Abstract
Nucleotide positions conserved on the 3′ side of the initiator codon ATG and the corresponding N-terminal amino acid residues in a number of highly abundant plant proteins were identified by computational analysis of a dataset of highly expressed plant genes. The reporter genes uidA and gfp were modified to introduce these features. Insertion of GCT TCC TCC after the initiator codon ATG augmented expression for both the reporter genes. The insertion of each successive codon improved the expression of β-glucuronidase (GUS) in an incremental fashion in transient transformation of tobacco (Nicotiana tabacum) leaves. The insertion of alanine-serine (Ser)-Ser resulted in about a 2-fold increase in the stability of GUS. However, this did not account for the 30- to 40-fold increase in GUS activity between the constructs coding for methionine-alanine-Ser-Ser-GUS and the native enzyme. Substitution of the codon for Ser at the third amino acid residue with synonymous codons reduced GUS expression. The results suggest a role for the conserved nucleotides in the +4 to +11 region in augmenting posttranscriptional events in the expression of genes in plants.
Sequences flanking the translation initiator codon ATG have been surveyed in vertebrate (Cavener, 1987; Kozak, 1987) and plant (Joshi et al., 1997; Sawant et al., 1999) gene databases to predict context requirements for translational initiation. Some of the predictions made by computational analysis have been substantiated experimentally by using in vitro translation systems. The current model (Kozak, 1989) for the initiation of eukaryotic translation broadly postulates that the first ATG codon is recognized by scanning of the mRNA sequence from the 5′ end by the 40S ribosomal initiation complex. The −3 and +4 positions (where the A of ATG is +1) are considered as the most important in determining a favorable context of initiator ATG (Sullivan and Green, 1993). The nucleotides upstream of the ATG are believed to comprise the context required for efficient initiation of translation. The modulation of gene expression by nucleotides upstream of the ATG has been studied in plant cells (Taylor et al., 1987; McElroy et al., 1991; Dinesh-Kumar and Miller, 1993; Johnston and Rochon, 1996; Marcin et al., 2000), though the functional roles of the preferred nucleotides at the −1, −2, and −3 positions are not known. Specific, “most preferred” nucleotides at these positions were reported to give a two-fold variation in gene expression, depending upon the plant species and the cell type (Marcin et al., 2000). Using in vitro translation, Lütcke et al. (1987) reported differences in the nucleotide requirement at the −3 position between mammalian (rabbit reticulocyte) and plant (wheat germ) systems. The role of the sequence context downstream of the ATG, especially beyond the +5 position, has not been examined in plants. The G at +4 and A at +5 positions were suggested to determine the efficient utilization of ATG initiation sites in in vitro translation experiments using the rabbit reticulocyte system (Grunert and Jackson, 1994). However, in disagreement with this study and that of Boeck and Kolakofsky (1994), Kozak (1997) reported a role of only up to the +4 position in correct recognition of the initiator codon. The extent of conserved positions further downstream of +4 and their function in in vivo gene expression have not been studied in detail. In none of the earlier studies was the context analysis done after classifying the gene database with respect to their level of expression.
The nucleotides following the initiator ATG also determine the type of amino acid residues at the N terminus of encoded proteins. This can additionally influence gene expression indirectly by altering the stability of proteins. A variety of mechanisms that affect the intracellular stability of proteins in yeast (Saccharomyces cerevisiae; Bachmair et al., 1986) and mammalian cells (Lèvy et al., 1999) have been reported to operate by recognizing the type of amino acids at the N-terminal end. We examined a dataset of highly expressed plant genes to see if they had features conserved downstream of the ATG initiator codon and if such features were in contrast to the genes expressed at low levels. The functional validity of the features noticed in computational analysis was established by determining their role on the level of in vivo gene expression. Keeping the ATG upstream context constant, this study reports the influence of nucleotide sequences conserved on the 3′ side of the initiator ATG on in vivo expression of uidA and gfp genes, using a transient assay involving biolistic transformation of tobacco (Nicotiana tabacum) leaves.
RESULTS
Features Conserved on the 3′ Side of the Initiator ATG in Highly Expressed Plant Genes
The underlying assumption of this study was that the features characteristic of the 3′ side of the initiator ATG codon found in a number of genes expressed at high levels in plants may reveal specialized architecture that facilitates the high level of their expression in vivo. Two features were noticed. First, there was a significant bias for specific nucleotides at positions +4, +5, +6, +8, +9, +10, and +11 in the dataset of highly expressed genes (Table I). This results in a second feature that characterizes the abundantly expressed plant proteins. The predominance of GCT at the +4 to +6 positions gives an Ala next to N-terminal Met in 95% of such proteins. Other conserved nucleotides in the second and third codon positions similarly result in the predominance of Ser as the third and the fourth residues in the highly expressed plant proteins (Table I). Features of the canonical ATG downstream sequence identified as conserved in the dataset of highly expressed plant genes remained the same, irrespective of whether, in the case of gene families, multiple members or orthologous representatives were considered in the analyses. When all members of the gene families were considered, the dataset comprised 236 entries. This dataset resulted in the nucleotide sequence on 3′ of the initiator ATG and the corresponding N-terminal amino acid sequence shown in Table I. Reconstruction of the database to include only one representative sequence per gene family gave a dataset comprising of 74 entries. This smaller dataset resulted in a similar conserved ATG 3′ nucleotide sequence, that is to say A100 T100 G100 G98 C92 T75 T40 C60 C40 (T37/A35) C41, and corresponding N-terminal amino acid sequence Met100 Ala92 Ser36 Ser30. Results from the complete dataset shown in Table I were used for detailed analyses.
Table I.
Modification of initiator ATG-downstream sequence in reporter genes
| Name of Construct | Insertion at ∗ | Name of Upstream Primer |
|---|---|---|
| uidA Constructs | ||
| (i) M-GUS | None | Primer 1 |
| (ii) MA-GUS | GCT | Primer 2 |
| (iii) MAS1-GUS | GCT TCC | Primer 3 |
| (iv) MAS1S-GUS | GCT TCC TCC | Primer 4 |
| (v) MDS1S-GUS | GAT TCC TCC | Primer 5 |
| (vi) MACS-GUS | GCT TGC TCC | Primer 6 |
| (vii) MAS2S-GUS | GCT TCG TCC | Primer 7 |
| (viii) MAS3S-GUS | GCT AGC TCC | Primer 8 |
| (ix) MAS4S-GUS | GCT AGT TCC | Primer 9 |
| gfp Constructs | ||
| (i) M-GFP | None | Primer 10 |
| (ii) MASS-GFP | GCT TCC TCC | Primer 11 |
Conserved ATG downstream features in highly expressed plant genes: (a) nucleotides to 3′ of initiator ATG, A100T100G100G98C94T75 NC62C40(T37/A35)C48, and (b) N-terminal amino acids, Met100 Ala95Ser32Ser31. uidA Constructs with modified ATG downstream sequence: upstream primers, 5′-AATTACATCTAGATAAACAATG∗TTACGTCCTGTAGAAACCCCAA-3′. gfp Constructs with modified ATG downstream sequence: upstream primers, 5′-ATGCATTCTAGATCAACAATG∗GGTAAAGGAGAAGAACTTTTCACTGGAGTT3′.
Conservation of Codons Following Initiator ATG in Highly Expressed Plant Genes
A high frequency of G at +4 and C at +5 noticed in our dataset of highly expressed plant genes has been documented earlier in vertebrate genes (Grunert and Jackson, 1994). It was also correlated with a high frequency of Ala as the corresponding amino acid at the second position in vertebrate (Grunert and Jackson, 1994) and plant (Luehrsen and Walbot, 1994) proteins. However, the advantage, if any, of an Ala next to the initiator Met has not been reported. The preferred occurrence of T at +6, which is the wobble position in the codon for Ala, is characteristic of the penultimate N-terminal codon in the dataset developed by us. In an earlier study (Murray et al., 1989), the frequency of GCT employed for Ala in plant gene database was calculated at 37%. Quite comparably, the dataset of highly expressed nuclear genes analyzed by us also showed GCT for Ala at 39% positions in the reading frame. However, the frequency of GCT usage at +4 to +6 position in the same dataset was 78%. This points to a possible role of U at +6 position over and above a possible advantage in having an Ala as the second amino acid in abundantly expressed proteins. Likewise, a preferred deployment of CC at the second and third codon positions, with Ser as the third amino acid residue, could be indicative of an architecture at +8 and +9 that facilitates a high level of gene expression. The UCC is used in only 20% of the cases for Ser elsewhere in the dataset of highly expressed nuclear genes in plants. However, Ser at the third amino acid position in the highly expressed genes is coded by UCC in 46% of the cases. The +10 and +11 positions show a preference for T/A and C, respectively, and a preference for Ser as the fourth N-end residue. Of the highly expressed proteins, 46% had Met-Ala, 18% had Met-Ala-Ser, 17% had Met-Ala-X-Ser, and 14% had Met-Ala-Ser-Ser as the N-terminal amino acids. The last class comprised members of the RuBP carboxylase small subunit family from different plants and forms part of the most abundant proteins in plants.
Effect of Conserved Features on Expression of the Reporter Genes
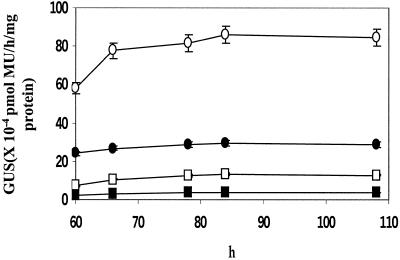
The functional validity of the conserved features was established by comparing the suitably mutated uidA genes in transient expression, following their bombardment on tobacco leaves. The uidA gene modified (Table I) to resemble the ATG downstream architecture of the highly expressed plant genes up to +11 position, i.e. MAS1S-β-glucuronidase (GUS), showed a 30- to 40-fold increase in GUS expression in tobacco leaves. The activity increased progressively (Fig. 1) as the nucleotide context in Met-Leu-Leu-Pro-GUS (M-GUS) was progressively extended to include the conserved motif up to +11 in MAS1S-GUS. In order to establish that the increase in GUS activity was the result of the specific ATG downstream nucleotide sequence and/or the corresponding amino acid residues and not merely the consequence of providing a 5′ extension to the mRNA, the expression of GUS was examined from constructs carrying point mutations of MAS1S-GUS. As shown in Table II, the substitution of C at +5 with A (MDS1S-GUS, results in Ala2→-Asp) and C at +8 with G (MACS-GUS, results in Ser3→-Cys) resulted in a significant decline in GUS activity. The decline was most severe in the case of the +5 mutation (MDS1S-GUS) and that corresponds with this position being conserved in 94% of the highly expressed genes (Table I). The importance of a specific nucleotide per se at a given position was further examined by making synonymous substitutions in TCC, the third N-terminal codon. The C→G mutation at +9 (MAS2S-GUS) gave a 5.5-fold reduction in expression (Table II), although both MAS1S- and MAS2S-GUS have Ser as the third N-terminal residue. A double mutation in the third codon, i.e. T→A and C→G at positions +7 and +8, respectively, in MAS3S, resulted in a six-fold reduction in expression level. When the TCC was replaced by the synonymous codon AGT (MAS4S), thereby mutating all three positions without altering the amino acid, the expression was reduced by seven-fold.
Figure 1.
Effect of ATG downstream modifications on in vivo GUS activity in cell-free extracts prepared at different time points after bombardment of tobacco leaves with gene constructs expressing M-GUS (▪), MA-GUS (□), MAS1-GUS (●), and MAS1S-GUS (○).
Table II.
Expression of reporter proteins with N-terminal modifications
| Name of Construct | Activity | Fold Activation |
|---|---|---|
| GUS constructs | ||
| (i) M-GUS | 14.6a (±1.2) | 1.0 |
| (ii) MA-GUS | 59.9 (±4.1) | 4.1 |
| (iii) MAS1-GUS | 181.6 (3.9) | 12.4 |
| (iv) MAS1S-GUS | 603.0 (±13.8) | 41.2 |
| (v) MDS1S-GUS | 4.2 (±0.3) | 0.3 |
| (vi) MACS-GUS | 44.4 (±2.6) | 3.1 |
| (vii) MAS2S-GUS | 110.2 (±2.8) | 7.6 |
| (viii) MAS3S-GUS | 100.6 (±1.5) | 6.9 |
| (ix) MAS4S-GUS | 86.3 (±3.6) | 5.9 |
| GFP constructs | ||
| (i) M-GFP | 16.3b (±1.1) | 1.0 |
| (ii) MAS1S-GFP | 345.5 (±9.7) | 21.2 |
GUS ± sd (glucuronidase activity expressed as ×103 pmol/h/mg protein).
GFP ± sd (GFP activity expressed as ×102 relative fluorescence/mg protein).
Validation of the results on the augmentation of GUS activity by the ATG 3′ motif in MAS1S-GUS was further substantiated by inserting the same motif after the ATG of a second reporter gene, in this case gfp. Insertion of the canonical +4 to +11 motif enhanced the expression of MAS1S-green fluorescent protein (GFP) also by 21-fold (Table II) as compared with the wild-type M-GFP.
Given the fact that the gene architecture upstream of the initiator codon is exactly similar in all the constructs, the increase in expression in the MAS1S-fusion constructs is likely to be due to the enhancement of one or more posttranscriptional events. It could be due to improved transcript stability, higher translational efficiency, an increase in activity of the proteins due to altered N-terminal conformation, decrease in their rate of intracellular degradation, or a combination of one or more of these aspects. The different GUS constructs were examined in detail to resolve these possibilities.
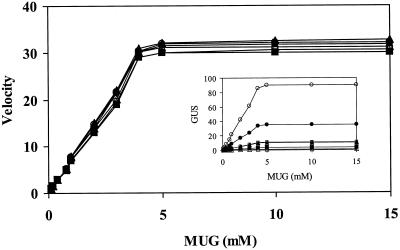
Substrate saturation curves (Fig. 2) showed that all four forms of GUS, that is to say M-GUS, MA-GUS, MAS1-GUS, and MAS1S-GUS, exhibited exactly similar reaction kinetics. Similar K0.5 values and saturation curves of the four forms suggested that the N-terminal modifications made in this study did not enhance the enzyme activity by imparting a catalytically favorable functional conformation to GUS. Similarly, the decline in GUS activities in MDS1S-GUS and MACS-GUS was not due to an effect on the catalytic site of the enzyme.
Figure 2.
Substrate saturation curves of GUS in cell-free extracts prepared from tobacco leaves, 36 h after bombardment with different ATG constructs. Using 4-methylumbelliferyl β-d-glucuronide as the variable substrate, the raw data on GUS-specific activities were plotted (shown in inset, with y axis as in Fig. 1) and then normalized to allow visual comparison of the shape of the curves. MDS1S-GUS (▵) and MACS-GUS (▴) are also included, besides the constructs given in Figure 1.
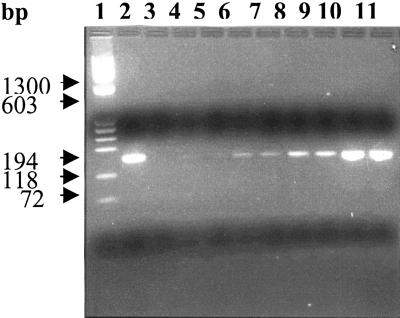
Figure 3 shows the results of reverse transcriptase (RT)-PCR, to compare relative steady state levels of GUS transcripts formed after bombardment with the constructs coding for M-GUS and MAS1S-GUS. From the 26th to 30th cycle, the amplification products showed a linear increase. The level of amplification achieved in different cases was comparable, suggesting that the different constructs transcribed uidA at similar levels. This excluded any substantial contribution of transcription and transcript stability on the 30- to 40-fold higher level of GUS expression from MAS1S-GUS as compared to M-GUS.
Figure 3.
Quantification of GUS transcripts by RT-PCR. One microgram of total RNA prepared from tobacco leaves bombarded with different constructs was subjected to RT-PCR. The amplification products in aliquots drawn at different stages were compared after electrophoresis. The agarose gel shows HaeIII ØX174 molecular weight standards (lane 1), PCR of standard GUS plasmid (lane 2), RT-PCR of control (non-bombarded; lane 3), and that of M-GUS (lanes 4, 6, 8, and 10) and MAS1S-GUS (lanes 5, 7, 9, and 11) on samples drawn after 26, 28, 30, and 40 cycles of amplification. Another control, comprising total RNA from MAS1S-GUS leaf but without adding the RT during the RT-PCR, showed no fragment after 40 cycles of amplification (lane not included).
Effect of Conserved N-Terminal Amino Acids on Protein Stability
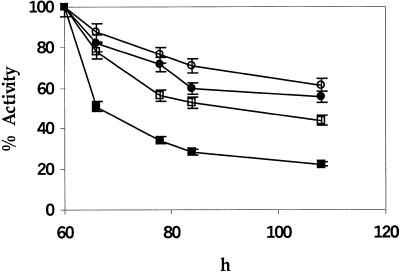
The cycloheximide treatment arrested the increase in GUS activity (Fig. 4). The half-life of GUS was calculated by using model y = α + βρx as the best fit. The activity of M-GUS declined with an average half-life of 4.26 h. Insertion of an Ala after the initiator Met gave a distinct advantage in reducing turnover of MA-GUS by 1.5-fold. The half life of GUS increased progressively in the constructs MA-GUS, MAS1-GUS, and MAS1S-GUS to 6.8, 7.6, and 9.3 h. However, the gain in intracellular stability was not sufficient to account for the nearly four-, eight-, and 30-fold higher in vivo activity seen, respectively (Fig. 1), in the case of MA-GUS, MAS1-GUS, and MAS1S-GUS, as compared with M-GUS.
Figure 4.
Relative activity of GUS in cell-free extracts prepared at different time points after transfer of leaves to medium containing cycloheximide. One set of leaf discs in each case was transferred to medium containing cycloheximide, 60 h after the bombardment. Each data point gives the average of six independent bombardment events. The symbols are the same as in Figure 1.
DISCUSSION
Our study shows that computational analysis of highly expressed plant genes can be used to identify features in gene architecture that possibly contribute toward determining their high level of expression. Though context sequences around the initiator ATG have been recognized earlier, such analyses have not been done after classifying the genes with respect to their level of expression or function. Following classification of genes into a dataset of highly expressed plant genes, novel features were identified downstream of the initiator ATG in this study. The validity of their function in augmenting the level of expression was supported by introducing such features in two reporter genes, uidA and gfp.
The results on the steady state of GUS transcripts in various constructs suggest that insertion of the conserved nucleotide sequence GCTNCC(T/A) CN following the initiator ATG does not enhance stability of the transcripts. A two-fold increase in intracellular stability of GUS protein suggests a modest role of the N-terminal amino acids in determining the half-life of proteins. However, a majority of the 30- to 40-fold increase in the expression of GUS following insertion of the ATG downstream conserved motif seems to be due to posttranscriptional events, although we can rule out protein and transcript stability. Though detailed steps in translational initiation and elongation are not understood, it is possible that nucleotides downstream of the ATG from +4 to +11 influence these events. Translation initiation factors (Raught et al., 2000) and ATG upstream nucleotides (Lütcke et al., 1987) specific to plants have been reported and may have distinct roles in determining translational efficiency. It is possible that efficient ribosomal recruitment at the initiator ATG involves an interaction between the +4 to +11 positions and the 48S pre-initiation complex in plants. The experiments with synonymous substitutions suggest that the TC at +7 to +8 and the C at +9 possibly influence translational efficiency favorably. Similarly, a C at +5 appears to be critical. In vitro studies on mRNAs with several substitutions in the ATG downstream region are required to understand the mechanism of such activation. The role of ATG downstream features in translational control is not known, though enhancer elements downstream of the ATG initiation codon have also been postulated in certain bacterial genes (O'Connor et al., 1999).
The results reported here show that the intracellular stability of GUS expressed in tobacco leaves was influenced by the specific nucleotide sequence and the corresponding amino acids at the N-terminal end. The stabilizing effect increased when the Met exposed at the N terminal of GUS was followed by Ala-Ser-Ser as the next three residues, with the maximum effect being observed when all three were present. Applicability of the result to GFP, used as a second reporter protein, further supports the validity of a purely computational analysis done by classifying genes by their level of expression. The results suggest that the specific nucleotide sequence and the corresponding amino acids at the N-terminal end of highly expressed proteins may have been evolutionarily selected to obtain high level of expression of these proteins. Interestingly, in Escherichia coli the presence of amino acids with smaller side chain lengths, i.e. Ala or Ser at the second position, was reported as particularly unfavorable to N-terminal stability of β-galactosidase (Hirel et al., 1989). The excision of N-terminal Met by Met aminopeptidase was reported to decrease in E. coli when amino acids with a longer side chain were inserted at the penultimate position of β-galactosidase. Thus, the length parameter rule for E. coli proteins beginning with Met does not apply to GUS and GFP stability in tobacco leaves.
In the case of processed proteins, different sets of amino acids at the non-Met N terminal impart stability in plants (Hondred et al., 1999), yeast, and rabbit reticulocytes (Tobias et al., 1991). Intracellular stability in such cases is determined by the ubiquitin-dependent N-end rule pathway. For instance, Met, Pro, Val, or Gly at the amino terminus stabilizes proteins in reticulocytes (Tobias et al., 1991). In addition to these, Thr, Ser, Ala, or Cys stabilizes proteins in yeast. In E. coli, any of the above with the exception of Pro and, in addition, Gln, Asn, Glu, Asp, Ile, or His, stabilize proteins. The principles of intracellular stability of ubiquitinylated proteins have recently been employed to enhance the accumulation of heterologous proteins in transgenic tobacco (Hondred et al., 1999). Our results suggest that Met-Ala-Ser-Ser, using the appropriate codons, may be a useful N-terminal tag for designing vectors for enhancing the stability of proteins in plants, provided their catalytic function is not affected by such modification.
MATERIALS AND METHODS
Computational Analysis
The EMBL gene database was screened manually to create a subset of angiosperm genes, potentially expressed at high level in plants, irrespective of their tissue or environmental specificities. The genes were classified as highly expressed, based on published information (Ruan et al., 1998; several other references cited in Sawant et al., 1999), on their expression level, various expressed sequence tags, and microarray analyses.
The selected dataset comprised highly expressed plant genes, several of which occur as gene families, that is to say chlorophyll a/b binding proteins, late embryogenesis abundant proteins, RuBP carboxylase small subunit, seed storage proteins (2S albumins, 7S albumins, 12S globulins, prolamins, napins, oleosins, 19-kD globulins, phaseolins, kafirins, zeins, and citrins), lectins, histones, photosystem related proteins (photosystem I proteins, photosystem II proteins, and cytochromes), nucleus coded mitochondrial proteins (RP15, citrate synthases, ATPases, and cytochromes), ribosomal proteins (L36, S10, S28, CL15, CL9, CS1, and CS70) Phe ammonia lyases, acyl carrier proteins, calmodulins, peroxidases, catalases, Pro- and Gly-rich proteins, extensins, polygalacturonases, cyclins (delta, 2A, 2B, 3A, and 3B), nodulins, translational elongation factors (TS, TU, and 1β), α-amylases, β-amylases, and amylase inhibitors. Other entries represented unique genes, that is to say actinidin protease from Actinidia delicosa, cinnamyl alchohol dehydrogenase, calcium binding protein, flavone isomerase, ferrodoxin, RNA binding protein from Arabidopsis, microspore specific protein from Brassica napus, fructokinase, phosphoglycerate kinase, ascorbate oxidase, nitrilase from tobacco (Nicotiana tabacum), allergen protein, glutathione reductase, lipoamide dehydrogenase from Pisum sativum, latex protein from Papaver somniferum, arabinogalactan, lipid transfer protein, Suc transfer protein from Pinus taeda, etc.
The sequences were manually aligned with respect to translational initiator ATG, as identified by the authors against individual entry in the database. Information about the ATG region of the 236 entries was employed in this analysis. A complete list of genes classified broadly with respect to their level of expression and used in this study is available on the Internet at www.geocities.com/rakeshtuli/PLNATG.htm, along with respective database references. The dataset of 236 entries was translated into nascent protein sequences to survey amino acid residues at the first 10 positions from the N-terminal region. The conserved nucleotide and the amino acid positions as determined by statistical tests of significance are given in Table I. A detailed analysis showing that several nucleotide sequence features in the highly expressed plant genes are in contrast to the features in genes expressed ubiquitously at low level has been published (Sawant et al., 1999). Differences in the levels of expression within gene families were ignored while creating the dataset since the objective was to identify features conserved in a majority of the members. As explained in “Results,” the conclusions did not change significantly if only one ortholog sequence representing a given gene family was included in the dataset instead of all the members reported from multiple plant species.
Construction of the Reporter Genes with Modified ATG 3′ Contexts
The uidA gene was amplified from pBI101.1, using nine different upstream primers designed to obtain the desired variants of the wild-type GUS mRNA. As given in Table I, the standard upstream primer (Primer 1) was used to amplify the wild-type uidA gene that comprises the native open reading frame that codes for M-GUS. The primers 2, 3, 4, 5, and 6 amplified GUS with Ala, Ala-Ser, Ala-Ser-Ser, Asp-Ser-Ser, and Ala-Cys-Ser, respectively, inserted between the first Met and the second Leu in the wild-type protein. The primers 7, 8, and 9 amplified variants of the Ala-Ser-Ser-coding mRNA in which the codon for the former Ser was substituted by synonymous codons. The nine constructs are referred to as M-, MA-, MAS1-, MAS1S-, MDS1S-, MACS-, MAS2S-, MAS3S-, and MAS4S-GUS, respectively. These were cloned into pUC19 by PCR-based ligation, in front of a highly expressed plant promoter (Sawant et al., 2001). The PCR was conducted using VentR DNA polymerase, which is known for a low error rate (5.7 × 10−5, which is very close to 4 × 10−5 for Klenow). The cloned fragments were sequenced in the ATG context region and found to be free of any errors. The different constructs, therefore, had exactly the same ATG-5′ translational context, untranslated leader, and promoter-regulatory region. They differed only with respect to the presence of additional codons inserted after the initiator Met as per the architecture identified in highly expressed plant genes. The primers 10 and 11 (Table I) were used (as upstream primers) to clone gfp (Reichel et al., 1996) to validate the effect of the conserved features on the expression of a second gene.
Analysis of Transient Expression of the Reporter Genes in Tobacco Leaves
The transient expression studies were carried out by microprojectile-mediated delivery of DNA in tobacco leaf discs using an improved method (Sawant et al., 2000). The modified protocol gives highly reproducible shot-to-shot results. Stability of the GUS in the bombarded leaf was measured by blocking de novo protein synthesis by placing the leaf discs on medium containing 300 μg mL−1 cycloheximide. Following bombardment with DNA, the leaf was cut into approximately 1-cm2 pieces and incubated on Murashige and Skoog-agar medium (in light). After 60 h of incubation, the leaf pieces were removed to determine GUS activity fluorometrically (Jefferson and Wilson, 1991). Endogenous GUS-like activity was suppressed by incorporating 20% (v/v) methanol in the reaction and treating the samples at 55°C for 10 min. At least six independent bombardments were done in each case to estimate GUS activity at each time point.
The single-point GUS and GFP activities were measured after incubating six replicates of the tissue on agar-Murashige and Skoog medium (in light) for 72 h after bombardment. The cell-free extracts for GFP measurements were prepared in 50 mm sodium phosphate buffer, pH 7. Emission was measured at 510 nm, following excitation at 475 nm (Reichel et al., 1996).
Analysis of GUS Transcript by RT-PCR
The relative amounts of GUS transcripts present in tobacco leaf discs were estimated in terms of the product formed in RT-PCR after bombardment of the discs with different gene constructs. One microgram of total RNA extracted in TRIZOL LS Reagent (Gibco-BRL, Cleveland) was treated with RNase-free DNase (1 unit of enzyme per microgram RNA; Amersham-Pharmacia, Uppsala), reverse transcribed by Superscript II (Gibco-BRL), and subjected to 40 cycles of PCR using primers that amplified a 175-bp fragment internal to the uidA gene. The phase of linear increase in PCR products was determined by drawing aliquots at different stages. The amount of amplified DNA was estimated both with Hoechst dye (H 33258, Sigma, St. Louis) and by scanning with the Flour-S (Bio-Rad Laboratories, Hercules, CA) documentation system after agarose gel electrophoresis.
ACKNOWLEDGMENTS
We thank Dr. S.K. Mandal of Central Drug Research Institute (Lucknow, India) for advice on statistical analysis of the data and Dr. Jaideep Mathur for providing the gfp construct.
Footnotes
This work was supported by fellowships (to S.V.S. and P.K.S.) and by a research grant from The Council of Scientific and Industrial Research, Government of India.
LITERATURE CITED
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-termianl residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Boeck R, Kolakofsky D. D Positions +5 and +6 can be major determinants of the efficiency of non-AUG initiation for protein synthesis. EMBO J. 1994;13:3608–3617. doi: 10.1002/j.1460-2075.1994.tb06668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavener DR. Comparison of the consensus sequence flanking translational start site in Drosophila and vertebrates. Nucleic Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Miller WA. Control of start codon choice on a plant viral RNA encoding overlapping genes. Plant Cell. 1993;5:679–692. doi: 10.1105/tpc.5.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunert S, Jackson RJ. The immediate downstream codon strongly influences the efficiency of utilization of eukaryotic translation initiation codons. EMBO J. 1994;13:3618–3630. doi: 10.1002/j.1460-2075.1994.tb06669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel Ph-H, Schmitter J-M, Dessen P, Fayat G, Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side- chain length of the penultimate amino acid. Proc Natl Acad Sci USA. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondred D, Walker JM, Mathews DE, Vierstra RD. Use of ubiquitin fusions to augment protein expression in transgenic plants. Plant Physiol. 1999;119:713–723. doi: 10.1104/pp.119.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Wilson KJ. The GUS gene fusion system. In: Gelvin S, Schilperoort R, editors. Plant Molecular Biology Manual. B14. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 1–33. [Google Scholar]
- Johnston JC, Rochon DM. Both codon context and leader length contribute to efficient expression of two overlapping open reading frames of a cucumber necrosis virus bifunctional subgenomic mRNA. Virology. 1996;221:232–239. doi: 10.1006/viro.1996.0370. [DOI] [PubMed] [Google Scholar]
- Joshi CP, Zhou H, Huang X, Chiang VL. Context sequences of translation initiation codon in plants. Plant Mol Biol. 1997;35:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lèvy F, Johnston JA, Varshavsky A. Analysis of a conditional degradation signal in yeast and mammalian cells. Eur J Biochem. 1999;259:244–252. doi: 10.1046/j.1432-1327.1999.00024.x. [DOI] [PubMed] [Google Scholar]
- Luehrsen KR, Walbot V. The impact of AUG start codon context on maize gene expression in vivo. Plant Cell Rep. 1994;13:454–458. doi: 10.1007/BF00231966. [DOI] [PubMed] [Google Scholar]
- Lütcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA. Selection of AUG codons differs in plants and animals. EMBO J. 1987;6:43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcin L, Marc F, Bènèdicte J, Arnaud S, Marc B. In vivo evaluation of the context sequence of the translation initiation codon in plants. Plant Science. 2000;154:89–98. doi: 10.1016/s0168-9452(00)00195-3. [DOI] [PubMed] [Google Scholar]
- McElroy D, Blowers AD, Jenes B, Wu R. Construction of expression vectors based on the rice actin 1 (Act1) 5′ region for use in monocot transformation. Mol Gen Genet. 1991;231:150–160. doi: 10.1007/BF00293832. [DOI] [PubMed] [Google Scholar]
- Murray EE, Lotzer J, Eberle M. Codon usage in plant genes. Nucleic Acids Res. 1989;17:477–498. doi: 10.1093/nar/17.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M, Asai T, Squires CL, Dahlberg AE. Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc Natl Acad Sci USA. 1999;96:8973–8978. doi: 10.1073/pnas.96.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Gingras A-C, Sonenberg N. Regulation of ribosomal recruitment in eukaryotes. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression, C.S.H. New York: Press; 2000. pp. 245–293. [Google Scholar]
- Reichel C, Mathur J, Eckes P, Langenkemper K, Koncz C, Schell J, Reiss B, Maas C. Enhanced green fluorescence by the expression of an Aequoreu victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proc Natl Acad Sci USA. 1996;93:5888–5893. doi: 10.1073/pnas.93.12.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Gilmore J, Conner T. Towards Arabidopsis genome analysis: monitoring expression profiles of 1400 genes using DNA microarays. Plant J. 1998;15:821–833. doi: 10.1046/j.1365-313x.1998.00254.x. [DOI] [PubMed] [Google Scholar]
- Sawant SV, Singh PK, Gupta SK, Madanala R, Tuli R. Conserved nucleotide sequence in highly expressed genes in plants. J Genet. 1999;78:1–8. [Google Scholar]
- Sawant SV, Singh PK, Madanala R, Tuli R. Designing of an artificial expression cassette for high level expression of transgenes in plants. Theor Appl Genet. 2001;102:635–644. [Google Scholar]
- Sawant SV, Singh PK, Tuli R. Pretreatment of microprojecticles to improve the delivery of DNA in plant transformation. BioTechniques. 2000;29:246–248. doi: 10.2144/00292bm09. [DOI] [PubMed] [Google Scholar]
- Sullivan ML, Green PJ. Post transcriptional regulation of nuclear-encoded genes in higher plants: the roles of mRNA stability and translation. Plant Mol Biol. 1993;23:1091–1104. doi: 10.1007/BF00042344. [DOI] [PubMed] [Google Scholar]
- Taylor JL, Jones JDG, Sandler S, Mueller GM, Bedbrook J, Dunsmuir P. Optimizing the expression of chimeric genes in plant cells. Mol Gen Genet. 1987;210:572–577. [Google Scholar]
- Tobias JW, Shrader TE, Rocap G, Varshavsky A. The N-end rule in bacteria. Science. 1991;254:1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- Tuli R (2000) List of highly expressed genes used for ATG region analysis. www.geocities.com/rakeshtuli/PLNATG.htm (December 28, 2000)