Abstract
Plants have evolved an intricate signaling apparatus that integrates relevant information and allows an optimal response to environmental conditions. For instance, the coordination of defense responses against pathogens involves sophisticated molecular detection and communication systems. Multiple protection strategies may be deployed differentially by the plant according to the nature of the invading organism. These responses are also influenced by the environment, metabolism, and developmental stage of the plant. Though the cellular signaling processes traditionally have been described as linear sequences of events, it is now evident that they may be represented more accurately as network-like structures. The emerging paradigm can be represented readily with the use of Boolean language. This digital (numeric) formalism allows an accurate qualitative description of the signal transduction processes, and a dynamic representation through computer simulation. Moreover, it provides the required power to process the increasing amount of information emerging from the fields of genomics and proteomics, and from the use of new technologies such as microarray analysis. In this review, we have used the Boolean language to represent and analyze part of the signaling network of disease resistance in Arabidopsis.
We learned from the last decades of molecular research that the genomic repertoire is not entirely expressed at any time in a cell. Organisms respond exquisitely to any given set of conditions by the production and activation of unique protein subsets from a wide number of possible combinations. To understand how these precise choices are made in a cell, research has focused on the study of the cellular perception and signal transduction mechanisms controlling gene expression and protein activity. With the use of molecular genetics, hundreds of genes and proteins responsible for perception have been identified and the precise sequence of signaling events starts to be unraveled. In parallel, biochemical analyses have described the nature of some of the transducing elements and their interactions, and key molecules now can be classified into various families of receptors or response regulators. Attempts are made to integrate the multiplicity of these results into a comprehensive description represented by hierarchical structures called “cascades” or tree-like series of reactions.
However, more recent findings suggest that signaling elements are not always operating in isolated pathways. Interactions between linear pathways during coincident activation have been termed crosstalks or interferences (Genoud and Métraux, 1999; Noselli and Perrimon, 2000). The physiological and molecular evidences of the cross talk phenomenon are numerous in plants, and add a new dimension to the study of signal processing (Genoud and Métraux, 1999; Feys and Parker, 2000).
STRUCTURE AND PLASTICITY OF NETWORKS
In eucaryotic systems, the organization of signaling events often depends upon a semistable structure composed of anchoring and scaffolding proteins (Pawson and Scott, 1997). These molecules typically display three functions: they selectively bind signal-processing elements to constitute a signaling complex and they may also target the modules of information transfer to a specific cellular location, or bridge the interaction between two partners. A classical example of a scaffolding protein is the Ste5 protein involved in the constitution of the mating pheromone's mitogen-activated protein kinase pathway in yeast (Faux and Scott, 1996; Garrington and Johnson, 1999).
Growing evidence suggests that the structure of signaling networks is rather plastic, whereby the process of perception often leads to modifications in the connections between elements, as well as to changes in their localization (Teruel and Meyer, 2000). For instance, the sensitivity and the specificity of a pathway may be increased or reduced upon perception, changing the qualitative properties of a signaling module. The reinforcement of a transduction pathway has been called “consolidation,” whereas the opposite corresponds to a “desensitization” phenomenon (Jordan et al., 2000). In higher plants, consolidation of signaling functions may occur in the perceptive process of several biotic and abiotic factors (Yamamoto et al., 1998). The activity of a pathway can also be modulated through direct or indirect intracellular crosstalk (Noselli and Perrimon, 2000).
The plasticity of a signaling network also ensues from relocation of elements, which may change potential interactions. It is known that proteins engaged in signaling are not necessarily active in a single cellular location (Teruel and Meyer, 2000). For instance, receptors can be translocated through different compartments of a cell. This is the case for the phytochrome A and B photoreceptors in plants that undergo a cytoplasm-to-nucleus translocation upon light perception (Kircher et al., 1999; Yamaguchi et al., 1999; Màs et al., 2000).
THE BOOLEAN FORMALISM
The emergent properties of the signaling process imply that our background formalism must be widened. We may select a new basic model that can include a connectionistic paradigm and the cross interactions among heterogeneous signals. For this purpose, a space of interconnected operators appears as a good starting framework. In an initial phase, a very reductive model, the standard Boolean network representation, may be applied (Genoud and Métraux, 1999). It has the advantage of eluding the statistical fundaments of stochastic neural networks and being strongly isomorphic to the classical genetic approach, which mostly divides altered characters in loss-of-function or gain-of-function mutations, and uses further dichotomic concepts such as dominant and recessive transmission, necessary or sufficient elements, positive and negative regulators, etc.
Another convenience provided by the Boolean framework resides in the possibility to perform simple simulations on a computer program, once the logical connections between constituents have been established. Also, if additional regulators are identified, they can easily be inserted into a current model to specify, for instance, a branch of the connected network. Although the results of computer simulation are still only qualitative in the present report, they may later include the product of quantitative algorithms, as soon as the biochemical parameters (structure, concentration, affinities, and localization) of the involved elements are known. It is clear, considering the achievements of computer engineering, that any quantitative set of data can be expressed in a Boolean (digital, numeric) language.
TRANSLATION OF GENETIC DATA INTO THE BOOLEAN LANGUAGE
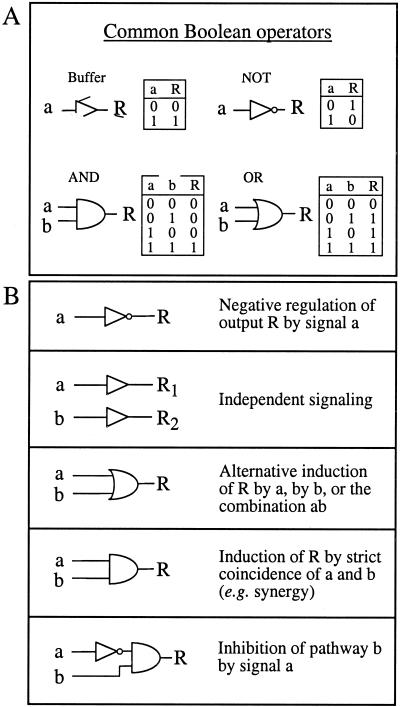
To represent genetic data in the form of a digital simulation program, several graphic softwares are freely available as shareware on the internet (e.g. the discrete time event simulator LogicSim created by Arnaud Masson; www.planete.net/∼amasson/logicsim.html). These applications require little knowledge in programming and can be directly converted into signal transduction simulators. To transform the function of signaling elements into logical operators, crosstalk switches have first to be postulated as elements located upstream of synergistic responses, or between multiple pathways regulating the expression of a similar gene or physiological response. The Boolean operators allowing the description of the majority of signal interactions are the NOT, AND, buffer, and OR gates (Fig. 1A). Combinations of these elements under the control of ON-OFF switches or clock-like input generators can model common signaling structures (see Fig. 2). The observed synergistic interactions are typically translated into AND operators, whereas negative interference can be expressed by the addition of a NOT operator to one of the two input connections of an AND gate (Fig. 1B). Isolated, cross talk-independent signal transducers can be represented as buffer gates, whereas elements used alternatively by two different pathways correspond to OR operators. Additional logical operators exist: the NAND (not and), NOR (not or), XOR (exclusive or), as well as XNOR (exclusive not or) switches, which add various specificities (Genoud and Métraux, 1999).
Figure 1.
Basic components and concepts of the Boolean formalism. A, The most frequent types of Boolean operators are the NOT, buffer, AND, and OR gates. They are shown with their corresponding truth table. Additional operators may be employed to translate integration steps in signaling (see text). Note that the inactive NOT gate produces a constitutive output of 1. B, Boolean representation of a few simple operations that may be found in a signal transduction network. a and b, Different input stimuli; R, R1, and R2, different responses.
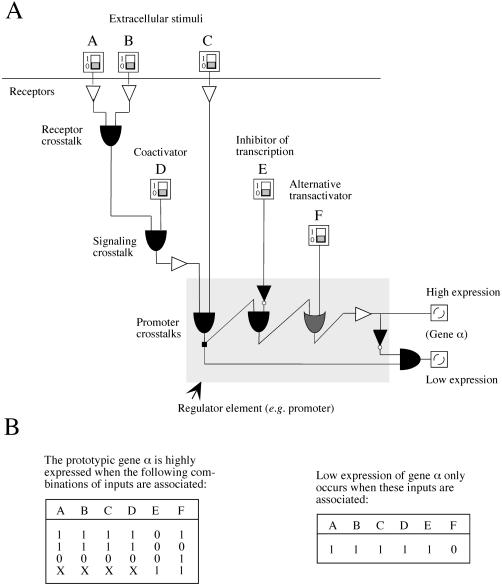
Figure 2.
Translation of signaling concepts into the Boolean formalism. A, Description of a signaling network regulating the expression of a model gene (α), combining several extracellular and intracellular parameters (represented as 1-0 switches). Final outputs are divided in three qualitative categories: no expression (no output), low expression, as well as high expression of gene α. Crosstalks may occur in different cellular compartments; they are depicted as OR and AND operators. Receptors and possible signaling components correspond to buffer elements (simple triangles). NOT operators (triangle followed by a small circle) correspond to signal inverters with a constitutive output of 1. Small squares indicate a branching of a line. A regulator element (shaded box), located in this instance at the level of the gene promoter, integrates signals through successive protein-DNA interactions leading to a specific level of transcription. B, Truth tables representing the various sets of input combinations that regulate high and low expression of gene α in A. In this particular example, low expression is only obtained by a single specific setting, whereas high expression may result from several settings. X values represent either 1 or 0. Any other combination leads to no expression.
TRANSLATION OF MICROARRAY DATA INTO THE BOOLEAN LANGUAGE
Large-scale microarrays have provided a first view on gene regulation in response to particular treatments in Arabidopsis (Maleck et al., 2000; Schenk et al., 2000). From such complex sets of information, qualitative categories of gene expression based on the relative transcript levels can be defined. Cutoff values may be rigorously selected to divide the transcription patterns into pragmatic qualitative classes, starting with the highly up-regulated and down-regulated genes. For each physiologically significant time point, one may select levels corresponding to an increase or decrease of four relative units, for instance. These first sets of genes can be considered as diagnostic markers for a specific treatment. In a second step, the expression profiles of diagnostic markers corresponding to two different treatments can be confronted. Genes that are up-regulated to the same level range in both treatments belong to a category associated to a logical OR operator (see Figs. 1B and 2) because both treatments produce the same qualitative effect; the related down-regulated genes are associated under an OR gate that switches a NOT operator (Fig. 3).
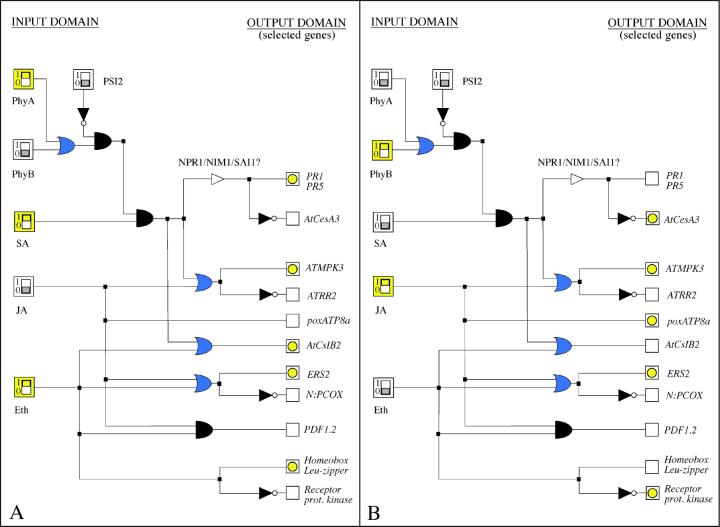
Figure 3.
Boolean representation of a signal transduction network as it may appear on the interface of a digital simulation program. A module of the signal transduction network controlling the plant's defense response against pathogens has been represented with a series of output genes selected among the classes deduced from microarray data corresponding to separate treatments of Arabidopsis with salicylic acid (SA), jasmonic acid (JA), and ethylene (E; Schenk et al., 2000). The chosen output genes present a variation in expression of at least four times the control level (except PDF1.2; for more details and gene nomenclature, see Schenk et al., 2000). Two of the various possible settings for the signal sources (input domain) have been represented (corresponding to A and B), along with their particular response profiles (output domain). Note that the deduced Boolean OR elements proposed in this working model, as well as the position of NPR1/NIM1/SAI1, need to be confirmed by further experiments. The activated switches are represented in yellow, whereas the yellow diode symbols indicate the induced genes. Empty squares correspond to no significant expression. Networks can be modeled by current digital simulators using graphic programs such as the discrete time event simulator LogicSim created by Arnaud Masson and available as (postcard) shareware on the internet (www.planete.net/∼amasson/logicsim.html).
GRAPHIC DYNAMICAL SIMULATION OF SIGNALING EVENTS: CROSSTALK INTERACTIONS AMONG SA-, JA-, AND E-MEDIATED PATHWAYS
Plants can deploy a number of different strategies to protect themselves against pathogen attack or potential microbial infection in wounded tissues. The defense system of the plant consists of a complex network of local and systemic signaling pathways, both general and specific, depending on the invading organism. The phytohormones SA, JA, and E are three of the most important signaling molecules involved in such defense-related responses. They mediate a variety of pathways that exhibit multiple crosstalks (for review, see Reymond and Farmer, 1998; Genoud and Métraux, 1999; Feys and Parker, 2000).
The hypersensitive response (HR) is an important nonspecific initial defense mechanism mounted against potentially pathogenic organisms; it consists of a syndrome of localized defense mechanisms that confines the intruders within a small area, slowing down the growth and progression of the pathogen through tissues, and resulting in localized cellular necroses (Greenberg et al., 1994; Durner et al., 1997). Necrotizing pathogens often elicit a systemic resistance in plants, mediated by a signal emitted from the invaded cells, which induces the expression of a long-lasting defense response in distal regions of the plant. Such systemic acquired resistance (SAR) provides strong immunity against a large spectrum of pathogens, and is characterized by an accumulation of several pathogenesis-related (PR) proteins (for review, see Sticher et al., 1997).
In Arabidopsis, several mutants with a defect in the regulation of PR gene activation have been isolated, some of which involve the interruption of the signal transduction downstream of SA. For instance, in the allelic mutants no PR gene expression, noninducible immunity, and SA-insensitive (npr1, Cao et al., 1994; nim1, Delaney et al., 1995; and sai1, Shah et al., 1997, respectively), SAR genes are not activated in response to applied SA or its functional analogs. As a consequence, these mutants display a higher susceptibility to infectious agents. The NPR1/NIM1/SAI1 gene encodes for a protein containing ankyrin repeats (Cao et al., 1997; Ryals et al., 1997) that interacts with a basic Leu zipper protein transcription factor in the nucleus (Zhang et al., 1999), which in turn binds sequences of the PR-1 gene promoter. Furthermore, it has been demonstrated recently that NPR1 accumulates in the nucleus in response to activators of SAR, and that nuclear localization of NPR1 is essential for the induction of PR genes (Kinkema et al., 2000).
Other mutants (lesion simulating disease resistance response [lsd], constitutive expression of PR proteins [cpr], and defense no cell death [dnd]) constitutively express the PR genes (for review, see Dangl et al., 1996; Dietrich et al., 1997); most of them also show an accumulation of SA. In contrast, mutants such as enhanced disease susceptibility (eds4; Rogers and Ausubel, 1997; Gupta et al., 2000), phytoalexin-deficient (pad4; Glazebrook et al., 1996), and SA induction-deficient (sid1 and sid2) have low levels of SA, show decreased pathogen resistance, and are affected in SA-mediated defense responses (Nawrath and Métraux, 1999).
The Arabidopsis pad4 mutant displays defects in defense responses, including camalexin synthesis and PR-1 gene expression, when infected by Pseudomonas syringae pv maculicola ES4326 but not when infected by an isogenic strain carrying the avirulence gene avrRpt2. In P. syringae pv maculicola ES4326-infected pad4 plants, SA synthesis is reduced and delayed compared with wild-type plants; moreover, SA treatment partially restores the wild-type camalexin production and PR-1 gene expression phenotypes (Zhou et al., 1998). In contrast, sid1 mutants have high levels of camalexin (Nawrath and Métraux, 1999), leaving open the question of the involvement of SA in camalexin biosynthesis. It is likely that PAD4 is required upstream from SA accumulation in regulating defense response expression upon infection with P. syringae pv maculicola ES4326; in contrast, PAD4 is not required for SA production upon infection with the avirulent strain, and in this case, PR-1 expression is independent of NPR1/NIM1/SAI1 (Zhou et al., 1998). The analysis of sid1 (allelic to eds5; Volko et al., 1998) and sid2 plants indicates that these mutations affect signaling steps upstream from SA biosynthesis, and confirms the existence of SA-independent compensation pathways (Nawrath and Métraux, 1999).
The study of the Arabidopsis mpk4 mutant has led recently to the discovery of a mitogen-activated protein kinase in the regulation of SAR. MPK4 kinase activity is required to repress SAR; moreover, SAR expression in mpk4 plants is dependent on elevated SA levels, but is independent of NPR1. It is interesting that induction of the JA-responsive genes PDF1.2 and THI2.1 was blocked in mpk4 expressing NahG (a hydroxylase that degrades SA to catechol), suggesting the requirement of MPK4 in JA-responsive gene expression (Petersen et al., 2000).
Recent studies with Arabidopsis and maize (Zea mays) mutants developing spontaneous HR lesions, and transgenic tomato expressing the R gene Pto, have suggested that light critically influences the HR in plants (Martienssen, 1997; Tang et al., 1999). Moreover, a light-hypersensitive mutant of Arabidopsis (phytochrome signaling [psi2]) has been shown to form HR-like lesions on leaves at high intensity of red light (Genoud et al., 1998). This strongly suggests that a crosstalk exists between the red light and PR expression signaling pathways (Fig. 3), a notion further confirmed by the observations that the light-hypersensitive Arabidopsis psi2 mutant exhibits a light fluence-dependent amplification of SA-induced PR-1a gene expression, and that the plants containing no detectable phytochrome A and B proteins (phyA and phyB double mutants) present a strong reduction in the expression of the PR genes elicited by either SA or benzothiadiazol (an SA agonist; T. Genoud and J.-P. Métraux, unpublished data).
Wounded plants express specific sets of genes, some of which are thought to have protective properties against microbial infection. JA and E are two of several signaling molecules involved in this phenomenon, which involves the expression of wound-responsive genes through diverse forms of cross talks (Genoud and Métraux, 1999). In Arabidopsis, for example, induction of the antifungal plant defensin gene PDF1.2 requires concomitant activation of the JA and E pathways (Penninckx et al., 1998).
The SA pathway also exhibits cross talk with JA/E pathways (for review, see Feys and Parker, 2000). The Arabidopsis mutants constitutive expression of PR mutants (cpr5 and cpr6), which have elevated levels of SA and express SAR constitutively, also express marker genes from the JA pathway (Bowling et al., 1997; Clarke et al., 1998). Further analysis indicates that CPR5 and CPR6 regulate resistance through distinct pathways, and that SA-mediated, NPR1-independent resistance involves components of the JA/E-mediated pathways (Clarke et al., 2000). In a similar manner, the ssi1 mutation, which bypasses the requirement of NPR1 for SAR function, renders the expression of PDF1.2 SA dependent (Shah et al., 1999). Furthermore, the eds4 and pad4 mutations, which cause reduced SA levels, lead to a heightened response to inducers of JA-dependent gene expression (Gupta et al., 2000). In contrast, resistance to turnip crinkle virus in Arabidopsis is mediated by a signaling pathway that is SA dependent, but NPR1, JA, and E independent (Kachroo et al., 2000).
JA and E are also involved in another type of defense response, mediating the so-called induced systemic resistance (ISR) elicited by root colonization of certain nonpathogenic Pseudomonas spp. strains (for review, see Pieterse and van Loon, 1999). ISR is, like SAR, a form of broad-spectrum systemic protection, though it is independent of SA and of PR gene expression (Pieterse et al., 1996, 1998). In Arabidopsis, both the SAR and ISR pathways are regulated by NPR1, and their activity may superimpose because their simultaneous activation produces an enhancement of disease resistance (van Wees et al., 2000).
A microarray analysis recently performed in Arabidopsis by Schenk et al. (2000) further emphasizes the complexity in the network of pathway interactions during plant defense responses. In this study, which involved 2,375 selected genes, a substantial change in the steady-state abundance of 705 mRNAs was observed in response to one or more of the following treatments: inoculation with an incompatible fungal pathogen, and exogenous application of SA, methyl-jasmonate (a biologically active JA derivative), or E. Out of these 705 mRNAS, 169 were regulated by multiple treatments, with the largest numbers of co-induced or corepressed genes observed in a class regulated by both SA and methyl-jasmonate.
In a similar experiment, gene expression in Arabidopsis was analyzed under 14 different SAR-inducing or SAR-repressing conditions with a DNA microarray representing approximately 7,000 genes (Maleck et al., 2000). These researchers found 413 expressed sequence tags showing differential expression equal to or greater than 2.5-fold in at least two SAR-relevant conditions. They used two different algorithms to generate a hierarchical “clustergram” and “self-organizing maps” (SOMs) to define groups of coregulated genes. The PR-1 gene clustered in SOM c1, which contained 45 expressed sequence tags (from a maximum of 31 different genes). Because PR-1 is a molecular marker for SAR, the genes in this PR-1 regulon are thought likely to function in SAR; moreover, they showed a unique expression profile, being strongly activated in secondary SAR tissue and dependent on NIM1/NPR1/SAI1. Upon analysis of the 26 available promoters from SOM c1, the authors found that the only cis regulatory element present in all of them is the binding site for WRKY transcription factors (W boxes: TTGAC). They propose that NIM1/NPR1/SAI1 may mediate WRKY-dependent derepression of PR-1 regulon genes, or alternatively, that it may drive early expression of a subset of WRKY proteins that subsequently regulate other WRKY-dependent SAR target genes. By describing the first map of the plant defense transcriptome during SAR in Arabidopsis, Maleck and coworkers illustrate the power that this type of approach provides for the analysis of complex signaling networks.
We have used the Boolean language to represent and analyze the plant defense signaling apparatus. A preliminary and simplified representation of currently available knowledge is shown in Figure 3.
CONCLUSION
As the experimental data continue to accumulate, it becomes more and more evident that the multiplicity of environmental stimuli is transduced via a complex intracellular signaling network and leads to the activation of multiple gene expression patterns. This system gives plants a remarkable flexibility in the utilization of their genome. To further understand the molecular mechanisms involved in the control of gene expression, new ways of representing and analyzing genetic and molecular data are required.
The elements that constitute perceptive systems are connected in networks, and function as signal receptors, transducers and/or integrators, generating patterns of gene expression and protein modification. To represent such complex structures, a connectionistic description seems a convenient paradigm. This provides the interesting possibility to operationally classify genes, create functional computer simulation, and make mechanistic predictions. Used as a starting framework, a logical translation of the genetic and microarray data into a digital formalism allows an immediate dynamic representation of signaling events through the use of a computer simulation program. From such a description, functional steps can be deduced and assigned to new signaling elements that may serve as targets for further genetic investigations. Though the Boolean interpretation of genetic networks can lead only to a qualitative description, it may, however, represent a good approximation of the cellular signaling process, resulting in pragmatic qualitative predictions.
Information on the biochemical properties of the elements, together with their quantitative occurrence and localization, will allow to further refine digital models. The existence of proteins functioning as signal coincidence detectors, and the occurrence of cellular signaling machines in diverse eucaryotic systems (Kennedy, 2000; Prehoda et al., 2000) confirm the broad idea that transduction elements may statistically display simple Boolean operations in network-like structures.
Microarray data obtained with two or more pathways activated in concert typically will show the effect of interferences on gene expression. The sites and roles of crosstalks thus may be identified and in turn this will reveal the activity of particular operators under certain conditions. Future comparisons between results from microarray analysis and classical genetics surely will add more resolution to the inferred model. Therefore, it is reasonable to use the same synthetic language to describe the results of these different approaches.
Because the Boolean formalism in principle can be used to process a practically unlimited amount of information, the limitations on its applicability will be a function of the quality and quantity of available data. In other words, the accuracy and precision of a modeled signaling network will be determined by the degree of similarity in relevant experimental conditions, the thoroughness of sampling (such as no. of time points and dosage levels) for a given stimulus, and/or combination of stimuli, and the selected criteria for data analysis (e.g. control values). Thus, although a digital modeling will be a powerful tool in the simulation of signaling networks, the diagnostic interpretation of the scientist will remain important in this field of research.
ACKNOWLEDGMENT
We thank Dominique Genoud for his help finding simulator programs.
Footnotes
This work was supported by the Swiss National Science Foundation (grant no. FN 3100–055662.98). M.B.T.S.C. is a postdoctoral fellow of the Swiss Federal Commission of Scholarships for Foreign Students.
LITERATURE CITED
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong C. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Clarke JD, Liu Y, Klessig DF, Dong X. Uncoupling PR gene expression from NPR1 and bacterial resistance: characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell. 1998;10:557–569. doi: 10.1105/tpc.10.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X. Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell. 2000;12:2175–2190. doi: 10.1105/tpc.12.11.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Dietrich RA, Richberg MH. Death don't have no mercy: cell death programs in plant-microbe interactions. Plant Cell. 1996;8:1793–1807. doi: 10.1105/tpc.8.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney T, Friedrich L, Ryals J. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL. A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell. 1997;88:685–694. doi: 10.1016/s0092-8674(00)81911-x. [DOI] [PubMed] [Google Scholar]
- Durner J, Shah J, Klessig DF. Salicylic acid and disease resistance in plants. Trends Plant Sci. 1997;2:266–274. [Google Scholar]
- Faux MC, Scott JD. Molecular glue: kinase anchoring and scaffold proteins. Cell. 1996;85:9–12. doi: 10.1016/s0092-8674(00)81075-2. [DOI] [PubMed] [Google Scholar]
- Feys BJ, Parker JE. Interplay of signaling pathways in plant disease resistance. Trends Genet. 2000;16:449–455. doi: 10.1016/s0168-9525(00)02107-7. [DOI] [PubMed] [Google Scholar]
- Garrington TP, Johnson GL. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr Opin Cell Biol. 1999;11:211–218. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- Genoud T, Métraux J-P. Crosstalks in plant cell signaling: structure and function of the genetic network. Trends Plant Sci. 1999;4:503–507. doi: 10.1016/s1360-1385(99)01498-3. [DOI] [PubMed] [Google Scholar]
- Genoud T, Millar AJ, Nishizawa N, Kay SA, Schäfer E, Nagatani A, Chua N-H. An Arabidopsis mutant hypersensitive to red and far-red light signals. Plant Cell. 1998;10:889–904. doi: 10.1105/tpc.10.6.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg J, Guo A, Klessig D, Ausubel F. Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell. 1994;77:551–563. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]
- Gupta V, Willits MG, Glazebrook J. Arabidopsis thaliana EDS4 contributes to salicylic acid (SA)-dependent expression of defense responses: evidence for inhibition of jasmonic acid signaling by SA. Mol Plant-Microbe Interact. 2000;13:503–511. doi: 10.1094/MPMI.2000.13.5.503. [DOI] [PubMed] [Google Scholar]
- Jordan JD, Landau EM, Iyengar R. Signaling networks: the origins of cellular multitasking. Cell. 2000;103:193–200. doi: 10.1016/s0092-8674(00)00112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo P, Yoshioka K, Shah J, Dooner HK, Klessig DF. Resistance to turnip crinkle virus in Arabidopsis is regulated by two host genes and is salicylic acid dependent but NPR1, ethylene, and jasmonate independent. Plant Cell. 2000;12:677–690. doi: 10.1105/tpc.12.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MB. Signal-processing machines at the postsynaptic density. Science. 2000;290:750–754. doi: 10.1126/science.290.5492.750. [DOI] [PubMed] [Google Scholar]
- Kinkema M, Fan W, Dong X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell. 2000;12:2339–2350. doi: 10.1105/tpc.12.12.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bogna L, Kim L, Adam E, Harter K, Schäfer E, Nagy F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- Martienssen R. Cell death: fatal induction in plants. Curr Biol. 1997;7:R534–R537. doi: 10.1016/s0960-9822(06)00274-0. [DOI] [PubMed] [Google Scholar]
- Màs P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408:207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
- Nawrath C, Métraux J-P. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noselli S, Perrimon N. Are there close encounters between signaling pathways? Science. 2000;290:68–69. doi: 10.1126/science.290.5489.68. [DOI] [PubMed] [Google Scholar]
- Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Penninckx IAMA, Thomma BPHJ, Buchala A, Métraux J-P, Broekaert WF. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell. 1998;10:2103–2113. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, van Loon LC. Salicylic acid-independent plant defense pathways. Trends Plant Sci. 1999;4:52–58. doi: 10.1016/s1360-1385(98)01364-8. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, van Wees SCM, Hoffland E, van Pelt JA, van Loon LC. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell. 1996;8:1225–1237. doi: 10.1105/tpc.8.8.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CML, van Wees SCM, van Pelt JA, Knoester M, Laan R, Gerrits N, Weisbeek PJ, van Loon LC. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:157–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehoda KE, Scott JA, Dyche Mullins R, Lim WA. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290:801–806. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- Reymond P, Farmer EE. Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol. 1998;1:404–411. doi: 10.1016/s1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- Rogers EE, Ausubel FM. Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell. 1997;9:305–316. doi: 10.1105/tpc.9.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner H-Y, Johnson J, Delaney TP, Jesse T, Vos P et al S. The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell. 1997;9:425–439. doi: 10.1105/tpc.9.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Kachroo P, Klessig DF. The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell. 1999;11:191–206. doi: 10.1105/tpc.11.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Tsui F, Klessig DF. Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol Plant-Microbe Interact. 1997;10:60–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- Sticher L, Mauch-Mani B, Métraux J-P. Systemic acquired resistance. Annu Rev Phytopathol. 1997;35:235–270. doi: 10.1146/annurev.phyto.35.1.235. [DOI] [PubMed] [Google Scholar]
- Tang X, Xie M, Kim YJ, Zhou J, Klessig DF, Martin GB. Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell. 1999;11:15–29. doi: 10.1105/tpc.11.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teruel MN, Meyer T. Translocation and reversible localization of signaling proteins: a dynamic future for signal transduction. Cell. 2000;103:181–184. doi: 10.1016/s0092-8674(00)00109-4. [DOI] [PubMed] [Google Scholar]
- van Wees SCM, de Swart EAM, van Pelt JA, van Loon LC, Pieterse CMJ. Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:8711–8716. doi: 10.1073/pnas.130425197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volko SM, Boller T, Ausubel FM. Isolation of new Arabidopsis mutants with enhanced disease susceptibility to Pseudomonas syringae by direct screening. Genetics. 1998;149:537–548. doi: 10.1093/genetics/149.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Matsui M, Deng X-W. Positive feedback in plant signaling pathways. Trends Plant Sci. 1998;3:374–375. [Google Scholar]
- Zhang Y, Fan W, Kinkema M, Li X, Dong X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci USA. 1999;96:6523–6528. doi: 10.1073/pnas.96.11.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell. 1998;10:1021–1030. doi: 10.1105/tpc.10.6.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]