodes a protein of 377 amino acids with a predicted molecular mass of 41.9661 kD. Sequence alignment using the John Hein method (Lasergene software package) revealed homology of 40.6% and 30% between the Arabidopsis, human, and yeast GGT-I β-subunit proteins, respectively (Fig. 1). In a similar manner, the homology between the tomato LeFTB protein and its mammalian and yeast homologs is 45% and 30%, respectively (Yalovsky et al., 1997). AtGGT-IB and AtFTB share 30% homology, similar to the homology between the Arabidopsis and the yeast GGT-I β-subunit proteins. This may suggest FTase and GGTase-I β-subunits diverged to form two separate enzymes very close to the time that plant and yeast diverged from a common ancestor.
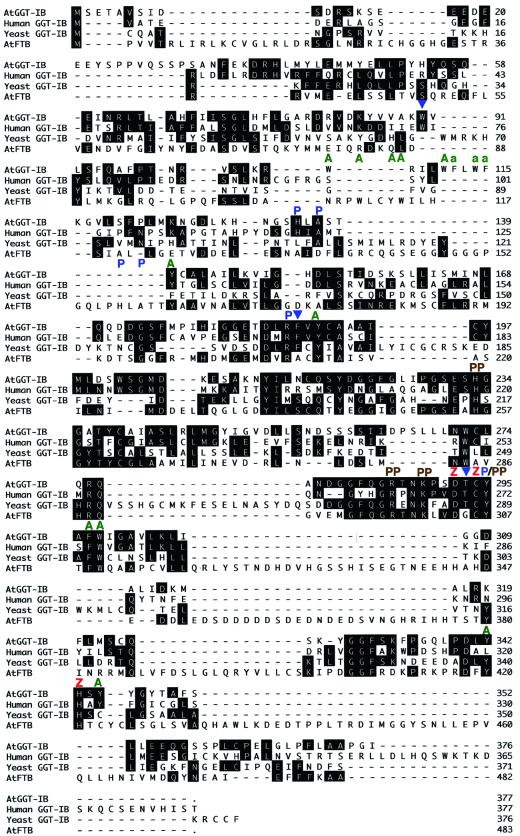
Figure 1.
Amino acid sequence alignment of protein geranylgeranyltransferase β-subunits and the Arabidopsis FTB. Sequence alignments were established by using the John Hein method (Lasergene). Amino acid identities between the Arabidopsis AtGGT-IB and AtFTB proteins, and GGT-I β-subunits from yeast and human, are indicated by black boxes. Dashes denote gaps formed by the alignment algorithm. In some regions the alignment program failed to align the AtFTB sequence correctly because of the divergence from the yeast sequence. Molecular functions were assigned to some of the residues using the crystal structure of the rat FTase as a model. Green A denotes residues in the hydrophobic pocket, blue P denotes residues that interact with the CaaX peptide, brown PP denotes residues that interact with the diphosphate group of FPP (or GGPP), red Z denotes the ligands of the catalytic zinc atom, and blue arrows denote residues that are conserved between all GGT-I but differ in FTBs. AtGGT-IB was isolated as a 1,351-bp clone with 1,128-bp open reading frame. Low stringency DNA-blot analysis indicates that AtGGT-IB exists in a single copy form in the Arabidopsis genome (data not shown). RNA-blot analysis (data not shown) and comparison to the genomic sequence of AtGGT-IB (GenBank accession no. ATAC004218) indicate that AtGGT-IB represents a full-length clone. The GenBank accession nos. for the aligned sequences are as follows: AtGGT-IB, AF311225; human GGT-IB, L25441; yeast (S. cerevisiae) GGT-IB, M74109; and Arabidopsis AtFTB, AF214106.
Using the structural models of Rat FTase with its isoprenoid and peptide substrates (Park et al., 1997; Long et al., 1998; Strickland et al., 1998) together with sequence alignments presented in Figure 1, it was possible to attribute specific functions to several amino acid residues in AtGGT-I. This analysis revealed that the core active site is conserved between GGT-I and FTase. The conservation includes the following amino acids: residues Asp-304, Cys-306, and His-421 in AtFTB that serve as ligands to the catalytic zinc atom correspond to residues Asp-292, Cys-294, and His-343 in AtGGT-IB (red Z in Fig. 1); residues His-255, Arg-298, Lys-301, and Tyr-307 in AtFTB, which make bonds with the diphosphate group of FPP, correspond to residues His-233, Arg-286, Lys-289, and Tyr-295 in AtGGT-IB (brown PP in Fig. 1); and residues His-157, Arg-210, and Tyr-307 in AtFTB, which make direct connection to the CaaX peptide, correspond to residues His-135, Arg-186, and Tyr-295 in AtGGT-IB (blue Ps in Fig. 1). Ala-159 in AtFTB and Ala-137 in AtGGT-IB are part of the peptide-binding pocket but do not bind the peptide. In addition, several conserved aromatic residues, which are likely constituents of the hydrophobic pocket that make the binding site for both FPP (or GGPP) and the CaaX peptide substrates, are dispersed throughout AtGGT-IB and AtFTB proteins (green As in Fig. 1). Both AtGGT-IB and AtFTB have a conserved aromatic domain that is absent from the yeast and human GGT-IB proteins. This domain stretches between residues 107 to 115 in AtGGT-IB, and 110 to 114 in AtFTB. In AtFTB, the domain contains three residues: Trp-110, Tyr-113, and Trp-114. These residues are conserved in FTBs of yeast, plants, and mammals (Yalovsky et al., 1997), and the cocrystal structure of rat FTase with FPP and CaaX peptide substrates shows that they are part of the hydrophobic binding site pocket for both substrates (Strickland et al., 1998). The homologous domain in AtGGT-IB contains five aromatic residues: Trp-107, Trp-111, Phe-112, Trp-114, and Phe-115, and these may enlarge the hydrophobic pocket to accommodate the 20-carbon GGPP as has been previously suggested (Long et al., 1998). The absence of this homologous domain in the yeast and human proteins may suggest a slightly different structure.
Several conserved residues of GGT-IB are occupied by different amino acids in FTBs (blue triangles in Fig. 1). Of particular interest is Thr-293 in AtGGT-IB. This Thr residue, which is located between the zinc atom ligands Asp-293 and Cys-294, is conserved in yeast plant and human GGT-IB proteins. In FTB proteins an invariant Gly residue occupies this position.
Expression Patterns of AtGGT-IB RNA and Protein
FTB RNA and proteins are ubiquitously expressed in both tomato and Arabidopsis (Schmitt et al., 1996; Ziegelhoffer et al., 2000) suggesting that FTase functions as a housekeeping protein. Reporter gene experiments in tobacco, using the pea PsFTB promoter fused to β-glucoronidase, revealed higher expression levels in meristematic zones and around vascular tissues (Zhou et al., 1997). These results, however, are not consistent with the RNA in situ data, which did not reveal higher expression levels of AtFTB in meristems (Ziegelhoffer et al., 2000). Because FTase and GGT-I are believed to be semiredundant, it was important to analyze the level and pattern of GGT-IB expression. To examine the possibility that expression levels of AtFTB and AtGGT-IB are coordinated, experiments were carried out in both Col-0 wt and era1-2 mutant Arabidopsis plants, which lack the entire gene encoding AtFTB (Cutler et al., 1996).
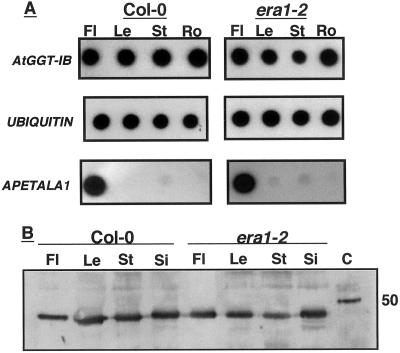
RNA levels in different tissues were determined by reverse transcriptase (RT)-PCR. Using this approach, no significant differences were detected in the level of AtGGT-IB RNA in flowers, leaves, stems, and root in both wt and era1-2 plants (Fig. 2A). Furthermore, comparison of AtGGT-IB RNA levels in wt and era1-2 plants failed to detect significant differences in GGT-IB RNA levels (Fig. 2A). As expected (Gustafson-Brown et al., 1994), the RNA of the floral transcription factor APETALA1 (AP1) was highly expressed in flower tissues (Fig. 2A), indicating that the conditions used in the RT-PCR experiments allowed detection of differences in level of gene expression.
Figure 2.
Expression patterns of AtGGT-IB RNA and protein. A, Dot-blot analysis of GGT-IB, Ubiquitin, and APETALA1 RT-PCR products prepared from wt Col-0 and era1-2 Arabidopsis plants. Fl, Flower; Le, leaf; St, stem; Ro, root. The RT-PCR was carried out as described in “Materials and Methods” B, Immunoblot analysis of AtGGT-IB expression in wt Col-0 and era1-2 Arabidopsis plants using α-AtGGT-IB polyclonal Abs (“Materials and Methods”). Fl, Flowers; Le, leaf; St, stem; Si, silique; C, recombinant GGT-IB control. The control protein appears larger because it contained additional residues that originated from the pFastBacHTa cloning vector (“Materials and Methods”).
The RT-PCR experiments are further supported by protein-blot analysis using α-AtGGT-IB polyclonal Abs (Fig. 2B). The results shown in Figure 2B clearly demonstrate that AtGG-IB is expressed approximately at the same level in all tissues tested, either in protein extracts prepared from wt or era1-2 plants.
Identification of GGT-I Substrates in Plants
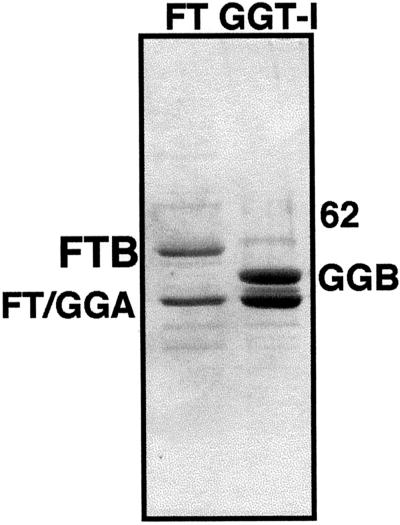
The expression pattern of GGT-IB (Fig. 2) cannot predict which signaling cascades this enzyme may affect in vivo. One approach to analyze GGT-I function is to identify its putative protein substrates. A number of proteins that terminate in a CaaL box motif were identified using computer-aided searches of the protein database. As a first step, the prenylation of CaaL boxes corresponding to some of these proteins was tested in vitro using recombinant purified GGT-I enzyme (Fig. 3). Efficient expression of (His)6-tagged versions of either FTase or GGT-I enzymes was achieved by co-infection of insect cells with baculoviruses harboring the common α-subunit and either of the β-subunits (FTB or GGT-IB). Purification of FTase and GGT-I was achieved by metal chellate chromatography (Fig. 3).
Figure 3.
Recombinant FTase and GGT-I proteins. Stained gel showing recombinant (His)6-tagged FTase (FT) and GGT-I that were expressed in baculovirus-infected insect cells, and purified over an Ni-NTA column (“Materials and Methods”). The Mr difference between FTB and GGT-I β-subunit (GGB) is visible. FT/GGA, The common α-subunit.
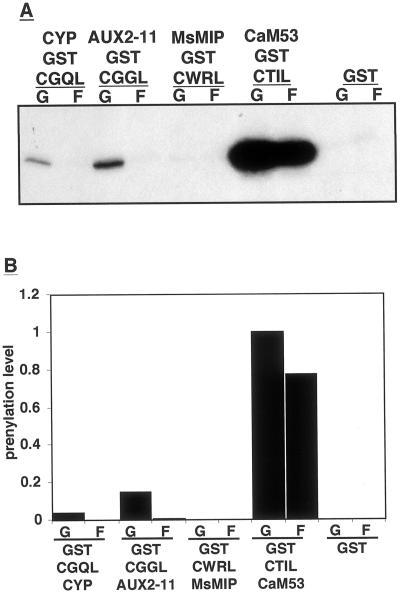
Figure 4 shows results of a prenylation experiment in which GST CaaL box fusion proteins were incubated with GGT-I and either [3H]GGPP or [3H]FPP. The results shown in Figure 4 demonstrate that the GST-CTIL CaaL box fusion protein, which corresponds to the petunia (Petunia hybrida) calmodulin CaM53, was prenylated much more efficiently by GGT-I than any of the other proteins tested. Only very low prenylation levels were detected for the GST-CWRL CaaL box, which corresponds to an alfalfa membrane channel protein (MIP; Fig. 4). A positively charged Arg residue occupies the a2 position in the CWRL box. In yeast, either positively or negatively charged residues have been shown to inhibit prenylation by FTase (Trueblood et al., 1997). The CGQL box of CYP was also poorly geranylgeranylated and no farnesylation could be detected. It is possible that the presence of the polar Gln residue at the a2 position makes this CaaL box an unfavorable substrate for prenylation. The GST-CGGL box of AUX2–11 was prenylated more efficiently than GST-CGQL box, indicating that a Gly residue at the a2 position is favorable over a Gln. To summarize, GGT-I can use either GGPP or FPP as prenyl group donors but prenylation with GGPP was more efficient. FPP, however, was a poor prenyl group donor when the protein substrate had an unfavorable CaaL sequence motif. The presence of charged or polar groups at the a2 position greatly reduced the prenylation efficiency, and it is still questionable whether CYP or Aux2-11 are prenylated in vivo.
Figure 4.
Prenylation efficiency of different gluthatione-S-transferase (GST)-CaaL box fusion proteins. Prenylation reactions were carried out with GST alone or with GST-CaaL fusion proteins, as indicated on the figure, with either GGPP (G) or FPP (F) as prenyl group donors. Reactions were terminated by denaturing proteins and separating them on SDS gels, which were in turn fluorographed and exposed to x-ray film (A). Bands in A were quantified and the band with the maximum intensity was given the value of 1 (B). Cyclophilin (CYP), CYP from Solanum commersonii (wild potato), GenBank accession no. U92087; AUX22-1 auxin-induced protein from Arabidopsis, GenBank accession no. L15450; MsMIP, membrane channel protein from alfalfa (Medicago sativa), EMBL accession no. L36881; CaM53 Ca2+, calmodulin from petunia, GenBank accession no. M80831.
The Role of the Carboxy-Terminal Polybasic Domain in Prenylation by GGT-I
Fusion proteins between the GFP and either the full-length CaM53 or the 34 C-terminal amino acids of CaM53 (BDCaM53) were localized to the plasma membrane, whereas a GFP-CTIL fusion protein (CTIL is the CaaL box of CaM53) was localized to the plasma membrane and the cytoplasm (Rodríguez-Concepción et al., 1999b). These data showed that other domains of CaM53, such as the calmodulin EF hands, are not required for prenylation (Rodríguez-Concepción et al., 1999b). The BDCaM53 domain is rich in basic amino acid residues, and the results suggested that it was required for efficient prenylation (Rodríguez-Concepción et al., 1999b). In vitro prenylation assays indicated that either CaM53 or a GST-BDCaM53 fusion protein were prenylated more efficiently than a GST-CTIL fusion protein (Rodríguez-Concepción et al., 1999b). However, due to technical difficulties in the expression of proteins, it was difficult to quantitatively determine the kinetic parameters of the prenylation reaction. BDCaM53 contains several stretches of Arg residues with at least three repetitions of the Arg codons AGG AGA in tandem. Codon usage prevents efficient translation of this sequence combination in bacterial cells resulting in substantial reduction in levels of full-length protein and contamination of samples with partially translated fragments (Brinkmann et al., 1989).
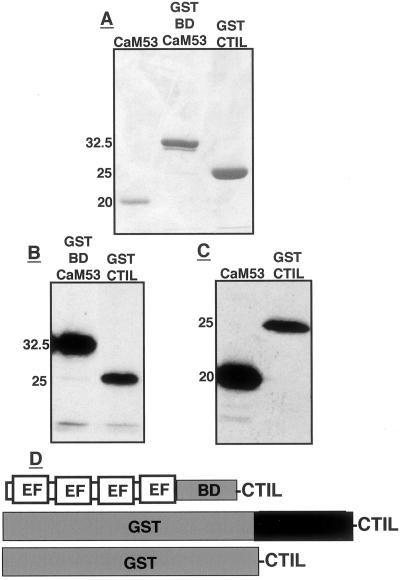
Expression in Escherichia coli in the presence of a plasmid that contained the appropriate tRNAs (Brinkmann et al., 1989; Schenk et al., 1995; see “Materials and Methods”) enabled the expression of full-length CaM53 and GST-BDCaM53 proteins (Fig. 5, A and D). Prenylation reactions with the purified proteins showed that both CaM53 and BDCaM53 were prenylated more efficiently than GST-CTIL (Fig. 5, B and C).
Figure 5.
CaM53, GST-BDCaM53, and GST-CTIL protein substrates and their prenylation. A, Stained SDS-PAGE of the purified protein substrates. B, Fluorogram showing prenylation of GST-BDCaM53 and GST-CTIL fusion recombinant protein. C, Fluorogram showing prenylation of CaM53 and GST-CTIL recombinant proteins. Numbers in A through C denote molecular mass in kilodaltons. D, Schematic presentation of CaM53, GST-BDCaM53, and GST-CTIL. BD, Basic domain; EF, Ca2+-binding EF hands.
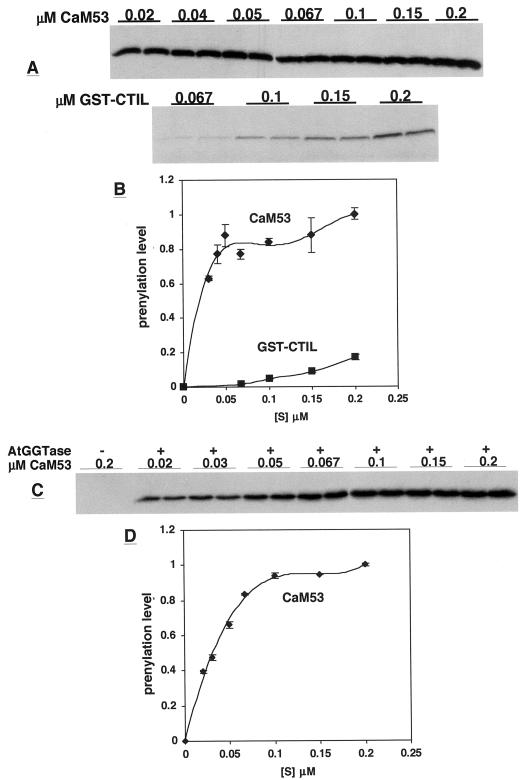
Substrate saturation kinetic analysis of prenylation of GST-CTIL, GST-BDCaM53, and CaM53 (Fig. 5D) were carried out (Figs. 6 and 7). Two different methods were used to separate between the prenylated proteins and the free unbound GGPP at the end of the reactions. One method was to denature proteins and separate them on SDS-gels, which were fluorographed and exposed to x-ray films for various lengths of time (Fig. 6, A and C). In the second method, the free GGPP was converted into GG-OH by an acidic-ethanol treatment (see “Materials and Methods”), followed by precipitation of proteins on glass filters and determination of the amount of radiolabled protein in a scintillation counter (Fig. 7). The results obtained by both methods were similar and confirmed the validity of each method.
Figure 6.
Substrate saturation curves for prenylation of CaM53 and GST-CTIL by GGT-I. Prenylation reactions were carried out with concentrations of CaM53 and GST-CTIL as indicated on the figure. All other conditions were as described in “Materials and Methods.” Reactions were terminated by separating the protein products on SDS gels, which were, in turn, fluorographed and exposed to x-ray films (A and C). The fluorograms in A were exposed for 60 h, and the fluorogram in C for 48 h. C, −, Denotes reactions carried out with boiled GGT-I enzyme; +, reactions carried out with active GGT-I. B and D, Quantification of the bands on fluorograms in A and C, respectively. Values for each band pair were averaged, and the maximal intensity was given a value of 1. The kinetics of the prenylation of CaM53 appeared more accurate following a shorter (48 h) exposure period of the film (compare A and B with C and D). However, the radiolabeled GST-CTIL was barely detectable when films were exposed for less then 60 h (data not shown).
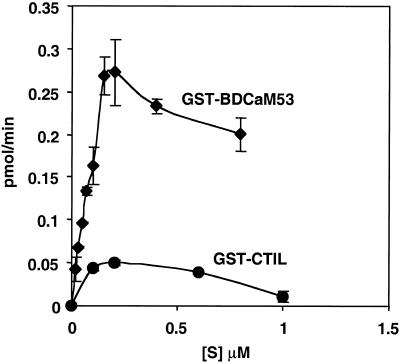
Figure 7.
Substrate saturation curves for prenylation of GST-BDCaM53 and GST-CTIL by GGT-I. Prenylation reactions were carried out with concentrations of GST-BDCaM53 and GST-CTIL as indicated in the figure. All other conditions were as described in “Materials and Methods.” Reactions were carried out in triplicates, and were terminated by separating between protein-incorporated and free GGPP using the acidic ethanol method (“Materials and Methods”). Each graph point represents the average value of each of the three reactions and bars are sd values. To express the incorporation of GGPP in values of pmol min−1, the radioactivity of known amount of [3H]GGPP was measured by scintillation counting.
Both GST-BDCaM53 and CaM53 were prenylated with Vmax values that were 5-fold greater than in the prenylation of GST-CTIL (Fig. 6, C and D and Fig. 7). The calculated Km values for the geranylgeranylation of CaM53 and GST-BDCaM53 ranged between 0.1 and 0.25 μm. In contrast, the calculated Km values for geranylgeranylation of GST-CTIL were an order of a magnitude higher and ranged between 2 and 3 μm. Thus, the polybasic domain of CaM53 increased the affinity of GGT-I toward its protein substrate. The results shown in Figures 4 through 7 demonstrate that the efficiency of prenylation by AtGGT-I depends on the composition of the amino acids in the CaaX box (Fig. 4), and a proximal polybasic domain (Figs. 5–7).
At concentrations higher than 0.5 μm, GST-CTIL inhibited its own prenylation (Fig. 7). The same holds true for GST-BDCaM53, although the inhibition was not as pronounced. Similar inhibition of the prenylation of H-Ras and K-RasB was formerly reported (James et al., 1995). These inhibitions in the prenylation reactions have not been explained.
The Regulation of Activity of AtGGT-I by Temperature and pH
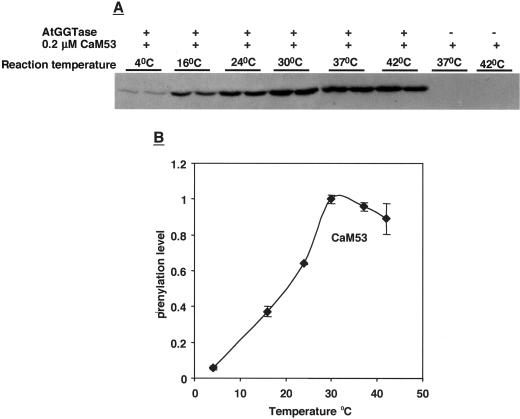
AtGGT-I was active in a wide range of temperatures (Fig. 8). The enzyme had maximal activity around 30°C to 37°C and it is stable up to 42°C. In contrast, prenylation activity quickly decreased at temperatures below 30°C. It was only 67% of the maximum at 24°C, 40% at 16°C, and 5% to 10% at 4°C. The experiment shown in Figure 8 was carried out using CaM53 as substrate protein, and similar results were obtained with GST-BDCaM53. The controls at 37°C and 42°C, which were carried out with boiled, inactive enzyme, indicate that the labeling of CaM53 by [3H]GGPP was due to AtGGT-I activity, and rule out the possibility of unspecific substrate binding.
Figure 8.
How does GGT-I activity depend on temperature? Prenylation reactions of CaM53 were carried out at different temperatures, as indicated on the figure. All other conditions were as described (“Materials and Methods”). Separation of the protein products on SDS gels terminated reactions, followed by fluorography of gels and exposure to x-ray films (A). Values for each band pair were averaged, and the maximal intensity was given a value of 1. −, Denotes reactions carried out with boiled GGT-I enzyme; +, reactions carried out with active GGT-I.
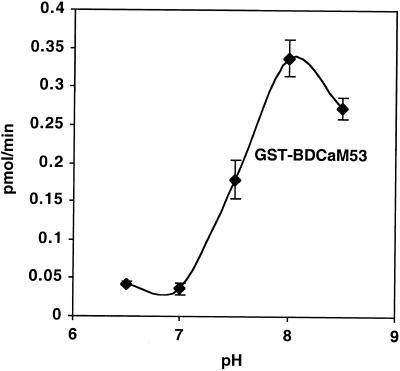
AtGGT-I had maximal activity at a relatively basic pH of 7.9 (Fig. 9). At a pH of 8.5 the activity was about 20% lower; however, at pH 7.5 the activity was only about 50% of the activity at pH 7.9 (Fig. 9). Similar results were obtained with other preparations of either GST-BDCaM53 or CaM53 substrate proteins (data not shown). Because the pH of the cytosol is usually around 7.5 (Sanders and Bethke, 2000), changes in cytoplasmic pH may modulate AtGGT-I activity.
Figure 9.
Regulation of GGT-I by pH. Prenylation reactions of GST-BDCaM53 were carried out at different pH values as indicated on the figure. All other conditions were as described (“Materials and Methods”). Reactions were carried out in triplicates, and were terminated by separating between protein-incorporated and free GGPP using the acidic ethanol method (“Materials and Methods”). Each graph point represents the average value of each of the three reactions and bars are sd values. The incorporation of GGPP into substrate protein was calculated as described in Figure 7.
Prenylation of CaM53 by GGT-I in Plants
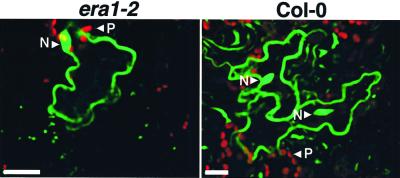
FTase can prenylate CaM53 in vitro but at a lower efficiency compared with GGT-I (Rodríguez-Concepción et al., 1999b). It is possible, however, that in vivo CaM53 is prenylated by both FTase and GGT-I as has been shown for RhoB in mammalian cells (Lebowitz et al., 1997). Because the possibility of changes in GGT-I activity in the absence of FTase in era1-2 plants could not be ruled out, we tested whether the absence of FTase may influence prenylation. GFP-BDCaM53 fusion protein was transiently expressed in wt and era1-2 Arabidopsis plants using biolistic bombardment (Fig. 10). The majority of the fusion protein was localized to the plasma membrane in bombarded cells from both the wt and era1-2 plants (Fig. 10). Non-prenylated forms of GFP-BDCaM53 accumulated in the nucleus due to the presence of the polybasic domain at the C-terminal end of the fusion protein as has already been shown in previous experiments (Rodríguez-Concepción et al., 1999b). This nuclear localization is observed in almost every experiment probably owing to overexpression of GFP-BDCaM53, which results in saturation of the GGT-I, and/or a depletion of the isoprenoid substrates (Rodríguez-Concepción et al., 1999b). The fact that the distribution of the fusion protein between the plasma membrane and the nucleus was similar in wt and era1-2 indicates that in vivo CaM53 is primarily prenylated by GGT-I. The results further indicate that GGT-I function was not influenced by the absence of FTase.
Figure 10.
The activity of GGT-I in era1-2 mutants. A GFP-BDCaM53 fusion protein was transiently expressed in leaves of wt Col-0 and era1-2 mutants following transformation by biolistic bombardments. The intracellular localization of the fusion protein was determined by imaging cell with confocal microscope. Prenylated GFP-BDCaM53 protein was located to the plasma membrane resulting in green fluorescence at the periphery of the cells. Un-prenylated fusion protein accumulated in the nucleus (N; Rodríguez-Concepción et al., 1999b). The red fluorescence came from plastids (P). Bars are 20 microns.
DISCUSSION
In this paper we report the cloning of the Arabidopsis AtGGT-IB gene. Purification of recombinant AtGGT-I from baculovirus-infected insect cells has enabled the biochemical characterization of the enzymatic activity, and to raise polyclonal Abs directed against the two subunits that comprise the enzyme. These tools facilitated the analysis of GGT-I at the molecular and biochemical levels and allowed an assessment of the functional relationship between FTase and GGT-I in plants.
Sequence comparison between the GGT-I β-subunits from yeast, plants, and mammals and the AtFTB revealed that the Arabidopsis FTase and GGT-I β-subunits are as closely related to one another as the yeast and plant GGT-I β-subunits. This finding may suggest that the two separate CaaX prenyl transferases evolved very close to the divergence of yeast and plants from a common ancestor.
In one region, between residues 283 and 298 of AtGGT-IB and 294 and 310 of AtFTB, the homology between the two proteins is strikingly high. Using sequence alignment tools together with the three-dimensional structure of rat FTase (Park et al., 1997; Long et al., 1998; Strickland et al., 1998) it became possible to assign putative functions to some of the residues in this region of the proteins. According to this comparison, the region is part of the core active site of the two CaaX prenyl transferases, containing ligands to the catalytic zinc ion, the diphosphate group of FPP, and to the CaaX peptide (Fig. 1). The combined sequence and structure analysis also revealed a residue, Thr 293 in AtGGT-IB, that is conserved between all GGT-I β-subunits and is occupied by a different residue, Gly 305, in all FTase β proteins (Fig. 1). Future structure function analysis in which the invariant Thr of GGT-IB will be converted into Gly should reveal whether this residue plays a role in substrate specificity.
The ubiquitous expression patterns of AtGGT-IB RNA and proteins (Fig. 2) suggest GGT-I activity is not regulated by differential expression, and that other factors regulate the enzymatic activity. In this context, expression levels of the protein substrate, and the abundance of GGPP could be such regulatory factors. The protein substrates of GGT-I may be expressed in a differential manner. For example, it has been shown that the floral transcription factor AP1, which is highly expressed in flowers (Gustafson-Brown et al., 1994), is a substrate of FTase (Yalovsky et al., 2000b). Prenylation might be regulated by changes in the level of FPP/GGPP that result from changes in HMGR activity and the levels of mevalonic acid. In trichome cells of Nicotiana benthamiana, a GFP-BDCaM53 fusion protein was not prenylated and accumulated in the nucleus (Rodríguez-Concepción et al., 1999b). Inhibition of HMGR activity by the drug lovastatin and incubation of leaf explants in the dark for an extended period of time resulted in similar nuclear accumulation of the fusion protein (Rodríguez-Concepción et al., 1999b).
The amino acid composition of the CaaL box and the adjacent region of the protein determine the affinity of GGT-I to its protein substrates (Figs. 4–7). Positively charged residues at the a2 position of the CaaL box greatly reduced the prenylation, whereas aliphatic residues enhanced it (Fig. 4). Yeast cells that expressed a-factor mutants in which the a2 Ile was substituted into either Gly, Lys, Arg, Asp, or Glu either failed or formed only very small growth inhibition halos when placed over a mat of sst2α cells (Trueblood et al., 2000), indicating that a-factor farnesylation was inhibited. These results provide further support to an earlier conclusion that the core active site of AtFTase and AtGGT-I are very similar.
The polybasic domain proximal to the CTIL CaaL box of CaM53 increased prenylation by GGT-I by an order of magnitude (Figs. 6 and 7). A polybasic region in the C-terminal end of K-RasB similarly increased the affinity of both FTase and GGT-I to the substrate protein (James et al., 1995).
The polybasic domain of CaM53 is comprised of 11 Arg and three Lys residues, whereas the polybasic domain of K-RasB is comprised of nine Lys residues. The polybasic domains of several Arabidopsis Rac-like GTPases (GenBank accession nos. U62746, AF107663, U41295, and U49971), which are putative substrates of GGT-I, are comprised of seven to eight Lys residues and in one case an additional Arg. These data suggest that charge rather than the specific composition of amino acid in the polybasic domain affect the efficiency of prenylation by GGT-I.
In K-RasB, the polybasic domain forms a type-I β-turn that binds along the rim of the hydrophobic cavity (Long et al., 2000). The polybasic domain of mammalian Cdc42 enlarges the binding pocket of RhoGDI for the geranylgeranylated Cys (Hoffman et al., 2000) and might have a similar role during the prenylation reaction itself. The differences in the prenylation efficiency of substrates with and without a polybasic domain raises the question: In vivo, must all the substrates of GGT-I have this domain to become modified? In an alternate manner, CaaL box-containing proteins that lack a polybasic domain should be expressed at higher level to become prenylated.
In the absence of a polybasic domain, GGT-I can only prenylate CaaX boxes in which the X residue is not Leu at a very low efficiency (Seabra et al., 1991; Trueblood et al., 1993). K-RasB terminates in a CVIM CaaX box, by itself a poor substrate for GGT-I. The efficient prenylation of K-RasB by GGT-I indicates that the polybasic domain changes the substrate specificity of GGT-I (James et al., 1995). Here, we showed that AtGGT-I inefficiently prenylated CaaL boxes in which the a2 position is occupied by a positively charged Arg residue (Fig. 4) and similar results were shown for yeast FTase (Trueblood et al., 2000). It should now be interesting to determine whether the presence of the polybasic domain in other proteins can facilitate prenylation of unfavorable CaaX boxes.
Because plants cannot change their location, adaptation to changes in their environment are crucial for their survival. AtGGT-I maintained high activity in a temperature range of 26°C, between 16°C and 42°C, with a maximum at 30°C to 37°C. Thus, the ability of certain enzymes to function at different temperatures may contribute to the ability of plant cells to survive in changing environments. From a structural point of view, it will be interesting to examine whether certain substitutions in the amino acid composition in GGT-I α- and β-subunits of plants have occurred to adapt this enzyme to function at a wide temperature range.
AtGGT-IB expression and activity remained unaltered in era1-2 plants, which lack AtFTB (Figs. 2 and 10). These results indicate that the expression of the two β-subunits is not coordinated. The results further suggest that AtGGT-I may prenylate some of FTase substrate proteins. In yeast ram1Δ cells that lack FTase, low efficiency prenylation of Ras proteins by GGT-I allows the cells to divide, albeit in slower rates (Trueblood et al., 1993; Yalovsky et al., 1997). Enhancement of geranylgeranylation of the mammalian RhoB protein in transformed tissue culture cells treated with FTase inhibitors have been correlated with the inhibition of cell proliferation (Lebowitz et al., 1997; Du et al., 1999). In analogy, AtGGT-I activity in era1-2 plants may act to suppress some phenotypes that might have resulted from the lack of farnesylation. On the other hand, geranylgeranylation rather than farnesylation of some proteins may change their function and induce new phenotypes.
Future studies to identify mutants in AtGGT-IB and AtFTA/GGT-IA, as well as identification and characterization of new prenylated proteins in plants will be required to identify the whether GGT-I is required for the survival of era1 plants, and whether proteins change their function when geranylgeranylated rather than farnesylated. Answers to these types of questions will be required to understand why eukaryotic cells have maintained two separate CaaX prenyl transferases.
MATERIALS AND METHODS
Yeast Two-Hybrid Screen
Bacterial and Yeast Strains
Escherichia coli DH5α and XL-1blue were used for plasmid propagation. E. coli MH4 was used to recover the Gal4 activation domain plasmid from yeast (Hall et al., 1984). Yeast Y190 was used as the host strain for the yeast two-hybrid screen (Harper et al., 1993).
Plasmid Construction
A full-length cDNA BamHI XhoI fragment of LeFTA was created by PCR and cloned into the yeast vector pGBT9.BS (Kim et al., 1997) to create pSY207.
Yeast Transformation
Yeast transformation was performed using the polyethylene glycol/LiAcetate method as described by Gietz et al. (1992). Yeast Y190 was first transformed with pSY207 to obtain strain Y190-1 followed by transformation of the λ-ACT cDNA library (Kim et al., 1997). When transforming the library, the following modifications were made in the transformation method: Two cultures of 500 mL Y190-1 cells were used for two independent transformations of 100 μg of λ-ACT library DNA. Following the incubation in 36 mL of polyethylene glycol/LiAcetate 3.9 mL of dimethyl sulfoxide (from a new bottle that was freshly opened, or stored at −80°C immediately after the bottle was opened) were added and then the cells were heat shocked as described (Gietz et al., 1992). An estimated number of 5 to 6 million transformants was obtained. Transformants were plated on synthetic complete media (Trp, Leu, and His containing 25 mm 3-aminotriazole [Sigma, St. Louis]) and incubated for 7 to 10 d at 30°C. 3-Aminotriazole was used to repress basal activity of HIS3 gene resulting in unspecific background (Kim et al., 1997). His+ colonies were spread on a secondary plate and assayed for β-gal activity using filter lift assay (Breeden and Nasmyth, 1985). pACT cDNA plasmids were rescued into E. coli MH4 cells (Kim et al., 1997). Plasmids, which gave strong β-gal reactions, were sequenced.
Protein Expression in Baculovirus-Infected Insect Cells
Bacterial Strains and Insect Cells
E. coli DH5α and XL-1blue were used for plasmid propagation. E. coli DH10Bac (Gibco BRL, Grand Island, NY) was used to obtain bacmids. Spugidera frugidera (Sf9) cell culture was used for protein expression.
AtFTA/GGT-IA Clone
An expressed sequence tag cDNA clone of ATFTA/GGT-IA (GenBank accession H37092) was obtained from The Arabidopsis Resource Center (Columbus, OH). The clone was sequenced to verify that it contains a full-length AtFTA/GGT-IA cDNA. The sequence of a full-length AtFTA/GGT-IA cDNA can be found in GenBank (accession no. AF064542).
Plasmid Construction
A full-length BamHI SalI fragment of AtGGT-IB was created by PCR and cloned into pFastBacHTa (Gibco) to create pHTaGGT-IB. A full-length HinDIII PstI fragment of AtGGT-IA was created by PCR and cloned into pFastBacHTa (Gibco) to create pHTaGGT-IA.
Insect Cell Infection and Protein Expression
To obtain bacmids, pHTaGGT-IA and pHTaGGT-IB were transformed into E. coli DH10Bac (Gibco). Recombinant baculovirus expressing the genes were prepared in the Bac to Bac system according to manufacturer's instructions (Gibco BRL). Recombinant proteins were purified on 1 mL Ni-NTA columns according to the manufacturer's instructions (Qiagen, Valencia, CA). Fractions containing purified proteins were pooled and dialyzed/concentrated under vacuum against 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 50 μm ZnCl2, and 1 mm dithiothreitol (DTT), on ice using a Collodion apparatus (Schleicher and Schuell, Dassel, Germany) with a dialysis membrane, molecular mass cutoff of 25 kD. FTase was expressed as previously described (Yalovsky et al., 2000b). The purified concentrated proteins were aliquotted, batch frozen in liquid N2, and kept at −80°C until further use.
Abs Production and Protein Immunoblots
Preparation of Abs
To separate the α- and β-subunits of AtGGT-I, purified protein was separated on SDS-PAGE (Laemmli, 1970). Protein bands were eluted from the gels and injected into rabbits.
Protein Immunoblots
Protein extracts were prepared as described (Yalovsky et al., 1996, 2000a). Equal amount of protein from each extract were loaded on SDS-PAGE and then electro-transferred onto nitrocellulose membranes (Schleicher and Schuell). Nitrocellulose membranes were first blocked with nonfat milk and subsequently incubated for 12 h at 4°C with the α-AtGGT-IB antibody (diluted 1:10,000), washed with TBST, and incubated for 1 h with a goat α-rabbit horseradish peroxidase-conjugated secondary Ab (diluted 1:30,000) for developing with Super Signal Substrate kit (Pierce, Rockford, IL).
Plant Material
Arabidopsis Col-0 and era1-2 were grown in 5-cm pots. Plants were grown on soil with vermiculite (Avi Saddeh mix, Pecka Hipper Gan) and were irrigated from below as described (Yalovsky et al., 2000a). Plants were grown in an environmental growth chamber under long days (16-h-light/8-h-dark cycles). Light intensity was 100 μE m−2 s−1.
RT-PCR
RNA Isolation
Total RNA was isolated from 50 to 100 mg of fresh or frozen tissues using SV total RNA isolation system (Promega, Madison, WI).
cDNA First Strand Synthesis
Five-hundred nanograms of total RNA was incubated with 500 ng of oligo dT primer and water was added to a final volume of 15 μL. The mixture was incubated at 70°C for 5 min and transferred to ice. The following mixture was then added and incubated at 42°C for 1 h.: 5 μL 5× M-MLV buffer (Promega), 2.5 μL of 5 mm dNTP mix, 1 μL RNAsine (25 units), 1 μL of M-MLV RT (200 units; Promega), and water to a final volume of 25 μL. The reaction was stopped by incubation at 95°C for 5 min.
PCR
For amplification of AtGGT-IB and AP1, 2 μL of concentrated first strand synthesis products were used as templates together with gene specific primers. For amplification of UBQ10, 2 μL of first strand synthesis products diluted 1:10 were used as template together with specific primers. All products were amplified using the following program: 1 cycle of 94°C, 5 min; 54°C, 5 min; 72°C, 2 min; followed by 25 cycles of 94°C, 1 min; 54°C, 1 min; 72°C, 2 min; and a final elongation step of 5 min at 72°C. Serial dilutions of the PCR products were applied onto a dot blot and hybridized with [32P]dCTP-labeled probes.
Protein Expression in E. coli
Bacterial Strains
E. coli DH5α and XL-1blue were used for DNA propagation and protein expression.
Plasmid Construction
The construction of pTA3CaM53 and pGEXBDCaM53 was described (Rodríguez-Concepción et al., 1999b). The pGEX-CaaL fusions were constructed by ligating annealed primer pairs into EcoRI XhoI sites of pGEX4T-1 (Pharmacia, Uppsala). The following primer pairs were used: CTIL (CaM53), AA TTG TGC ACA ATA CTG TGA and C ACG TGT TAT GAC ACT AGCT; CGGL (AUX-2.11), AA TTG TGC GGT GGA CTG TGA and C ACG CCA CCT GAC ACT AGCT; CGQL (CYP), AA TTG TGC GGT CAG CTG TGA and C ACG CCA GTC GAC ACT AGCT; and CWRL (MIP), AA TTG TGC TGG AGA CTG TGA and C ACG ACC TCT GAC ACT AGCT.
Protein Expression
Full-length CaM53 and GST-BDCaM53 were expressed successfully by cotransforming cells with the protein expression plasmids and the tRNA Arg plasmid pUBS520 (Brinkmann et al., 1989; Schenk et al., 1995). The pUBS520 plasmid suppresses misexpression of proteins that contain AGG AGA Arg codons in tandem (Brinkmann et al., 1989). Expression of all proteins was induced with 1 mm isopropyl thio glactoside. Purification of CaM53 was as described (Rodríguez-Concepción et al., 1999b). Purification of GST fusion proteins was carried out according to the manufacturer (Pharmacia). Purified proteins were dialyzed/concentrated under vacuum against 50 mm Tris-HCl pH 7.5 and 1 mm DTT, on ice, using a Collodion apparatus (Schleicher and Schuell) with a dialysis membrane of a 10-kD molecular mass cutoff. The concentrated proteins were aliquotted, batch frozen in liquid N2, and kept at −80°C until further use.
Prenylation Reactions
Protein Quantitation
Two methods were used to measure the concentration of purified GGT-I and substrate proteins. Optical density of the protein solution was measured from A240 to A320 (optical absorbance at 240 to 320 nm). The values obtained for A280 were used to determine protein concentration using the “Protparam” tool site on the Expasy Molecular Biology server (www.expasy.ch/sprot/protparam.html). Protein concentration was measured using the BCA kit (Pierce). In short, equal volumes (usually 500 μL) of protein solution (in water) and BCA reagents were mixed and incubated at 60°C for 1 h. Following the incubation, the optical density was measured at A562.
Reactions
Unless indicated otherwise, each reaction contained reaction buffer {50 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-NaOH, pH 7.8, 5 mm MgCl2, 50 μm ZnCl2, 2.5 mm DTT, and 0.3% [v/v] NP40}, 0.2 μm protein substrate, 0.8 μm all-trans [3H] GGPP or [3H]FPP 30 Ci mmol−1 (American radiolabeled), and 0.5 pmol (0.01 μm) AtGGT-I, in a final volume of 25 μL. All ingredients except the enzyme were mixed and kept on ice. Reactions were initiated by adding the enzyme to each mix and transferring to incubation at 30°C (unless indicated otherwise) for either 15 or 30 min. Reactions were carried out in triplicates or in a few cases in duplicates.
Quantitation of Protein Incorporated GGPP
To determine the amount of prenylated proteins, reactions were terminated by adding 1 mL of HCL:ethanol (1:9, v/v; acidic ethanol) solution and incubation for 30 min at room temperature (this treatment breaks the diphosphate group off GGPP and enables the separation of geranylgeranylated proteins from unbound GG-OH by additional washes in ethanol [Seabra et al., 1993]). Following the incubation, the solution was filtered through glass filters (GF/C, Whatman, www.Whatman.com) to capture precipitated radiolabeled proteins, the tubes were washed with 2 mL of 100% (v/v) ethanol, and the filters were washed with an additional 3 mL of 100% (v/v) ethanol. The amounts of radiolabeled proteins were estimated by scintillation counting. In an alternate manner, reactions were terminated by denaturing proteins in SDS-PAGE denaturing buffer, separating equal amount of each reaction by SDS-PAGE (Laemmli, 1970), after which gels were fixed, treated with Amplify reagent (Amersham, Little Chalfont, UK), and exposed to x-ray films. The x-ray films were scanned to quantify the band intensity using ImageMaster 1D software (Pharmacia LKB).
Transient Expression of GFP Fusion Proteins
GFP-BDCaM53 construct (Rodríguez-Concepción et al., 1999b) was transformed into leaves of Col-0 and era1-2 plants using biolistic bombardments (Rodríguez-Concepción et al., 1999b). Protein expression in the epidermal cell layer was analyzed with confocal laser scanning microscope (RS 510, Zeiss, Jenna, Germany) as previously described (Rodríguez-Concepción et al., 1999b; Yalovsky et al., 2000a, 2000b).
ACKNOWLEDGMENTS
The plasmid pUBS520 was a gift from Prof. Ralf Mattes (University of Stuttgart, Germany). We would like to thank Prof. Loy Volkmann (University of California, Berkeley) and Dr. Sasson Dorri (Tel Aviv University) for technical support, and members of the Yalovsky and Gruissem laboratories for their support and advice.
Footnotes
This research was supported by The Israel Science Foundation (grant no. 571/99 to S.Y.), by the Binational Science Foundation (grant no. 1999423 to S.Y.), and by the U.S. Department of Energy (grant no. 85ER13375 to W.G.). D.C. was supported by a fellowship from the Swiss National Science Foundation, and M.R.C. had support from the Spanish Ministry of Education and Culture.
LITERATURE CITED
- Armstrong SA, Hannah VC, Goldstein JL, Brown MS. CAAX geranylgeranyl transferase transfers farnesyl as efficiently as geranylgeranyl to RhoB. J Biol Chem. 1995;270:7864–7868. doi: 10.1074/jbc.270.14.7864. [DOI] [PubMed] [Google Scholar]
- Bonetta D, Bayliss P, Sun S, Sage T, McCourt P. Farnesylation is involved in meristem organization in Arabidopsis. Planta. 2000;211:182–190. doi: 10.1007/s004250000283. [DOI] [PubMed] [Google Scholar]
- Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Brinkmann U, Mattes RE, Buckel P. High-level expression of recombinant genes in Escherichia coli is dependent on the availability of the dnaY gene product. Gene. 1989;85:109–114. doi: 10.1016/0378-1119(89)90470-8. [DOI] [PubMed] [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. A protein farnesyl transferase involved in ABA signal transduction in Arabidopsis. Science. 1996;273:1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- Del Villar K, Mitsuzawa H, Yang W, Sattler I, Tamanoi F. Amino acid substitutions that convert the protein substrate specificity of farnesyltransferase to that of geranylgeranyltransferase type I. J Biol Chem. 1997;272:680–687. doi: 10.1074/jbc.272.1.680. [DOI] [PubMed] [Google Scholar]
- Díaz M, Sanchez Y, Bennet T, Sun CR, Godoy C, Tamanoi F, Duran A, Perez P. The Schizosaccharomyces pombe Cwg2+ gene codes for the β-subunit of a geranylgeranyltransferase type I required for β-glucan synthesis. EMBO J. 1993;12:5245–5254. doi: 10.1002/j.1460-2075.1993.tb06220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Lebowitz PF, Prendergast GC. Cell growth inhibition by farnesyltransferase inhibitors is mediated by gain of geranylgeranylated RhoB. Mol Cell Biol. 1999;19:1831–1840. doi: 10.1128/mcb.19.3.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykema PE, Sipes PR, Marie A, Bierman BJ, Crowell DN, Randall SK. A new class of proteins capable of binding transition metals. Plant Mol Biol. 1999;41:139–150. doi: 10.1023/a:1006367609556. [DOI] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acid Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman LE, Perou CM, Fujiyama A, Tamanoi F. Structure and expression of yeast DPR1, a gene essential for the processing and intracellular localization of ras proteins. Yeast. 1988;4:271–281. doi: 10.1002/yea.320040405. [DOI] [PubMed] [Google Scholar]
- Gustafson-Brown C, Savidge B, Yanofsky MF. Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell. 1994;76:131–143. doi: 10.1016/0092-8674(94)90178-3. [DOI] [PubMed] [Google Scholar]
- Hall MN, Hereford L, Herskowitz I. Targeting of E. coli beta-galactosidase to the nucleus in yeast. Cell. 1984;36:1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Hoffman GR, Nassar N, Cerione R. Structure of the Rho family GTP-binding protein Cdc42 in complex with multifunctional regulator RhoGDI. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- James GL, Goldstein JL, Brown MS. Polylysine and CVIM sequences of K-RasB dictate specificity of prenylation and confer resistance to benzodiazepine peptidomimetic in vitro. J Biol Chem. 1995;270:6221–6226. doi: 10.1074/jbc.270.11.6221. [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lebowitz PF, Casey PJ, Prendergast GC, Thissen JA. Farnesyltransferase inhibitors alter the prenylation and growth-stimulating function of RhoB. J Biol Chem. 1997;272:15591–15594. doi: 10.1074/jbc.272.25.15591. [DOI] [PubMed] [Google Scholar]
- Li H, Wu G, Ware D, Davis KR, Yang Z. Arabidopsis Rho-related GTPases: differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 1998;118:407–417. doi: 10.1104/pp.118.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wang Y, Zhu J-K, Yang Z. Localization of a Rho GTPase implies a role in tip growth and movement of the generative cell in pollen tubes. Plant Cell. 1996;8:293–303. doi: 10.1105/tpc.8.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SB, Casey PJ, Beese LS. Cocrystal structure of protein farnesyltransferase complexed with a farnesyl diphosphate substrate. Biochemistry. 1998;37:9612–9618. doi: 10.1021/bi980708e. [DOI] [PubMed] [Google Scholar]
- Long SB, Casey PJ, Beese LS. The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 A resolution ternary complex structures. Structure Fold Des. 2000;8:209–222. doi: 10.1016/s0969-2126(00)00096-4. [DOI] [PubMed] [Google Scholar]
- Ohya Y, Goebl M, Goodman LE, Peterson-Bjorn S, Friesen JD, Tamanoi F, Anraku Y. Yeast CAL1 is a structural and functional homologue to the DPR1(RAM1) gene involved in ras processing. J Biol Chem. 1991;266:12356–12360. [PubMed] [Google Scholar]
- Park HW, Boduluri SR, Moomaw JF, Casey PJ, Beese LS. Crystal structure of protein farnesyltransferase at 2.25 angstrom resolution. Science. 1997;275:1800–1804. doi: 10.1126/science.275.5307.1800. [DOI] [PubMed] [Google Scholar]
- Randall SK, Marshall MS, Crowell DN. Protein isoprenylation in suspension-cultured tobacco cells. Plant Cell. 1993;5:433–442. doi: 10.1105/tpc.5.4.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss Y, Brown MS, Goldstein JL. Divalent cation and prenyl pyrophosphate specificities of the protein farnesyltransferase from rat brain, a zinc metalloenzyme. J Biol Chem. 1992;267:6403–6408. [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Yalovsky S, Gruissem W. Protein prenylation in plants: old friends and new targets. Plant Mol Biol. 1999a;39:865–870. doi: 10.1023/a:1006170020836. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Yalovsky S, Zik M, Fromm H, Gruissem W. The prenylation status of a novel plant calmodulin directs plasma membrane or nuclear localization of the protein. EMBO J. 1999b;18:1996–2007. doi: 10.1093/emboj/18.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Bethke P. Membrane transport. In: Buchanan BB, Gruissem W, Jones RL, editors. Biochemistry and Molecular Biology of Plants. Ed 1. Rockville, MD: American Society of Plant Physiologists; 2000. pp. 110–158. [Google Scholar]
- Schafer WR, Rine J. Protein prenylation: genes, enzymes, targets and functions. Annu Rev Genet. 1992;30:209–237. doi: 10.1146/annurev.ge.26.120192.001233. [DOI] [PubMed] [Google Scholar]
- Schafer WR, Trueblood CE, Yang C-C, Mayer MP, Rosenberg S, Poulter CD, Kim S-H, Rine J. Enzymatic coupling of cholesterol intermediates to a mating pheromone precursor and to the Ras protein. Science. 1990;249:1133–1139. doi: 10.1126/science.2204115. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Baumann S, Mattes R, Steinbiss HH. Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare Arg tRNAs. Biotechniques. 1995;19:196–198. [PubMed] [Google Scholar]
- Schmitt D, Callan K, Kim S-H, Gruissem W. Molecular and biochemical characterization of tomato farnesyl-protein transferase. Plant Physiol. 1996;112:767–777. doi: 10.1104/pp.112.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabra MC, Brown MS, Goldstein JL. Retinal degeneration in choroidermia: deficiency of Rab geranylgeranyl transferase. Science. 1993;259:377–381. doi: 10.1126/science.8380507. [DOI] [PubMed] [Google Scholar]
- Seabra MC, Reiss Y, Casey P, Brown MS, Goldstein JL. Protein farnesyltransferase and geranylgeranyltransferase share a common α-subunit. Cell. 1991;65:429–434. doi: 10.1016/0092-8674(91)90460-g. [DOI] [PubMed] [Google Scholar]
- Sinensky M. Recent advances in the study of prenylated proteins. Biochem Biophys Acta. 2000;1484:93–106. doi: 10.1016/s1388-1981(00)00009-3. [DOI] [PubMed] [Google Scholar]
- Strickland CL, Windsor WT, Syto R, Wang L, Bond R, Wu Z, Schwartz J, Le Hung V, Beese LS, Weber PC. Crystal structure of farnesyl protein transferase complexed with a CaaX peptide and farnesyl diphosphate analogue. Biochemistry. 1998;37:16601–16611. doi: 10.1021/bi981197z. [DOI] [PubMed] [Google Scholar]
- Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- Trainin T, Shmuel M, Delmer DP. In vitro prenylation of the small GTPase Rac13 in cotton. Plant Physiol. 1996;112:1491–1497. doi: 10.1104/pp.112.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood CE, Boyartchuk VL, Picologlou EA, Rozema D, Poulter D, Rine J. The CaaX proteases, Afc1p and Rce1p, have overlapping but distinct substrate specificities. Mol Cell Biol. 2000;20:4381–4392. doi: 10.1128/mcb.20.12.4381-4392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood CE, Boyartchuk VL, Rine J. Substrate specificity determinants in the farnesyltransferase beta-subunit. Proc Natl Acad Sci USA. 1997;94:10774–10779. doi: 10.1073/pnas.94.20.10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood CE, Ohya Y, Rine J. Genetic evidence for in vivo cross-specificity of the CaaX box protein prenyltransferases farnesyltransferase and geranylgeranyltransferase-I in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:4260–4275. doi: 10.1128/mcb.13.7.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Kulukian A, Rodríguez-Concepción M, Young CA, Gruissem W. Functional requirement of plant farnesyltransferase during development in Arabidopsis. Plant Cell. 2000a;12:1267–1278. doi: 10.1105/tpc.12.8.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Loraine AE, Gruissem W. Specific prenylation of tomato Rab proteins by geranylgeranyl type-II transferase requires a conserved cysteine-cysteine motif. Plant Physiol. 1996;110:1349–1359. doi: 10.1104/pp.110.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Rodríguez-Concepción M, Bracha K, Toledo-Ortiz G, Gruissem W. Prenylation of the floral transcription factor APETALA1 modulates its function. Plant Cell. 2000b;12:1257–1266. doi: 10.1105/tpc.12.8.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Rodríguez-Concepción M, Gruissem W. Lipid modification of proteins-slipping in and out of membranes. Trends Plant Sci. 1999;4:429–438. doi: 10.1016/s1360-1385(99)01492-2. [DOI] [PubMed] [Google Scholar]
- Yalovsky S, Trueblood CE, Callan KL, Narita JO, Jenkins SM, Rine J, Gruissem W. Plant farnesyltransferase can restore yeast ras signaling and mating. Mol Cell Biol. 1997;17:1986–1994. doi: 10.1128/mcb.17.4.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K, McGeady P, Gelb M. Mammalian protein geranylgeranyltransferase-I: substrate specificity, kinetic mechanism, metal requirement and affinity labeling. Biochemistry. 1995;34:1344–1354. doi: 10.1021/bi00004a029. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, Zimmerman K, Scholten J, Gelb MH. Differential prenyl pyrophosphate binding to mammalian protein geranylgeranyltransferase-I and protein farnesyltransferase and its consequence on the specificity of protein prenylation. J Biol Chem. 1997;272:3944–3952. doi: 10.1074/jbc.272.7.3944. [DOI] [PubMed] [Google Scholar]
- Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- Zhou D, Qian D, Carmer CL, Yang Z. Developmental and environmental regulation of tissue- and cell-specific expression for a pea protein farnesyltransferase gene in transgenic plants. Plant J. 1997;12:921–930. doi: 10.1046/j.1365-313x.1997.12040921.x. [DOI] [PubMed] [Google Scholar]
- Ziegelhoffer EC, Medrano LJ, Meyerowitz EM. Cloning of the Arabidopsis WIGGUM gene identifies a role for farnesylation in meristem development. Proc Natl Acad Sci USA. 2000;97:7633–7638. doi: 10.1073/pnas.130189397. [DOI] [PMC free article] [PubMed] [Google Scholar]