Abstract
The expression of alternative oxidase (Aox) and uncoupling proteins (Ucp) was investigated during ripening in mango (Mangifera indica) and compared with the expression of peroxisomal thiolase, a previously described ripening marker in mango. The multigene family for the Aox in mango was expressed differentially during ripening. Abundance of Aox message and protein both peaked at the ripe stage. Expression of the single gene for the Ucp peaked at the turning stage and the protein abundance peaked at the ripe stage. Proteins of the cytochrome chain peaked at the mature stage of ripening. The pattern of protein accumulation suggested that increases in cytochrome chain components played an important role in facilitating the climacteric burst of respiration and that the Aox and Ucp may play a role in post-climacteric senescent processes. Because both message and protein for the Aox and Ucp increased in a similar pattern, it suggests that their expression is not controlled in a reciprocal manner but may be active simultaneously.
The plant mitochondrial electron transport chain displays a remarkable plasticity with respect to the flow of electrons from a substrate. Oxidation of NADH can occur via the rotenone sensitive multisubunit, proton-pumping NADH ubiquinone reductase (complex I) or via the rotenone insensitive internal or external NAD(P)H dehydrogenases. Oxidation of ubiquinol can occur via the so-called cytochrome chain and terminate at cytochrome c oxidase, which is sensitive to cyanide. In addition, ubiquinol may be oxidized by the alternative oxidase (Aox; Moore and Siedow, 1991). A more recent addition to the complexity of the plant mitochondrial respiratory chain is the presence of uncoupling proteins (Ucp), which are proposed to allow the reentry of protons from the intermembrane space to the matrix, bypassing the ATP synthase complex (Vercesi et al., 1995; Laloi et al., 1997). Hence, although by different mechanisms, both Aox and Ucp are termed non-phosphorylating bypasses of respiration. These activities are not unique to plants (Douce and Neuburger, 1989; Ricquier and Bouillaud, 2000) but their expression profile in various plant tissues during development suggests differential regulation (Laloi et al., 1997; Vanlerberghe and McIntosh, 1997; Kowaltowski et al., 1998; Maia et al., 1998; McCabe et al., 1998; Ito, 1999; Nantes et al., 1999; Casolo et al., 2000; Pastore et al., 2000). The function of these activities in plant respiration and metabolism is still largely unknown.
The Aox is the best characterized of the non-phosphorylating bypasses in plants. It consists of an approximately 34-kD protein that is active as a dimer (Vanlerberghe and McIntosh, 1997). Extensive resources include: (a) a monoclonal antibody that cross-reacts with all known Aox proteins (Elthon et al., 1989); (b) cDNA and genomic clones from a variety of plants and fungi (Vanlerberghe and McIntosh, 1997); (c) activation via α-keto acids and redox state (Millar et al., 1993; Umbach and Siedow, 2000); (d) expression of various mutants in Escherichia coli, yeast, and plants (Albury et al., 1998; Berthold, 1998; Djajanegara et al., 1999); and (e) antisense and overexpressing plants have all been and are being used in efforts to understand the role of this activity (Vanlerberghe et al., 1997; Maxwell et al., 1999). Except in thermogenic plants, where the Aox activity assists pollination (Moore and Siedow, 1991), a defined role does not exist. Its capacity to prevent the production of reactive oxygen species (ROS) that could result from a variety of stresses has been proposed by several independent studies (Wagner, 1995; Millar and Day, 1996, 1997; Maxwell et al., 1999).
The Ucp of plant mitochondria is an approximately 32-kD protein, with six transmembrane regions. The gene sequence was first reported from plants in 1997 (Laloi et al., 1997), but its role has not been defined in plants. On the basis of sequence homology to the well-characterized mammalian Ucp1 of brown adipose tissue, it is proposed to be an uncoupling protein. Such proteins uncouple ATP synthesis from respiration and such an activity has been reported from plant mitochondria (Vercesi et al., 1995; Jezek et al., 1997; Laloi et al., 1997). It is ostensibly stimulated by free fatty acids that inhibit Aox and is proposed to supplement Aox activity when Aox is inhibited or absent (Sluse et al., 1998; Almeida et al., 1999; Jarmuszkiewicz et al., 2000a). Like Aox, its capacity to reduce the production of ROS in wheat (Triticum aestivum), potato (Solanum tuberosum), and pea (Pisum sativum) and its induction by cold in potato, Arabidopsis, and skunk cabbage (Symplocarpus foetidus) have been reported (Laloi et al., 1997; Kowaltowski et al., 1998; Maia et al., 1998; Ito, 1999; Nantes et al., 1999; Casolo et al., 2000; Murayama and Handa, 2000; Pastore et al., 2000).
Climacteric fruits are defined by a respiratory burst, which can be as high as 6-fold in mango (Mangifera indica) and other tropical fruits (Rhodes, 1970; Biale and Young, 1981). Such respiration has been linked to the non-phosphorylating bypasses due to its insensitivity to cyanide (Solomos and Laties, 1974; Kumar et al., 1990) and typically occurs at the mature and turning stages of fruit development (Biale and Young, 1981; Jobin-Decor, 1988; Holmes et al., 1990). In mango, it has been shown that abundance of the Aox protein increases during ripening, as does expression at the level of mRNA (Cruz-Hernandez and Gomez-Lim, 1995). However, previous reports suggest that during postharvest ripening in tomato (Lycopersicon esculentum Mill.), the Aox protein and then the Ucp protein decrease early in the ripening process (Almeida et al., 1999; Costa et al., 1999). It is well known that tomato is climacteric (Rhodes, 1970; Biale and Young, 1981; Almeida et al., 1999), but such results appear to contradict the presence of a respiratory burst.
Because mango undergoes a large and well-defined respiratory burst, we investigated the expression of the Aox and Ucp at both a message level and protein abundance during fruit ripening. In addition, we analyzed the presence of several other respiratory chain components by western-blot analysis. We used mangoes that were naturally ripened on the plant, quantitative analysis of message levels using a real-time PCR approach and of protein levels using digital detection of signal, which overcomes the inherent problems of the narrow range of x-ray film (Laskey, 1980; Murcha et al., 1999).
RESULTS
Identification of Aox and Ucp Genes from Mango
A priority in studying the expression of Aox and Ucp during mango fruit ripening was to determine the copy number of each of these genes. Using primers designed to regions conserved in plant, fungal, and protist Aox and previously used to characterize a multigene family in soybean (Glycine max L. Merr.; Whelan et al., 1996), we identified four distinct sequences in mango. In total, 40 Aox inserts of 234 to 432 bp (excluding primers) depending on the primer combination, were sequenced from mango, so it is unlikely that any other Aox genes are present. Comparison with other Aox sequences indicated that three were of the Aox1 type, entitled MnAox1a, b, and c (accession nos. AF329895, AF329896, and AF329897, respectively). The fourth gene was identical to that previously described and clearly an Aox2 type, entitled MnAox2 (accession no. AF329898, Cruz-Hernandez and Gomez-Lim, 1995).
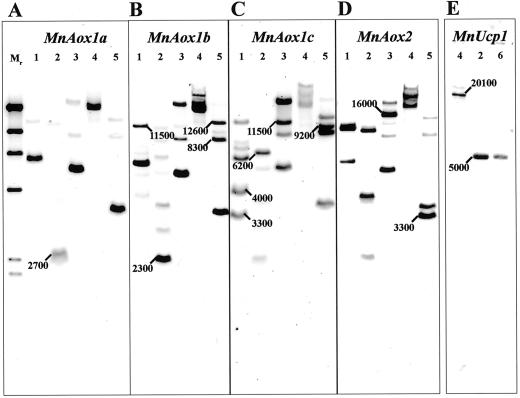
To confirm the gene copy number, Southern hybridization was carried out with the four Aox fragments against mango DNA cut with various restriction enzymes. To ensure that the results were comparable, each digest was set up in a master reaction and loaded onto an agarose gel in quadruplicate. The gel was blotted, cut into four strips, and each probed with an equal amount of digoxigenin-11-dUTP (DIG)-labeled probe. Although washed at a high stringency, cross-hybridizations between the different fragments were evident (Fig. 1, A–D). This was not unexpected because the homology between the Aox1-type inserts was 68% to 83%. However, the pattern and intensity of bands gave each insert a unique profile. The most similar profile was between MnAox1a and MnAox1b, which were distinguished by a double digest (Fig. 1, A and B, lane 5).
Figure 1.
Southern-blot analyses of mango genomic DNA hybridized with DIG-labeled DNA clones of MnAox1a, 1b, 1c, and 2 (A–D) or MnUcp1 (E). Genomic DNA isolated from mango leaves was digested (3 μg per lane but in a pooled sample for MnAox) with the restriction enzyme(s): 1, Eco RI; 2, Hind III; 3, Eco RV; 4, Bam HI; 5, Eco RV/Bam HI; and 6, Bam HI/Hind III. DNA was electrophoresed on 0.8% (w/v) agarose gel against a DIG-labeled molecular mass marker (Mr). After blotting to Nylon+ membrane and hybridization with a known amount of the relevant DIG-labeled clone, blots were washed at a high stringency (0.1× SSC and 0.1% [w/v] SDS at 68 °C), detected, and then visualized using an LAS-1000. Diagnostic restriction fragments are indicated with their apparent molecular mass (base pairs).
A similar approach, with primers designed to the putative Ucp, yielded only a single gene from mango. A sequence-invariable genomic fragment of 1,596 bp was obtained in all of the 10 inserts sequenced, entitled MnUcp1 (accession no. AF329899). The use of primers designed to other regions of the Ucp failed to amplify further Ucp homologs from mango. Southern analysis of mango genomic DNA with this fragment yielded results consistent with a single gene copy (Fig. 1E). Multiple genes have been identified in Arabidopsis with 64% identity (Watanabe et al., 1999). Because the primers used would predictably amplify these genes and did amplify multiple genes from soybean (M.J. Considine and J. Whelan, unpublished data), we concluded that only a single locus exists in mango. MnUcp1 shared 78% identity with the potato StUcp1 (Laloi et al., 1997).
Validation of the Real-Time PCR Method to Measure Relative Transcript Abundance
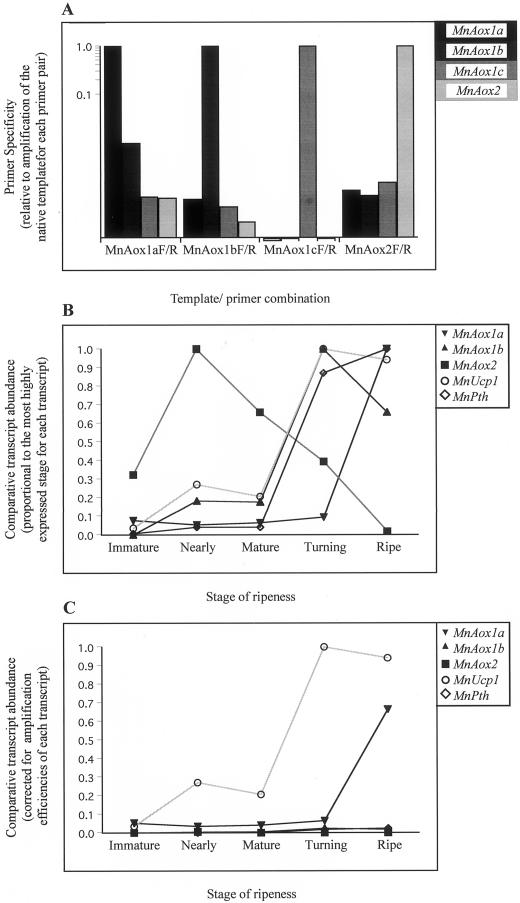
The lack of cross-reactivity between MnAox primers was established by amplification from the nonspecific templates, calculated relative to a standard curve of the specific template and subjected to the appropriate analysis of variance. The magnitude of the resulting variance ratio (vr, also known as an F statistic) represents a confidence in the treatment effect. {If the null hypothesis were true, vr would equal 0, whereas the more evidence [data] we have that disproves the null hypothesis, the closer vr would be to infinity. Given the degrees of freedom [df] concerning the treatment and residual effects, we can estimate the probability [p] of obtaining our particular results [vr] if the null hypothesis were true. The significant p that is conventionally used is P < 0.05, so variables were quoted as significant [P < 0.05] or not. Regarding this experiment, dftemplate = 3 (ntemplate − 1) and dfresidual = 8 [(nreplicates − 1) × ntemplate)], where nreplicates = 3.} For each given MnAox primer set, vr was >1 × 104, which was significant at P < 0.05 (data not shown). Hence, the probability that relative amplification from each MnAox template was equal to 1 (null hypothesis) was <0.05. With this confidence in the data, greater than a 1,000-fold discrimination was observed between most MnAox genes (Fig. 2A). The exception was between MnAox1a and MnAox1b, where greater than 100-fold discrimination was achieved.
Figure 2.
Validation and summary of results of the real-time PCR approach to assay developmental expression of select genes throughout ripening of mango. A, Lack of cross-reactivity between MnAox primers was first established and the data analyzed for significance (refer to the text). B, Total RNA (1 μg) from mango fruit of five stages of ripeness was then reverse transcribed two independent times. Standard curves of cDNA from the stage of ripeness with greatest expression for each transcript were used to calculate relative abundance at the other stages. Unedited data was analyzed by the appropriate analysis of variance to calculate variance ratios (vr) and t statistics (refer to Table IB). The data was presented individually for each transcript where the stage of ripening that displayed maximal expression was set to 1. C, Corrections were made for differences in amplification efficiency (refer to Table IA) between primer sets for the various transcripts and then data were expressed relative to the greatest corrected expression value.
Determination of the efficiency of each primer set enabled the comparison between transcripts. MnAox2 was set to 1 because it was amplified most efficiently and all other transcripts were calculated relative to this value (Table IA). Analysis of variance showed a vr of 78.0 (dftranscript = 5 and dfresidual = 18), which was significant at P < 0.05. Large differences in amplification efficiency were observed, due to small differences in amplification being amplified exponentially. The values obtained were used to calculate relative transcript abundance between the genes studied.
Table I.
A summary of statistical data used in comparing the expression of MnAox, MnUcp, and MnPth genes throughout the ripening of mango fruit
| Introduction | MnAox1a | MnAox1b | MnAox1c | MnAox2 | MnUcp1 | MnPth |
|---|---|---|---|---|---|---|
| A | ||||||
| Ar | 0.006 | 0.176 | 0.012 | 1.000 | 0.004 | 0.173 |
| B | ||||||
| vr (rn) | 12.56a | 0.01 | 3.69 | 5.75a | 0.00 | |
| vr (stg) | 124.45a | 16.74a | 164.67a | 91.93a | 116.13a | |
| sed | 0.05 | 0.14 | 0.04 | 0.07 | 0.07 | |
| df (rn; stg; ɛ) | 1; 4; 14 | 1; 3; 9 | 1; 4; 13 | 1; 4; 12 | 1; 4; 13 |
A, The amplification efficiency (Ar) of each primer set from the specific template was calculated relative to the most efficient one. PCR using the LightCycler was performed on 0.01 fmol each template standard using specific primers. The amplicon from MnAox2 reached exponential amplification at the earliest cycle. Amplification efficiencies of the other primer sets were calculated relative to a standard curve of MnAox2. Analysis of variance, using a completely randomized design was performed before normalising data so that the mean of MnAox2 was equal to 1. The variance ratio (vr = 78.0), with 5 and 18 degrees of freedom (df) concerning the primer set and residual, respectively, was statistically significant (P < 0.05). The se of the difference between means (sed) was 0.066. B, Unedited data from the gene expression experiments were treated as having a randomized block design, with PCR-run (rn) as the block factor, stage of ripening (stg) as the treatment factor, and relative transcript abundance as the response variate. The residual of the model was denoted ɛ. Given the se of the difference of means (sed) and the relevant degrees of freedom (df), t statistics can be calculated to test for difference between individual means.
From the analysis of variance, significant variance ratios (vr with P < 0.05).
Expression of Aox and Ucp Genes during Fruit Ripening
We designed primers to amplify a transcript of the mango peroxisomal thiolase (MnPth), which is up-regulated throughout mango and tomato ripening and thus provides a suitable marker for ripening (Bojorquez and Gomez-Lim, 1995). The vr showed a significant effect of stage of ripeness for each transcript (Table IB). Transcript abundance of the mango peroxisomal thiolase (MnPth, Fig. 2B) was comparable to that previously described (Bojorquez and Gomez-Lim, 1995). This validated both the real-time PCR approach and the fruit tissue used, affirming that the fruit underwent a normal ripening pattern and were scored for stage of ripeness appropriately.
When the changes were normalized to the maximum expression for each gene, it was observed that the MnAox genes changed differentially during ripening (Fig. 2B). MnAox2, which has previously been reported to be up-regulated, was seen to peak at the nearly stage and decline steadily toward a minimum at the ripe stage. MnAox1a was very low from immature to turning and then increased almost 10-fold by the ripe stage. Likewise, MnAox1b increased 5-fold during ripening but peaked at the turning stage before decreasing slightly at the ripe stage. MnAox1c was not expressed at any stage. MnUcp1 showed a very similar profile to MnAox1b, increasing 5-fold from mature to turning with a small decrease seen at the ripe stage. When gene expression was corrected for amplification efficiency, as outlined in Table IA, MnUcp1 was the most abundant gene transcript followed by MnAox1a (Fig. 2C). These two genes were expressed at a 10-fold higher level than the other genes examined (Fig. 2C). Both were up-regulated but MnUcp1 peaked at the turning stage, whereas MnAox1a increased at the ripe stage (Fig. 2C). As a comparison, we carried out a similar analysis of leaf RNA whereby it was apparent that MnAox2 expression was much higher in leaf compared with ripening fruit but MnAox1c remained undetectable (data not shown).
Abundance of Aox, Ucp, and Other Respiratory Proteins in Ripening
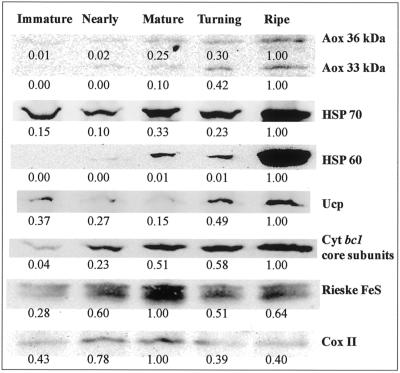
Several mitochondrial proteins were analyzed by western-blot analysis during ripening using total protein extracts from fruit tissue and a typical pattern of protein abundance is shown (Fig. 3). For Aox and Ucp, the observed amount of protein followed the transcript abundance by peaking at the ripe stage. Analysis of the respiratory chain components for subunit 2 of cytochrome c oxidase and the Rieske FeS protein of the cytochrome bc1 complex indicated that these proteins peaked at the mature stage and had decreased 2-fold by the ripe stage. The core subunits of the cytochrome bc1 complex peaked at the ripe stage. The difference in the pattern of abundance between the Rieske FeS protein, and the core subunits of the cytochrome bc1 complex may be due to the different roles of these proteins in mitochondria. The Rieske FeS protein is directly involved in electron transport, whereas the core subunits are necessary for the structural integrity of the cytochrome bc1 and are involved in the processing of mitochondrial precursor protein upon import into mitochondria (Glaser and Dessi, 1999). The pattern observed with the Rieske FeS was similar to cytochrome oxidase subunit 2, another electron carrying component of the cytochrome chain. The core proteins of the cytochrome bc1 complex peak at the ripe stage, as do Aox and Ucp. This may be due to the fact that the core subunits of cytochrome bc1 are necessary to remove the mitochondrial targeting signal of Aox and other mitochondrial proteins that may be accumulating at this stage (with the exception of Ucp, which has no cleavable N-terminal presequence; Bouillaud et al., 1986; Laloi, 1999). HSP-60 and HSP-70 were also seen to peak at the ripe stage, most dramatically for HSP-60, which displayed a 100-fold increase between the turning and ripe stages. However, the HSP-60 and HSP-70 antibodies are not specific to the mitochondrial isoforms and may have cross-reacted with the cytosolic and plastid isoforms.
Figure 3.
Immunoblots of total protein from ripening mango fruit using antibodies raised against various mitochondrial proteins. Protein was loaded in lanes for SDS-PAGE on an equal mass (wet weight) basis. Proteins cross-reacting with the various antibodies were visualized using an LAS-1000 and quantitated digitally, where the highest intensity band of the profile was set to 1 and others calculated relative to that value.
DISCUSSION
The roles of Aox and Ucp in plant metabolism are unclear. Both reduce, by different means, the amount of ATP synthesized by mitochondria per oxidation of substrate. In the present investigation, we have demonstrated the up-regulation of both Aox and Ucp after the climacteric ripening phase in mango fruit that were ripened and scored according to industry standards and displayed similar patterns in Aox and peroxisomal thiolase expression as previously reported (Bojorquez and Gomez-Lim, 1995; Cruz-Hernandez and Gomez-Lim, 1995). In contrast, electron transport components of the cytochrome chain peaked at the mature, preclimacteric stage. We suggest that the respiratory burst may be supported by an increase in cytochrome chain components, with some uncoupling possibly facilitated by Ucp. The role of Aox and Ucp may be to maintain respiration after a respiratory burst in the presence of high levels of ATP, and thus allow senescence to proceed.
Independent reports previously have suggested the induction of Aox and Ucp by several different stress conditions, leading to the proposal that they are involved in preventing the production of ROS (Wagner, 1995; Millar and Day, 1996, 1997; Kowaltowski et al., 1998; Maia et al., 1998; Maxwell et al., 1999; Nantes et al., 1999; Casolo et al., 2000; Pastore et al., 2000). However, using cold as a stress it was reported that Aox was induced in mung bean (Vigna radiata L. R. Wilcz) but not in soybean (Gonzalez-Meler et al., 1999). Likewise, induction of Ucp by cold treatment has been reported in potato, Arabidopsis, and skunk cabbage but in wheat, Ucp was unaffected by cold (Laloi et al., 1997; Maia et al., 1998; Ito, 1999; Murayama and Handa, 2000). No consensus on the role of these activities has been drawn across species.
Climacteric respiration in ripening fruit has been traditionally linked to the alternative pathway, due to its insensitivity to cyanide, which inhibits the cytochrome c oxidase (Solomos and Laties, 1974; Kumar et al., 1990). However, it is known now that the phosphorylating and non-phosphorylating pathways can switch in the presence of inhibitors (Day et al., 1996). In fact, the climacteric in mango appears to be facilitated by up-regulation of some cytochrome chain components. Although Aox and Ucp proteins were significantly up-regulated at the post-climacteric stages, possibly reflecting the prior increases in message levels observed and contributing to maintaining the respiratory burst, they may also play other roles and facilitate senescence. One possible role is the maintenance of electron transport when the cytochrome chain may be declining in activity due to developmental regulation as evidenced by the abundance of various proteins, or limited by adenylate control produced in the respiratory burst or due to a decreased demand for ATP during senescence.
The up-regulation of respiratory chain components reported here contrasts with a previous analysis of ripening in postharvest tomato fruit. The abundance of a single 36-kD tomato Aox decreased dramatically from the green stage and that of Ucp (32 kD) declined steadily from the yellow stage (Almeida et al., 1999). Likewise, ATP synthesis-sustained respiration decreased markedly from the green stage. It is possible that some molecular events in mango and tomato differ throughout the climacteric respiratory. It is established that postharvest ripening can invoke an altered ripening process (Rhodes, 1970; Biale and Young, 1981). Previous researchers have suggested that free fatty acids could activate Ucp while inhibiting Aox (Sluse et al., 1998; Almeida et al., 1999; Jarmuszkiewicz et al., 2000a). We have observed similar up-regulations of Aox and Ucp message and protein. An increase in free fatty acids throughout mango ripening is well documented (Lizada, 1993). If a similar system of biochemical regulation is operating in mango, it indicates that the signals that lead to the up-regulation of both message and protein for these activities in mango are distinct to those that regulate their biochemical activity. Alternatively, the differences observed, compared with those of postharvest-ripened tomato, may be due to the fact that mango undergoes a large climacteric, displaying a 4- to 6-fold increase in respiration, whereas in tomato the respiratory climacteric represents only a 2-fold increase (Rhodes, 1970; Biale and Young, 1981).
The precise nature of the activity of Ucp proteins remains uncertain in plant tissues (Ricquier and Bouillaud, 2000). These proteins have been designated uncouplers based on sequence homology to the classical Ucp of brown adipose tissue. Several other isoforms of Ucp have now been identified in mammalian and other systems (Boss et al., 1997; Fleury et al., 1997; Gong et al., 1997; Sanchis et al., 1998; Jarmuszkiewicz et al., 1999, 2000b; Mao et al., 1999; Stuart et al., 1999). Much experimental effort has been applied to determine how the activity of mammalian Ucp1-homologs relates to the classical uncoupling activity of mammalian Ucp1. Studies based on knockout mice and ectopic expression in yeast have concluded that only Ucp1 of mammalian mitochondria displays a physiologically relevant uncoupling, in that Ucp2 and Ucp3 could not compensate for the uncoupling or thermogenic activity of Ucp1 (Matthias et al., 1999; Nedergaard et al., 1999, 2001; Matthias et al., 2000; Klingenberg and Echtay, 2001; Stuart et al., 2001). Other isoforms identified in mammals (Ucp2–5) and other organisms have not been reported to support uncoupling in vivo and lack one or both of the essential His residues required for uncoupling activity supported by mammalian Ucp1 (Bienengraeber et al., 1998; Laloi, 1999; Nedergaard et al., 1999; Ricquier and Bouillaud, 2000). Given that the plant Ucp sequences display greater homology with the mammalian isoforms other than Ucp1 and do not contain either of the essential histidines (Bienengraeber et al., 1998; Laloi, 1999; Nedergaard et al., 1999; Ricquier and Bouillaud, 2000), it is premature to definitively link these proteins with the uncoupling activity measured with isolated plant mitochondria. Transgenic approaches using both antisense and overexpressing genetics, where a regulated uncoupling activity can be correlated to protein abundance, are needed to further establish the role of plant Ucp in mitochondrial energy metabolism.
In conclusion, the climacteric respiratory burst in mango is accompanied by changes in several respiratory chain proteins. In comparison with other respiratory chain components, the expression profile and protein accumulation of Aox and Ucp increased in a similar, post-climacteric manner. Further research, including transgenic plants that overexpress or underexpress these proteins, particularly Ucp, is required before a consensus can be reached on their physiological relationship. However, in regards to fruit, they are more likely to play a role in senescence after ripening rather than during the climacteric phase, as was previously believed.
MATERIALS AND METHODS
Plant Material
Mango (Mangifera indica L. cv Kensington Pride) fruit were harvested and scored for stage of ripeness according to mesocarp color standards, as described by the Queensland Department of Primary Industry (Jobin-Decor, 1988; Holmes et al., 1990), before being snap frozen in liquid nitrogen and stored at −70°C. Because mango fruit ripen differentially on the tree, all fruit was harvested within 1 week, whereas the profile of immature to ripe actually represents a period of approximately 14 d (Biale and Young, 1981). Leaves (not yet fully expanded) were also collected and snap frozen.
Nucleic Acid Isolation and Preparation
Genomic DNA from mango leaves was isolated by a modified CsCl gradient procedure (Jofuku and Goldberg, 1988). Total RNA from mango leaves and fruit was extracted three separate times by a modified hot-borate procedure developed for tissues rich in polysaccharides (Wilkins and Smart, 1996). Residual DNA was removed by diluting 10 to 100 μg total RNA to 400 μL in DNase I buffer, adding 40 units RNase-free DNase I and incubating at 37°C for 30 min (Roche, Sydney). DNase I was removed by phenol extraction. The concentration and integrity of RNA was assessed by spectrophotometry and agarose gel electrophoresis. cDNA from 1 μg total RNA was prepared using Expand Reverse Transcriptase according to the manufacturer's instructions (Roche) and desalted through a QIAquick PCR Purification Kit (Qiagen, Adelaide, Australia) with the final elution in water before being used for amplification.
DNA Amplification and Sequencing
For each amplicon, the forward and reverse primers were: mango Aox DNA, AoxF2 (MGNTAYGGNTGYMGNGCNATGATG), or AoxF3 (GARWSNRTNGCNGSNKKNCCNGG); AoxR2 (GCYTCYTCYTCNARRTANCCNAC) or AoxR3 (TGRTTNACRTCNCKRTGRTG); mango Ucp DNA, UcpF1 (GNCTNGAYACNGCNAARGTNMG); UcpR1 (GGNTCNTGGAAYGTNATHATG), MnAox1a cDNA, MnAox1aF (GCTGTTACACTGCAAGTCAC); MnAox1aR (CGTGTTCTTCAACGCCTACTT); MnAox1b cDNA, MnAox1bF (GGTGGGCGGCATGCTGTTACACTG); MnAox1bR (GAAGAATACTCCTTGGACTGC); MnAox1c cDNA, MnAox1cF (GGAGGCATGCTGTTACACTT); MnAox1cR (AAATGCATACTCTTTGGGC); MnAox2 cDNA, MnAox2F (TTHCTHCATCTHAAHTCTCTCC); MnAox2R (AAAGAAGACTCCCTGCACAGCA); MnUcp1 cDNA, MnUcp1F (CCCAGGCTTTACAGATAATG); MnUcp1R (AGCCCTTGTAAAATGCAAGC); MnPth cDNA, MnPthF (GCCTACCAAGATTGTTGACCC); and MnPthR (CAGCAAAAGTCCTGAATACACC).
Aox and Ucp were amplified from mango cv Kensington Pride genomic DNA using primers outlined above with 200 ng template DNA by Expand High Fidelity PCR system and reagents suggested by the manufacturer (Roche). The peroxisomal thiolase (Pth) was amplified from mango cDNA derived from leaf RNA. Amplification conditions for Aox were: one cycle of 94°C, 2 min; 40 cycles of 94°C, 30 s, 55°C, 30 s, and 72°C, 1 min; one cycle of 72°C, 5 min. Amplification conditions of Ucp were: one cycle of 94°C, 2 min; 40 cycles of 94°C, 45 s, 59°C, 45 s, 68°C, 2 min; and one cycle of 68°C, 5 min. Amplification conditions for Pth were: one cycle of 94°C, 2 min; 40 cycles of 94°C, 30 s, 80°C to 50°C, 30 s with a step of −2°C cycle−1, 72°C, 45 s; and one cycle of 72°C, 10 min. Fragments from the amplification were purified either from low-melting point agarose using the QIAquick Agarose Gel Extraction Kit (Qiagen) or directly using the QIAquick PCR Purification Kit (Qiagen). Fragments were subcloned into pCR2.1 plasmid vector before transformation into INVαF' competent cells as described by the TA Cloning Kit (Invitrogen, Adelaide, Australia). Plasmid DNA was purified using the HighPure Plasmid Preparation Kit for minipreparations (Roche) and inserts sequenced using an ABI PRISM BigDye Terminator sequencing kit analyzed on an ABI PRISM 310 Genetic Analyzer according to the manufacturer's instructions (Perkin-Elmer, Melbourne, Australia).
Genomic Southern Hybridization and Analysis
Three micrograms of mango genomic DNA was digested with 20 units of each restriction enzyme at 37°C for 2 h before adding a further 20 units of enzyme and repeating the incubation. The digested DNA was electrophoresed on a 0.8% (w/v) agarose gel, photographed, transferred to Nylon+ membrane (Roche; Southern, 1975; Brown, 1998), and fixed by baking at 120°C for 30 min. DNA probes were labeled with DIG-11-dUTP by the PCR-DIG Labeling Mix and quantitated according to the DIG System User's Guide for Filter Hybridization (Roche) using the LAS-1000 (Fuji, Tokyo) and Image Gauge v3.0 software (Fuji). Southern hybridization was performed using the recommended concentration of DIG-labeled DNA diluted in DIG Easy Hyb and washed at high stringency (0.1× SSC and 0.1% [w/v] SDS at 68°C) according to the manufacturer's instructions (Roche). The membranes were incubated with the anti-DIG alkaline phosphatase conjugate before adding the CDP-Star chemiluminescent substrate and visualizing the hybridized DNA fragments using the LAS-1000 (Fuji). The DNA fragments were sized relative to the DIG-labeled Molecular Weight Marker II (Roche).
Raising the Anti-SoyUcp Polyclonal Antibody
Soybean (Glycine max L. Merr.) RNA was prepared and reverse transcribed as described for mango. A partial cDNA of the soybean Ucp was amplified using gene-specific primers and subcloned into pCR2.1 (Invitrogen). A restriction enzyme was introduced to the 5′ end of the fragment to facilitate subcloning into a bacterial expression vector pQE-31 (Qiagen) such that the cDNA would be in frame under the vectors' promoter system. The 6× His-tagged soybean Ucp protein was expressed and purified using the QIAexpressionist system (Qiagen). The purified antigen was injected into two rabbits, the anti-soyUcp antibody raised, and the serum collected according to standard procedures (Cooper and Paterson, 1996).
Isolation, Blotting, and Immunodetection of Total Protein
Two grams (wet weight) of frozen mango fruit at each development stage was ground in a mortar and pestle before adding 2 mL of extraction buffer (125 mm Tris-Cl, 7.75% [w/v] SDS, and 10% [v/v] mercaptoethanol, pH 7.0) and grinding further. The mixture was transferred to a centrifuge tube, thawed to room temperature, and centrifuged in a swing-out rotor at 3,500g for 10 min. Several extraction and blots were carried out to optimize the extraction method and to ensure the pattern of protein observed was representative. The supernatant was mixed with 6× (v/v) sample buffer (0.1 m Tris-Cl, 12% [w/v] SDS, 9% [v/v] glycerol, and 60% [v/v] mercaptoethanol, pH 6.8) before loading 60 μL to be separated by SDS-PAGE (Laemmli, 1970). After electrophoresis, the separated proteins were transferred to a Hybond-C Extra nitrocellulose membrane (Amersham Pharmacia Biotech, Sydney) using a Multiphor II semi-dry blotting apparatus (Amersham Pharmacia Biotech) according to the manufacturer's instructions. The detection of western hybridization was carried out using the BM Chemiluminescence Blotting Substrate POD system (Roche). The primary antibodies were used at the following dilutions: Aox, 1:50 (Dr. Thomas E. Elthon, University of Nebraska, Lincoln); cytochrome oxidase subunit 2, 1:7,500 (D.O. Daley and J. Whelan, unpublished data); cytochrome bc1 (core subunits), 1:10,000 (Dr. Hans-Peter Braun, University of Hannover, Germany); HSP-60, 1:2,500 (Stressgen, Canada); HSP-70, 1:2,500 (Prof. Elzbieta Glaser, Stockholm University, Sweden); Rieske FeS, 1:5,000; and anti-soyUcp, 1:10,000. Proteins cross-reacting with the various antibodies were visualized using an LAS-1000 (Fuji) and quantitated using the Image Gauge v3.0 software (Fuji), where the highest intensity band of the profile was set to 1 and others calculated relative to that value.
Real-Time PCR Analysis of Gene Expression
Gene expression was assayed using the LightCycler (Roche) and FastStart DNA Master SYBR Green I kit (Roche). Reaction conditions (10-μL volume) were optimized according to the manufacturer's instructions to minimize primer-dimer formation, and 0.008% (w/v) bovine serum albumin was also included. To compare the many different transcripts, primers were designed within a confined range of melting temperature and amplicon size so that a single thermocycling program could be used. Thermocycling conditions were: one cycle of 95°C, 5 min; 50 cycles of 85°C to 50°C, 5 s with a step of −2°C cycle−1, 72°C, 15 s, single acquisition; one cycle of 95°C, 0 s, 70°C, 1 min, 95°C, 0 s with a transition rate of 0.1°C s−1, continuous acquisition; and one cycle of 40°C, 30 s. The lack of primer-dimer or nonspecific product accumulation was checked by melt-curve analysis and affirmed by agarose gel electrophoresis. After standardizing the baseline, relative template abundance was quantified using the second derivative maximum method of the LightCycler version 3.0 software (Roche) to determine the theoretical cycle at which each PCR reached exponential amplification, a requirement for quantitative PCR. Data were exported and analyzed by the appropriate analysis of variance using Genstat 5 version 4.1 (Rothamsted Agricultural Station, Harpenden, UK).
Before analyzing cDNA samples, we: (a) demonstrated the lack of contaminating genomic DNA in each RNA sample using control reverse transcription PCR that lacked the reverse transcriptase, (b) demonstrated the specificity of primers, and (c) measured the efficiency of each set of primers. The amplification specificity and relative efficiency of each primer pair were optimized using standard amounts of the genomic clones as template. To demonstrate specificity, the amplification of each gene was carried out with specific primers, e.g. MnAox1a with MnAox1aF/R primer set, and compared with amplification with this primer set from all other MnAox clones. This procedure was carried out for all MnAox genes and primer sets. Data were analyzed as a completely randomized experiment by analysis of variance, with template as the treatment factor and relative amplification as the response variate. Analysis of variance tested the null hypothesis that relative amplification from each MnAox template, with a given MnAox primer set equaled 1 (i.e. equal to amplification of the specific amplicon). To assess relative amplification efficiency of each amplification reaction, PCR was performed on 1 × 10−15 molecules of each template using the specific primers. The template that reached exponential amplification at the earliest cycle was titrated to generate a standard curve from which to calculate the relative efficiencies. Analysis of variance was based on a completely randomized experiment, with transcript as the treatment factor and relative amplification as the response variate.
Total RNA from representative tissue samples of each stage of ripeness was isolated separately three times. Two separate reverse transcription assays were carried out on total RNA and were assayed separately, in duplicate for each transcript, by real-time PCR. Using 1-μL 10−1 dilutions of cDNA, we determined the stage of ripeness that displayed the highest expression for each gene. From that stage, a series of three 5-fold cDNA dilutions were carried out and transcript abundance at the other stages was measured relative to that standard curve. The data were analyzed as a randomized block experiment by analysis of variance, where PCR-run was the block factor, stage of ripeness was the treatment factor, and relative transcript abundance was the response variate. Using a block factor reduced the effect of variation between PCR-runs on the response. Therefore, the expression data was expressed relative to the highest value for each gene and then corrected for amplification efficiencies. This allowed comparisons of each gene between the stages of development and also of relative amounts of each gene.
ACKNOWLEDGMENTS
We are especially grateful to Dr. Nick Galwey for his guidance on statistical analysis and to Prof. David Day, Dr. Harvey Millar, and Tulene McCabe for their manuscript revision and comments offered. We also appreciate access to plant material granted by the Gascoyne Research Station and Dr. Sujit Dey.
Footnotes
This research was supported by the Australian Research Council (grant to J.W.). M.J.C. was sponsored by AgWEST.
LITERATURE CITED
- Albury MS, Affourtit C, Moore AL. A highly conserved glutamate residue (Glu-270) is essential for plant alternative oxidase activity. J Biol Chem. 1998;273:30301–30305. doi: 10.1074/jbc.273.46.30301. [DOI] [PubMed] [Google Scholar]
- Almeida AM, Jarmuszkiewicz W, Khomsi H, Arruda P, Vercesi AE, Sluse FE. Cyanide-resistant, ATP-synthesis-sustained, and uncoupling-protein-sustained respiration during postharvest ripening of tomato fruit. Plant Physiol. 1999;119:1323–1329. doi: 10.1104/pp.119.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold DA. Isolation of mutants of the Arabidopsis thaliana alternative oxidase (ubiquinol:oxygen oxidoreductase) resistant to salicylhydroxamic acid. Biochim Biophys Acta. 1998;1:73–83. doi: 10.1016/s0005-2728(98)00015-2. [DOI] [PubMed] [Google Scholar]
- Biale JB, Young RE. Respiration and ripening in fruits: retrospect and prospect. In: Friend J, Rhodes MJC, editors. Recent Advances in the Biochemistry of Fruits and Vegetables. London: Academic Press; 1981. pp. 1–40. [Google Scholar]
- Bienengraeber M, Echtay KS, Klingenberg M. H+ transport by uncoupling protein (UCP-1) is dependent on a histidine pair, absent in UCP-2 and UCP-3. Biochemistry. 1998;37:3–8. doi: 10.1021/bi972463w. [DOI] [PubMed] [Google Scholar]
- Bojorquez G, Gomez-Lim MA. Peroxisomal thiolase mRNA is induced during mango fruit ripening. Plant Mol Biol. 1995;28:811–820. doi: 10.1007/BF00042067. [DOI] [PubMed] [Google Scholar]
- Boss O, Samec S, Paoloni GA, Rossier C, Dulloo A, Seydoux J, Muzzin P, Giacobino JP. Uncoupling protein-3: a new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997;408:39–42. doi: 10.1016/s0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- Bouillaud F, Weissenbach J, Ricquier D. Complete cDNA-derived amino acid sequence of rat brown fat uncoupling protein. J Biol Chem. 1986;261:1487–1490. [PubMed] [Google Scholar]
- Brown T. Analysis of DNA sequences by blotting and hybridization. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. Vol. 1. New York: John Wiley and Sons; 1998. pp. 2.9.1–2.9.2. [Google Scholar]
- Casolo V, Braidot E, Chiandussi E, Macri F, Vianello A. The role of mild uncoupling and non-coupled respiration in the regulation of hydrogen peroxide generation by plant mitochondria. FEBS Lett. 2000;474:53–57. doi: 10.1016/s0014-5793(00)01576-3. [DOI] [PubMed] [Google Scholar]
- Cooper HM, Paterson Y. Production of polyclonal antisera. In: Coligan JE, Kruisbeek AE, Margulies DH, Shevach EM, Strober W, editors. Current Protocols in Immunology. Vol. 1. New York: John Wiley and Sons; 1996. pp. 2.4.1–2.4.2. [Google Scholar]
- Costa ADT, Nantes IL, Jezek P, Leite A, Arruda P, Vercesi AE. Plant uncoupling mitochondrial protein activity in mitochondria isolated from tomatoes at different stages of ripening. J Bioenerg Biomembr. 1999;31:527–533. doi: 10.1023/a:1005408809619. [DOI] [PubMed] [Google Scholar]
- Cruz-Hernandez A, Gomez-Lim MA. Alternative oxidase from mango (Mangifera indica L.) is differentially regulated during fruit ripening. Planta. 1995;197:569–576. doi: 10.1007/BF00191562. [DOI] [PubMed] [Google Scholar]
- Day DA, Krab K, Lambers H, Moore AL, Siedow JN, Wagner AM, Wiskich JT. The cyanide-resistant oxidase: to inhibit or not to inhibit, that is the question. Plant Physiol. 1996;110:1–2. doi: 10.1104/pp.110.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djajanegara I, Holtzapffel R, Finnegan PM, Hoefnagel MHN, Berthold DA, Wiskich JT, Day DA. A single amino acid change in the plant alternative oxidase alters the specificity of organic acid activation. FEBS Lett. 1999;454:220–224. doi: 10.1016/s0014-5793(99)00808-x. [DOI] [PubMed] [Google Scholar]
- Douce R, Neuburger M. The uniqueness of plant mitochondria. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:371–414. [Google Scholar]
- Elthon TE, Nickels RL, McIntosh L. Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol. 1989;89:1311–1317. doi: 10.1104/pp.89.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury C, Neverova M, Collins S, Raimbault S, Champigny O, Levi Meyrueis C, Bouillaud F, Seldin MF, Surwit RS, Ricquier D. Uncoupling protein-2: a novel gene linked to obesity and hyperinsulinemia. Nat Genet. 1997;15:269–272. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- Glaser E, Dessi P. Integration of the mitochondrial-processing peptidase into the cytochrome bc1 complex in plants. J Bioenerg Biomembr. 1999;31:259–274. doi: 10.1023/a:1005475930477. [DOI] [PubMed] [Google Scholar]
- Gong DW, He Y, Karas M, Reitman M. Uncoupling protein-3 is a mediator of thermogenesis regulated by thyroid hormone, beta-3-adrenergic agonists, and leptin. J Biol Chem. 1997;272:24129–24132. doi: 10.1074/jbc.272.39.24129. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Meler MA, Ribas-Carbo M, Giles L, Siedow JN. The effect of growth and measurement temperature on the activity of the alternative respiratory pathway. Plant Physiol. 1999;120:765–772. doi: 10.1104/pp.120.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes R, Campbell T, Ledger S. Mango Picking Guide. Brisbane, Queensland: Queensland Department of Primary Industries; 1990. [Google Scholar]
- Ito K. Isolation of two distinct cold-inducible cDNAs encoding plant uncoupling proteins from the spadix of skunk cabbage (Symplocarpus foetidus) Plant Sci. 1999;149:167–173. [Google Scholar]
- Jarmuszkiewicz W, Almeida AM, Vercesi AE, Sluse FE, Sluse-Goffart CM. Proton re-uptake partitioning between uncoupling protein and ATP synthase during benzohydroxamic acid-resistant state 3 respiration in tomato fruit mitochondria. J Biol Chem. 2000a;275:13315–13320. doi: 10.1074/jbc.275.18.13315. [DOI] [PubMed] [Google Scholar]
- Jarmuszkiewicz W, Milani G, Fortes F, Schreiber AZ, Sluse FE, Vercesi AE. First evidence and characterization of an uncoupling protein in fungi kingdom: CpUCP of Candida parapsilosis. FEBS Lett. 2000b;467:145–149. doi: 10.1016/s0014-5793(00)01138-8. [DOI] [PubMed] [Google Scholar]
- Jarmuszkiewicz W, Sluse-Goffart CM, Hryniewiecka L, Sluse FE. Identification and characterization of a protozoan uncoupling protein in Acanthamoeba castellanii. J Biol Chem. 1999;274:23198–23202. doi: 10.1074/jbc.274.33.23198. [DOI] [PubMed] [Google Scholar]
- Jezek P, Costa ADT, Vercesi AE. Reconstituted plant uncoupling mitochondrial protein allows for proton translocation via fatty acid cycling mechanism. J Biol Chem. 1997;272:24272–24278. doi: 10.1074/jbc.272.39.24272. [DOI] [PubMed] [Google Scholar]
- Jobin-Decor MP. Mango Ripening Guide. Queensland Agric J. 1988;114:369–371. [Google Scholar]
- Jofuku KD, Goldberg RB. Analysis of plant gene structure. In: Shaw CH, editor. Plant Molecular Biology: A Practical Approach. Oxford: IRL Press; 1988. pp. 37–42. [Google Scholar]
- Klingenberg M, Echtay KS. Uncoupling proteins: the issues from a biochemists point of view. Biochim Biophys Acta. 2001;1504:128–143. doi: 10.1016/s0005-2728(00)00242-5. [DOI] [PubMed] [Google Scholar]
- Kowaltowski AJ, Costa ADT, Vercesi AE. Activation of the potato plant uncoupling mitochondrial protein inhibits reactive oxygen species generation by the respiratory chain. FEBS Lett. 1998;425:213–216. doi: 10.1016/s0014-5793(98)00231-2. [DOI] [PubMed] [Google Scholar]
- Kumar S, Patil BC, Sinha SK. Cyanide resistant respiration is involved in temperature rise in ripening mangoes. Bioch Biophys Res Commun. 1990;168:818–822. doi: 10.1016/0006-291x(90)92394-f. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laloi M. Plant mitochondrial carriers: an overview. Cell Mol Life Sci. 1999;56:918–944. doi: 10.1007/s000180050484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi M, Klein M, Riesmeier JW, Muller-Rober B, Fleury C, Bouillaud F, Ricquier D. A plant cold-induced uncoupling protein. Nature. 1997;389:135–136. doi: 10.1038/38156. [DOI] [PubMed] [Google Scholar]
- Laskey RA. The use of intensifying screens or organic scintillators for visualising radioactive molecules resolved by gel electrophoresis. Methods Enzymol. 1980;65:363–371. doi: 10.1016/s0076-6879(80)65047-2. [DOI] [PubMed] [Google Scholar]
- Lizada C. Mango. In: Seymour GB, Taylor JE, Tucker GA, editors. Biochemistry of Fruit Ripening. London: Chapman & Hall; 1993. pp. 255–271. [Google Scholar]
- Maia IG, Benedetti CE, Leite A, Turcinelli SR, Vercesi AE, Arruda P. Atpump: an Arabidopsis gene encoding a plant uncoupling mitochondrial protein. FEBS Lett. 1998;429:403–406. doi: 10.1016/s0014-5793(98)00634-6. [DOI] [PubMed] [Google Scholar]
- Mao WG, Yu XX, Zhong A, Li W, Brush J, Sherwood SW, Adams SH, Pan GH. UCP4, a novel brain-specific mitochondrial protein that reduces membrane potential in mammalian cells. FEBS Lett. 1999;443:326–330. doi: 10.1016/s0014-5793(98)01713-x. [DOI] [PubMed] [Google Scholar]
- Matthias A, Jacobsson A, Cannon B, Nedergaard J. The bioenergetics of brown fat mitochondria from UCP1-ablated mice: UCP1 is not involved in fatty acid-induced de-energization (“uncoupling”) J Biol Chem. 1999;274:28150–28160. doi: 10.1074/jbc.274.40.28150. [DOI] [PubMed] [Google Scholar]
- Matthias A, Ohlson KBE, Fredriksson JM, Jacobsson A, Nedergaard J, Cannon B. Thermogenic responses in brown fat cells are fully UCP1-dependent: UCP2 or UCP3 do not substitute for UCP1 in adrenergetically or fatty acid-induced thermogenesis. J Biol Chem. 2000;275:25073–25081. doi: 10.1074/jbc.M000547200. [DOI] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L. The alternative oxidase lowers mitochondria reactive oxygen production in plant cells. Proc Natl Acad Sci USA. 1999;96:8271–8276. doi: 10.1073/pnas.96.14.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe TC, Finnegan PM, Millar AH, Day DA, Whelan J. Differential expression of alternative oxidase genes in soybean cotyledons during postgerminative development. Plant Physiol. 1998;118:675–682. doi: 10.1104/pp.118.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Day DA. Nitric oxide inhibits the cytochrome oxidase but not the alternative oxidase of plant mitochondria. FEBS Lett. 1996;398:155–158. doi: 10.1016/s0014-5793(96)01230-6. [DOI] [PubMed] [Google Scholar]
- Millar AH, Day DA. Alternative solutions to radical problems. Trends Plant Sci. 1997;2:289–290. [Google Scholar]
- Millar AH, Wiskich JT, Whelan J, Day DA. Organic acid activation of the alternative oxidase of plant mitochondria. FEBS Lett. 1993;329:259–262. doi: 10.1016/0014-5793(93)80233-k. [DOI] [PubMed] [Google Scholar]
- Moore AL, Siedow JN. The regulation and nature of the cyanide-resistant alternative oxidase of plant mitochondria. Biochim Biophys Acta. 1991;1059:121–140. doi: 10.1016/s0005-2728(05)80197-5. [DOI] [PubMed] [Google Scholar]
- Murayama S, Handa H. Isolation and characterization of cDNAs encoding mitochondrial uncoupling proteins in wheat: wheat UCP genes are not regulated by low temperature. Mol Gen Genet. 2000;264:112–118. doi: 10.1007/s004380000289. [DOI] [PubMed] [Google Scholar]
- Murcha MW, Huang T, Whelan J. Import of precursor proteins into mitochondria from soybean tissues during development. FEBS Lett. 1999;464:53–59. doi: 10.1016/s0014-5793(99)01674-9. [DOI] [PubMed] [Google Scholar]
- Nantes IL, Fagian MM, Catisti R, Arruda P, Maia IG, Vercesi AE. Low temperature and aging-promoted expression of PUMP in potato tuber mitochondria. FEBS Lett. 1999;457:103–106. doi: 10.1016/s0014-5793(99)01017-0. [DOI] [PubMed] [Google Scholar]
- Nedergaard J, Golozoubova V, Matthias A, Asadi A, Jacobsson A, Cannon B. UCP1: the only protein able to mediate adaptive non-shivering thermogenesis and metabolic inefficiency. Biochim Biophys Acta. 2001;1504:82–106. doi: 10.1016/s0005-2728(00)00247-4. [DOI] [PubMed] [Google Scholar]
- Nedergaard J, Matthias A, Golozoubova V, Jacobsson A, Cannon B. UCP1: The original uncoupling protein: and perhaps the only one? New perspectives on UCP1, UCP2, and UCP3 in the light of the bioenergetics of the UCP1-ablated mice. J Bioenerg Biomembr. 1999;31:475–491. doi: 10.1023/a:1005400507802. [DOI] [PubMed] [Google Scholar]
- Pastore D, Fratianni A, Di Pede S, Passarella S. Effects of fatty acids, nucleotides and reactive oxygen species on durum wheat mitochondria. FEBS Lett. 2000;470:88–92. doi: 10.1016/s0014-5793(00)01292-8. [DOI] [PubMed] [Google Scholar]
- Rhodes MJC. The climacteric and ripening of fruits. In: Hulme AC, editor. The Biochemistry of Fruits and Their Products. Vol. 1. London: Academic Press; 1970. pp. 521–536. [Google Scholar]
- Ricquier D, Bouillaud F. The uncoupling protein homologues: UCP1, UCP2, UCP3, StUCP and AtUCP. Biochem J. 2000;345:161–179. [PMC free article] [PubMed] [Google Scholar]
- Sanchis D, Fleury C, Chomiki N, Goubern M, Huang Q, Neverova M, Gregoire F, Easlick J, Raimbault S, Levi-Meyrueis C. BMCP1, a novel mitochondrial carrier with high expression in the central nervous system of humans and rodents, and respiration uncoupling activity in recombinant yeast. J Biol Chem. 1998;273:34611–34615. doi: 10.1074/jbc.273.51.34611. [DOI] [PubMed] [Google Scholar]
- Sluse FE, Almeida AM, Jarmuszkiewicz W, Vercesi AE. Free fatty acids regulate the uncoupling protein and alternative oxidase activities in plant mitochondria. FEBS Lett. 1998;433:237–240. doi: 10.1016/s0014-5793(98)00922-3. [DOI] [PubMed] [Google Scholar]
- Solomos T, Laties GG. Similarities between the actions of ethylene and cyanide In initiating the climacteric and ripening of avocados. Plant Physiol. 1974;54:506–511. doi: 10.1104/pp.54.4.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stuart JA, Cadenas S, Jekabsons MB, Roussel D, Brand MD. Mitochondrial proton leak and the uncoupling protein homologues. Biochim Biophys Acta. 2001;1504:144–158. doi: 10.1016/s0005-2728(00)00243-7. [DOI] [PubMed] [Google Scholar]
- Stuart JA, Harper JA, Brindle KM, Brand MD. Uncoupling protein 2 from carp and zebrafish, ectothermic vertebrates. Biochim Biophys Acta. 1999;1413:50–54. doi: 10.1016/s0005-2728(99)00081-x. [DOI] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN. The cyanide-resistant alternative oxidases from the fungi Pichia stipitis and Neurospora crassa are monomeric and lack regulatory features of the plant enzyme. Arch Biochem Biophys. 2000;378:234–245. doi: 10.1006/abbi.2000.1834. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L. Alternative oxidase: from gene to function. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:703–734. doi: 10.1146/annurev.arplant.48.1.703. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, Vanlerberghe AE, McIntosh L. Molecular genetic evidence of the ability of alternative oxidase to support respiratory carbon metabolism. Plant Physiol. 1997;113:657–661. doi: 10.1104/pp.113.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercesi AE, Martins IS, Silva MAP, Leite HMF, Cuccovia IM, Chaimovich H. PUMPing plants. Nature. 1995;375:24. [Google Scholar]
- Wagner AM. A role for active oxygen species as second messengers in the induction of alternative oxidase gene expression in Petunia hybrida cells. FEBS Lett. 1995;368:339–342. doi: 10.1016/0014-5793(95)00688-6. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Nakazono M, Tsutsumi N, Hirai A. AtUCP2: a novel isoform of the mitochondrial uncoupling protein of Arabidopsis thaliana. Plant Cell Physiol. 1999;40:1160–1166. doi: 10.1093/oxfordjournals.pcp.a029501. [DOI] [PubMed] [Google Scholar]
- Whelan J, Millar AH, Day DA. The alternative oxidase is encoded in a multigene family in soybean. Planta. 1996;198:197–201. doi: 10.1007/BF00206244. [DOI] [PubMed] [Google Scholar]
- Wilkins TA, Smart LB. Isolation of RNA from plant tissue. In: Krieg PA, editor. A Laboratory Guide to RNA: Isolation, Analysis and Synthesis. New York: Wiley-Liss; 1996. pp. 21–42. [Google Scholar]