Abstract
Phytic acid (myo-inositol hexakisphosphate) is the major storage form of phosphorus in plant seeds. During germination, stored reserves are used as a source of nutrients by the plant seedling. Phytic acid is degraded by the activity of phytases to yield inositol and free phosphate. Due to the lack of phytases in the non-ruminant digestive tract, monogastric animals cannot utilize dietary phytic acid and it is excreted into manure. High phytic acid content in manure results in elevated phosphorus levels in soil and water and accompanying environmental concerns. The use of phytases to degrade seed phytic acid has potential for reducing the negative environmental impact of livestock production. A phytase was purified to electrophoretic homogeneity from cotyledons of germinated soybeans (Glycine max L. Merr.). Peptide sequence data generated from the purified enzyme facilitated the cloning of the phytase sequence (GmPhy) employing a polymerase chain reaction strategy. The introduction of GmPhy into soybean tissue culture resulted in increased phytase activity in transformed cells, which confirmed the identity of the phytase gene. It is surprising that the soybean phytase was unrelated to previously characterized microbial or maize (Zea mays) phytases, which were classified as histidine acid phosphatases. The soybean phytase sequence exhibited a high degree of similarity to purple acid phosphatases, a class of metallophosphoesterases.
Adequate levels of phosphorus are critical to the growth and development of all organisms for a range of functions such as macromolecular structure, energy generation, and metabolic regulation. The demand for phosphorus increases dramatically during periods of rapid cell growth and division, such as seed germination. Phosphorus is stored in plant seeds as phytic acid (myo-inositol hexakisphosphate) during seed development (Lott et al., 1995; Raboy, 1997). In soybeans (Glycine max L. Merr.), phytic acid is deposited in protein bodies (protein storage vacuoles) as a complex of chelated minerals and protein known as phytin (Prattley and Stanley, 1982). Maximal phytic acid levels are achieved at seed maturity immediately preceding desiccation (Raboy and Dickinson, 1987) and represent approximately 1% of the total weight in dry seeds.
Upon seed germination, hydrolysis of phytin reserves provides nutrients for the rapidly growing seedling. Dephosphorylation of phytic acid to a series of myo-inositol esters and inorganic phosphate is catalyzed by the enzyme phytase (Loewus and Murthy, 2000). Phytases have been purified from the seeds of various monocot and dicot species (Wodzinski and Ullah, 1996). Only partial purification has been achieved in most examples, which has prevented amino acid sequencing and subsequent phytase gene isolation. In soybean seed germination, a pronounced increase in phytase activity accompanies a concomitant decrease in phytic acid, with maximal phytase activity attained at approximately 10 d after germination (Gibson and Ullah, 1988).
Animal feeds are comprised primarily of plant seed components, typically from corn and soybean. For example, high protein poultry diets contain up to 50% soybean meal. Seed phytic acid is largely unavailable to monogastric animals, including poultry, swine, fish, and humans (Reddy et al., 1989; Ravindran et al., 1995). Excretion of undigested phytic acid in manure leads to the redistribution of phosphorus to the soil. An undesirable side effect of high soil phosphorus levels is the loss of this important nutrient, due to its entry into watersheds via runoff. As a limiting nutrient in aquatic environments, elevated phosphorus levels can lead to eutrophication and water quality issues (Sharpley et al., 1994). Numerous studies have indicated that the use of phytases as feed supplements can improve phosphorus availability and reduce the phosphorus content in manure (Simons et al., 1990; Cromwell et al., 1993; Jackson et al., 1996; Yi et al., 1996).
Maize (Zea mays) is the only plant for which phytase gene isolation has been reported (Maugenest et al., 1997, 1999). Two phytase genes in maize are highly homologous and tightly linked on the long arm of chromosome 3. The maize phytase cDNA was isolated by screening a seedling expression library using a polyclonal antibody. The maize phytase genes encode a homodimeric protein, with a subunit molecular mass of 38 kD, pH optimum of 4.5, and temperature optimum of 55°C. The maize coding sequence contains the amino acid sequence motif, RHGXRXP, a hallmark of histidine acid phosphatases (Ostanin et al., 1992), including fungal phytases (Ullah and Dischinger, 1993; Mitchell et al., 1997). The maize phytase lacks any additional regions of sequence homology to fungal phytases. Sequence homology to the histidine acid phosphatase active site motif was used to identify a potential candidate phytase from the Arabidopsis database (Mullaney and Ullah, 1998). The function of the Arabidiopsis sequence has yet to be defined experimentally. Here, we report the isolation of a phytase gene from soybean using a biochemical approach. Use of the isolated gene for transformation studies confirmed its identity as a phytase.
RESULTS
Purification of a Phytate-Degrading Enzyme from Cotyledons of Germinated Soybeans
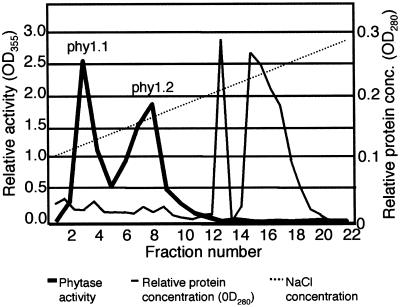
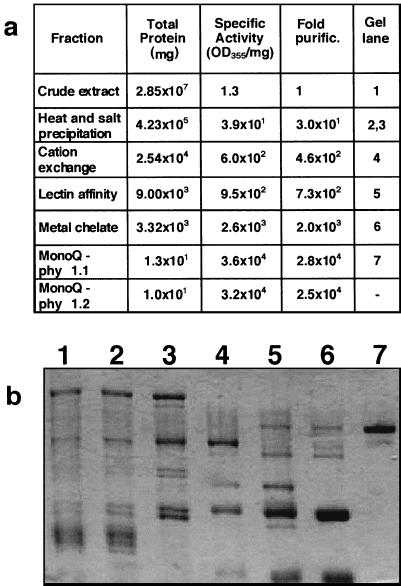
A seven-step procedure was developed for purification of a soybean phytase from cotyledons of 10-d-old seedlings. The protein was subjected to heat and salt precipitation prior to column chromatography. In the first chromatography step, nonspecific acid phosphatase activity (Ullah and Gibson, 1988) was effectively separated from the phytase peak on the cation exchange column. A concanavalin-A lectin affinity chromatography step in the phytase purification procedure took advantage of the presence of high-Man glycan side chains. This resulted in separation of the phytase from major non-glycosylated proteins with otherwise similar physical properties, including β-amylase (Totsuka and Fukazawa, 1993). This protein copurified as a contaminant in previous reports of soybean phytase isolation (Gibson and Ullah, 1990). The ability of phytase to bind to metal ions facilitated further purification of the protein using a Cu2+- charged metal chelate column. Purification to electrophoretic homogeneity was achieved with the final high-resolution anion exchange (monoQ) column (Fig. 1). Two distinct peaks of phytase activity were eluted at NaCl concentrations of approximately 128 mm (phy1.1) and 165 mm (phy1.2). The final phytase purification ranged from 22,000- to 28,000-fold in three large-scale preparations. A purification table and polyacrylamide gel of protein samples from each purification step for one representative purification are illustrated in Figure 2.
Figure 1.
Soybean phytase protein purification by monoQ anion-exchange chromatography. Two phytase activity peaks, phy1.1 and phy1.2, were observed. The heavy line shows phytase activity, the narrow line represents relative protein concentration, and the dotted line indicates the linear salt gradient used for elution.
Figure 2.
Purification of soybean phytase. a, Purification table. b, SDS-polyacrylamide electrophoresis of protein samples from protein purification steps. 1, Crude extract; 2, heat shock precipitation; 3, ammonium sulfate precipitation; 4, cation exchange chromatography; 5, lectin affinity chromatography; 6, metal chelate chromatography; 7, phy 1.1 from monoQ chromatography. Approximately 1 μg of protein was loaded in each lane.
Phy1.1 and phy1.2 were very similar in molecular mass as determined by denaturing SDS-PAGE (not shown). Edman degradation confirmed that the N-terminal amino acid sequences (15 residues) of phy1.1 and phy1.2 were identical, suggesting that the two peaks likely represent different modifications, such as glycosylation, of the same gene product. Staining with periodic acid-Schiff reagent confirmed that the purified protein was glycosylated (data not shown). The major monoQ peak, phy1.1, was the focus of further analysis and internal amino acid sequencing. The molecular mass of the purified protein was estimated to be 70 to 72 kD on an SDS polyacrylamide gel. Sephacryl-300 gel filtration provided an estimate of 130 kD for the native protein, suggesting that it may exist as a homodimer (data not shown).
Characteristics of the Purified Soybean Phytase
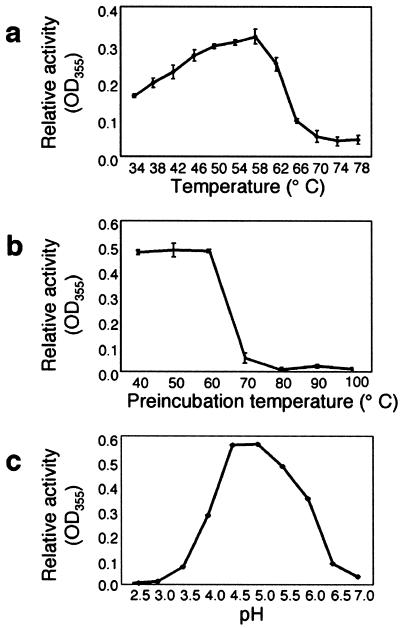
To optimize conditions for detection of phytase activity of the purified protein, assays were conducted over a range of temperature and pH. Under our assay conditions, the highest enzymatic activity was recorded at 58°C (Fig. 3a). Thermostability of the purified enzyme was tested by conducting a 10-min pre-incubation at temperatures ranging from of 40°C to 100°C prior to the standard activity assay (Fig. 3b). The results indicated that soybean phytase is stable up to 60°C, but does not regain activity after heat denaturation at temperatures greater that 60°C. The pH optimum of soybean phytase was estimated between pH 4.5 and 5.0 (Fig. 3c), similar to phytase activity data previously reported for soybean and maize (Sutardi and Buckle, 1986; Gibson and Ullah, 1988; Labore et al., 1993). The low pH optimum of phytase is characteristic of a vacuolar protein.
Figure 3.
Effects of temperature and pH on soybean phytase activity. a, Activity of purified soybean phytase was assayed at temperatures ranging from 34°C to 78°C. b, Thermostability of the purified phytase was determined by preincubating samples at temperatures ranging from 40°C to 100°C for 10 min and subsequently assaying remaining phytase activity. c, Activity of the purified soybean phytase was assayed from pH 2.5 to 7.0.
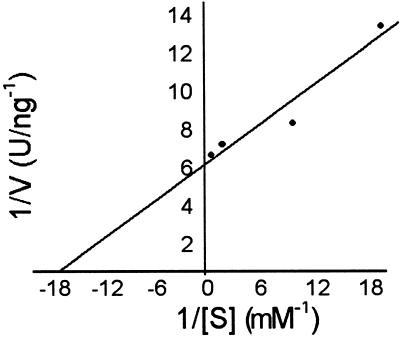
To determine the most likely in vivo substrate for the phytase enzyme, activity of the purified soybean phytase was assayed with several phosphorylated substrates at different concentrations. The Km value for phytate was 61 μm (Fig. 4), which is significantly lower than estimates obtained for ATP (1,700 μm), polyphosphate (3,300 μm), or p-nitrophenyl phosphate (pNPP; 4,300 μm). Phytase was inhibited by phytate, ATP, and polyphosphate at substrate concentrations greater than 5 mm. pNPP was not inhibitory at concentrations up to 10 mm.
Figure 4.
Kinetic analysis of soybean phytase. Activity of the purified phytase was measured for phytic acid over a range of substrate concentrations. Km was estimated from the y intercept on the Lineweaver-Burke double reciprocal plots.
Amino Acid Sequencing and Isolation of the Soybean Phytase cDNA
The N terminus of the purified protein and four internal peptides were sequenced by Edman degradation (N terminus = HIPSTLEGPFDPVTV, peptide 1 = EVGDQIYIVRQPDICPIHQRR, peptide 2 = WLERDLENVDRSITP, peptide 3 = FCWDRQPDYSAFRESSFGYGILEVK, and peptide 4 = TVSSVVQYGTSRFELVHE ARGQSLIYNQLYPFEGLQXYTSGII). Degenerate oligonucleotide primers were designed from two internal peptides for amplification of the phytase gene from soybean cDNA. Overlapping regions of the cDNA corresponding to the purified soybean phytase subsequently were amplified using multiple PCR-based strategies involving both reverse transcriptase (RT)-PCR and genomic DNA amplification (see “Materials and Methods”). The resulting soybean phytase (GmPhy) sequence contained a 1,644-bp open reading frame that could encode a protein with a predicted molecular mass of 62.3 kD.
The phytase sequence was examined for the presence of a predicted signal sequence using the program SignalP (Nielsen et al., 1997). A cleavage site was predicted between amino acids 28 and 29, which concurs with the sequence data from the N terminus of the purified protein. The presence of a signal peptide indicates that the protein should be directed to the endomembrane system for secretion or further subcellular sorting.
Phylogenetic Analysis of the GmPhy Sequence
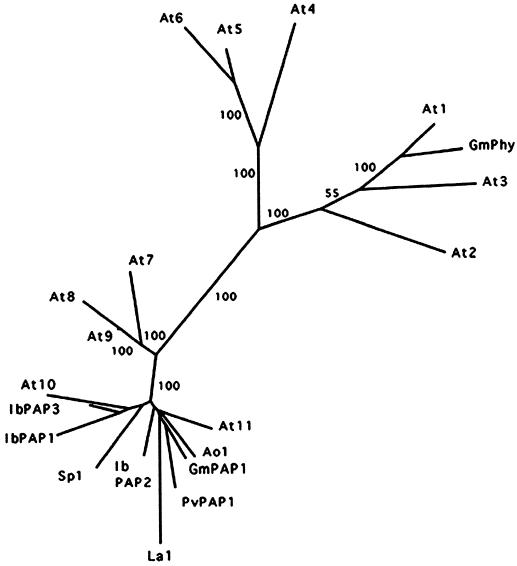
GmPhy was used as the query sequence in a BLASTX search (Altschul et al., 1997) for similarities to other sequences in GenBank. No homology was revealed to any of the previously reported phytase sequences from maize or microbes. The top scoring results included known purple acid phosphatases (PAPs) from several plants (Klabunde et al., 1994; Durmas et al., 1999; Schenk et al., 1999), as well as plant genomic and expressed sequence tag sequences predicted to encode PAPs. An alignment of the plant sequences was performed using ClustalW (Thompson et al., 1994) and the data were used to generate an un-rooted tree by the neighbor joining method in the PHYLIP program (Felsenstein, 1989). Figure 5 shows that GmPhy and three predicted PAP-like genes from Arabidopsis (At1–At3) cluster on a separate branch of the tree, which is clearly distinct from the other PAPs or PAP-like sequences. No experimentally defined plant PAPs are included in the cluster with GmPhy. The three closely related Arabidopsis sequences share 51% to 74% identity with the soybean phytase, whereas the remaining PAP and PAP-like sequences located on other branches of the tree show only 21% to 33% identity to the phytase.
Figure 5.
Phylogenetic relationships among the soybean phytase, known plant PAPs, and plant PAP-like sequences. PAP designator is used for sequences encoding proteins previously identified as PAPs. Other sequences are abbreviated by genus and species. The consensus tree was generated from 1,000 bootstrap replicates using the neighbor joining method in the PHYLIP program. Bootstrap values are indicated for major branches as percentages. GmPhy (accession no. AF272346) and three sequences from the Arabidopsis genome sequencing project (At1–At3) are located together on a separate branch of the tree, distinct from the remaining sequences. Clustering of the soybean phytase and three Arabidopsis sequences was also observed using the Protein Parsimony method in PHYLIP (not shown). The plant sequences, accession numbers, and percentage identities to the GmPhy sequence are: At1, AAF20233, 74.1%; At2, AAC04486, 51.6%; At3, CAB36834, 51.3%; At4, CAB89239, 32.3%; At5, CAB89243, 32.9%; At6, CAB89242, 32.5%; At7, CAA18136, 26.8%; At8, AAD31353, 23.4%; At9, AAF30342, 24.5%; At10, AAD26885, 27.9%; At11, AAD22297, 27.1%; IbPAP1 (from Ipomea batatas), AAF19821, 25.9%; IbPAP2, AAF19822, 28.5%; IbPAP3, CAA07280, 30.5%; PvPAP1 (from Phaseolus vulgaris), CAA04644, 27.5%; GmPAP1 (from soybean) AAF19820, 27.1%; Sp1 (Spirodela punctata), BAA92365, 26.8%; La1 (Lupinus albinus), BAA82130, 20.6%; and Ao1 (Anchusa officinalis), AAD20634, 25.7%. Where multiple identical Arabidopsis sequences were identified from the database, a single representative accession number was used in the phylogenetic tree.
The soybean phytase lacks the active site motif, RHGXRXP, previously reported for fungal and maize phytases (Ullah and Dischinger, 1993; Maugenest et al., 1999). Instead, the soybean phytase contains motifs characteristic of a large group of phosphoesterases, including the PAPs (Koonin, 1994; Zhou et al., 1994; Lohse et al., 1995). Five sequence blocks comprising two motifs (D*X[G/H*] − (Xñ) − GD*XX[Y/X] − (Xñ) − GN*H[E/D], and VXXH* − (Xñ) − GH*XH*) contain conserved metal-ligating residues (asterisks) required for enzyme function, which have been identified based on sequence comparison and structural analysis of the active site of the kidney bean (Phaseolus vulgaris) PAP (Sträter et al., 1995; Klabunde et al., 1996; Schenk et al., 2000). The complete sequences of At1 and At3 are very similar to soybean phytase and share the conserved motifs; however, At2 is divergent in critical amino acids in the metallophosphoesterase motifs. Table I shows a comparison of the signature metallophosphoesterase domains from the soybean phytase and plant PAP or PAP-like sequences.
Table I.
Comparison of GmPhy with homologous plant sequences
| Source | Accession No. | Identity | Consensus
Motifsa
|
||||
|---|---|---|---|---|---|---|---|
| I DXG | II GDXXY | III GNH(E,D) | IV VXXH | V GHXH | |||
| % | |||||||
| GmPhy | AF272346 | 100.0 | DLG | GDVTY | GNHE | VTWH | GHVH |
| At1 | AC012395 | 74.1 | DLG | GDVSY | GNHE | ASWH | GHVH |
| At2 | AC003974 | 51.6 | DLG | GGFSY | GEHE | ATWS | SHVD |
| At3 | AL035528 | 51.3 | DLG | GDLTY | GNHE | ATMH | GHVH |
| At4 | AL353912 | 32.3 | DLG | GDLSY | GNHE | AVVH | GHVH |
| At5 | AL353912 | 32.9 | DLG | GDLSY | GNHE | VLLH | GHVH |
| At6 | AL353912 | 32.5 | DLG | GDLSY | GNHE | VVMH | GHVH |
| At7 | AL022141 | 26.8 | DLG | GDLSY | GNHE | VMVH | GHVH |
| At8 | AC007212 | 23.4 | DLG | GDLSY | GNYE | VLVH | GHVH |
| At9 | AC019018 | 24.5 | DLG | GDLSY | GNHE | VLVH | GHVH |
| At10 | AC007290 | 27.9 | DLG | GDLSY | GNHE | VLVH | GHVH |
| At11 | AC007047 | 27.1 | DLG | GDISY | GNHE | VLMH | GHVH |
| IbPAP1 | AF200825 | 25.9 | DIG | GDLSY | GNHE | VLVH | GHVH |
| IbPAP2 | AF200826 | 28.5 | DLG | GDLSY | GNHE | VLMH | GHVH |
| IbPAP3 | AJ006870 | 30.5 | DLG | GDLSY | GNHE | VLMH | GHVH |
| PvPAP1 | AJ001270 | 27.5 | DLG | GDLSY | GNHE | VLMH | GHVH |
| GmPAP1 | AF200824 | 27.1 | DLG | GDLSY | GNHE | VLMH | GHVH |
| Sp1 | AB039746 | 26.8 | DLG | GDLSY | GNHE | VLMH | GHVH |
| La1 | AB023385 | 20.6 | DLG | GDLSY | GNHE | VLVH | GHVH |
| Ao1 | AF126255 | 25.7 | DLG | GDLSY | GNHE | VLMH | GHVH |
The source and accession no. of the top scoring sequences from the BLAST search, and percentage identity of the predicted amino acid sequences to the soybean phytase sequence (as determined by the Megalign sequence alignment program in DNAStar) are shown. The amino acids sequences from regions corresponding to the conserved motifs are listed for each plant sequence.
Motifs as described by Schenk et al. (2000).
Temporal Regulation of GmPhy Transcript in Soybean Cotyledons
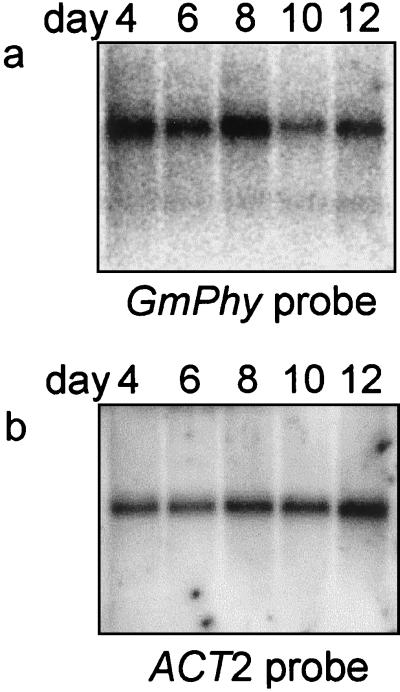
RNA-blot analysis was performed to analyze the expression of GmPhy in soybean cotyledons at 4, 6, 8, 10, and 12 d after germination (Fig. 6). A band at approximately 1,700 nucleotides was detected by hybridization at all time points tested. The highest steady state RNA levels were detected at 8 d after germination, which precedes maximal enzyme activity. The increase in phytase RNA levels after germination was not as dramatic as the increase in enzyme activity. This may indicate that phytase synthesized early in germination persists for several days in the cotyledons.
Figure 6.
Phytase RNA expression in cotyledons from germinating soybean seedlings. a, Poly(A+) RNA from soybean cotyledons (2.5 μg) was probed with a 950-bp phytase probe amplified from the 3′ end of the phytase coding sequence. b, The blot was stripped and reprobed with a labeled Arabidopsis ACT2 actin gene (An et al., 1996) to test for equivalent RNA loading.
Functional Expression of GmPhy in Transgenic Cell Cultures
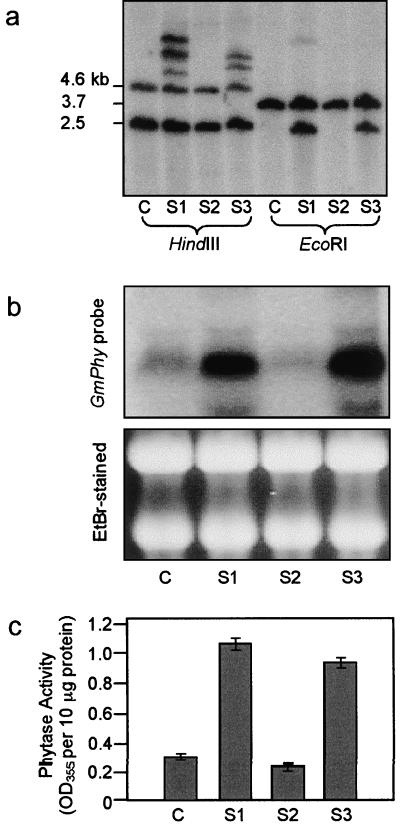
The soybean phytase coding region was cloned into a plant transformation plasmid for constitutive expression in tissue culture. Following bombardment and hygromycin selection, small calli were distinguishable from the background of dying cells. The majority of the calli failed to thrive, but a total of three transgenic culture lines (S1, S2, and S3) were recovered and analyzed. Non-transformed, wild-type cultures and transformed cell suspension cultures were analyzed for the presence of the phytase transgene using DNA gel-blot analysis (Fig. 7a). The endogenous phytase gene was observed in the control culture probed with the phytase cDNA. In the transformed cell cultures S1 and S3, multiple copies of the phytase gene were present in addition to the endogenous copy. Although the culture S2 survived hygromycin selection, no ectopic copies of the phytase sequence were detected, indicating that the sequence was lost during or following plasmid integration.
Figure 7.
Soybean phytase transgene incorporation and expression in transformed cells. a, DNA from control (C) and transgenic soybean cell cultures (S1–S3) was digested with HindIII or EcoRI and analyzed by DNA gel-blot analysis using a 950-bp probe amplified from the 3′ end of the phytase coding sequence. b, Total RNA was isolated from soybean cell cultures and subjected to RNA blot analyses using the same phytase probe as above. RNA quality and equivalent loading were assessed by loading of duplicate samples for ethidium bromide staining. c, Phytase assays were performed on extracts from control (C) and transformed (S1–S3) soybean cell cultures. Phytase activity was measured as phosphate released (absorbance at 355 nm per 10 μg of protein extract). Each data point represents the mean of three assays. se is indicated.
RNA-blot analysis was performed to compare levels of phytase expression in transformed and control cell suspension cultures (Fig. 7b). In control cells, a faint band was detected at the predicted size of approximately 1,700 nucleotides. Increased steady state levels of the phytase transcript were observed in the transformed cell cultures S1 and S3, which contained multiple copies of the recombinant phytase gene. Activity assays confirmed expression of phytase in transformed cultures S1 and S3. Cell extracts from S1 and S3 exhibited levels of phytase activity 2- to 3-fold higher than the control (Fig. 7c). In the media of these cultures, phytase levels were only slightly elevated compared with the control (data not shown), suggesting that the phytase protein was localized within the cells. As expected, only background levels of mRNA or phytase activity were detected in the culture S2, which lacked the phytase transgene.
DISCUSSION
Cloning of the soybean phytase sequence was made possible by successful purification and partial amino acid sequencing of the protein. Biochemical approaches to phytase gene discovery have been limited because of relatively few reports of purification to homogeneity for plant phytases. Although phytase activity was detectable in cotyledons from germinating soybean, the protein was not abundant. Low-phytase mRNA levels, as evidenced by RNA-blot analyses, may explain why the phytase cDNA sequence has not appeared in the soybean expressed sequence tag databases. Considering the similarity of the soybean phytase to PAPs, appearance of GmPhy in the database would have gone unrecognized as a phytase by annotation programs. A recent patent application (Morgan et al., 1997) reported purification and partial peptide sequencing of a soybean enzyme with phytase activity. Although the patent application did not report a corresponding phytase DNA sequence, the partial amino acid sequences are located in the coding sequence predicted from GmPhy. This indicates that the two enyzme activities most likely correspond to the same phytase gene. Lacking the complete DNA sequence, sequence similarities to PAPs were not identified in the patent application.
Previously reported phytases from fungi and maize are histidine acid phosphatases. Our sequence data and experimental evidence indicate that the soybean phytase belongs to an entirely different class of phosphatases. The soybean phytase exhibits a high degree of sequence similarity to PAPs. PAPs contain binuclear Fe(III)-Me(II) centers where Me is Fe, Mn, or Zn (Sträter et al., 1995; Klabunde et al., 1996; Schenk et al., 1999). The metal ions are coordinated by seven invariant amino acids, which are essential features of the active site motif found in all PAP sequences. The soybean phytase sequence harbors this motif in conserved positions relative to the kidney bean PAP, which has been characterized extensively at the structural level. In addition to PAPs, the core motif appears in a large group of diverse enzymes including Ser/Thr phosphatases, exonucleases, nucleotidases, and other phosphoesterases (for review, see Lohse et al., 1995). The ubiquitous nature of this catalytic motif suggests that the hydrolytic mechanism is very effective and, for that reason, has been preserved throughout evolution (Lohse et al., 1995).
The biological roles of PAPs are not well defined in plants or animals. Kidney bean PAP exhibited phosphatase activity on a broad range of substrates, including polyphosphate as the preferred substrate, but lacked any phytase activity (Cashikar et al., 1997). Because polyphosphate is not found naturally in kidney bean, no physiological function could be inferred from activity studies. Duff et al. (1994) suggested that nonspecific PAPs may function in phosphate transport or acquisition. A nonspecific PAP was purified from soybean (SBPAP; Lebansky et al., 1992; Schenk et al., 1999). Sequence data confirmed that the SBPAP protein is distinct from the phytase we characterized. Compared with soybean phytase, the SBPAP had a 340-fold greater affinity for ATP and a 540-fold greater affinity for pNPP (based on Km). SBPAP hydrolyzed all substrates tested; however, no physiologically relevant substrate was identified. Although our soybean phytase demonstrated activity with a variety of substrates, phytate was clearly preferred. The soybean phytase represents the first account of a PAP-like protein with an obvious metabolic function.
The Arabidopsis genome contains several sequences that are related to the soybean phytase. On a phylogenetic tree, the soybean phytase groups with three Arabidopsis database entries that are putative PAPs. Based on the sequence similarity to the soybean phytase, it is likely that one or more of these putative proteins may possess the ability to hydrolyze phytate. Phytases would be expected to play the same physiological roles in Arabidiopsis as in other plant seeds and should be required for germination and development. We currently are characterizing At1, a strong candidate ortholog of GmPhy.
Expression of the soybean phytase cDNA in plant cell culture confirmed that the isolated sequence encodes a enzyme that catalyzes phytic acid degradation. However, transgenic cell lines expressing phytase were recovered at a much lower frequency than transformed lines bombarded with the GUS reporter gene. A possible explanation could be that constitutive expression of high intracellular levels of soybean phytase may be lethal. In the surviving transgenic cultures, phytase activity levels were only two to three times higher than endogenous levels. However, soybean cell cultures expressing high phytase activity levels were previously obtained using the fungal phytase transgene (Li et al.,1997). Differential viability may be due to intracellular retention of the soybean phytase, as opposed to extracellular localization of the fungal phytase. Recovery of transgenic plants expressing the soybean phytase will provide additional insight into roles for the enzyme in the soybean plant.
The use of a fungal phytase as a feed supplement has proven effective in alleviating the negative effects of phytate in livestock diets. The fungus Aspergillus niger produces an extracellular phytase (Shieh and Ware, 1968), which is widely used in Europe as a commercial feed supplement. Numerous feeding studies with poultry, swine, and fish have demonstrated the efficacy of phytase supplementation for improving phosphorus and mineral availability (Simons et al., 1990; Cromwell et al., 1993; Jackson et al., 1996; Yi et al., 1996). However, the added expense of feed supplements can be significant. Supplementation is not currently profitable for the average livestock producer in the United States.
Reduction of phytic acid levels is an alternative strategy for improving nutrient management in animal production. Isolation and manipulation of the soybean phytase gene may provide the opportunity to alter seed phytic acid metabolism. Phytic acid-rich seeds such as corn and soybean are major components of commercial livestock feed preparations. Expression of a phytase during soybean seed development to improve phosphorus availability may alleviate problems with high levels of phosphorus excretion and the associated impact on water quality.
MATERIALS AND METHODS
Purification of a Soybean (Glycine max L. Merr. cv Williams 82) Phytase Enzyme
Phytase was purified from coyledons of germinated soybean seeds. For enzyme purification, phytase activity was assayed by measuring the amount of phosphate released by a modification of the method of Heinonen and Lahti (1981). Protein samples were assayed in a 500-μL reaction volume at 55°C in 50 mm NaOAc, pH 4.5 for 30 min using 0.5 mm phytate (dipotassium salt) as the substrate. After incubation, 1.0 mL of a freshly prepared solution containing 50% (v/v) acetone, 2.5 mm ammonium molybdate, and 1.25 n sulfuric acid was added to allow color development. Absorbance was measured at 355 nm to quantify the release of inorganic phosphate from phytate.
To prepare a crude extract for protein purification, each batch of cotyledons from 10- to 11-d-old seedlings (8 × 250 g) was homogenized in 750 mL ice-cold extraction buffer (100 mm NaOAc, pH 5.5, 20 mm CaCl2, 1 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride). Following centrifugation to remove cell debris, the supernatant was subjected to ammonium sulfate precipitation. The pellet (50%–80% [w/w] precipitation) was resuspended in buffer A (50 mm NaOAc, pH 4.3) and dialyzed overnight. Precipitate formed during dialysis was removed by centrifugation (10,000g, 20 min, 4°C) and filtered through a 1.2-μm membrane.
Chromatography steps for further purification were performed using Fast Protein Liquid Chromatography (Amersham Pharmacia Biotech, Piscataway, NJ). The dialyzed sample was applied to a carboxymethyl-sepharose cation exchange column that was equilibrated with buffer A. Bound protein was eluted with a linear gradient of 30 to 285 mm NaCl in buffer A. Active fractions were pooled and concentrated using Centriplus spin-concentrator units (Millipore, Bedford, MA). To yield a working concentration of buffer B (0.1 m NaOAc, pH 5.6, 0.5 m NaCl, 1 mm MgCl2, 1 mm MnCl2, and 1 mm CaCl2), a 5× concentrated stock was added to the sample. The sample was loaded onto a concanavalin A sepharose column equilibrated with buffer B. Bound glycoproteins were eluted with 0.5 m α-l-methylglucopyranoside in buffer B and were collected as a single fraction, which was concentrated in buffer C (50 mm NaOAc, pH 5.5, and 0.5 m NaCl) as described above. Iminodiacetic acid metal chelate resin (Amersham Pharmacia Biotech) was charged with Cu2+ ions and equilibrated with buffer C prior to application of the concanavalin A eluant. Bound protein was eluted by decreasing the pH with a gradient of 0% to 100% buffer D (50 mm NaOAc, pH 4.0, and 0.5 m NaCl) in metal chelate buffer C (pH 5.5). Active fractions were pooled and concentrated in buffer E (20 mm Tris-Acetate, pH 7.4) as described above. The phytase-containing fractions from the metal chelate column were loaded onto a mono-Q anion exchange column (HR 5/5, Amersham Pharmacia Biotech) equilibrated with buffer E. Bound protein was eluted with a linear gradient of 105 to 285 mm NaCl in buffer E. Purity of the active fractions was determined by SDS-PAGE. Protein concentration was determined by the method of Bradford (1976) using a protein assay kit (Bio-Rad, Hercules, CA). A Protein Dot-Metric kit (GenoTech, Maplewood, MO) was used to quantify 1- to 2-μL aliquots of the concentrated mono-Q fractions.
Biochemical Characterization of Phytase
Phytase activity assays were performed as described above to determine optimal pH and temperature conditions. For temperature studies, reactions were incubated at temperatures ranging from 38°C to 78°C at 4°C intervals for 30 min at pH 4.5 with 0.5 mm phytate as substrate. To measure the effect of heat treatment on activity, purified samples were preincubated for 10 min at 10°C intervals from 40°C to 100°C. Following pre-incubation, the enzyme samples were cooled to room temperature. Sodium phytate (in 50 mm NaOAc, pH 4.5) was added to a final substrate concentration of 0.5 mm and assays were performed at 55°C as described above. To achieve a pH range range from 2.5 to 7, three different buffer systems were used at 50 mm: Gly buffer for pH 2.0 and 2.5, acetate buffer for pH 3.5 to 5.5, and MES [2-(N-morpholino)-ethanesulfonic acid] for pH 6.0 to pH 7.0. For kinetic studies, activity of purified phytase was assayed using pNPP, polyphosphate, ATP, and phytate. The final substrate concentrations for each set of assays were 10, 5, 2.5, 1, 0.5, 0.1, and 0.05 mm. For determination of catalytic parameters, Lineweaver-Burke double reciprocal plots were generated (S−1 v. V−1) using the program SigmaPlot (SPSS Science, Chicago, IL).
Amino Acid Sequencing
Protein from the mono-Q fraction exhibiting the highest specific activity was sent to the University of Virginia Biomolecular Research Facility (Charlottesville) for microsequencing. Amino acid sequence data were obtained for the N terminus of the intact protein and the N termini of four internal peptides generated by digestion with lysyl peptidase. Edman chemistry was performed on a Procise protein sequencer (Applied Biosystems, Foster City, CA).
Isolation of the Soybean Phytase cDNA
The first step in the isolation of a soybean phytase cDNA was the amplification of a portion of the sequence by RT-PCR. Total RNA was extracted from cotyledons of 9-d-old 100-mg seedlings using the RNAeasy kit (Qiagen, Valencia, CA). Oligo-dT-primed cDNA was synthesized from 5 μg total RNA template using Superscript II RT (Life Technologies, Rockville, MD). For PCR amplification, the degenerate upstream primer 1 (5′-GC GAY YTN GAR AAY GTT GA-3′) and the degenerate downstream primer 2 (5′-TC NGG YTG NCK ATC CCA ACA-3′) were synthesized based on the amino acid sequences of internal peptides (peptides 2 and 3). A 406-bp soybean phytase fragment was amplified from the cDNA template using Taq Master Mix (Qiagen). The RACE procedure was used to amplify the 5′ and 3′ ends of the soybean phytase cDNA. Amplification using the 5′ RACE system (Life Technologies) yielded a truncated product. To obtain the complete sequence of the 5′ end of the soybean phytase coding region, fragments were amplified from genomic DNA. Inverse PCR was used to amplify the region upstream from the soybean phytase coding region. Soybean genomic DNA (2 μg) was digested for 1 h with 40 units XbaI (Promega, Madison, WI). After heat inactivation of the enzyme, the DNA was self-ligated in a 1-mL volume with 80 units of T4 ligase (20 units μL−1, Promega) at 16°C overnight. The ligation reaction was used as template for inverse PCR with the primers oriented in opposite directions. Taq extender (Stratagene, La Jolla, CA) was added to the reaction to facilitate synthesis of long PCR products. The resulting 1,198-bp product contained the 5′-coding region as well as 986 bp of upstream promoter sequence.
Construction of a Soybean Phytase Plant Expression Plasmid
A clone containing the entire soybean phytase coding sequence was generated by high-fidelity PCR. To circumvent apparent secondary structure near the 5′ end of the cDNA, a small 5′ portion of the coding sequence was amplified from genomic DNA and joined at a unique EcoRV site to the remainder of the coding sequence, which was amplified from cDNA. The fragment was cloned into a unique KpnI site of the final transformation vector, p35SD, a modification of plasmid construct pPHY35P (Li et al., 1997).
Particle Bombardment and Recovery of Transgenic Cultures
Soybean cell suspension cultures (courtesy of Dr. Jack M. Widholm, University of Illinois, Urbana) were maintained in Murashige and Skoog medium (basal Murashige and Skoog, Sigma Chemical Co., St. Louis, supplemented with 30 g L−1 Suc and 0.4 mg L−1 2,4-dichlorophenoxyacetic acid) in darkness in an orbital shaker at 120 rpm. Aliquots of suspension culture cells (0.6 mL) were dispersed onto sterile 3.0-cm No. 1 filters (Whatman, Clifton, NJ) placed on Murashige and Skoog plates. M10 tungsten particles (Bio-Rad) were coated with plasmid DNA and bombardment was performed as previously described (Finer et al., 1992). After bombardment, cells were incubated in a growth chamber at 28°C for 2 d. Cells were transferred weekly to medium supplemented with 50 μg mL−1 hygromycin B (Calbiochem, La Jolla, CA) for a total of 6 weeks. Surviving cell foci were transferred weekly until calli were of a sufficient size to reinitiate suspension cultures.
Analysis of Transgenic Lines
Transformed suspension culture cells were analyzed for integration and expression of the phytase transgene. DNA and RNA were isolated from exponentially growing suspension cultures 3 d after subculture. DNA was extracted as described by Dellaporta et al. (1983) and samples were digested with either HindIII or EcoRI. Total RNA was isolated from 100 mg of cells using the RNAeasy Plant Kit (Qiagen). Total RNA was isolated from cotyledons 4, 6, 8, 10, and 12 d after germination using the same kit and the Oligtex kit (Qiagen) was used to further purify poly(A+) mRNA. Electrophoresis and blotting of DNA and RNA were performed according to standard procedures. The blots were probed with a 950-bp fragment from the 3′ end of the phytase cDNA labeled with α-32P-dATP by random priming (Random Primers Kit, Life Technologies). Hybridizations and washes were carried out in UltraHyb hybridization buffer according to the manufacturer's specifications (Ambion, Austin, TX). To verify equal loading and sample integrity, the mRNA blot was stripped and reprobed with the labeled Arabidopsis ACT2 actin gene (An et al., 1996). For phytase assays, cells and culture media were collected 5 d after transfer by vacuum filtration. Cells (0.5 g) were homogenized in 2 mL cold extraction buffer (50 mm NaOAc, pH 4.5, 1 mm dithiothreitol, and 20 mm CaCl2) for 1 min on ice. Homogenates were centrifuged for 15 min at 10,000 rpm in a microcentrifuge to remove debris. Phytase assays were performed as described above.
ACKNOWLEDGMENTS
We thank Regina Hanlon for technical assistance and Peter Kennelly, John McDowell, and Brenda Winkel for critical review of the manuscript.
Footnotes
This work was supported in part by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program.
LITERATURE CITED
- Altschul S, Madden T, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y-Q, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB. Strong, constitutive expression of Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 1996;10:107–121. doi: 10.1046/j.1365-313x.1996.10010107.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cashikar AG, Kumaresan R, Rao NM. Biochemical characterization and subcellular localization of the red kidney bean purple acid phosphatase. Plant Physiol. 1997;114:907–915. doi: 10.1104/pp.114.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromwell GL, Stahly TS, Coffey RD, Monegue HJ, Randolph JH. Efficacy of low-activity, microbial phytase in improving the bioavailability of phosphorus in corn-soybean meal diets for pigs. J Anim Sci. 1993;71:1831–1840. doi: 10.2527/1993.7171831x. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minpreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Duff SMG, Sarath G, Plaxton WC. The role of acid phosphatases in plant phosphorus metabolism. Physiol Plant. 1994;90:791–800. [Google Scholar]
- Durmas A, Eicken C, Sift BH, Kratel A, Kappl R, Huttermann J, Krebs B. The active site of purple acid phosphatase from sweet potatoes (Ipomoea batatas): metal content and spectroscopic chracterization. Eur J Biochem. 1999;260:709–716. doi: 10.1046/j.1432-1327.1999.00230.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- Finer JJ, Vain P, Jones MW, McMullen MD. Development of particle inflow gun for DNA delivery to plant cells. Plant Cell Rep. 1992;11:323–328. doi: 10.1007/BF00233358. [DOI] [PubMed] [Google Scholar]
- Gibson DM, Ullah AHJ. Purification and characterization of phytase from germinating soybean seeds. Arch Biochem Biophys. 1988;260:503–513. doi: 10.1016/0003-9861(88)90475-4. [DOI] [PubMed] [Google Scholar]
- Gibson DM, Ullah AHJ. Phytases and their action on phytic acid. In: Morré DJ, Boss WF, Loewus FA, editors. Inositol Metabolism in Plants. New York: Wiley-Liss; 1990. pp. 77–92. [Google Scholar]
- Heinonen JK, Lahti RJ. A new and convenient colorimetric determination of inorganic orthophosphate and its application to the assay of inorganic pyrophosphate. Anal Biochem. 1981;113:313–317. doi: 10.1016/0003-2697(81)90082-8. [DOI] [PubMed] [Google Scholar]
- Jackson LS, Li MH, Robinson EH. Use of microbial phytase in channel catfish Ictalurus punctatusdiets to improve utilization of phytate phosphorus. J World Aquaculture. 1996;27:309–313. [Google Scholar]
- Klabunde T, Stahl B, Suerbaum H, Hahner S, Karas M, Hillenkamp F, Krebs B, Witzel H. The amino acid sequence of the red kidney bean Fe(III)-Zn(II) purple acid phosphatase. Eur J Biochem. 1994;226:369–375. doi: 10.1111/j.1432-1033.1994.tb20061.x. [DOI] [PubMed] [Google Scholar]
- Klabunde T, Sträter N, Fröhlich R, Witzel H, Krebs B. Mechanism of Fe(III)-Zn(II) purple acid phosphatase based on crystal structures. J Mol Biol. 1996;259:737–748. doi: 10.1006/jmbi.1996.0354. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Conserved sequence pattern in a wide variety of phosphoesterases. Protein Sci. 1994;3:356–358. doi: 10.1002/pro.5560030218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labore AM, Gagnon J, Lescure AM. Purification and characterization of a phytase (myo-inositol-hexakisphosphate phosphohydrolase) accumulated in maize (Zea mays) seedlings during germination. Biochem J. 1993;295:413–419. doi: 10.1042/bj2950413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hegeman CE, Hanlon RW, Lacy GH, Denbow DM, Grabau EA. Secretion of active recombinant phytase from soybean cell-suspension cultures. Plant Physiol. 1997;114:1103–1111. doi: 10.1104/pp.114.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebansky BR, McKnight TD, Griffing LR. Purification and characterization of a secreted purple phosphatase from soybean suspension cultures. Plant Physiol. 1992;99:391–395. doi: 10.1104/pp.99.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus FA, Murthy PPN. Myo-Inositol metabolism in plants. Plant Sci. 2000;150:1–19. [Google Scholar]
- Lohse DL, Denu JM, Dixon JE. Insights derived from the structures of the Ser/Thr phosphatases calcineurin and protein phosphatase 1. Structure. 1995;3:987–990. doi: 10.1016/s0969-2126(01)00234-9. [DOI] [PubMed] [Google Scholar]
- Lott JNA, Greenwood JS, Batten BD. Mechanisms and regulation of mineral nutrient storage during seed development. In: Kigel J, Galili G, editors. Seed Development and Germination. New York: Marcel Dekker; 1995. , 215–235. [Google Scholar]
- Maugenest S, Martinez I, Godin B, Perez P, Lescure A-M. Structure of two maize phytase genes and their spatio-temporal expression during seedling development. Plant Mol Biol. 1999;39:503–514. doi: 10.1023/a:1006131506193. [DOI] [PubMed] [Google Scholar]
- Maugenest S, Martinez I, Lescure A-M. Cloning and characterization of a cDNA encoding a maize seedling phytase. Biochem J. 1997;322:511–517. doi: 10.1042/bj3220511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DB, Vogel K, Weimann BJ, Pasamontes L, van Loon APBM. The phytase subfamily of histidine acid phosphatases: isolation of genes for two novel phytases from the fungi Aspergillus terreus and Myceliophthora thermophila. Microbiology. 1997;143:245–252. doi: 10.1099/00221287-143-1-245. [DOI] [PubMed] [Google Scholar]
- Morgan AJ, Hessing M, Sleijster-Selis HE, inventors. May 14, 1998. Phytase from germinated soybeans. International Patent Application No. WO 98/20139
- Mullaney EJ, Ullah AJH. Identification of a histidine acid phosphatase (phyA)-like gene in Arabidopsis thaliana. Biochem Biophys Res Commun. 1998;251:252–255. doi: 10.1006/bbrc.1998.9452. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, Heijne GV. Identification of prokaryotic and eukaryotic signal pepetides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Ostanin K, Harms EH, Stevis PE, Kuciel R, Zhou MM, Van Etten RL. Overexpression, site-directed mutagenesis, and mechanism of Escherichia coliacid phosphatase. J Biol Chem. 1992;32:22830–22836. [PubMed] [Google Scholar]
- Prattley CA, Stanley DW. Protein-phytate interactions in soybean: I. Localization of phytate in protein bodies and globoids. J Food Biochem. 1982;6:243–253. [Google Scholar]
- Raboy V. Accumulation and storage of phosphate and minerals. In: Larkins B A, Vasil IK, editors. Cellular and Molecular Biology of Plant Seed Development. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 441–477. [Google Scholar]
- Raboy V, Dickinson DB. The timing and rate of phytic acid accumulation in developing soybean seeds. Plant Physiol. 1987;85:841–844. doi: 10.1104/pp.85.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran V, Bryden WL, Kornegay ET. Phytates: occurrence, bioavailability and implications in poultry nutrition. Poultry Avian Biol Rev. 1995;6:125–143. [Google Scholar]
- Reddy NR, Pierson MD, Sathe SK, Salunkhe DK. Phytates in Cereals and Legumes. Boca Raton, FL: CRC Press; 1989. [Google Scholar]
- Schenk G, Ge Y, Carrington LE, Wynne CJ, Searle IR, Carroll BJ, Hamilton S, deJersey J. Binuclear metal centers in plant purple acid phosphatases: Fe-Mn in sweet potato and Fe-Zn in soybean. Arch Biochem Biophys. 1999;370:183–189. doi: 10.1006/abbi.1999.1407. [DOI] [PubMed] [Google Scholar]
- Schenk G, Guddat LW, Ge Y, Carrington LE, Hume DA, Hamilton S, de Jersey J. Identification of mammalian-like purple acid phosphatases in a wide range of plants. Gene. 2000;250:117–125. doi: 10.1016/s0378-1119(00)00186-4. [DOI] [PubMed] [Google Scholar]
- Sharpley AN, Chapra SC, Wedepohl R, Sims JT, Daniel TC, Reddy KR. Managing agricultural phosphorus for protection of surface waters: issues and options. J Environ Qual. 1994;23:437–451. [Google Scholar]
- Shieh TR, Ware JH. Survey of microorganisms for the production of extracellular phytase. Appl Microbiol. 1968;16:1348–1351. doi: 10.1128/am.16.9.1348-1351.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons PCM, Versteegh AJ, Jongbloed AW, Kemme PA, Slump P, Bos KD, Wolters GE, Buedeker RF, Verschoor GJ. Improvement of phosphorus availability by microbial phytase in broilers and pigs. Br J Nutr. 1990;64:525–540. doi: 10.1079/bjn19900052. [DOI] [PubMed] [Google Scholar]
- Sträter N, Klabunde T, Tucker P, Witzel H, Krebs B. Crystal structure of a purple acid phosphatase containing a dinuclear Fe(III)-Zn(II) active site. Science. 1995;268:1489–1492. doi: 10.1126/science.7770774. [DOI] [PubMed] [Google Scholar]
- Sutardi, Buckle KA. The characteristics of soybean phytase. J Food Biochem. 1986;10:197–216. [Google Scholar]
- Thompson JD, Higgins D, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsuka A, Fukazawa C. Expression and mutation of soybean beta-amylase in Escherichia coli. Eur J Biochem. 1993;214:787–794. doi: 10.1111/j.1432-1033.1993.tb17981.x. [DOI] [PubMed] [Google Scholar]
- Ullah AHJ, Dischinger HC., Jr Aspergillus ficuumphytase: complete primary structure elucidation by chemical sequencing. Biochem Biophys Res Commun. 1993;192:747–753. doi: 10.1006/bbrc.1993.1477. [DOI] [PubMed] [Google Scholar]
- Ullah AHJ, Gibson DM. Purification and characterization of acid phosphatase from cotyledons of germinating soybean seeds. Arch Biochem Biophys. 1988;260:514–520. doi: 10.1016/0003-9861(88)90476-6. [DOI] [PubMed] [Google Scholar]
- Wodzinski RJ, Ullah AHJ. Phytases. Adv Appl Microbiol. 1996;42:263–302. doi: 10.1016/s0065-2164(08)70375-7. [DOI] [PubMed] [Google Scholar]
- Yi Z, Kornegay ET, Ravindran V, Denbow DM. Improving phytate phosphorus availability in corn and soybean meal for broilers using microbial phytase and calculation of phosphorus equivalency values for phytase. Poultry Sci. 1996;75:240–249. doi: 10.3382/ps.0750240. [DOI] [PubMed] [Google Scholar]
- Zhou S, Clemens JC, Stone RL, Dixon JE. Mutational analysis of a Ser/Thr phosphatase. J Biol Chem. 1994;269:26234–26238. [PubMed] [Google Scholar]