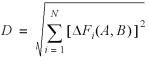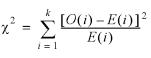Abstract
Based on searches for disabled homologs to known proteins, we have identified a large population of pseudogenes in four sequenced eukaryotic genomes—the worm, yeast, fly and human (chromosomes 21 and 22 only). Each of our nearly 2500 pseudogenes is characterized by one or more disablements mid-domain, such as premature stops and frameshifts. Here, we perform a comprehensive survey of the amino acid and nucleotide composition of these pseudogenes in comparison to that of functional genes and intergenic DNA. We show that pseudogenes invariably have an amino acid composition intermediate between genes and translated intergenic DNA. Although the degree of intermediacy varies among the four organisms, in all cases, it is most evident for amino acid types that differ most in occurrence between genes and intergenic regions. The same intermediacy also applies to codon frequencies, especially in the worm and human. Moreover, the intermediate composition of pseudogenes applies even though the composition of the genes in the four organisms is markedly different, showing a strong correlation with the overall A/T content of the genomic sequence. Pseudogenes can be divided into ‘ancient’ and ‘modern’ subsets, based on the level of sequence identity with their closest matching homolog (within the same genome). Modern pseudogenes usually have a much closer sequence composition to genes than ancient pseudogenes. Collectively, our results indicate that the composition of pseudogenes that are under no selective constraints progressively drifts from that of coding DNA towards non-coding DNA. Therefore, we propose that the degree to which pseudogenes approach a random sequence composition may be useful in dating different sets of pseudogenes, as well as to assess the rate at which intergenic DNA accumulates mutations. Our compositional analyses with the interactive viewer are available over the web at http://genecensus.org/pseudogene.
INTRODUCTION
We have identified pseudogenes in several completely sequenced eukaryotic genomes, mapping their positions on the chromosomes using BLAST (1) and related programs to search against protein databases (2–6). Here, we define pseudogenes as disabled copies of genes that do not produce a functional, full-length copy of a protein (7). Operationally, these are identified as regions of the chromosome that are similar to known proteins but contain obvious disablements (such as stop codons or frameshifts) mid-domain. There are two types of pseudogenes: (i) duplicated pseudogenes, which arise from duplication of a gene followed by an initial disablement (usually a premature stop-codon or frameshift mutation); and (ii) processed pseudogenes, which arise from reverse transcription of mRNA transcripts followed by reintegration into the genome and subsequent disablement (8). Pseudogenes are of special interest in the study of genomic evolution; since they are no longer functional, their subsequent degradation through accumulation of further coding disablements is generally not subject to selective pressure. Therefore, their sequence composition reflects the biochemical requirements of the gene to which they are related (as they are clearly similar to functional proteins) and the accumulation of mutations inherent in non-coding DNA. Consequently, they serve as markers to measure the overall mutation processes and the stability of the genomic sequence (9).
In an earlier large-scale survey of pseudogenes in the worm Caenorhabditis elegans (2), we examined a number of near-extinct gene families with large numbers of pseudogenes. We found that the amino acid composition of these pseudogenes is at an intermediate level between genes and non-coding DNA—the amino acid composition of non-coding regions simply being translations of randomly selected intergenic regions. This analysis clearly indicated that this is the result of incremental mutations causing a drift towards overall chromosomal composition rather than a statistical artifact. Here we have greatly expanded our analysis to include the yeast, fly and human chromosomes 21 and 22, and widened the scope to include codon and nucleotide as well as amino acid composition.
It has been demonstrated that the dinucleotide composition of eukaryotic chromosomes is homogeneous within an organism, even for regions with high coding content (10). However, the composition of genes is strongly affected by evolutionary constraints and therefore may be statistically ‘unfavorable’ in the context of the genome as a whole, for example, in codon use. Thus, pseudogenes might be expected to drift towards a make-up similar to ‘random’ intergenic DNA, even though they remain detectable by sequence alignment. Although the mechanisms and rates of random mutation (as well as gene loss) may differ across organisms—leading to pseudogene populations that differ greatly in size and composition—it is likely that dimer and trimer frequencies will, across the entire set, change from that favored by genes to the chromosomal bias. We have, in fact, observed this intermediate composition in multiple organisms, both on the nucleotide and implied amino acid level, in multiple organisms.
Our work follows upon a number of recent analyses of proteome composition, especially in the context of evolutionary relationships between organisms and in the broader functional implications of sequence make-up (2,9–18). Although intergenic DNA does not have an amino acid composition in the strict biological sense, this view is one of the most useful for examining different features, since it results from several different biases in sequence composition.
MATERIALS AND METHODS
Data sets
The yeast genome and proteome sequences (19) were obtained from the Saccharomyces Genome Database (ftp://genome-ftp.Stanford.edu/pub/yeast/SacchDB). For the worm genome (20), we used the Wormpep18 database and the complete chromosome sequences (December 1999 versions) available from the Sanger Centre ftp site (ftp://ftp.sanger.ac.uk). For the fly, Release 2 of the fly chromosomes (21) and protein predictions was obtained from the Berkeley Drosophila Genome project website (http://www.fruitfly.org). The Mycoplasma genitalium and Escherichia coli genomes (22,23) were obtained from GenBank (April 2001). The composition statistics for the human genome were taken from chromosomes 21 and 22 (24,25). Genes were predicted using the program GenomeScan (26); we have also used the Ensembl set for some comparisons. GenomeScan finds 279 and 648 genes for chromosomes 21 and 22, respectively, while Ensembl lists 290 and 711 genes (see Sequence Analysis for a detailed description). We also use the most current releases of the human genome, including the April 1, 2001 assemblies (http://genome.ucsc.edu) and release 1.0 of the Ensembl Project gene annotations (http://www.ensembl.org and ftp://ftp.sanger.ac.uk). The release files that we use in the current study are available from our website; it is important to note that data files are constantly changing given the ongoing nature of sequencing projects.
Pseudogene annotations
We derived sets of pseudogene sequences for the worm, fly and yeast genomes and for human chromosomes 21 and 22 (2–4). Pseudogenes were predicted by searching for homologies with disablements in the genomic DNA aligned against the entire SWISS-PROT (27) database plus the proteome of the organism studied. This was done using the BLAST (1) and FASTX/Y (28) alignment programs, with repeats masked using RepeatMasker (29), and low-complexity regions masked with SEG (30) (settings ‘25 3.0 3.3’ and ‘45 3.4 3.75’). Specifically, for yeast, pseudogenic sequences were extended at either end into the optimal disabled open reading frame. A summary of all the pseudogenes we annotated is shown in Table 1. Note that the finding of all these pseudogenes involves a massive amount of calculation. On average, one assignment of pseudogenes to each 20 Mb of genomic sequence took at least 10 CPU days (on a 1.2 GHz processor). Thus, the full set of assignments reported here involved many months of sequence comparisons. It is also worth noting that since most pseudogenes have multiple disablements in the homologous region (for humans, >90%), the possibility that assignments may in fact be functional genes is negligible.
Table 1. Statistics for pseudogene sets.
| Organism | No. of pseudogenes (no. of residues) | No. of genes (no. of residues) | Chromosome size (Mb) | A/T content (%) | Source |
|---|---|---|---|---|---|
| Worm | 1836 (215 995) | 18 680 (8 140 673) | 99 | 63.1 | (2) |
| Yeast | 166 (34 099) | 6280 (2 974 116) | 12.3 | 61.7 | (3) |
| Fly | 114 (40 505) | 14 332 (7 177 167) | 116 | 56.2 | (5) |
| Human | 325 (77 031) | 927 (447 410) | 68.5 | 54.5 | (4); chromosomes 21 and 22 only |
Pseudogene identification used the process described in Harrison et al. (2), except for yeast. Some of the sets described here are from intermediate data sets, so the total number may differ slightly from published results.
We assigned pseudogenes as processed or duplicated using two criteria: first, we picked as candidate processed pseudogenes, all those matches that comprise >70% of the length of the closest matching human Ensembl or SWISS-PROT database sequence in a continuous segment. We allowed for any obvious surrounding exon structure, and for known single-exon human genes. Secondly, we assigned any genes with evidence for a polyadenine tail as candidate processed pseudogenes. Pseudogenes were separately split into roughly equal proposed modern and ancient subsets based on percent identity to the closest Ensembl protein, divided around the median score (FASTA identity of 79%).
Sequence analysis
Coordinates for introns and exons for human genes were extracted using the predictions from GenomeScan (26). Gene composition statistics were taken from assembled exons or full amino acid predictions. Amino acid statistics for pseudogenes were made using the FASTA-generated alignment; codons were extracted from the chromosome based on start coordinates and implied positions from alignments, skipping over frameshifts and partial codons.
The implied composition of intergenic DNA was calculated using the first frame of the forward strand only (other frames yield identical results). Dinucleotide predictions (and trinucleotide, where distinct from codons and residues) used every frame. No masking for other features (including repeats) was used for intergenic statistics, but gaps (indicated by ‘N’ in the raw sequence) were passed over. Masking does not produce significantly different statistics, even in the human chromosomes where removing repeats nearly halves the total sequence length. Codon frequencies in all cases refer to the usage compared with synonymous codons, so that Trp and Met codons will have the same frequency in all features, no matter what the trinucleotide composition.
RESULTS
Statistical characterization of composition
We have used a formula independent of scale to quantitatively evaluate the compositional difference of two sets of features (where a feature could be such things as ‘genes in the worm’ or ‘intergenic DNA in the human’). The similarity in residue frequencies between any two features was determined by treating each set as an N-dimensional vector and calculating the ‘distance’ between these vectors. For a given sequence element indexed by i (either amino acid, codon or dinucleotide), we use the following notation for the absolute difference in frequency between features A and B:
ΔFi(A,B) = │Fi(A) – Fi(B)│
Then distance can thus be expressed as:
where N is 20 for amino acids (21 if stop codons are included), 64 for codons and 16 for dinucleotides. This calculation is most useful in determining the divergence of pseudogene composition from the presumed original level, but as part of a larger matrix can be used to define relationships across a larger set. Since the D vectors here all have a sum of 1, distance is always relative to this number, and average change for each amino acid or codon depends on the value of N. We have converted all distances to percent values here.
Amino acids in any group can also be treated individually: in a large enough set of features, the standard deviation for the frequency of each residue can be calculated. We have scaled this figure to be comparable across the entire set by dividing by the mean frequency. This allows the relative variability or ‘spread’ of each residue in a set of features to be expressed quantitatively as well as qualitatively. Cysteine provides an example; it has the third highest spread value due to the low mean frequency but wide difference in frequency across the genes. However, in the comparisons of pseudogenes, standard deviation is less useful. For the purposes of plotting the relationships between pseudogenes, genes and chromosomes, we have simply sorted by values of ΔF(genes,intergenic), as will be discussed in the results for pseudogenes. This quantity is the difference in composition between genes and translated intergenic DNA for each amino acid (or codon). Typically, this value ranges from nearly zero (especially for Ser) to as much as 0.04 for Ala, Asp, Glu or Phe, and 0.06 for the stop codon.
Genes versus non-coding DNA
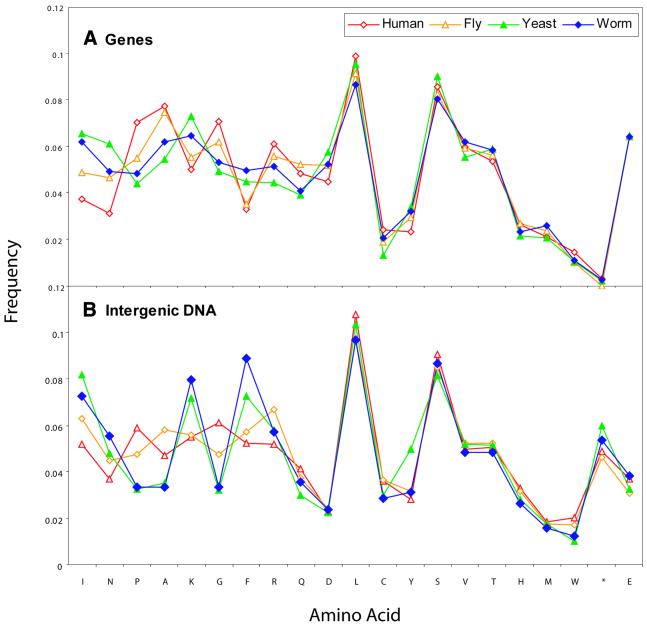
We compare amino acid composition of genes and translated intergenic regions in Figure 1. Several amino acids, in particular Ile, Lys and Asn are extremely variable, and Cys usage in human is twice that in yeast. Glu, on the other hand, is virtually constant. Within a genome, different chromosomes tend to have a very similar composition for both amino acids and dinucleotides (10). Human chromosomes vary most for the codons encoding Gly, Pro, Ile, Tyr and Ala, but frequencies for most other amino acids are quite close, even for relatively short chromosomes. We discuss the differences across the human genome below.
Figure 1.
(A) Gene and (B) intergenic region composition for the 20 amino acids and stop signal (*) in the four eukaryotes. Residues are sorted in decreasing order by standard deviation of gene frequencies across the organisms. Human genes are taken from GenomeScan predictions along chromosomes 21 and 22; for other organisms the available complete proteomes have been used. Some gene sequences may include the terminating stop codon, thus there is some variation in the frequency shown for this signal.
A striking result is the influence of the A/T content of genomes (Table 1) on the distribution of amino acid compositions for genes (Fig. 1). This is apparent when we compare the data in Figure 1 and Table 1. We observe that the ordering of the organisms by their A/T content (Table 1) is also maintained in the distributions of amino acid compositions (Fig. 1). Thus, genes in worm and yeast—two genomes with relatively high A/T content in the data set—have similar amino acid compositions. The same applies to genes in the fly and human genomes, which have relatively low A/T content. A similar observation has previously been reported for less complex genomes (31), and we propose that the main (but not exclusive) reason for this is the effect of A/T content on codon usage.
Implied amino acid composition of pseudogenes
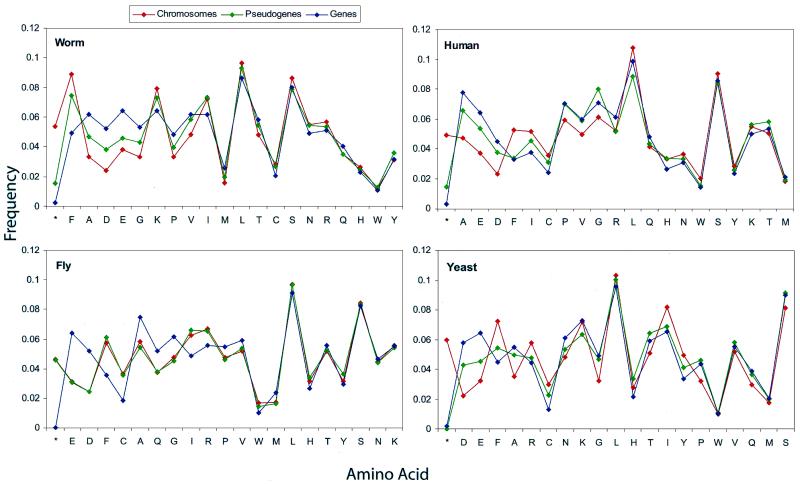
Figure 2 shows plots of the amino acid composition of pseudogenes from each organism versus the corresponding genes and intergenic DNA; Table 2 lists the distances obtained from these sets of statistics. The composition of most of the sets of pseudogenes found is intermediate between that of genes and non-coding regions. This is evident in the worm, both human chromosomes, and the yeast genome; however, the fly pseudogenes have almost exactly the same composition as intergenic DNA. The latter result may stem from a high underlying rate of point mutations in the fly genome. In addition, the small number of fly pseudogenes is consistent with the observed high rate of genomic DNA loss in the fly (32) and is not necessarily indicative of an especially ancient pseudogene population.
Figure 2.
Compositon of ΨG in the eukaryotes. The amino acid content of pseudogene predictions is compared with the implied translation of unmasked chromosomes and identified genes. For the human, only chromosomes 21 and 22 are used in the plot shown. In each case, residues have been sorted in order of the difference in frequency between genes and chromosomes [ΔF(genes,intergenic)].
Table 2. Distances for features in eukaryotic genomes (without stop codon).
| Intergenic regions (%) | Genes (%) | |
|---|---|---|
| Worm genes |
7.04 |
– |
| Worm pseudogenes |
3.31 |
4.49 |
| Human genes |
6.00 |
– |
| Human pseudogenes |
4.92 |
2.91 |
| Fly genes |
6.30 |
– |
| Fly pseudogenes |
0.96 |
6.67 |
| Yeast genes |
7.33 |
– |
| Yeast pseudogenes | 4.92 | 3.56 |
Column headings apply only to the named feature within the same organism.
Overall, 16 out of 21 amino acid types (including stop codons) in the worm have occurrences between that of intergenic DNA and that of genes. This is reduced to 14 residues in both human and yeast (sometimes less when individual chromosomes are used instead of all chromosomes). There are very few cases in any organism where pseudogene frequencies differ greatly from this intermediate range. More importantly, in Figure 2 we have sorted residues by ΔF(genes,intergenic), showing that virtually all of the amino acids with large overall divergence have an intermediate frequency for pseudogenes.
From the distances (without stop codons) shown in Table 2 and from the plots it is obvious that the worm pseudogenes are overall closer in composition to intergenic DNA, while the opposite largely holds for human; yeast is nearly exactly intermediate. We have not plotted pseudogenes from multiple organisms together because although the ‘direction’ of compositional drift is towards non-coding DNA in each organism, their exact composition depends on their age or the amount of mutation in each species. Therefore, we cannot directly compare pseudogenes from different organisms. The type of pattern shown in Figure 1 does not appear, since organismal genome composition has little relation to mutation patterns and rates on this scale.
Classes of pseudogenes
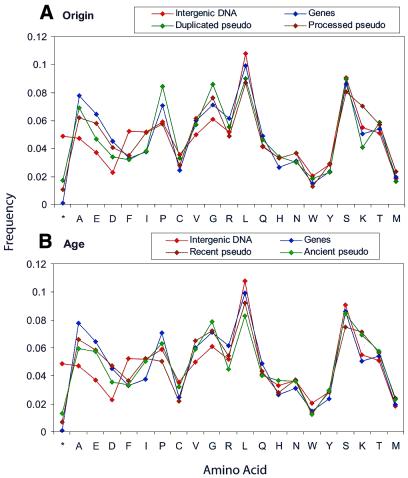
We divided human pseudogenes two different ways into subsets: (i) We classified human pseudogenes as processed or duplicated (Fig. 3A); these sets have distinct compositions but do not have a consistent pattern relative to genes and intergenic DNA. Some variations can be explained by the implied functional characteristics of these sets; the processed pseudogenes have a high number of disabled ribosomal proteins, whose higher Lys content contributes to the peak frequency for this residue on the plot. However, we cannot draw any conclusions about the overall degree of mutation (and the corresponding relative age) of the processed and duplicated subsets of pseudogenes. (ii) We also then subdivided the processed human pseudogenes into hypothetical ‘ancient’ and ‘modern’ subsets based on their similarity to the closest matching human gene. Figure 3B shows the composition of these groups relative to genes and chromosomes, again sorted by ΔF(genes,intergenic). Modern pseudogenes are generally closer to genes than intergenic DNA, and again this relationship is clearest in the residues with highest variability (only two out of the top 10 are anomalous). The pattern seen in Figure 2 appears to be the cumulative effect of both classes, and with smaller sets of pseudogenes there are more cases of erratic frequencies for individual residues.
Figure 3.
Classifications of pseudogenes. Residues are sorted as above by ΔF(genes,intergenic). (A) Pseudogenes divided into putative processed and duplicated sets. (B) Processed pseudogenes divided into recent and ancient sets based on a median FASTA identity value of 79%.
Codon and dinucleotide usage
Pseudogene codon usage in both worm and human also tends to lie between that of genes and intergenic regions. Of the 64 codons in worm pseudogenes, 47 have intermediate frequencies and seven are within 5% of the frequency for another feature. In human chromosomes 21 and 22, 49 and 53 codons, respectively, are intermediate. Several codons in chromosome 21 have highly elevated frequencies—in particular, CAC, CGT, GGA and GGT. If codon biases for individual amino acids are examined instead, 56 and 59 codons (excluding Met and Trp) in chromosomes 21 and 22 have a composition intermediate between genes and chromosomes.
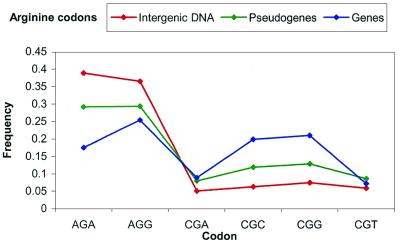
We have plotted the biases for the set of the codons forming Arg in Figure 4, which best characterizes the pattern. Applying the distance formula with N = 64 again indicates that human pseudogenes are slightly closer to genes, while worm pseudogenes are closer to intergenic DNA. We have not attempted to quantify codon bias in this manner, since it is dependent on overall implied amino acid frequencies, and is only relevant for individual sets of codons.
Figure 4.
Arginine codon bias in human chromosomes 21 and 22. Frequency is out of all Arg codons, or Fcodon/FArg.
Statistics for pseudogene subsets again display the expected distinction between ancient and modern sets. Whether all codons or only Arg codons are sampled, the modern set tends to have a bias closer to genes than less homologous pseudogenes. Processed versus duplicated pseudogenes again do not follow any trend, rather varying by chromosome and scope of comparison—though in Arg codons alone, processed pseudogenes are more closely related to genes (especially in chromosome 21).
Relationship to dinucleotide relative abundance
Gentles and Karlin (10) have shown in a large set of both prokaryotic and eukaryotic organisms that dinucleotide frequencies represent a distinct ‘signature’, consistent across all chromosomes of a genome, and apparently distinct from preferences towards individual base pairs. This is most evident in the extremely low occurrence of the CG dimer in the human, which we have calculated to comprise only 1.6% of all dimers in chromosomes 21 and 22 (versus, e.g., 5.7% for GC). A predictable result of this is the corresponding depletion of the four Arg codons starting with CG in raw genomic DNA; AGA and AGG instead comprise ∼80% of the full set. Human genes maintain considerably higher levels of Arg CG codons, perhaps since these are less susceptible to non-synonymous substitution. However, other CG-containing codons are used much more often in genes than their random frequency would suggest, and CG levels are thus elevated overall throughout human exons, at 4.2% in chromosome 22.
Worm does not have any example of this kind of discrepancy; Arg codons in genes follow a considerably different pattern. Though both the human and worm genomes are rich in adenine and thymine, human genes tend to use CGC/G more than CGA/T. Percent CG in pseudogenes increases relative to chromosomes to 2.2 and 2.6% in 21 and 22, respectively, again much closer than expected from the amino acid distributions.
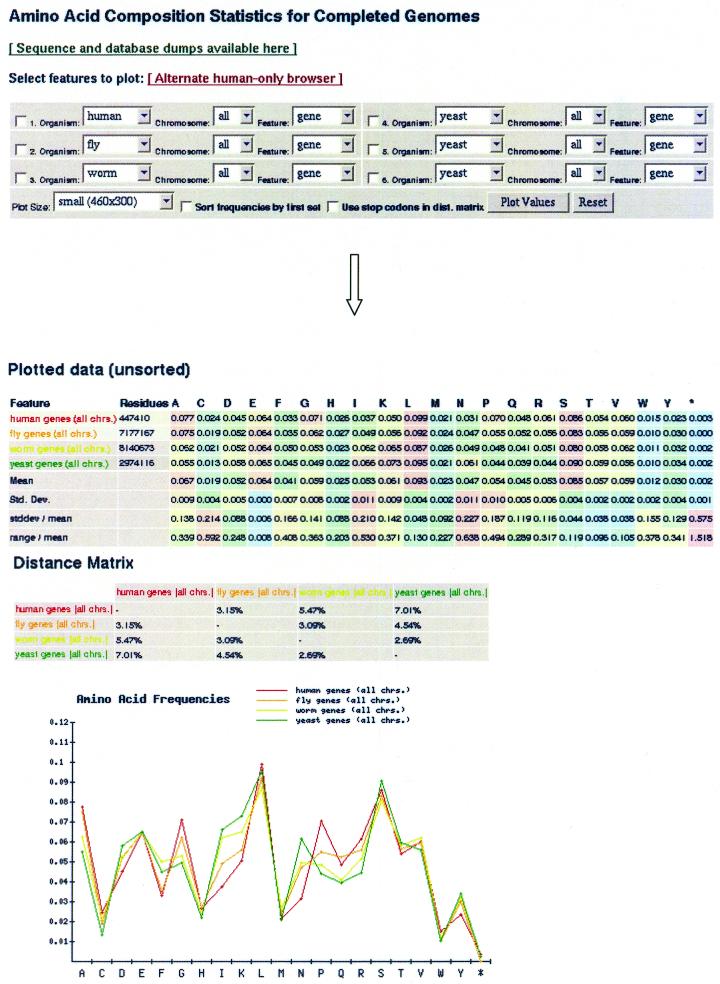
Web composition browser
We have made available an online browser (Fig. 5) that summarizes and builds upon all of our results relating to amino acid composition and eukaryotic pseudogenes, described at http://genecensus.org/pseudogene. Data from other organisms and features not discussed here are viewable along with primary results through several plots and charts created based on user input. Most of the data presented here is directly replicated at this site in both numerical and graphical form. Results from additional completed microbial genomes have been included in the database for comparison. We have also created an additional page focusing on the expanded human genome with a larger presentation of the various sets of features now available.
Figure 5.
Sample screen of the online composition browser. The database is accessible through a form that allows selection of any combination of features for which amino acid composition has been determined. Included in the display is a plot of the compositions and statistics for each feature.
DISCUSSION
We have surveyed nucleotide and amino acid composition across pseudogenes, genes and intergenic DNA, the relationship between coding and non-coding sequences and patterns in mutations in disabled genes. Our analysis suggests a trend for both genes and pseudogenes to assume the underlying composition of the genome, to the extent that this does not interfere with biochemical function or result in potentially much less stable codon usage. Though genes have amino acid compositions distinct from that of translated intergenic DNA, they clearly reflect the underlying levels of nucleotide usage in the genomes as a whole. Pseudogenes can be viewed as genes removed from selective constraints, and our results show that the cumulative effect of mutations is to yield a sequence approaching homogeneity with the surrounding non-coding DNA. Our results can be affected by several factors, particularly the size and statistical relevance of the data sets and issues of compositional bias in the portion of the human genome studied.
Literal statistical significance
The usefulness of the annotations relied upon here depends on the size of our data sets. The scale of this analysis is large enough to ensure that compositional biases are real rather than random. For the composition of any pair of features, the chi-squared statistic can be calculated by mutiplying the frequency in either set of each amino acid by the total number of amino acids in the observed set, and summing as shown:
The summation is for the 20 amino acids, where k = 20, O(i) is the count for a particular amino acid in the observed set, and E(i) is the count for a particular amino acid in the expected set. For samples of the size used here, e.g. approximately 150 000 amino acids for genes in human chromosome 21 and 82 000 for human pseudogenes, the chi-squared value will be extremely high even with only slight compositional differences, with a correspondingly low P-value. Even for features that are nearly indistinguishable on a plot, the number of amino acids is so large that the chi-squared statistic is always highly significant.
In light of this, the distance calculations used here are more indicative of the distinction between two composition figures. Thus, the proteins in human and yeast have a distance of 7%, while human and fly proteins have a distance of 3%. A distance of 1% is usually negligible—this is about the average difference in implied amino acid composition between worm chromosomes’ intergenic regions. However, this is affected by the selection of particular features and thus by any distinct composition they possess, discussed in the next section.
Expansion to the entire human genome
We have evaluated two sets of annotations for chromosomes 21 and 22. The set of predicted human genes in chromosomes 21 and 22, assigned using the GenomeScan algorithm, is close in size and composition to the current Ensembl Project gene predictions for these chromosomes. Using GenomeScan allowed us to assign a large and relatively complete set of genes at an early date, before the Ensembl predictions were completed. More importantly, it gives a uniform set of predictions for genes—in contrast to Ensembl, which combines gene identified by a variety of methods.
We have chosen here to concentrate only on chromosomes 21 and 22, the earliest and most thoroughly completed chromosomes, to ensure full coverage by pseudogene predictions and accurate composition statistics. Incomplete sequences for other chromosomes make it difficult to rely on homology searches as undertaken here. For instance, as of September 2001, only 70% of chromosome 1 is currently represented as actual nucleotides rather than null bases (though these numbers are continuously changing).
However, it is useful to evaluate how representative chromosomes 21 and 22 are of the rest of the draft genome sequence. We have looked at the April 2001 assemblies of the Golden Path to evaluate the similarity in composition of the smaller chromosomes to the complete set. Human intergenic DNA tends to vary most for the trinucleotides encoding Gly, Pro, Ile, Tyr and Ala, but frequencies for most other amino acids are quite close, even for the relatively short chromosomes we studied. Distances between gene and intergenic amino acid frequencies vary from 4.5 to 7%, with chromosomes 21 and 22 being close to opposite extremes. Although chromosome 22 tends to be most divergent for the variable amino acids, both chromosomes have compositions that follow those of longer chromosomes. Interestingly, however, in their intergenic DNA they differ more from each other than most other pairs, with a distance of 5.4%.
Though the genes used here and those from Ensembl have similar compositions (distances of 1.2% for either chromosome), both are distinctly biased in composition relative to the entire Ensembl set of approximately 27 000 genes. To evaluate this bias (33), we have randomly sampled with replacement sets of 500 genes from Ensembl (compared with totals of approximately 300 and 700 for chromosomes 21 and 22, respectively), and examined the distribution of amino acid frequencies for these sets. Genes in chromosomes 21 and 22 have frequencies that often fall outside or at one extreme of the random distribution. The residues Ala, Pro, Arg, Trp and Tyr are all enriched, while Asp, Glu, Phe, Ile, Lys and Asn are depleted. It is reasonable to assume that this corresponds to functional differences in these genes. Nevertheless, substituting the full Ensembl predictions for the chromosome 21 and 22 GenomeScan annotations in Figure 1A results in a substantially similar plot (data not shown). This means that while there are real differences in composition between the genes on chromosomes 21 and 22 and those in the full human genome, on the scale of our analysis (particularly with regard to trends with respect to other organisms) these differences are not significant.
Classification and dating by composition
One interesting application we see for such composition statistics is dating of pseudogenes. We have tried to make this distinction based on the degree of similarity to human proteins, and simply divided ancient and modern sets around the median. We would like to find a more precise method of classifying pseudogenes by age, instead of depending entirely on the results from BLAST and FASTA similarity. Our treatment of amino acid composition in non-coding features relies on shifts in frequency as representative of the amount of mutation overall. This is not necessarily the case for an individual sequence, which ideally needs to be examined on the scale of codons—that is, taking into account synonymous as well as non-synonymous mutations. Furthermore, it seems reasonable to expect that codon bias will tend to reach that of chromosomes for individual homologies, but this may involve so great an amount of mutation that non-synonymous changes will make the homology undetectable by our methods. It is also difficult to guess at the exact original sequence of the pseudogene, especially in cases that are obviously duplicated and those most closely matching proteins in other species. Lastly, codon bias in genes as described here is again a cumulative effect, not applicable to individual genes where nucleotide make-up is used as an altogether different measure, as in the codon adaptation index for prediction of expression level (34).
We do believe that it is possible and practical to determine the overall age of a population of pseudogenes taken as a whole, given that our search characteristics are largely similar in all the eukaryotes used. Though individual pseudogenes do not comprise a large enough sample size for this to work on a smaller scale, except in the few cases where the exact original gene sequence is preserved elsewhere, it may still be feasible to segregate pseudogenes into smaller sets based on predicted age. If one accepts the make-up of genes and non-coding DNA—both codons and residues—as relatively constant over time, an average figure for the origin of disablements or reverse transcription might be calculated based on rates of mutation and the divergence from genes.
Acknowledgments
ACKNOWLEDGEMENTS
M.G. thanks the Keck Foundation and the NIH (P50 HG02357-01) for support. N.M.L. is sponsored by the Anna Fuller Fund.
REFERENCES
- 1.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison P.M., Echols,N. and Gerstein,M.B. (2001) Digging for dead genes: an analysis of the characteristics of the pseudogene population in the Caenorhabditis elegans genome. Nucleic Acids Res., 29, 818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison P., Kumar,A., Lan,N., Echols,N., Snyder,M. and Gerstein,M. (2002) A small reservoir of disabled ORFs in the yeast genome and its implications for the dynamics of proteome evolution. J. Mol. Biol., 316, 409–419. [DOI] [PubMed] [Google Scholar]
- 4.Harrison P.M., Hegyi,H., Balasubramanian,S., Luscombe,N.M., Bertone,P., Echols,N., Johnson,T. and Gerstein,M. (2002) Molecular fossils in the human genome: identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res., 12, 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison P.M. and Gerstein,M. (2002) Studying genomes through the aeons: protein families, pseudogenes and proteome evolution. J. Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]
- 6.Harrison P.M., Kumar,A., Lang,N., Snyder,M. and Gerstein,M. (2002) A question of size: the eukaryotic proteome and the problems in defining it. Nucleic Acids Res., 30, 1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mighell A.J., Smith,N.R., Robinson,P.A. and Markham,A.F. (2000) Vertebrate pseudogenes. FEBS Lett., 468, 109–114. [DOI] [PubMed] [Google Scholar]
- 8.Vanin E.F. (1985) Processed pseudogenes: characteristics and evolution. Annu. Rev. Genet., 19, 253–272. [DOI] [PubMed] [Google Scholar]
- 9.Gojobori T., Li,W.H. and Graur,D. (1982) Patterns of nucleotide substitution in pseudogenes and functional genes. J. Mol. Evol., 18, 360–369. [DOI] [PubMed] [Google Scholar]
- 10.Gentles A.J. and Karlin,S. (2001) Genome-scale compositional comparisons in eukaryotes. Genome Res., 11, 540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ophir R. and Graur,D. (1997) Patterns and rates of indel evolution in processed pseudogenes from humans and murids. Gene, 205, 191–202. [DOI] [PubMed] [Google Scholar]
- 12.Kreil D.P. and Ouzounis,C.A. (2001) Identification of thermophilic species by the amino acid compositions deduced from their genomes. Nucleic Acids Res., 29, 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White S.H. and Jacobs,R.E. (1993) The evolution of proteins from random amino acid sequences. I. Evidence from the lengthwise distribution of amino acids in modern protein sequences. J. Mol. Evol., 36, 79–95. [DOI] [PubMed] [Google Scholar]
- 14.Qian J., Stenger,B., Wilson,C.A., Lin,J., Jansen,R., Teichmann,S.A., Park,J., Krebs,W.G., Yu,H., Alexandrov,V., Echols,N. and Gerstein,M. (2001) PartsList: a web-based system for dynamically ranking protein folds based on disparate attributes, including whole-genome expression and interaction information. Nucleic Acids Res., 29, 1750–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin J. and Gerstein,M. (2000) Whole-genome trees based on the occurrence of folds and orthologs: implications for comparing genomes on different levels. Genome Res., 10, 808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen R. and Gerstein,M. (2000) Analysis of the yeast transcriptome with structural and functional categories: characterizing highly expressed proteins. Nucleic Acids Res., 28, 1481–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerstein M. (1998) Patterns of protein-fold usage in eight microbial genomes: a comprehensive structural census. Proteins, 33, 518–534. [DOI] [PubMed] [Google Scholar]
- 18.Gerstein M. and Levitt,M. (1997) A structural census of the current population of protein sequences. Proc. Natl Acad. Sci. USA, 94, 11911–11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherry J.M., Ball,C., Weng,S., Juvik,G., Schmidt,R., Adler,C., Dunn,B., Dwight,S., Riles,L., Mortimer,R.K. and Botstein,D. (1997) Genetic and physical maps of Saccharomyces cerevisiae. Nature, 387, 67–73. [PMC free article] [PubMed] [Google Scholar]
- 20. The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 21.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 22.Fraser C.M., Gocayne,J.D., White,O., Adams,M.D., Clayton,R.A., Fleischmann,R.D., Bult,C.J., Kerlavage,A.R., Sutton,G., Kelley,J.M. et al. (1995) The minimal gene complement of Mycoplasma genitalium. Science, 270, 397–403. [DOI] [PubMed] [Google Scholar]
- 23.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F., Gregor,J., Davis,N.W., Kirkpatrick,H.A., Goeden,M.A., Rose,D.J., Mau,B. and Shao,Y. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 24.Hattori M., Fujiyama,A., Taylor,T.D., Watanabe,H., Yada,T., Park,H.S., Toyoda,A., Ishii,K., Totoki,Y., Choi,D.K. et al. (2000) The DNA sequence of human chromosome 21. Nature, 405, 311–319. [DOI] [PubMed] [Google Scholar]
- 25.Dunham I., Shimizu,N., Roe,B.A., Chissoe,S., Hunt,A.R., Collins,J.E., Bruskiewich,R., Beare,D.M., Clamp,M., Smink,L.J. et al. (1999) The DNA sequence of human chromosome 22. Nature, 402, 489–495. [DOI] [PubMed] [Google Scholar]
- 26.Yeh R.F., Lim,L.P. and Burge,C.B. (2001) Computational inference of homologous gene structures in the human genome. Genome Res., 11, 803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson W.R., Wood,T., Zhang,Z. and Miller,W. (1997) Comparison of DNA sequences with protein sequences. Genomics, 46, 24–36. [DOI] [PubMed] [Google Scholar]
- 29.Smit A.F. (1999) Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev., 9, 657–663. [DOI] [PubMed] [Google Scholar]
- 30.Wootton J.C. and Federhen,S. (1996) Analysis of compositionally biased regions in sequence databases. Methods Enzymol., 266, 554–571. [DOI] [PubMed] [Google Scholar]
- 31.Singer G.A. and Hickey,D.A. (2000) Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol. Biol. Evol., 17, 1581–1588. [DOI] [PubMed] [Google Scholar]
- 32.Petrov D.A., Lozovskaya,E.R. and Hartl,D.L. (1996) High intrinsic rate of DNA loss in Drosophila. Nature, 384, 346–349. [DOI] [PubMed] [Google Scholar]
- 33.Gerstein M. (1998) How representative are the known structures of the proteins in a complete genome? A comprehensive structural census. Fold Des., 3, 497–512. [DOI] [PubMed] [Google Scholar]
- 34.Sharp P.M. and Li,W.H. (1987) The codon Adaptation Index—a measure of directional synonymous codon usage bias and its potential applications. Nucleic Acids Res., 15, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]