Abstract
Glutathione (GSH) and homo-GSH (hGSH) are the major low-molecular weight thiols synthesized in Medicago truncatula. Two M. truncatula cDNAs (gshs1 and gshs2) corresponding to a putative GSH synthetase (GSHS) and a putative hGSH synthetase (hGSHS) were characterized. Heterologous expression of gshs1 and gshs2 cDNAs in an Escherichia coli strain deficient in GSHS activity showed that GSHS1 and GSHS2 are a GSHS and an hGSHS, respectively. Leucine-534 and proline-535 present in hGSHS were substituted by alanines that are conserved in plant GSHS. These substitutions resulted in a strongly stimulated GSH accumulation in the transformed E. coli strain showing that these residues play a crucial role in the differential recognition of β-alanine and glycine by hGSHS. Phylogenetic analysis of GSHS2 and GSHS1 with other eukaryotic GSHS sequences indicated that gshs2 and gshs1 are the result of a gene duplication that occurred after the divergence between Fabales, Solanales, and Brassicales. Analysis of the structure of gshs1 and gshs2 genes shows they are both present in a cluster and in the same orientation in the M. truncatula genome, suggesting that the duplication of gshs1 and gshs2 occurred via a tandem duplication.
Glutathione (γ-glutamyl-cysteinyl-Gly; GSH) is the major low-Mr thiol in most eukaryotic organisms, where it is involved in the control of redox status, the detoxification of xenobiotics, and the protection against oxidative stress. In plants, GSH also acts as a storage form of reduced sulfur and is the precursor of phytochelatins, which participate in the sequestration of heavy metals (Noctor et al., 1998). There are at least two higher plant phylogenetic groups in which other γ-glutamyl-Cys-tripeptides are found either in addition to, or instead of, GSH. γ-Glutamyl-cysteinyl-Ser was identified in the Poaceae family (Klapheck et al., 1992) and γ-glutamyl-cysteinyl-Glu was detected in maize (Zea mays) seedlings upon exposure to cadmium (Meuwly et al., 1993). Homo-GSH (hGSH) is a low-Mr thiol that has been detected only in leguminous plants. This tripeptide was first found in leaves of Phaseolus vulgaris and Glycine max (Price, 1957) and was characterized as γ-glutamyl-cysteinyl-β-Ala (Carnegie, 1963). The level of hGSH and the ratio of hGSH to GSH vary dramatically between different genera of the family Fabaceae (Klapheck, 1988; Matamoros et al., 1999). hGSH is not detected in legumes like Vicia faba or Lupinus albus. In contrast, hGSH is the major low-molecular thiol in G. max, P. vulgaris, and Phaseolus coccineus. Moreover, in legumes where both GSH and hGSH are abundant, such as in Pisum sativum or in Medicago truncatula, the ratio of hGSH to GSH varies between the different organs of a given plant species (Frendo et al., 1999; Matamoros et al., 1999).
Many of the roles ascribed to GSH are also performed by hGSH. As such, it is the major storage form of reduced sulfur in Vigna radiata (Macnicol and Bergmann, 1984). hGSH has also been implicated in the plant defense against heavy metals acting as a precursor in the biosynthetic pathway of the metal-complexing phytochelatins (Grill et al., 1986; Klapheck et al., 1995) and against xenobiotics via the GSH-S-transferases (Skipsey et al., 1997). Dalton et al. (1986) also have suggested that hGSH is implicated in the scavenging of reactive oxygen species.
Synthesis of GSH and hGSH is a two-step process. In the first step, γ-glutamyl-Cys synthetase produces the dipeptide γ-glutamyl-Cys (γ-GC) from l-Glu and l-Cys. The formation of GSH and hGSH is determined by the substrate specificity of the enzyme catalyzing the second step. GSH synthetase (GSHS; E.C. 6.3.2.3) catalyzes the addition of Gly to γ-GC, whereas hGSH synthetase (hGSHS; E.C. 6.3.2.23) catalyzes the addition of β-Ala to γ-GC. GSHS from mammals (Rathbun et al., 1977; Oppenheimer et al., 1979), yeast (Mooz and Meister, 1967), and tobacco (Nicotiana tabacum; Hell and Bergmann, 1988) are highly specific for Gly and do not accept β-Ala as a substrate. In contrast, the hGSHS partially purified from shoots of V. radiata and from leaves of P. coccineus demonstrated a much higher affinity for β-Ala than for Gly (Macnicol, 1987; Klapheck et al., 1988). To our knowledge, no hGSHS has been purified yet to homogeneity and the specificity of these enzymes remains to be determined.
In M. truncatula, a model plant for the study of plant-Rhizobium sp. symbioses (Barker et al., 1990; Cook et al., 1997), we have shown previously that hGSH can only be detected in the underground parts of the mature plants (Frendo et al., 1999). This localization of hGSH was correlated with the presence of hGSHS activity that was detected in roots of mature plants (Frendo et al., 1999). Two partial cDNAs gshs1 and gshs2 showing high identity with Arabidopsis GSHS were isolated. Based on their expression, they were putatively ascribed to be gshs and hgshs, respectively (Frendo et al., 1999). Here, we show by heterologous expression of gshs1 and gshs2 in Escherichia coli that GSHS1 and GSHS2 are a GSHS and an hGSHS, respectively. Using phylogenetic analysis, we conclude that gshs1 and gshs2 result from a gene duplication that occurred after the divergence between the Fabales, Solanales, and Brassicales. Site-directed mutagenesis of GSHS2 shows that Leu534 and Pro535 play a crucial role in the differential recognition of β-Ala and Gly by hGSHS.
RESULTS
Isolation and Characterization of a cDNA Clone Encoding hGSHS in M. truncatula
Northern-blot analysis showed that gshs2 is expressed preferentially in roots and nodules rather than in leaves and flowers of mature M. truncatula plants (Frendo et al., 1999). Therefore, to isolate a full-length gshs2 cDNA, an M. truncatula cDNA library constructed with RNAs extracted from roots infected with Sinorhizobium meliloti was screened using gshs2 partial cDNA as a probe. Twenty-one positive clones were obtained from 5 105 clones in the first round of screening, out of which 10 were purified through the second and third rounds of screening.
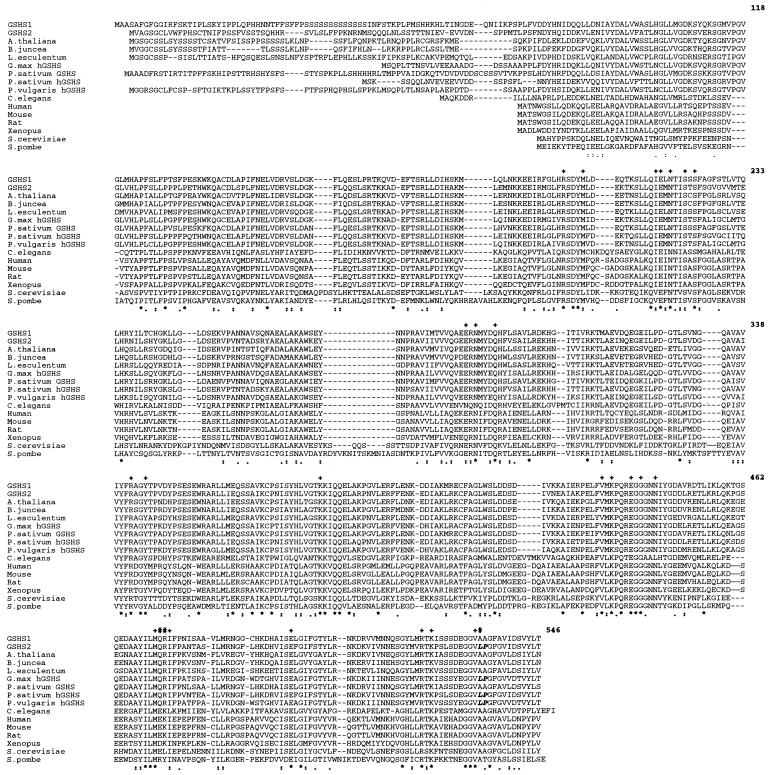
The nucleotide and the deduced amino acid sequences of the longest clone (phGSHS 2.3; accession no. AF194422) insert were determined. 5′-RACE was performed to obtain the gshs2 full-length cDNA sequence. An ATG (nucleotide 69) was detected on the translated sequence obtained from the RACE procedure. The presence of a stop codon (nucleotide 60) upstream and in frame with this ATG indicates that the encoded protein is complete. Protein analysis of the translated sequence using PSORT and CHLOROP computer programs predicted the presence of a plastid transit peptide. Comparison of GSHS2 with plant hGSHS and eukaryotic GSHS sequences is presented in Figure 1. All these proteins are strongly conserved among distantly related eukaryotic taxa (from plants to yeast and animals). However, all the plant sequences presented an N-terminal sequence extension, suggesting the presence of transit peptide on all the deduced protein sequences. GSHS2 shows higher identity with the P. sativum putative hGSHS (87%), the G. max putative hGSHS (76%), and the P. vulgaris putative hGSHS (72%) than with GSHS1 from M. truncatula (68%).
Figure 1.
Multiple alignment of eukaryotic GSHS and hGSHS. SWISSPROT-TREMBL accession nos. are as follows: Q21549 (M176.2, Caenorhabditis elegans), P46413 (GSHS rat [Rattus norvegicus]), P51855 (GSHS mouse [Mus musculus]), P48637 (GSHS human), P35668 (GSHS African clawed frog [Xenopus laevis]), Q08220 (GSHS baker's yeast [Saccharomyces cerevisiae]), P35669 (GSHS fission yeast [Schizosaccharomyces pombe]), O22494 (GSHS tomato [Lycopersicon esculentum]), O23732 (GSHS leaf mustard [Brassica juncea]), AJ243812 (GSHS Arabidopsis), AJ272035 (hGSHS G. max), AF194421 (GSHS1 M. truncatula), AF194422 (GSHS2 M. truncatula), AF258320 (hGSHS P. vulgaris), AF258319 (hGSHS P. sativum), and AF231137 (GSHS P. sativum). Amino acid residues corresponding to GSHS2 are numbered on the right. Stars represent identical amino acids and dots represent conserved amino acids (below). Amino acids involved in the catalytic site of human GSHS and conserved in GSHS2 are represented by + and amino acids located in the catalytic site of human GSHS and changed in GSHS2 are represented by # (above). Leu534 and Pro535 are represented in bold letters.
To determine the activity of GSHS2, the cDNA was expressed in an E. coli mutant deficient in GSHS activity. A PCR fragment corresponding to the protein sequence conserved among the GSHS (Met70 to Thr546 of GSHS2) was cloned in the pBluescript SK plasmid (pSK) in frame with the β-galactosidase coding sequence under the control of the lac promoter (phGSHS.exp). The phGSHS.exp was transfected in the gshsB-deficient E. coli strain 830 (strain hgshs830). As shown in Table I, a high level of GSH and a very low level of hGSH were detected in the wild-type strain AB1157. In comparison, the GSHS-deficient E.coli strain 830 carrying the pSK vector (strain 830sk) accumulated γ-GC and contained a very diminished level of GSH. hGSH was below the limit of detection in the 830sk strain, even when β-Ala was added to the growth medium. Accumulation of GSH and hGSH was observed in the hgshs830 strain and addition of β-Ala to the growth medium increased the level of hGSH 9-fold. Furthermore, both hGSHS (0.32 ± 0.03 nmol hGSH min−1 mg protein−1) and GSHS (0.12 ± 0.02 nmol GSH min−1 mg protein−1) activities were found in the hgshs830 strain, whereas no GSHS and hGSHS activities were detected in the 830sk strain. The specificity of the expressed protein with respect to β-Ala and Gly was estimated by steady-state kinetic measurements in crude bacterial extracts. The recombinant enzyme displayed a much higher affinity for β-Ala [Km(app.) 28 mm, Vmax 1;1 nmol hGSH min−1 mg−1 protein] than for Gly [Km(app.) 260 mm, Vmax 0; 26 nmol GSH min−1 mg−1 protein]. Thus, the specificity constant (Vmax/Km) of GSHS2 for β-Ala (3, 9 10−2) is 40-fold higher than that for Gly (1 10−3). Taken together, these results indicate that GSHS2 is an hGSHS.
Table I.
γ-GC, GSH, and hGSH content measured in E. coli extracts
| Strains | γ-GC | GSH | hGSH |
|---|---|---|---|
| nmol per 5.109 cells | |||
| AB1157 | nd | 44 ± 5 | 0.3 ± 0.2 |
| 830sk | 63 ± 6 | 1 ± 0.2 | nd |
| 830sk + β-ala | 61 ± 7 | 1.3 ± 0.1 | nd |
| hgshs 830 | 47.7 ± 1.3 | 2.2 ± 0.4 | 0.5 ± 0.2 |
| hgshs 830 + β-ala | 45.4 ± 3.2 | 2 ± 0.3 | 4.3 ± 0.4 |
Extracts prepared from GSHS-deficient E. coli strain 830 containing the pSK vector (830sk) or the phGSHS exp vector (hgshs830) were grown with 1 mm isopropyl β-d-thiogalactopyranoside (IPTG). β-Ala was added to the growth medium where indicated. Nos. represent the means of three independent experiments ± se. nd, Not detected.
Phylogenetic and Genomic Analysis of gshs1 and gshs2
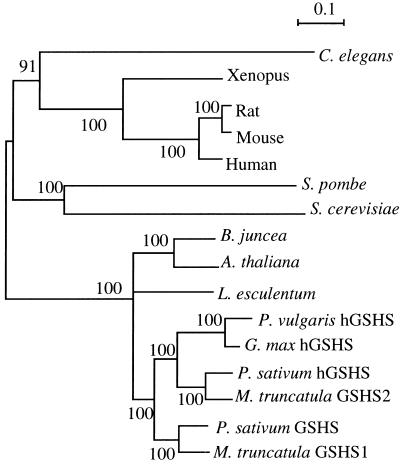
Multiple sequence comparison of GSHS revealed a high identity among all eukaryotic GSHS. The phylogenetic tree of eukaryotic GSHS protein family, shown in Figure 2, clearly indicates that gshs1 and gshs2 were produced by gene duplication that occurred in the Fabale lineage after the divergence between Fabales, Solanales (tomato), and Brassicales (Arabidopsis and leaf mustard) and before the divergence between the different legume families.
Figure 2.
Unrooted phylogenetic tree of the GSHS and hGSHS protein family. Branch lengths are proportional to sequence divergence. Branch labels record the stability of the branches over 500 bootstrap replicates. SWISSPROT-TREMBL accession nos. are indicated in Figure 1.
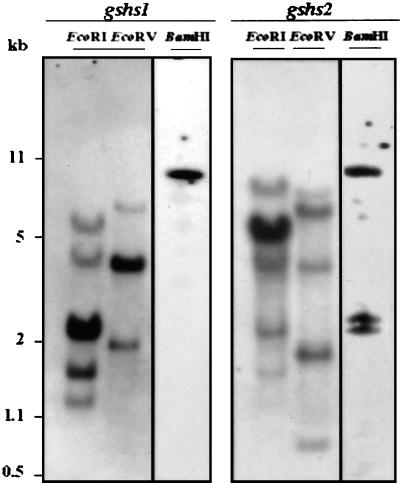
Genomic Southern-blot analysis was used to estimate the copy number of gshs1 and gshs2 in the M. truncatula genome. As shown in Figure 3, multiple restriction fragments from each digestion hybridized to the gshs2 probes, suggesting that hGSHS is encoded by a small gene family. However, based on the identity observed between the different GSHS and hGSHS cDNAs, we cannot exclude that some of the hybridizing fragments correspond to other gshs genes. Only one BamHI restriction fragment hybridized to the gshs1 probe, suggesting that this enzyme is encoded by a single gene. It is interesting that for all other lineages for which data are available, GSHS appears to be encoded by a single gene. We did not find any other gshs-related genes in baker's yeast, C. elegans, and Arabidopsis genomes that have been entirely sequenced. Compilation of DNA sequence databases and Southern-blot analyses also suggest that there is a single GSHS gene in the human genome (Webb et al., 1995).
Figure 3.
Genomic southern analysis of M. truncatula gshs1 and gshs2. Total genomic M. truncatula DNA was digested with EcoRI, EcoRV, and BamHI. The filters were probed with gshs1 cDNA or gshs2 cDNA as described in the experimental procedures. Size markers are indicated on the left.
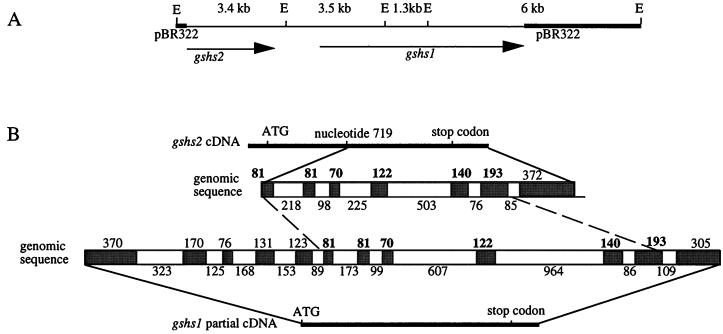
Screening of a M. truncatula genomic library using the gshs2 cDNA (Frendo et al., 1999) as a probe was performed to characterize the structure of gshs2 gene. The nucleotide sequence of a 9.5-kb genomic clone G1-gshs (accession no. AF194421) was determined and analyzed (Fig. 4). gshs1 and partial gshs2 gene sequences were both present in the same orientation in the genomic DNA clone (Fig. 4A). This result strongly suggests that the gene duplication of gshs1 and gshs2 has occurred via a tandem duplication due to an unequal crossing over. The gshs2 partial genomic sequence contains seven exons ranging in size from 70 to 305 bp and six introns that range from 76 to 503 bp (Fig. 4B).
Figure 4.
Genomic organization of gshs1 and gshs2. A, Restriction map of the genomic clone G1-gshs showing the positions of EcoRI sites (E) and the sizes (in kilobase pairs) of EcoRI fragments. The arrows indicate the regions of the clone complementary to gshs1 and gshs2 cDNAs and the direction of transcription. B, The intron/exon structure of gshs1 and gshs2 genes. Size of exons (above) and introns (below) is indicated in base pairs. Conserved exons between gshs1 and gshs2 are indicated in bold.
Reverse transcriptase (RT)-PCR experiments were performed to determine the 5′ end of gshs1 coding sequence. The gshs1 genomic sequence corresponding to the gshs1 cDNA contains 12 exons ranging from 70 to 370 bp and 11 introns that range from 86 to 964 bp. Structural comparison of gshs1 and gshs2 shows that the exon size has been conserved in the two genes. In contrast, intron size has not been maintained between the two genes even if intron sequences bordering the splicing sites are highly similar. Comparison of gshs1 with the Arabidopsis gshs (accession no. AJ243812) indicates that the structure of the two genes presents a high similarity. The number and the size of the exons are conserved between the two genes. These results demonstrate that the genomic structure of these genes is highly conserved in plants.
Characterization of GSHS1 Activity
Phylogenetic analysis of the eukaryotic GSHS shows that gshs1 and gshs2 are the result of a gene duplication. After duplication, gshs1 may have retained its original GSHS function, whereas gshs2 has acquired the ability to synthesize hGSH. This hypothesis is in agreement with our previous results showing that the expression of gshs1 in leaves is correlated with the detection of GSHS activity in this tissue and the lack of hGSHS activity (Frendo et al., 1999). Nevertheless, to rule out the possibility that gshs1 is an alternative hGSHS, GSHS1 was expressed in the E. coli strain 830 deficient in GSHS activity. A PCR fragment corresponding to the protein sequence conserved among the GSHS (Pro78 to Thr554 of GSHS1) was cloned in the pSK plasmid in frame with the β-galactosidase coding sequence under the control of the lac promoter (pGSHS.exp). In the gshsB-deficient E. coli strain 830 carrying the pGSHS.exp (strain gshs830), an accumulation of GSH (44 ± 1 nmol GSH/5 109cells) was observed and no hGSH was detected even when β-Ala was added to the growth medium. These results clearly show that GSHS1 is a GSHS and indicate that it does not accept β-Ala as a substrate. Thus, GSHS1 has retained its GSHS activity after the gene duplication that occurred in the Fabale lineage.
Substitution of Leu534 and Pro535 by Alas Resulted in a Change of GSHS2 Activity
The amino acids involved in the human GSHS catalytic site (Polekhina et al., 1999) are highly conserved in hGSHS (Fig. 1). The amino acids that are part of the magnesium ion binding site (Glu-216, Asn-218, and Glu-439) and that interact with the γ-GC moiety (Arg-200, Ser-221, Ser-223, Glu-278, Asn-280, Gln-284, Arg-332, and Tyr-345) are fully conserved. The amino acids contributing to the ATP-binding site (Met-204, Iso-215, Lys-381, Val-433, Lys-435, Gly-441, Asn-444, Tyr-446, Met-471, Ileu-474, Glu-498, and Lys-525) are conserved, whereas Gln-472 and Arg-473 replace Glu and Lys, respectively. These two conservative changes in amino acids were observed in all the plant GSHS and hGSHS sequences reported so far. Likewise, the Arg-522 and Val-533 present in the glycyl moiety-binding site of GSHS are conserved in hGSHS.
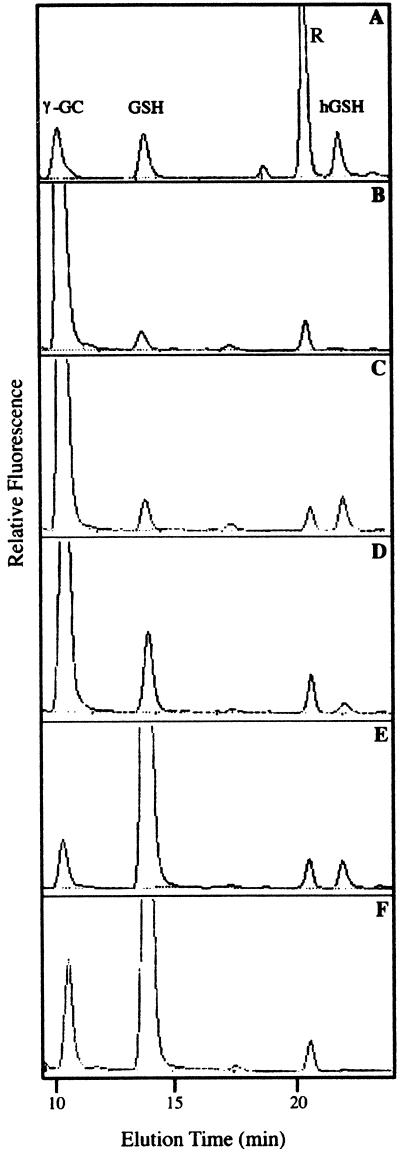
The major difference in the amino acids involved in the catalytic site between hGSHS and GSHS is the replacement of Leu-534 by Ala. This residue has been proposed to be implicated in the glycyl-binding site of the human GSHS (Polekhina et al., 1999). It is interesting that the Pro-535, present in hGSHS, is also replaced by an Ala in the plant GSHS. To test whether these two amino acids play a role in the differential recognition of Gly and β-Ala by GSHS2, site-directed mutagenesis was performed to substitute Leu-534 by Ala (phgshsL/A) and to substitute Leu-534 and Pro-535 by two Alas (phgshsLP/AA) on the recombinant hGSHS. The production of hGSH and GSH was tested in the GSHS-deficient E.coli strain 830 transformed with the phgshsL/A (strain hgshsL/A) or the phgshsLP/AA (strain hgshsLP/AA; Fig. 5). As shown in Table II, an increased production of GSH and a reduced formation of hGSH were observed in the hgshsL/A strain as compared with the hgshs830 strain. A ratio (hGSH to GSH) of 0.21 was obtained in the hgshsL/A strain compared with the ratio of 6.14 for the hgshs830 strain. Thus, the Leu-534 residue appears to be important for the differential recognition of β-Ala/Gly by hGSHS. The substitution of Leu-534 and Pro-535 by Alas led to a further increased formation of GSH in the hgshsLP/AA strain as compared with the hgshs830 strain; formation of hGSH was also detected and the ratio of hGSH to GSH produced in this strain was lowered to 0.08.
Figure 5.
HPLC Analysis of thiol compounds in E. coli mutant strain expressing the different constructs. A, Elution profile of thiol standard containing γ-GC, GSH, and hGSH. Elution profile of thiol compounds present in the E. coli 830sk strain extract (B), the E. coli hgshs830 strain extract (C), the E. coli hgshsL/A strain extract (D), the E. coli hgshsLP/AA strain extract (E), and the E. coli gshs830 strain extract (F). R represents a reagent peak. β-Ala was added to the growth medium.
Table II.
GSH and hGSH content measured in E. coli extracts
| Strains | GSH | hGSH |
|---|---|---|
| nmol per 5.109 cells | ||
| 830sk | 1.3 ± 0.2 | nd |
| hgshs 830 | 2 ± 0.3 | 4.3 ± 0.4 |
| hgshsL/A | 6 ± 0.7 | 1 ± 0.1 |
| hgshsLP/AA | 47 ± 6 | 3.5 ± 0.6 |
| gshs830 | 44 ± 1 | nd |
Extracts prepared from GSHS-deficient E. coli strain 830sk, hgshs830, hgshsL/A, hgshsLP/AA, and gshs830 were grown with 1 mm IPTG and β-Ala. Nos. represent the means of three independent experiments ± se. nd, Not detected.
DISCUSSION
hGSHS Specificity
There has been an open question about whether the formation of hGSH is catalyzed by a specific hGSHS or by a GSHS with a broad substrate specificity in plants that contain both GSHS and hGSH activities. In P. sativum (Bergmann and Rennenberg, 1993; Matamoros et al., 1999) and in M. truncatula (Frendo et al., 1999) where both tripeptides are present, studies of GSHS and hGSHS activities indicated that both plants have a specific hGSHS in addition to a GSHS. cDNA sequences corresponding to legume putative hGSHS and GSHS have been described very recently (Moran et al., 2000). However, to our knowledge, no biochemical data have been reported about the activities of the proteins encoded by these cDNAs. Here, we show for the first time the hGSHS activity of a recombinant protein by heterologous expression of a cDNA in E. coli. This protein, named GSHS2, has a high identity with putative hGSHS from other plants. Heterologous expression of gshs2 cDNA in E. coli shows that recombinant GSHS2 has hGSHS activity. GSHS2 also shows GSHS activity but with an approximately 10-fold higher Km(app) for Gly than that for β-Ala. This differential substrate recognition by the recombinant GSHS2 is similar to the 6-fold ratio observed for the hGSHS of P. sativum, a plant that contains both hGSH and GSH (Bergmann and Rennenberg, 1993). However, this ratio is much lower than the 73-fold ratio observed in partial purified protein extracts from leaves of P. coccineus, a legume plant that contains only hGSH (Klapheck et al., 1988). With respect to the M. truncatula GSHS1, heterologous expression of gshs1 cDNA in E. coli shows that GSHS1 is a GSHS and has a high specificity for Gly. This result is in agreement with the results observed for the other eukaryotic GSHS (Mooz and Meister, 1967; Rathbun et al., 1977; Oppenheimer et al., 1979; Hell and Bergmann, 1988).
Comparison of the amino acids involved in the catalytic site of human GSHS with those of plant GSHS and hGSHS shows that they are very conserved among these proteins. This observation is in accordance with the in vitro biochemical data obtained on plant GSHS and hGSHS. Both plant GSHS and hGSHS showed maximum activity at pH 8.0 to 9.0, an absolute requirement for magnesium and slight stimulation by potassium (Bergmann and Rennenberg, 1993). Moreover, both enzymes showed similar apparent Km for γ-GC (Bergmann and Rennenberg, 1993). Thus, GSHS and hGSHS are very similar enzymes that differ only by the differential affinity for their substrates Gly and β-Ala. The substitution of Leu534 by Ala in GSHS2 results in a 70% decrease in the production of hGSH in E. coli, indicating that this Leu residue plays a major role in the recognition of β-Ala. The hgshsLP/AA strain produces the same amount of GSH as the strain expressing GSHS1. This indicates that the replacement of the Leu and Pro residues by Alas results in the restoration of a high GSHS activity. On the other hand, in contrast with the strain expressing GSHS1, the hgshsLP/AA strain keeps an hGSHS activity, indicating that other amino acids are involved in the Gly/β-Ala specificity.
Acquisition of hGSHS Activity in Legumes
Phylogenetic analysis of the eukaryotic GSHS shows that gshs1 and gshs2 are the result of a gene duplication that occurred after the divergence between Fabales, Solanales, and Brassicales. Sequencing of genomic DNA reveals that gshs1 and gshs2 are present in a cluster and in the same orientation on the M. truncatula genome, strongly suggesting that this gene duplication has occurred via a tandem duplication due to an unequal crossing over. Gene duplications have played a major role in the acquisition of new functions during evolution (Ohno, 1970; Li and Graur, 1991). After duplication, one copy may retain its original function, whereas the other accumulates sequence changes such that it may be modified in its expression pattern and/or in the biochemical activity of its protein product. The evolution of gshs1 and gshs2 genes seems to have followed this scenario: gshs1 has retained the original GSHS activity, whereas gshs2 has acquired a distinct expression pattern and the ability to synthesize hGSH. Consistent with this model, GSHS1 is slightly more conserved than GSHS2 with respect to their common ancestral gene. The Fabales are the only plants in which hGSH has been detected. Thus, it is tempting to propose that this feature results directly from the acquisition of hGSHS activity in the Fabale lineage after duplication of the ancestral gshs gene. The differential localization of hGSH and various hGSH/GSH ratios observed in different legume plants (Klapheck, 1988; Matamoros et al., 1999) suggests that the hgshs and gshs genes have evolved in various manners in the different genera of the Fabaceae family after the gene duplication.
It will now be of interest to understand why this activity has been specifically developed in leguminous plants and why the expression patterns of hgshs and gshs have evolved in various manners in the different genera of the Fabaceae family.
MATERIALS AND METHODS
Plants, Bacterial Strains, and Libraries
Seeds of Medicago truncatula cv Jemalong genotype J5 were grown as previously described (Frendo et al., 1999). Escherichia coli parental strain AB1157 (F−, thr−, leu−, proA−, his−, arg−, thi−, and strA−) and the GSHS-deficient gshB mutant strain 830 (F−, thr−, leu−, proA−, his−, arg−, thi−, strA−, and gshB−) were obtained from the Delft University of Technology (Delft, The Netherlands). The M. truncatula cDNA library, constructed with RNAs extracted from roots 6 to 48 h after infection with Sinorhizobium meliloti, and the genomic library were generous gifts from Dr. Pascal Gamas and from Dr. David Barker, respectively (Laboratoire de Biologie Moléculaire des Relations Plantes-Microorganismes, Castanet-Tolosan, France).
Cloning and Analysis of cDNA and Genomic Clones
The 1-kb gshs2 partial cDNA (Frendo et al., 1999) was radiolabeled with [α32P] dCTP by random priming (Prime-a-gene labeling system, Promega, Charbonnières, France) and used as a probe to screen M. truncatula cDNA and genomic libraries. Hybridization was performed at 65°C and filters were washed twice in 6× SSC and twice in 1× SSC (1× SSC: 150 mm NaCl and 15 mm sodium citrate, pH 7.0) at 65°C.
DNA sequencing was carried out on both strands by the dideoxy chain termination method. Sequence data were analyzed using the UWGCG software (Devereux et al., 1984). Deduced GSHS1 and GSHS2 protein sequences were compared with all sequences available in the National Center for Biotechnology Information databases with BLASTP and BLASTN programs (Altschul et al., 1997). Homologous sequences were aligned with CLUSTALW (Thompson et al., 1994). The phylogenetic tree was derived from this multiple alignment (306 sites, gap excluded), using the neighbor-joining method (Saitou and Nei, 1987) with percentage of accepted point mutations distances. Analysis of possible transit peptide was performed using PSORT (Nakai and Kanehisa, 1992) and CHLOROP programs (Emanuelsson et al., 1999).
Characterization of gshs1 and gshs2 5′ cDNA Ends
The characterization of the 5′ part of the gshs1 open reading frame was achieved by RT-PCR amplification. Total RNA was purified from leaves according to Shirzadegan et al. (1991). Reverse transcription reaction was performed using the Omniscript RT kit (Qiagen, Courtaboeuf, France). PCR amplification was achieved using a genomic clone- (G1-gshs) specific primer, 5′-GCAATAGGCAATG-GCTGCTTC-3′, and a gshs1 cDNA clone-specific primer, 5′-CCATGGCAAGTAAGTATGTACCTG-3′, during 35 cycles of sequential incubations at 95°C for 0.5 min, 60°C for 1 min, and 72°C for 1 min in a 50-μL reaction mixture containing 10 pmol of each primer and 5 units of Taq DNA polymerase (Appligene-Oncor, Illkirch, France).
The rapid amplification of the 5′ gshs2 cDNA end (5′-RACE) was performed using the Marathon cDNA amplification kit (CLONTECH, Palo Alto, CA). PCR amplification was performed using the adaptor-specific primer (AP1) 5′-CATAACGAATCTGGAAAGCAGATGG- 3′ and a gshs2-specific primer (GSHS2.1) 5′-TGAGTTTCAGGAA-GAGGGGGAGGAG- 3′ during 35 cycles of sequential incubations at 95°C for 0.5 min and 68°C for 2 min in a 50-μL reaction mixture containing 10 pmol of each primer and 5 units of Taq DNA polymerase (Appligene-Oncor). The reaction product was purified from agarose gel using QIAEX II kit (Qiagen) and used as template for a nested PCR using a second gshs2-specific primer (GSHS2.2) 5′-AAGATGCACCAACCCTACACCAGGAACTC-3′ with the same reaction parameters.
The PCR-amplified DNA fragments were inserted in the pGEM-T vector (Promega) and three independent transformants from each subcloning were sequenced and found to be identical.
Southern-Blot Analysis
Genomic DNA was extracted from leaves of M. truncatula genotype J5 with Genomic-tip 500/G kit (Qiagen). DNA samples (10 μg) were digested, fractionated by electrophoresis on agarose gel, and transferred by capillary flow onto Hybond N membranes (Amersham, Orsay, France). A filter carrying EcoRI and EcoRV DNA digests was probed at 65°C with the gshs1-partial cDNA (1,133 bp; Frendo et al., 1999) and gshs2 full-length cDNA radiolabeled with [α32P]dCTP. Filters were washed twice in 6× SSC and twice in 1× SSC at 65°C. The filter carrying BamHI DNA digest was probed at 55°C with the gshs1- and gshs2-partial cDNAs radiolabeled with [α32P]dCTP and washed twice in 6× SSC at 55°C.
Expression of gshs1 and gshs2 cDNAs in the GSHS Activity-Deficient Mutant Strain of E. coli
To express GSHS2, a DNA fragment (1,450 bp) was amplified by PCR using 0.5 μg of phGSHS-2.3 as template and 50 pmol each of primers: sense, 5′-TTCT-GGATCCATGACGCTTCCCAGCTT-3′ (BamHI site is underlined); and antisense, 5′-AATTCCTCGAGTTGGTTTC-ATCATGTGAGG-3′ (XhoI site is underlined). DNA was amplified during 10 cycles of sequential incubations at 95°C for 0.5 min, 60°C for 1 min, and 72°C for 2 min, in a final 50-μL reaction mixture containing 5 units of Taq DNA polymerase (Appligene-Oncor).
To express GSHS1, RT-PCR amplification was performed, essentially as described previously in this paper. PCR amplification was realized using 2.5 units of pfuturbo DNA polymerase (Stratagene Europe, Amsterdam) and the following oligonucleotides: sense, 5′-AAAGGATCCTTC-TCCTCTATTCGTTGATG-3′ (BamHI site is underlined); and antisense, 5′-AAACTCGAGCAATGTGGCCTCCCTTTC -3′ (XhoI site is underlined). DNA amplification was performed during 30 cycles of sequential incubations at 95°C for 0.5 min, 60°C for 1 min, and 72°C for 2 min in a final 50-μL reaction volume.
The PCR reaction products were purified from agarose gel using QIAEX II kit (Qiagen) and inserted in the pGEM-T vector (Promega). The PCR inserts were digested with BamHI and XhoI and the restricted DNA fragments were cloned in the pBS phagemid expression vector (Stratagene, Europe), in frame with the β-galactosidase gene. The constructions (phGSHS.exp and pGSHS.exp) were verified by sequencing. gshsB-deficient E. coli strain was transformed with each construction and grown in Luria broth medium containing 100 μg ml−1 ampicillin, 1 mm IPTG, and 400 μg ml−1 β-Ala.
Site-Directed Mutagenesis of GSHS2
Substitution of Gly534 and Pro535 by Alas was introduced using the quick change site-directed mutagenesis kit (Stratagene Europe). PCR was achieved using the phGSHS.exp as template and the following oligonucleotide primers: sense, 5′-GATGAAGGTGGGGTTGCGCCTGG-3′ (substituted nucleotides are underlined); antisense, 5′- CCAG-GCGCAACCCCACCTTCATC-3′ for Gly/Ala substitution and sense, 5′-GATGAAGGTGGGGTTGCG- GCTGG-3′ (substituted nucleotides are underlined); and antisense, 5′-CCAGCCGCAACCCCACCTTCATC-3′ for Gly-Pro/Ala-Ala double substitution. DNA amplification was performed during 35 cycles of sequential incubations at 95°C for 1 min, 58°C for 1 min, and 72°C for 6 min, in a 50-μL reaction mixture containing 0.2 μg of phGSHS.exp, 50 pmol of each primer, and 2.5 units of pfuturbo DNA polymerase (Stratagene Europe). The construction was verified by sequencing (MWG-Biotech, Courtaboeuf, France). gshsB-deficient E. coli strain was transformed and grown in Luria broth medium containing 100 μg ml−1 ampicillin and 1 mm IPTG and 400 μg ml−1 β-Ala.
Quantification of GSH and hGSH and Determination of GSHS and hGSHS Activities
Thiols were extracted with perchloric acid, derivatized with monobromobimane, and quantified after separation on reverse phase HPLC as described previously (Frendo et al., 1999). Commercial GSH (Sigma, Saint Quentin Fallarier, France) and α-GC (Promochem, Molsheim, France) were used as standards. The hGSH used as a standard was synthesized by Neosystem (Strasbourg, France). Results are the mean of three independent experiments ± se.
For the enzymatic activity determination, cells were resuspended in 1% of the original volume in the extraction buffer (100 mm Tris-HCl [pH7.8], 5 mm EDTA, 20 mm MgCl2, and 0.4 mm dithiothreitol), disrupted by two passages through a French press (8,500 lb/in2) and centrifuged at 33,000g for 30 min at 4°C. Endogenous low-Mr thiols were eliminated by gel filtration of the resulting supernatant on Sephadex G15 columns (Pharmacia, Orsay, France) using the extraction buffer. Protein containing fractions were concentrated with a Diaflow cell fitted with a YM5-type membrane (MERCK-EUROLAB, Illkirch, France). Proteins were quantified according to the method of Bradford (Bradford, 1976) with bovine serum albumin as a standard. Activities of GSHS and hGSHS were determined as described (Frendo et al., 1999). Results are the mean of two independent experiments ±se. Apparent Km and Vm values were determined using Lineweaver-Burk plot.
ACKNOWLEDGMENTS
We gratefully acknowledge Prof. Bruce Demple, Dr. Dominique Job, and Dr. Judith Harrison for critical reading of the manuscript. We thank Dr. Anuradha Alahari, Dr. Cecile Bladier, Dr. Emmanuel Baudouin, and Dr. Sophie Moreau for helpful discussions. We acknowledge Dr. Pascal Gamas and Dr. David Barker for providing us with the M. truncatula cDNA library and the genomic library, respectively. We are indebted to Dr. Muriel Sagan and Dr. Gérard Duc for providing us with the M. truncatula seeds and to Mr. Alain Gilabert for technical assistance.
Footnotes
This work was funded in part by the European Communities' Human Capital and Mobility program (contract no. CT94–0605), by the Ministère de l'Education Nationale et de la Recherche, and by the Centre National de la Recherche Scientifique. M.J.H.J. is the recipient of a postdoctoral fellowship from the European Communities' Training and Mobility of Researchers program.
LITERATURE CITED
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DG, Bianchi S, Blondon F, Dattée Y, Duc G, Flament P, Galusci P, Génier G, Guy P, Muel X. Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol Biol Rep. 1990;8:40–49. [Google Scholar]
- Bergmann L, Rennenberg H. Glutathione metabolism in plants. In: De Kok LJ, editor. Sulfur Nutrition and Assimilation in Higher Plants. The Hague, The Netherlands: SPB Academic Publishing; 1993. pp. 109–123. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carnegie PR. Structure and properties of a homologue of glutathione. Biochem J. 1963;89:471–478. doi: 10.1042/bj0890471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR, VandenBosch K, de Bruinj FJ, Huguet T. Model legumes get the nod. Plant Cell. 1997;9:275–280. [Google Scholar]
- Dalton DA, Russell SA, Hanus FJ, Pascoe GA, Evans HJ. Enzymatic reactions of ascorbate and glutathione that prevent peroxide damage in soybean root nodules. Proc Natl Acad Sci USA. 1986;83:3811–3815. doi: 10.1073/pnas.83.11.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendo P, Gallesi D, Turnbull R, Van de Sype G, Hérouart D, Puppo A. Localization of glutathione and homoglutathione in Medicago truncatula is correlated to a differential expression of genes involved in their synthesis. Plant J. 1999;17:215–219. [Google Scholar]
- Grill E, Gekeler W, Winnaker E-L, Zenk MH. Homo-phytochelatins are heavy metal-binding peptides of homo-glutathione containing Fabales. FEBS Lett. 1986;205:47–50. [Google Scholar]
- Hell R, Bergmann L. Glutathione synthetase in tobacco suspension cultures: catalytic properties and localization. Physiol Plant. 1988;72:70–76. [Google Scholar]
- Klapheck S. Homoglutathione: isolation, quantification and occurrence in legumes. Physiol Plant. 1988;74:727–732. [Google Scholar]
- Klapheck S, Chrost B, Starke J, Zimmermann H. γ-Glutamylcysteinylserine: a new homologue of glutathione in plants of the family Poaceae. Bot Acta. 1992;105:174–179. [Google Scholar]
- Klapheck S, Schlunz S, Bergmann L. Synthesis of phytochelatins and homophytochelatins in Pisum sativum L. Plant Physiol. 1995;107:515–521. doi: 10.1104/pp.107.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapheck S, Zopes H, Levels H-G, Bergmann L. Properties and localization of the homoglutathione synthetase from Phaseolus coccineus leaves. Physiol Plant. 1988;74:733–739. [Google Scholar]
- Li WH, Graur D. Fundamentals of molecular evolution. Sunderland, MA: Sinauer Associates, Inc.; 1991. [Google Scholar]
- Macnicol PK. Homoglutathione and glutathion synthetases of legumes seedlings: partial purification and substrate specificity. Plant Sci. 1987;53:229–235. [Google Scholar]
- Macnicol PK, Bergmann L. A role for homoglutathione in organic sulfur transport to the developing mung bean seed. Plant Sci. 1984;36:219–223. [Google Scholar]
- Matamoros MA, Moran JF, Iturbe-Ormaetxe I, Rubio MC, Becana M. Glutathione and homoglutathione synthesis in legume root nodules. Plant Physiol. 1999;121:879–888. doi: 10.1104/pp.121.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwly P, Thibault P, Rauser WE. γ-Glutamylcysteinylglutamic acid-a new homologue of glutathione in maize seedlings exposed to cadmium. FEBS Lett. 1993;336:472–476. doi: 10.1016/0014-5793(93)80858-r. [DOI] [PubMed] [Google Scholar]
- Mooz ED, Meister A. Tripeptide (glutathione) synthetase: purification, properties, and mechanism of action. Biochemistry. 1967;6:1722–1734. doi: 10.1021/bi00858a022. [DOI] [PubMed] [Google Scholar]
- Moran JF, Iturbe-Ormaetxe I, Matamoros MA, Rubio MC, Clemente MR, Brewin NJ, Becana M. Glutathione and homoglutathione synthetases of legumes nodules: cloning, expression, and subcellular localization. Plant Physiol. 2000;124:879–888. doi: 10.1104/pp.124.3.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Arisi M-CM, Jouanin L, Kunert KJ, Rennenberg H, Foyer CH. Glutathione: biosynthesis, metabolism and relationship to stress tolerance explored in transformed plants. J Exp Bot. 1998;49:623–647. [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Berlin: Springer-Verlag; 1970. [Google Scholar]
- Oppenheimer L, Wellner VP, Griffith OW, Meister A. Glutathione synthetase: purification from rat kidney and mapping of the substrate binding sites. J Biol Chem. 1979;254:5184–5190. [PubMed] [Google Scholar]
- Polekhina G, Board PG, Gali RR, Rossjohn J, Parker MW. Molecular basis of glutathione synthetase deficiency and a rare gene permutation event. EMBO J. 1999;18:3204–3213. doi: 10.1093/emboj/18.12.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. A new thiol in legumes. Nature. 1957;180:148–149. doi: 10.1038/180148a0. [DOI] [PubMed] [Google Scholar]
- Rathbun WB, Sethna SS, Van Buskirk GE. Purification and properties of glutathione synthetase from bovine lens. Exp Eye Res. 1977;24:145–158. doi: 10.1016/0014-4835(77)90255-x. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shirzadegan M, Christie P, Seemann JR. An efficient method for isolation of RNA from tissue cultured plant cells. Nucleic Acids Res. 1991;19:6055. doi: 10.1093/nar/19.21.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skipsey M, Andrews CJ, Townson JK, Jepson I, Edwards R. Substrate and thiol specificity of a stress-inducible glutathione transferase from soybean. FEBS Lett. 1997;409:370–374. doi: 10.1016/s0014-5793(97)00554-1. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb GC, Vaska VL, Gali RR, Ford JH, Board PG. The gene encoding human glutathione synthetase (GSS) maps to the long arm of chromosome 20 at band 11.2. Genomics. 1995;30:617–619. doi: 10.1006/geno.1995.1287. [DOI] [PubMed] [Google Scholar]