Abstract
Nucleotide excision repair in Escherichia coli involves formation of the UvrB–DNA complex and subsequent DNA incisions on either site of the damage by UvrC. In this paper, we studied the incision of substrates with different damages in varying sequence contexts. We show that there is not always a correlation between the incision efficiency and the stability of the UvrB–DNA complex. Both stable and unstable UvrB–DNA complexes can be efficiently incised. However some lesions that give rise to stable UvrB–DNA complexes do result in a very low incision. We present evidence that this poor incision is due to sterical hindrance of the damage itself. In its C-terminal region UvrC contains two helix–hairpin–helix (HhH) motifs. Mutational analysis shows that these motifs constitute one functional unit, probably folded as one structural unit; the (HhH)2 domain. This (HhH)2 domain was previously shown to be important for the 5′ incision on a substrate containing a (cis-Pt)·GG adduct, but not for 3′ incision. Here we show that, mainly depending on the sequence context of the lesion, the (HhH)2 domain can be important for 3′ and/or 5′ incision. We propose that the (HhH)2 domain stabilises specific DNA structures required for the two incisions, thereby contributing to the flexibility of the UvrABC repair system.
INTRODUCTION
Nucleotide excision repair (NER) is responsible for the removal of a vast array of structurally different lesions from the DNA. This repair process involves specific damage recognition, dual incision of the damaged strand, removal of the lesion followed by gap filling and finally strand ligation (reviewed in 1–3). In Escherichia coli, damage recognition is accomplished by the heterotrimeric UvrA2B complex. After the damage has been located, the UvrA dimer dissociates from the complex to form the UvrB–DNA complex. Binding of ATP to the UvrB–DNA complex induces a conformational change in the DNA that is a prerequisite for the UvrC endonuclease to catalyse the first incision on the fourth or fifth phosphodiester bond 3′ from the lesion (4). This incision is immediately followed by an incision on the eighth phosphodiester bond 5′ from the damage. Both incisions are made by UvrC, which contains the catalytic sites for both incisions (5,6). Recently, a second endonuclease named Cho has been identified that is involved in NER as well (7). This nuclease contains the catalytic domain found in UvrC that is responsible for the 3′ incision and is indeed capable of performing the 3′ incision on a damaged substrate although the incision position is shifted 4 nt away from the lesion (7).
Several studies have shown that different substrates are incised with varying efficiencies (8–12). Since the UvrABC repair reaction is a multi-step process, the efficiency of the reaction can be influenced by various determinants. A correlation between the rate of UvrA binding and the incision efficiency has been suggested (10). In addition the rates of UvrB– (9) and/or UvrBC–DNA complex (11) formation or the 3′ incision event itself have been proposed to be rate limiting (12). On the other hand, an inverse correlation between incision efficiency and stability of the UvrB–DNA complex has been proposed (8). Recently it has been shown that some bulky lesions that are poorly incised by UvrC are very efficiently incised by Cho, which cuts the DNA 4 nt further away from the damage, suggesting that the 3′ incision by UvrC can be hampered by sterical hindrance (7).
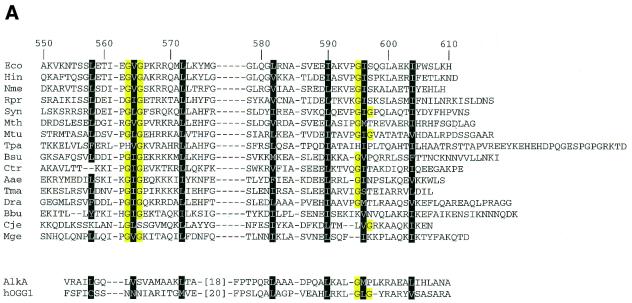
In the C-terminal region of UvrC a helix–hairpin–helix (HhH) motif was found and this region has been shown to be important for the 5′ incision on a substrate carrying a cis-diamminedichloroplatinum(II) (cis-Pt)·GG adduct but not for the 3′ incision (13). The proposed consensus sequence of the HhH motif is hxxhxGhGxxxAxxhh (14) and recently it has been shown that most HhH motifs are integrated as part of a five-helical domain, termed the (HhH)2 domain (15). This structural domain consists of two consecutive HhH motifs linked by a connector helix. The (HhH)2 domain is a structurally compact unit with a well defined hydrophobic core composed of seven residues from the two individual HhH motifs (Fig. 1A). Co-crystallisation studies have shown that in proteins containing the (HhH)2 domain, both DNA strands are contacted and the two HhH motifs each contact a different strand (15). The HhH motif itself is formed by two anti-parallel helices connected by a hairpin-like loop (14,15). HhH motifs are found in many DNA-processing proteins (16), where they are involved in non-sequence-specific DNA binding (14). All co-crystal structures known so far of these proteins show that the conformations of their respective DNA substrates in the complex deviate from normal B-DNA (17–19). For instance, the HhH motifs in RuvA are involved in binding and recognition of the Holliday junction (17) and in the BER enzymes AlkA and hOGG1 they are involved in stabilisation of the protein-induced conformational changes in the DNA that lead to flipping of the damaged nucleotide (18,19). These structures suggest that HhH motifs are important for binding and/or stabilisation of specific DNA structures.
Figure 1.
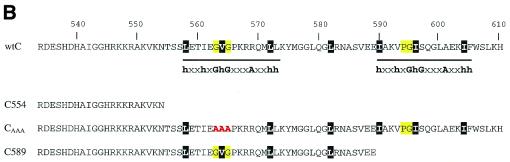
(A) Sequence alignment of the putative (HhH)2 motifs of UvrC proteins from a number of different bacteria with the structurally determined (HhH)2 domain found in AlkA and hOGG1 (Eco, E.coli; Hin, Haemophilus influenzae; Nme, Neisseria meningitidis; Rpr, Rickettsia prowazekii; Syn, Synechocystis; Mth, Methanobacterium thermoautotrophicum; Mtu, Mycobacterium tuberculosis; Tpa, Treponema pallidum; Bsu, Bacillus subtilis; Ctr, Chlamydia trachomatis; Aae, Aquifex aeolicus; Tma, Thermatoga maritima; Dra, Deinococcus radiodurans; Bbu, Borrelia burgdorferi; Cje, Campylobacter jejuni; Mge, Mycoplasma genitalum). The conserved glycine residues of the hairpin in the HhH motif are highlighted in yellow. The residues highlighted in black form the hydrophobic core of the proposed (HhH)2 domain as observed in the crystal structures of AlkA and hOGG1. (B) Overview of mutants used in this study. The HhH motifs are underlined and the consensus sequence for the HhH motif is indicated. The conserved glycine residues are highlighted in yellow and the hydrophobic core of the (HhH)2 domain in black. The mutated residues are in red.
Sequence alignment shows that UvrC contains the seven conserved hydrophobic residues of the (HhH)2 domain in its C-terminal region as well (Fig. 1A). This strongly suggests that UvrC contains two HhH motifs that are probably folded like the (HhH)2 domain. We show here that indeed the two HhH motifs of UvrC form one functional unit involved in DNA binding. In addition, we show that the HhH motifs can be important for the 3′ and/or the 5′ incision and that the importance of the HhH motifs for both incisions mainly depends on the sequence context and only to a minor extent on the lesion itself.
MATERIALS AND METHODS
Oligonucleotides and plasmids
Bio421 (GACCGCGCTTGCCAGCGTGTTGAAATTGCCGG) encodes UvrC residues 381–391. Bio1048 (TCAAGTCTTGACATATGTCATTCCTCGACGCTGGCGTTAC) contains a NdeI restriction site, a stopcodon and encodes UvrC residues 583–589. Bio1308 (CGACGTTTTGGCGCCGCGGCTTCAATGCTTTCC) corresponds to UvrC residues 569–559, but residues 565–563 (GVG) are changed to AAA. pBL12 expresses the UvrC gene from the tac-promoter (20).
Construction of mutant proteins
All mutant proteins are schematically shown in Figure 1B. The mutants were constructed in pBL12. Construction of the UvrC554 mutant has been described before (13). The C-terminal deletion mutant UvrC589 was constructed via PCR using primers Bio421 (encoding UvrC residues 381–391) and Bio1048 (encoding UvrC residues 583–589 immediately followed by a stopcodon). The resulting PCR fragment was inserted in pBL12 via NcoI and NdeI digestion. Mutant UvrCAAA was constructed via site-directed mutagenesis using the Gene-editor kit II (Promega) and oligo Bio1308, which changes residues GVG (563–565) into AAA. All constructs were verified by sequence analysis.
Protein purification
The purification of wild-type (wt) UvrC (21), Cho (7) and the UvrC554 mutant (13) has been described. Mutant UvrCAAA was purified according to the protocol for wt UvrC and behaved like UvrC during the purification procedure. Mutant UvrC589 did not bind to the ssDNA cellulose column during the purification and therefore this column was replaced by a 20-ml Blue Agarose column. The fractions containing UvrC589 were diluted to a concentration of 150 mM KCl and loaded onto the Blue Agarose column. The protein was eluted with a 0.15–1.5 M KCl gradient in buffer C. The protein was further purified according to the purification procedure for wt UvrC.
Construction of DNA substrates
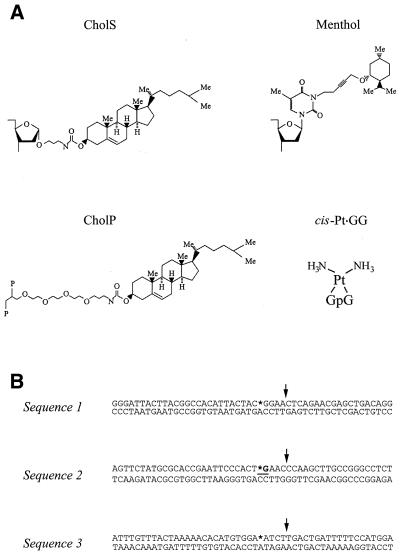
The 50 bp fragment containing a cis-Pt·GG adduct was constructed similar to the 96 bp fragment described previously (22). Ligation of the same central 18 nt oligo used for the 96 bp fragment to two 16mer flanking oligos resulted in a 50 nt damaged top strand, which was isolated from a denaturing acrylamide gel followed by hybridisation to the complementary 50 nt bottom strand. The CholS and CholP lesions consist of cholesterol adducts at the sugar ring or the phosphodiester backbone, respectively. The menthol lesion contains a menthol group attached to a thymine at the N3 position (Fig. 2A). The oligonucleotides containing the CholP lesion were all provided by Eurogentec (Seraing, Belgium) (23) and gel purified. The CholS lesion was synthesised as a building block and incorporated into an oligonucleotide using a DNA synthesiser (24). Synthesis of the N3 menthol lesion is described below. The different nucleotide sequences used in this study are shown in Figure 2B. The CholS, CholP and Menthol lesions were placed in sequence 1. Sequence 2 carries a cis-Pt·GG adduct or a CholP lesion and sequence 3 contains a CholP or Menthol damage.
Figure 2.
DNA substrates used in this study. (A) The structures of the different lesions used. The CholS lesion consists of a cholesterol moiety attached to the sugar ring of DNA and has been described before (24). The menthol damage is a thymine with a menthol group attached to the N3 position. The CholP lesion contains a cholesterol moiety attached to the phoshodiester backbone (23). The cis-Pt·GG lesion is a intra-strand crosslink of two adjacent guanines linked at the N7 position (22). (B) DNA sequences of the substrates. Sequence 1 contains the CholS, CholP or Menthol damages, sequence 2 contains the cis-Pt·GG or CholP lesion and sequence 3 carries a CholP or Menthol lesion. The asterisk indicates the position of the damage. In the case of the cis-Pt·GG damage, the asterisk represents a guanine and the two crosslinked residues are underlined. The arrows indicate the position of the nick in the nicked substrates.
For 5′ labelling 2 pmol of the appropriate oligo was incubated with 10 U T4 polynucleotide kinase in 70 mM Tris–HCl (pH 7.6), 10 mM MgCl2, 5 mM DTT and 1.8 pmol of [γ-32P]ATP (Amersham). After incubation at 37°C for 45 min, the reaction was terminated by adding 20 mM EDTA and incubation at 80°C for 10 min. The different substrates were constructed by hybridising 2 pmol of the appropriate oligos in the presence of 50 mM Tris–HCl, 50 mM NaCl and 1 mM EDTA. The substrates were purified from non-incorporated nucleotides by G50 gel filtration.
Synthesis of the N3 menthol building block
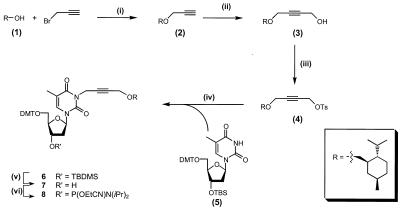
Synthesis of the N3 menthol building block is schematically shown in Figure 3.
Figure 3.
Reaction scheme for N3 menthol synthesis. Reagents and conditions. (i) 0.2 eq. TBAI, 1.1 eq. NaH, 0°C, 1 h then 4 eq. propargyl bromide, DMF, 4 days. (ii) 1.1 eq. nBuLi, –50°C, THF, 1 h, then 1.2 eq. (CH2O)n, reflux, 3 h. (iii) 1.5 eq. TosCL, 1.5 eq. TEA, 0.1 eq. DMAP, 0°C, DCM, 6 h. (iv) 1.1 eq. compound 4, 1.1 eq. NaH, 0.1 eq. Bu4NI, DMF, overnight. (v) TEA·3HF, pyridine, overnight. (vi) 1.3 eq. (iPr)2NP(Cl)OEtCN, 4 eq. DiPEA, DCE, 1 h.
(i) 3-(1R,2S,5R)-(–)-menthyloxy-1-propyne (2) (25). l-Menthol (1) (2.37 g, 15 mmol) and tBAI (1.13 g, 3 mmol) were co- evaporated with dry toluene a few times and dissolved in dry DMF (20 ml). The reaction mixture was cooled to 0°C, NaH (60% dispersion in mineral oil, 0.72 g, 18 mmol) was added and the reaction was left stirring at room temperature for 1 h. Propargyl bromide [80% (w/w) solution in toluene, 7 ml, 62 mmol] was added to the mixture and the reaction was stirred at room temperature for 4 days. The excess propargyl bromide was quenched with methanol and after partial evaporation of the solvent the reaction mixture was diluted with diethyl ether, washed with water, 5% NaHCO3 and brine. The organic phase was dried over anhydrous MgSO4, filtered and evaporated under reduced pressure. The crude product was purified via silica gel column chromatography using a mixture of petroleum ether/diethyl ether 98:2 as an eluent. The product was obtained as an oil. Yield, 32%.
(ii) 4-[1-(1R,2S,5R)-(–)-menthyloxy]-2-butynyl-1-ol (3). Compound 2 (0.474 g, 2.4 mmol) was dissolved in freshly distilled THF (3 ml). The solution was cooled at –10°C and a solution of nBuLi (1.6 M in hexane, 1.7 ml, 2.6 mmol) was added, followed by (CH2O)n (80 mg, 2.7 mmol). The reaction was stirred at room temperature for 30 min and refluxed for 3 h. The reaction was quenched with water at 0°C. The water layer was washed a few times with Et2O and the organic layer was subsequently washed with saturated NH4Cl, dried over anhydrous MgSO4, filtered and evaporated under reduced pressure. The crude product was purified via silica gel column chromatography using a mixture of EtOAc/petroleum ether/MeOH 5:30:1 as an eluent. The product was obtained as an oil. Yield, 50%.
(iii) 1-tosyl-4-[1-(1R,2S,5R)-(–)-menthyloxy]-2-butynyl-1-ol (4). TosCl (857 mg, 4.5 mmol), TEA (625 µl, 4.5 mmol) and DMAP (18.3 mg, 0.15 mmol) were dissolved in dry DCM (15 ml) and cooled at 0°C. Compound 3 (673 mg, 3 mmol) was also dissolved in dry DCM (15 ml) and this solution was slowly dropped into the previous one. After 4 h at 0°C the mixture was dissolved in more DCM and washed with brine at 0°C. The organic phase was dried over anhydrous MgSO4, filtered and evaporated under reduced pressure (temperature water bath 30°C). The crude product was purified via silica gel column chromatography using a mixture of EtOAc/petroleum ether 1:8 as an eluent. Yield, 42%.
5′-(4,4′-dimethoxytrityl)-3′-tert-butyldimethylsilyl-2′-deoxyuridine (5) (26). 2′-Deoxythymidine (3.63 g, 15 mmol) was dissolved in dry pyridine and DMTCl (6.22 g, 18 mmol) was added. The reaction was stirred at room temperature for 2 h. The pyridine was partially evaporated and the residue was dissolved in DCM. The organic phase was washed with 5% NaHCO3 and brine, dried over anhydrous MgSO4, filtered and evaporated. The product was obtained by reverse precipitation from petroleum ether/Et2O/DCM. The crude product was dissolved in DMF (40 ml) and dry imidazole (4.7 g, 70 mmol) and TBSCl (2.95 g, 19 mmol) were added. The reaction mixture was stirred at room temperature overnight. The reaction mixture was diluted with Et2O. The organic phase was washed several times with water, 5% NaHCO3 and brine, dried over anhydrous MgSO4, filtered and evaporated under reduced pressure. The crude product was purified via silica gel column chromatography using a gradient 33→40% of EtOAc in petroleum ether. The product was obtained as an oil. Yield, 90% (over two steps).
(iv) 3-N-{4-[1-(1R,2S,5R)-(–)-menthyloxy]-2-butyn-1-yl} 5′-(4,4′-dimethoxytrityl)-3′-tert-butyldimethylsilyl-2′-deoxyuridine (6). Compound 5 (0.63 g, 1 mmol) and tBAI (38 mg, 0.1 mmol) were co-evaporated a few times with dry toluene and dissolved in dry DMF (7 ml). Compound 4 (0.41 g, 1.08 mmol) was also dissolved in DMF and added to the reaction mixture. The reaction was stirred at room temperature overnight. The reaction mixture was diluted with Et2O, washed several times with water, 5% NaHCO3 and brine. The organic phase was dried over anhydrous MgSO4, filtered and evaporated under reduced pressure. The crude product was purified via silica gel column chromatography using a gradient 16→18% of EtOAc in petroleum ether containing 1% of TEA. The product was obtained as a foam. Yield, 78%.
(v) 3-N-{4-[1-(1R,2S,5R)-(–)-menthyloxy]-2-butyn-1-yl}-5′-(4,4′-dimethoxytrityl)-2′-deoxyuridine (7). Compound 6 was dissolved in a mixture 1:3 (v/v) of TEA·HF/pyridine (8 ml) and the reaction mixture was stirred at room temperature for 3 h. Afterwards, EtOAc was added and the organic phase washed a few times with brine, dried over anhydrous MgSO4, filtered and evaporated under reduced pressure. The crude product was purified via silica gel column chromatography using a mixture of EtOAc/petroleum ether 1:2 containing 1% of MeOH and TEA. The product was obtained as a white foam. Yield, 83%.
(vi) 3-N-{4-[1-(1R,2S,5R)-(–)-menthyloxy]-2-butyn-1-yl} 5′-(4,4′-dimethoxytrityl)-2′-deoxyuridinyl-3′-(2-cyanoethyl-N,N,-diisopropyl)phosphoramidite (8). Compound 7 (0.2 mmol) was co-evaporated twice with dry pyridine and twice with dry DCE. It was dissolved in dry DCE (2 ml) and DiPEA (0.8 mmol) and N,N-diisopropylamino-cyanoethoxychlorophsosphine (0.26 mmol) were added. The reaction was stirred under argon atmosphere for 1.5 h. The reaction mixture was diluted with DCM and the organic phase washed with 5% NaHCO3, brine, dried over anhydrous MgSO4, filtered and evaporated under reduced pressure. The crude product was purified via silica gel column chromatography using a mixture of EtOAc/petroleum ether 1:3.5 containing 1% of MeOH and 3% of TEA. Yield, 90%.
All products were verified using 1H-NMR and ESI–MS.
Incision and gel retardation assays
Incision assay. The different DNA substrates (2 fmol) were incubated with 2.5 nM UvrA, 100 nM UvrB and 25 nM of wt UvrC, mutant UvrC or Cho in 20 µl Uvr-endo buffer (50 mM Tris–HCl pH 7.5, 100 mM KCl, 10 mM MgCl2, 0.1 µg/µl BSA and 1 mM ATP). After 20 min the reaction was stopped by adding 2 µl of 2 µg/ml glycogen followed by ethanol precipitation. The incision products were visualised on a 15% denaturing acrylamide gel. To determine the incision efficiencies, the reaction products were loaded onto an 8% denaturing polyacrylamide gel, which was subsequently dried after which the incision products were quantified using a PhosphorImager.
Gel retardation assay. The different DNA substrates (2 fmol) were incubated in 10 µl Uvr-endo buffer with 2.5 nM UvrA, 100 nM UvrB and 50 nM of wt or mutant UvrC as indicated for 5 min at room temperature to prevent excessive incision. The samples were loaded on a 3.5% native polyacrylamide gel containing 1 mM ATP and 10 mM MgCl2 (21). The gel system was cooled by continuous circulation of ice-cold water.
RESULTS AND DISCUSSION
The C-terminal region of UvrC containing the putative (HhH)2 motif can be important for both 3′ and 5′ incisions
We have previously shown that the C-terminal region of UvrC containing the putative (HhH)2 domain is important for the 5′ incision on a 96 bp substrate containing a cis-Pt·GG adduct but not for the 3′ incision (13). Similarly, when the cis-Pt·GG adduct is placed in a 50 bp substrate with the same sequence context as the previously described 96mer (Pt-2), we found that UvrC554 lacking the putative (HhH)2 domain is disturbed in 5′ but not in 3′ incision (Fig. 4, panel 1). Incubation of substrate Pt-2 with UvrAB and wt UvrC gives rise to a 19 nt incision product resulting from the 3′ incision which is immediately followed by the 5′ incision. This incision product is subsequently a substrate for the damage-independent UvrBC incision (23) resulting in a 12 nt incision product (Fig. 4, panel 1). Incubation of Pt-2 with UvrC554 results in the 31 nt 3′ incision product only.
Figure 4.
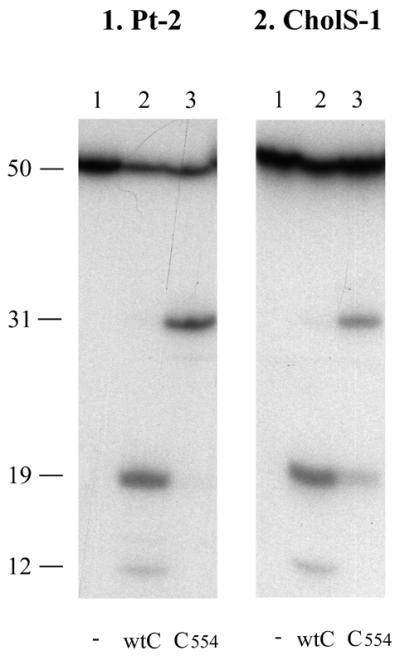
Incision of substrate containing a cis-Pt·GG damage or CholS lesion by wt UvrC and UvrC554. The double-stranded substrates were incubated with UvrABC or UvrABC554 as indicated. The DNA fragments of 31 nt correspond to incision at the 3′ site. The fragments of 19 nt correspond to incision at the 5′ site. The fragments of 12 nt result from the damage-independent UvrBC incision.
However, when we tested the activity of UvrC554 on another substrate, CholS-1, the mutant protein appeared to be disturbed in both 3′ and 5′ incision (Fig. 4, panel 2; Fig. 5, panels 1 and 2, lane 3). The resulting activities of UvrC554 with respect to the wt protein on this substrate are 45.8% for 3′ incision and 37.9% for 5′ incision (Table 1C). Substrates CholS-1 and Pt-2 not only differ in the type of damage but also in base composition (Fig. 2B). To study whether the differential effect of UvrC554 is due to the damage itself or to the DNA sequence surrounding the damage we constructed a variety of substrates with different lesions in different sequence contexts. Four different types of damages were used, i.e. the cis-Pt·GG, CholS, CholP and menthol lesions (Fig. 2A), which were placed in sequence 1 (CholS-1, CholP-1 and Menthol-1), sequence 2 (Pt-2 and CholP-2) or sequence 3 (CholP-3 and Menthol-3) (Fig. 2B). In addition, to study 5′ incision independent of the 3′ incision, a similar set of substrates carrying a nick at the 3′ side of the lesion was made (nicked CholS-1, etc.).
Figure 5.
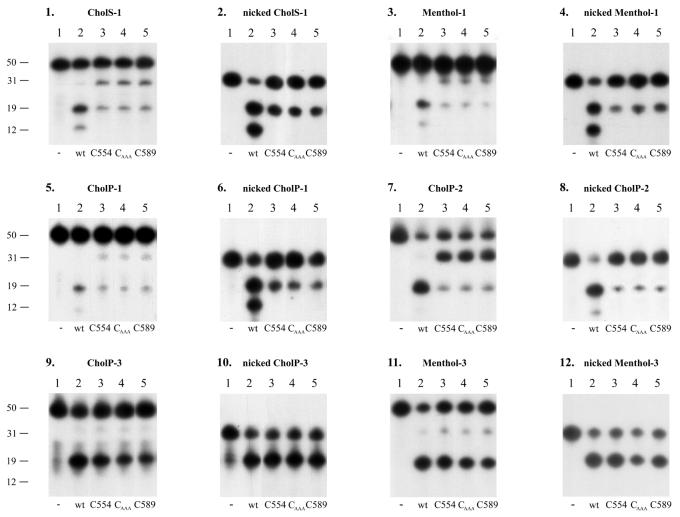
Incision by the different UvrC mutants. The substrates used are indicated above each panel. The different substrates were incubated with UvrA, UvrB and the different UvrC mutants as indicated. The DNA fragments of 31 nt correspond to incision at the 3′ site. The fragments of 19 nt correspond to incision at the 5′ site. Fragments of 12 nt result from the damage-independent UvrBC incision.
Table 1. Incision percentages of wt UvrC and UvrC554.
| Damages | CholS sequence 1 | Menthol sequence 1 | CholP sequence 1 | Pt·GG sequence 2 | CholP sequence 2 | CholP sequence 3 | Menthol sequence 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (A) Percentage 3′ incision | |||||||||||||
| wt UvrC | 20.3 ± 3.1 | 8.8 ± 1.5 | 5.3 ± 0.8 | 51.3 ± 6.5 | 62.5 ± 3.8 | 42.5 ± 1.5 | 75.3 ± 5.6 | ||||||
| UvrC554 | 9.3 ± 1.4 | 3.0 ± 0.4 | 3.2 ± 1.1 | 49.2 ± 1.0 | 53.8 ± 3.5 | 19.9 ± 4.8 | 51.6 ± 5.8 | ||||||
| (B) Percentage of total 3′ incision resulting in 5′ incision | |||||||||||||
| wt UvrC | 96.5 ± 0.7 | 92.7 ± 1.3 | 95.6 ± 3.3 | 95.4 ± 1.0 | 95.2 ± 0.7 | 90.8 ± 2.7 | 90.4 ± 1.8 | ||||||
| UvrC554 | 36.6 ± 1.1 | 37.8 ± 2.8 | 43.0 ± 5.4 | 3.6 ± 1.7 | 15.8 ± 3.4 | 94.7 ± 1.2 | 95.2 ± 0.8 | ||||||
| (C) Percentage UvrC554 incision relative to wt UvrC | |||||||||||||
| 3′ Incision | 45.8 | 34.1 | 60.4 | 95.9 | 86.1 | 46.8 | 68.5 | ||||||
| 5′ Incision | 37.9 | 40.8 | 45.0 | 3.8 | 16.6 | 104 | 105 | ||||||
The data represent the mean of three independent experiments. Incision was analysed on an 8% denaturing polyacrylamide gel and quantified using a PhosphorImager. (A) The 3′ incision percentage was determined by adding the total amount of 3′ and 5′ incision products and comparing this value with the amount of the remaining uncut substrate. (B) The values were determined by comparing the total amount of 5′ incision products with the total amount of 3′ incision as mentioned in (A). (C) The amount of 3′ and 5′ incisions by UvrC554 was determined as described in (A) and (B) and subsequently compared with the amount of the two incisions by wt UvrC.
The incision efficiency of wt UvrC is not correlated with the stability of the UvrB–DNA complex
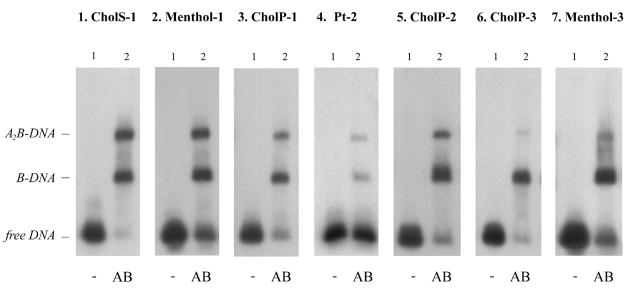
First, we tested the incision efficiencies of wt UvrC on the different double-stranded DNA fragments. On these substrates a large variation in incision efficiencies is observed (Fig. 5). Different types of damage within the same sequence are differently incised, as can be seen by comparing CholS-1 (20.3%) and CholP-1 (5.3%) or CholP-3 (42.5%) and Menthol-3 (75.3%) (Fig. 5, panels 1, 5, 9 and 11, and Table 1A). On the other hand the incision of CholP and Menthol in sequence 3 (42.5 and 75.3%, respectively) is much more efficient than the incision of the same lesions in sequence 1 (5.3 and 8.8%, respectively), showing that also the sequence surrounding the lesion influences the incision efficiency. To test whether these differences are due to a difference in damage recognition on the various substrates we analysed UvrB–DNA complex formation in a gel retardation assay (Fig. 6). The results show that, with the exception of Pt-2 (lane 2), all substrates give similar amounts of UvrB–DNA complexes. Interestingly, the incision of Pt-2 is very efficient, showing that proficient UvrB–DNA complexes are formed, but apparently these complexes are less stable than the complexes formed on the other substrates. It has been proposed that there is an inverse correlation between the stability of a UvrB–DNA complex and the incision efficiency (8). This was concluded from the observation that a DNA fragment with an AAF lesion in a specific sequence context resulted in a very stable UvrB–DNA complex and a very poor incision, whereas the same lesion in a different sequence context gave rise to an unstable UvrB–DNA complex and a very efficient incision. However, on our substrates we did not observe such an inverse correlation. All substrates with CholS, CholP or Menthol lesions result in stable UvrB–DNA complexes, but some of these complexes are poorly incised (e.g. CholP-1) whereas others are incised even better than the unstable complex on Pt-2 (e.g. Menthol-3). Apparently, the stability of a UvrB–DNA complex on a specific substrate in a gel retardation assay is no measure for the subsequent incision by UvrC.
Figure 6.
Complex formation on double-stranded substrates with UvrA and UvrB. All substrates were incubated with the different Uvr proteins as indicated. Incubation mixtures were run on a native gel containing ATP. The positions of the different Uvr–DNA complexes in the gel are shown.
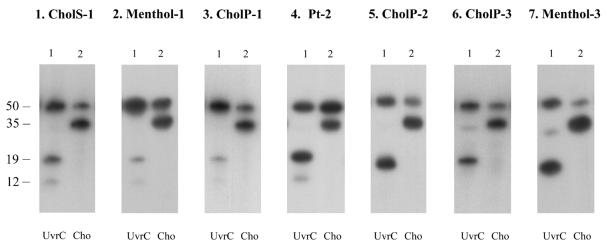
Why are some substrates that form stable UvrB–DNA complexes poorly incised by UvrC? The answer to this question can be given by testing the incision of the different substrates by the second endonuclease for NER present in E.coli, Cho. Recently we have shown that this Cho protein, which is homologous to the N-terminal half of UvrC, can induce 3′ incision on a UvrB–DNA complex. For this incision, which takes place 4 nt further away from the damage than incision by UvrC, the Cho protein binds to a different domain of UvrB than UvrC (7). As shown before (7), incision of substrate Pt-2 by Cho is somewhat reduced compared with incision of the same substrate by UvrC (Fig. 7, panel 4). All other substrates used in this study are very efficiently incised by Cho (Fig. 7, lane 2). Taken together, these results show that indeed all seven substrates do give rise to proper UvrB–DNA preincision complexes. The fact that nevertheless some of these complexes are not efficiently incised by UvrC is most likely the result of a sterical hindrance of the lesion itself. We have recently proposed a model where upon binding of UvrB, the damaged nucleotide is flipped out of the DNA helix, away from the UvrB protein. The exposed adduct, especially when it is very bulky, might in some cases interfere with a proper binding of the UvrC protein. The Cho protein, which is smaller and which binds to a different domain of UvrB, can circumvent this sterical hindrance (7). Since identical damage in different DNA sequences result in different numbers of UvrC-mediated incisions, our model implies that the way a lesion is exposed as a result of nucleotide flipping depends on the sequences surrounding the lesion. As proposed before (7), in the case of the cisplatin damage the flipped nucleotide(s) seem to cause the opposite effect, obstruction of Cho instead of UvrC, explaining the more efficient incision by UvrC in this case.
Figure 7.
Incision of different substrates by UvrC and Cho. The substrates used are indicated above each panel. The different substrates were incubated with UvrA, UvrB and UvrC or Cho as indicated. The fragments of 19 nt correspond to 5′ incision by UvrC, whereas the 35 nt incision products correspond to the 3′ incision by Cho. The fragments of 12 nt result from the damage-independent UvrBC incision.
Incubation of all prenicked substrates with wt UvrABC leads to very efficient 5′ incisions (Fig. 5, even panels, lanes 2). This confirms that all damages in the three different sequences give rise to proficient UvrB–DNA complex formation. Moreover, it shows that (at least on the substrates tested here) the lesion does not form a sterical hindrance for the 5′ incision by wt UvrC. The only difference observed with the nicked substrates is the absence of the extra 5′ incision for CholP-3 and Menthol-3 (Fig. 5, panels 10 and 12, lanes 2). Probably, the very A/T-rich 19 nt oligo that results from the 5′ incision (Tm of 38°C) dissociates from the substrate before the extra incision can occur.
The requirement of the putative (HhH)2 motif is mainly dependent on the sequence context of the lesion
Next, the activity of UvrC554 was compared with that of wt UvrC on the different substrates (Fig. 5, lanes 3 of all panels; Table 1C). On the substrates with the CholS, CholP or Menthol lesions in sequence 1, deletion of the putative (HhH)2 domain affects both 3′ and 5′ incisions. The remaining activity slightly varies for the different lesions, e.g. the 3′ incision activity of UvrC554 with respect to wt UvrC is 60.4% on CholP-1 and 34.1% on Menthol-1, but the effect on the 3′ and 5′ incisions is more or less similar. On the substrates with the lesions in sequence 2 (Pt-2 and CholP-2), however, the 5′ incision by UvrC554 is much more disturbed (residual activities of 3.8 and 16.6%, respectively) whereas the 3′ incision in this sequence is only slightly affected (remaining activities 95.9 and 86.1%). The base composition of sequence 3 has yet another effect on the activity of UvrC554. For CholP-3 and Menthol-3 the 5′ incision activity is like that of wt UvrC, whereas the 3′ incision is reduced (residual activities of 46.8 and 68.5%, respectively). Taken together, these results show that, mainly depending on the sequence context of the lesion, three classes of DNA substrates can be distinguished. In the sequence 1 substrates the HhH motifs are involved in both 3′ and 5′ incision. In the sequence 2 substrates the HhH motifs are essential for 5′ incision, but they play only a minor role in 3′ incision. Finally, in the sequence 3 substrates the HhH motifs are only required for 3′ incision and not for 5′ incision. The differential effect of the putative (HhH)2 domain on the two incisions shows that this domain has a different function in each of the two incision reactions.
Several studies have shown that the DNA in the UvrB–DNA preincision complex is severely distorted. The binding of ATP by UvrB, which is essential for formation of the preincision complex and the subsequent 3′ incision (4), results in a wrapping of the DNA around the UvrB protein (27) and a very strong DNase I hypersensitive site at the 5′ side of the lesion (22,28). In addition, there are strong indications that in the preincision complex the damaged nucleotide is flipped out of the helix (29). Finally, it has been shown that binding of UvrB induces DEPC sensitive sites at the 3′ side of the lesion (30,31) and that introduction of unpaired bases around the lesion significantly stimulates the 3′ incision (32). All of these findings indicate that a specific conformation of the DNA in the UvrB–DNA complex is induced which is required for the 3′ incision. It is conceivable that the stability of this DNA conformation is influenced by the base pair composition of the substrate. We propose that the putative (HhH)2 domain of UvrC stabilises the specific DNA structure required for 3′ incision, most likely by binding the DNA at the 3′ side of the damage. In some sequences (like sequence 2) the conformational changes in the DNA induced by UvrB alone lead to the proper DNA structure for 3′ incision and hence further stabilisation by the HhH motifs of UvrC is no longer needed. In other sequences (like sequences 1 and 3) the required DNA conformation is more unstable and the HhH motifs become important.
After the 3′ incision, the DNA conformation in the UvrB–DNA complex will change. It has been shown that the introduction of a nick at the 3′ incision site releases the stress in the DNA (4). This conformational change seems to be required to trigger the 5′ incision, because on a ‘normal’ damaged substrate 5′ incision can only occur after the 3′ nick has been introduced. It has been shown that the introduction of 10–12 unpaired bases 3′ to the damage leads to uncoupled 5′ incision, without any 3′ incision (32). Taken together these results suggest that the conformational change in the DNA after the 3′ incision involves strand opening around the 3′ incision site.
We propose that the putative (HhH)2 domain also stabilises this new DNA conformation required for 5′ incision, most likely by binding the DNA at the 3′ side of the damage. It was shown that the same UvrC molecule that makes the 3′ incision also induces the 5′ incision (4). Therefore, we do not expect that the DNA region that interacts with the HhH motifs is different for the two incisions. The effect of this interaction on the DNA conformations required for the two incisions, however, is apparently different. It should be noted that certain sequences (e.g. sequence 2), which in the absence of the HhH motifs support the 3′ incision very well, are very poor substrates for the 5′ incision when this DNA binding motif is lacking. For other sequences (e.g. sequence 3) the opposite effect is observed. This means that the respective DNA conformations required for 3′ and 5′ incisions are influenced by the base composition of different regions of the substrate. Identification of these regions awaits a systematic base pair change of the DNA substrates.
It has been shown before that the damage-independent incision by UvrBC, which gives rise to the extra 5′ incision product, is dependent on the C-terminal region of UvrC (23). Indeed, none of the double-stranded or nicked substrates gives rise to the 12 nt extra incision product with UvrC554 (Fig. 5, lane 3 of all panels). Apparently the requirement of the putative (HhH)2 domain for this extra 5′ incision is independent of the sequence context. Recent data strongly suggest that the damage-independent UvrBC incision is the equivalent of the 5′ incision on a damaged substrate (29). The nick in the DNA that results from the 5′ incision is recognised and processed like a ‘normal’ damage by UvrB. However, whereas the damage-independent incision is fully dependent on the putative (HhH)2 domain, this domain is dispensable for 5′ incision on certain damaged substrates. This suggests that the UvrB–DNA complexes on damaged or undamaged DNA are not identical. Very likely the presence of the damage provides additional protein–DNA interactions, either directly or indirectly, thereby stabilising the complex and/or the DNA conformation required for 5′ incision. Apparently, the DNA interactions provided by the putative (HhH)2 domain of UvrC become crucial for stabilisation of a proficient incision complex when UvrB lacks these damage-specific protein–DNA contacts.
The two HhH motifs of UvrC form one functional unit
The two HhH motifs in the C-terminal part of UvrC are probably folded like the recently described (HhH)2 domain (15), since sequence alignment of this region shows the presence of seven conserved residues that are known to constitute the core of the (HhH)2 domain in proteins like AlkA or Ogg1 (Fig. 1A). To study whether the two HhH motifs indeed form one functional unit, we have constructed two mutants (Fig. 1B) in which one of the HhH motifs is disrupted. In the UvrCAAA mutant, hairpin residues 563–565 (GVG) are replaced by AAA thereby disrupting the first HhH motif. The second HhH motif is deleted in the UvrC589 protein. Like UvrC554, lacking both HhH motifs, the two mutant genes fail to complement a ΔuvrC strain for survival after UV irradiation (data not shown). We have tested the incisions of the different damaged substrates by the two mutant proteins. In all assays, the UvrCAAA and the UvrC589 mutants behave exactly like UvrC554 (Fig. 5, lanes 4 and 5 of all panels). This strongly suggests that both HhH motifs are one functional unit as was suggested for a (HhH)2 domain (15).
It has been proposed that the presence of two HhH motifs in a (HhH)2 domain provides symmetric binding to both DNA strands, via interaction between the hairpin loop of either motif with the phosphate backbone of either DNA strand (15). During the purification of UvrC589, it became clear that the mutant protein does not bind to a ssDNA column as has been observed for the UvrC554 protein (13), indicating a defect in DNA interaction of this protein. In contrast, the UvrCAAA mutant with substitutions in the loop of the first motif is still capable of binding the ssDNA column. Most likely the ssDNA binding observed during affinity chromatography requires the completely folded (HhH)2 domain but interaction with the loop of only the second motif is sufficient. For the function of the (HhH)2 domain during incision, however, apparently both HhH motifs are essential, most likely because productive binding requires interaction with the two DNA strands.
In summary, we have shown that damage recognition and subsequent UvrB–DNA complex formation are not always sufficient to induce the proper DNA conformations required for 3′ and 5′ incisions. The (HhH)2 domain can help to stabilise the DNA conformation in the respective incision complexes thereby contributing to the flexibility of the UvrABC repair system.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the J. A. Cohen Institute for Radiopathology and Radiation Protection (IRS) and an EC grant on ‘Quality of Life and Management of Living Resources’ (QLG1-CT-1999-00008).
REFERENCES
- 1.Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- 2.Goosen N., Moolenaar,G.F., Visse,R. and Van de Putte,P. (1998) In Eckstein,F. and Lilley,D.M.J. (eds), Nucleic Acids and Molecular Biology: DNA Repair. Springer Verlag, Berlin.
- 3.Van Houten B. and McCullough,A. (1994) Nucleotide excision repair in Escherichia coli. Ann. N. Y. Acad. Sci., 726, 236–251. [DOI] [PubMed] [Google Scholar]
- 4.Moolenaar G.F., Herron,M.F., Monaco,V., van der Marel,G.A., van Boom,J.H., Visse,R. and Goosen,N. (2000) The role of ATP binding and hydrolysis by UvrB during nucleotide excision repair. J. Biol. Chem., 275, 8044–8050. [DOI] [PubMed] [Google Scholar]
- 5.Lin J.-J. and Sancar,A. (1992) Active site of (A)BC excinuclease. I. Evidence for 5′ incision by UvrC through a catalytic site involving Asp399, Asp438, Asp466 and His538 residues. J. Biol. Chem., 267, 17688–17692. [PubMed] [Google Scholar]
- 6.Verhoeven E.E.A., van Kesteren,M., Moolenaar,G.F., Visse,R. and Goosen,N. (2000) Catalytic sites for 3′ and 5′ incision of Escherichia coli nucleotide excision repair are both located in UvrC. J. Biol. Chem., 275, 5120–5123. [DOI] [PubMed] [Google Scholar]
- 7.Moolenaar G.F., van Rossum,S., van Kesteren,M. and Goosen,N. (2002) Cho, a second endonuclease involved in Escherichia coli nucleotide excision repair. Proc. Natl Acad. Sci. USA, 99, 1467–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delagoutte E., Bertrand-Burggraf,E., Dunand,J. and Fuchs,R.P. (1997) Sequence-dependent modulation of nucleotide excision repair: the efficiency of the incision reaction is inversely correlated with the stability of the pre-incision UvrB-DNA complex. J. Mol. Biol., 266, 703–710. [DOI] [PubMed] [Google Scholar]
- 9.Hoare S., Zou,Y., Purohit,V., Krishnasamy,R., Skorvaga,M., Van Houten,B., Geacintov,N.E. and Basu,A.K. (2000) Differential incision of bulky carcinogen-DNA adducts by the UvrABC nuclease: comparison of incision rates and the interactions of Uvr subunits with lesions of different structures. Biochemistry, 39, 12252–12261. [DOI] [PubMed] [Google Scholar]
- 10.Luo C., Krishnasamy,R., Basu,A.K. and Zou,Y. (2000) Recognition and incision of site-specifically modified C8 guanine adducts formed by 2-aminofluorene, N-acetyl-2-aminofluorene and 1-nitropyrene by UvrABC nuclease. Nucleic Acids Res., 28, 3719–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mekhovich O., Tang,M. and Romano,L.J. (1998) Rate of incision of N-acetyl-2-aminofluorene and N-2-aminofluorene adducts by UvrABC nuclease is adduct- and sequence-specific: comparison of the rates of UvrABC nuclease incision and protein–DNA complex formation. Biochemistry, 37, 571–579. [DOI] [PubMed] [Google Scholar]
- 12.Visse R., van Gool,A.J., Moolenaar,G.F., de Ruijter,M. and van de Putte,P. (1994) The actual incision determines the efficiency of repair of cisplatin-damaged DNA by the Escherichia coli UvrABC endonuclease. Biochemistry, 33, 1804–1811. [DOI] [PubMed] [Google Scholar]
- 13.Moolenaar G.F., Uiterkamp,R.S., Zwijnenburg,D.A. and Goosen,N. (1998) The C-terminal region of the Escherichia coli UvrC protein, which is homologous to the C-terminal region of the human ERCC1 protein, is involved in DNA binding and 5′-incision. Nucleic Acids Res., 26, 462–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doherty A.J., Serpell,L.C. and Ponting,C.P. (1996) The helix–hairpin–helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res., 24, 2488–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao X. and Grishin,N.V. (2000) Common fold in helix–hairpin–helix proteins. Nucleic Acids Res., 28, 2643–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aravind L., Walker,D.R. and Koonin,E.V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ariyoshi M., Nishino,T., Iwasaki,H., Shinagawa,H. and Morikawa,K. (2000) Crystal structure of the holliday junction DNA in complex with a single RuvA tetramer. Proc. Natl Acad. Sci. USA, 97, 8257–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruner S.D., Norman,D.P. and Verdine,G.L. (2000) Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature, 403, 859–866. [DOI] [PubMed] [Google Scholar]
- 19.Hollis T., Ichikawa,Y. and Ellenberger,T. (2000) DNA bending and a flip-out mechanism for base excision by the helix–hairpin–helix DNA glycosylase, Escherichia coli AlkA. EMBO J., 19, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zwetsloot J.C., Barbeiro,A.P., Vermeulen,W., Arthur,H.M., Hoeijmakers,J.H. and Backendorf,C. (1986) Microinjection of Escherichia coli UvrA, B, C and D proteins into fibroblasts of xeroderma pigmentosum complementation groups A and C does not result in restoration of UV-induced unscheduled DNA synthesis. Mutat. Res., 166, 89–98. [DOI] [PubMed] [Google Scholar]
- 21.Visse R., de Ruijter,M., Moolenaar,G.F. and van de Putte,P. (1992) Analysis of UvrABC endonuclease reaction intermediates on cisplatin-damaged DNA using mobility shift gel electrophoresis. J. Biol. Chem., 267, 6736–6742. [PubMed] [Google Scholar]
- 22.Visse R., de Ruijter,M., Brouwer,J., Brandsma,J.A. and van de Putte,P. (1991) Uvr excision repair protein complex of Escherichia coli binds to the convex side of a cisplatin-induced kink in the DNA. J. Biol. Chem., 266, 7609–7617. [PubMed] [Google Scholar]
- 23.Moolenaar G.F., Bazuine,M., van Knippenberg,I.C., Visse,R. and Goosen,N. (1998) Characterization of the Escherichia coli damage-independent UvrBC endonuclease activity. J. Biol. Chem., 273, 34896–34903. [DOI] [PubMed] [Google Scholar]
- 24.Monaco V., van de Wetering,K.I., Meeuwenoord,N.J., van den Elst,H.A., Stuivenberg,H.R., Visse,R., van der Kaaden,J.C., Moolenaar,G.F., Verhoeven,E.E.A., Goosen,N., van der Marel,G.A. and van Boom,J.H. (1999) Synthesis and biological evaluation of modified DNA fragments for the study of nucleotide excision repair in E. coli. Nucl. Nucl., 18, 1339–1341. [DOI] [PubMed] [Google Scholar]
- 25.Almansa C., Moyano,A., Pericas,M.A. and Serratosa,F. (1988) A convenient procedure for the synthesis of propargyl ethers derived from secondary alcohols. Synthesis, 9, 707–709. [Google Scholar]
- 26.Sakurai M., Wirsching,P. and Janda,K.D. (1996) Synthesis of a nucleoside hapten with a [P(O)-O-N] linkage to elicit catalytic antibodies with phosphodiesterase activity. Bioorg. Med. Chem. Lett., 6, 1055–1060. [Google Scholar]
- 27.Verhoeven E.E.A., Wyman,C., Moolenaar,G.F., Hoeijmakers,J.H. and Goosen,N. (2001) Architecture of nucleotide excision repair complexes: DNA is wrapped by UvrB before and after damage recognition. EMBO J., 20, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertrand-Burggraf E., Selby,C.P., Hearst,J.E. and Sancar,A. (1991) Identification of the different intermediates in the interaction of (A)BC excinuclease with its substrates by DNase I footprinting on two uniquely modified oligonucleotides. J. Mol. Biol., 219, 27–36. [DOI] [PubMed] [Google Scholar]
- 29.Moolenaar G.F., Höglund,L. and Goosen,N. (2001) Clue to damage recognition by UvrB: residues in the beta-hairpin structure prevent binding to non-damaged DNA. EMBO J., 20, 6140–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J.-J., Phillips,A.M., Hearst,J.E. and Sancar,A. (1992) Active site of (A)BC excinuclease. II. Binding, bending and catalysis mutants of UvrB reveal a direct role in 3′ and an indirect role in 5′ incision. J. Biol. Chem., 267, 17693–17700. [PubMed] [Google Scholar]
- 31.Visse R., King,A., Moolenaar,G.F., Goosen,N. and van de Putte,P. (1994) Protein–DNA interactions and alterations in the DNA structure upon UvrB–DNA preincision complex formation during nucleotide excision repair in Escherichia coli. Biochemistry, 33, 9881–9888. [DOI] [PubMed] [Google Scholar]
- 32.Zou Y. and Van Houten,B. (1999) Strand opening by the UvrA(2)B complex allows dynamic recognition of DNA damage. EMBO J., 18, 4889–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]