Abstract
Uterine Corpus Endometrial Carcinoma (UCEC) represents a common malignant neoplasm in women, with its prognosis being intricately associated with available therapeutic interventions. In the past few decades, there has been a burgeoning interest in the role of mitochondria within the context of UCEC. Nevertheless, the development and application of prognostic models predicated on mitochondrial-related genes (MRGs) in UCEC remains in the exploratory stages. This study utilized RNA sequencing data and clinical information from the TCGA database to identify differentially expressed MRGs (DEMRGs) between UCEC and normal groups that are associated with overall survival (OS). Patients were randomly assigned to training and testing cohorts in a 1:1 ratio. In the training cohort, a risk model based on DEMRGs was developed using Lasso Cox regression analysis. Subsequently, patients in both cohorts were stratified into high-risk and low-risk groups based on their median risk scores. The prognostic performance of the model was validated through Kaplan-Meier survival analysis, ROC curves, and nomograms. Additionally, further analyses including functional enrichment, immune landscape assessment, prediction of response to ICB therapy, mutation profiling, and drug sensitivity analysis elucidated biological distinctions between the identified risk groups. We established a risk model incorporating eight MRGs. Patients classified within he high-risk group exhibited significantly poorer prognoses relative to those in the low-risk group. Functional enrichment analysis identified substantial differences in biological processes and signaling pathways between the high-risk and low-risk cohorts. Immune landscape analysis showed that patients with elevated risk scores exhibited significant immunosuppressive and immune evasion mechanisms. Conversely, low-risk patients exhibited higher expression of human leukocyte antigen (HLA) family members and immune checkpoint genes (ICGs) compared to their high-risk counterparts.Consequently, low-risk patients showed greater responsiveness to immunotherapy and potential small molecule drugs, whereas high-risk patients were more susceptible to chemotherapy. The mitochondrial-related risk model formulated in this study demonstrates efficacy in predicting both prognosis and response to immunotherapy in patients with UCEC, thereby providing a scientific basis for personalized treatment strategies. Future research endeavors should focus on further validating the clinical utility of this model and investigate the specific mechanisms of the identified MRGs in UCEC.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-85537-7.
Keywords: Uterine corpus endometrial cancer, Mitochondrial, Prognosis, Immunotherapy, Tumor microenvironment
Subject terms: Cancer models, Tumour biomarkers, Tumour immunology, Cancer, Immunology, Biomarkers, Oncology
Introduction
Uterine corpus endometrial carcinoma (UCEC) is one of the most common malignant tumors within the female reproductive system. According to statistical data, the number of new cases of UCEC in 2022 surpassed 420,0001, and the global incidence rate during the period from 2007 to 2016 was approximately 1.3%2. UCEC is responsible for approximately 97,000 female deaths globally each year, and it has a five-year survival rate of approximately 80%1,2. Owing to early detection and intervention, the five-year survival rate for patients diagnosed with stage I UCEC can approachreach approximately 95%3. However, individuals diagnosed with advanced stages, recurrent conditions, or distant metastasis of UCEC encounter a markedly poor prognosis, with a five-year survival rate falling of less than 20%4. The principal risk factors for UCEC are complex and multifactorial, including age, endocrine influences, genetic predispositions, and lifestyle choices5. Notably, diseases related to energy metabolism abnormalities, such asinsulin resistance, obesity, and diabetes emerge as pivotal metabolic factors that significantly elevate the risk of developing UCEC6. Over the past few decades, the intricate relationship between aberrant energy metabolism and cancer has emerged as a pivotal focus in oncological research. The earliest identified tumor-specific metabolic alteration is the reprogramming of energy metabolism, also known as the Warburg effect, wherein cancer cells tend to metabolize glucose into lactate even under aerobic conditions7. This phenomenon underscores a fundamental shift in cellular energy production pathways, favoring glycolysis over oxidative phosphorylation despite the presence of oxygen, thus facilitating rapid tumor growth and proliferation7. The metabolic processes of cancer cells exhibit significant deviations from those of normal cells. This metabolic reprogramming not only underpins the rapid proliferation of malignant cells but also furnishes the requisite energy and molecular substrates essential for tumor growth and metastasis.
However, despite glycolysis being the primary energy source, the mitochondria of cancer cells remain active and participate in metabolic reprogramming to support cell survival and growth8. Furthermore, the disruption of Ca2 + homeostasis within mitochondria, the increased levels of reactive oxygen species (ROS), alterations in mitochondrial dynamics, and mitochondrial genetic defects collectively impair mitochondrial functionality, thereby fostering the initiation and progression of tumors9–11. As research into the mechanisms of tumorigenesis linked to mitochondrial dysfunction advances, the role of mitochondria in UCEC has garnered considerable attention. Research has shown that the expression of MCU and VDAC1 is significantly upregulated in UCEC tissues, and MCU-induced mitochondrial calcium uptake plays a crucial role in UCEC progression through its interaction with VDAC112. Additionally, research has reported that FBXO7, as a novel tumor suppressor factor in UCEC, inhibits the occurrence and development of UCEC by negatively regulating the INF2-DRP1 axis-related mitochondrial fission13. This provides a new theoretical basis for the pathogenesis and clinical treatment of UCEC.
Throughout the evolution of research on UCEC, it is evident that prognosis and treatment decisions were predominantly based on staging in the past. However, staging alone is insufficient for prognosis, as distinct molecular subtypes significantly influence both prognosis and therapeutic choices. Bokhman categorized UCEC into two types based on endocrine and metabolic functions, with Type I estrogen-dependent tumors having a significantly better prognosis than Type II non-estrogen-dependent tumors14. This method, however, suffers from high subjectivity and low reproducibility, posing substantial limitations. With advancements in molecular medicine, The Cancer Genome Atlas (TCGA), by considering tumor mutation burden (TMB) and multi-omic characteristics, more precisely classified UCEC into four categories15. This advancement has provided crucial insights for prognosis assessment and adjuvant therapy in endometrial cancer. Despite significant strides in molecular typing, numerous challenges persist. For example, integrating molecular typing with immunotherapy and refining molecular classifications to address the specific needs of different patients remain unresolved issues. In this study, leveraging the TCGA database, we constructed a novel risk model closely associated with the prognosis of UCEC patients using eight mitochondrial-related genes (MRGs). We stratified patients into high-risk and low-risk groups based on their prognostic characteristics, and conducted a comprehensive analysis between these groups. In summary, our mitochondrial-related risk model has proven effective in predicting the prognosis and immunotherapy responsiveness of UCEC patients, thereby offering a foundation for prognosis assessment and personalized treatment strategies.
Methods
Data collection and processing
We obtained RNA-seq data (FPKM) and clinical information from the TCGA (http://cancergenome.nih.gov/) database. After excluding patients with ambiguous prognostic information, we included 543 UCEC samples and 35 normal samples. The list of MRGs was collected from the MitoCarta 3.0 database (https://www.broadinstitute.org/mitocarta/mitocarta30-inventory-mammalian-mitochondrial-proteins-and-pathways) and Gene Set Enrichment Analysis (http://www.gsea-msigdb.org/gsea/index.jsp)16 (Supplementary Table S1). Subsequently, we utilized the “limma” package to identify differentially expressed MRGs (DEMRGs) between UCEC and normal tissues, using criteria of |Log fold change| >1 and adjusted P-value < 0.05. Furthermore, we conducted univariate COX regression analysis to screen for DEMRGs associated with overall survival (OS). Finally, for risk model construction and validation, we used stratified random sampling to divide the 543 UCEC samples into a training set and a testing set in a 1:1 ratio.
Construction of mitochondrial-related risk model
Based on the Cox regression model, we utilized the R package “glmnet” to implement LASSO regression for variable selection in the training set17–19. We conducted 10-fold cross-validation to determine the optimal regularization parameter, identifying variables with non-zero coefficients. These selected variables are regarded as having a significant impact on survival outcomes. Ultimately, we established a risk model based on MRGs, and the risk score for each UCEC sample was calculated using the following formula: Risk score = (Coef1 * expression mRNA1) + (Coef2 * expression mRNA2) + … + (Coefn * expression mRNAn). Coef represents the coefficient index of the LASSO regression coefficient for each mRNA. Then We divided the UCEC patients from the training and testing cohorts into high-risk and low-risk groups based on the median value of the risk score.
Survival and clinical characteristics analyses
We evaluated the differences in OS and disease specific survival (DSS) between high-risk and low-risk patient groups using Kaplan-Meier (KM) survival curves and the log-rank test. We utilized the “survival” and “timeROC” packages to plot receiver operating characteristic (ROC) curves to assess the model’s predictive value for 1-year, 3-year, and 5-year survival rates. Furthermore, we analyzed the relationship between the risk score and clinical characteristics by categorizing patients based on various clinical features.
Nomogram model construction
Based on the risk score and various clinical characteristics, we evaluated whether the risk score is an independent prognostic factor for predicting OS through univariate and multivariate regression analyses. Subsequently, using the “rms” R package, we constructed a nomogram model by integrating risk scores and clinical characteristics, assigning values to these variables within the model. The total score for each patient was obtained by summing the scores of the included predictive factors. Finally, the 1-year, 3-year, and 5-year OS probabilities for patients were predicted based on the total score and survival outcome probabilities. The nomogram model’s discrimination and accuracy were assessed using ROC curves and calibration curves.
Functional enrichment analysis
In this study, we employed the “clusterProfiler” and “org.Hs.eg.db” R packages20–24 to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses on DEMRGs between the UCEC group and the normal group, as well as differentially expressed genes (DEGs) between the high-risk and low-risk groups, with a corrected P-value threshold of less than 0.05. DEGs between the high-risk and low-risk groups in the training cohort were screened using “limma” package, applying criteria of |Log fold change| >1 and adjusted P-value < 0.05. Furthermore, we conducted Gene Set Enrichment Analysis (GSEA)25 for differential pathway expression analysis between the high-risk and low-risk groups to explore enriched pathways.
Immune landscape analysis and prediction of immunotherapy response
We presented the infiltration abundance of 24 immune cell types in UCEC samples with the single sample GSEA (ssGSEA) method, which assesses immune cell infiltration by quantifying the relative enrichment of specific immune cell gene sets within each sample’s gene expression profile26. Subsequently, we scrutinized the disparities in immune infiltration between high-risk and low-risk groups, as well as the differential expression of human leukocyte antigen (HLA) family members and immune checkpoint genes (ICGs). The ESTIMATE algorithm analyzes transcriptomic data to assess the infiltration levels of immune and stromal cells in malignant tumors27. Using the ESTIMATE algorithm, we calculate the Stromal Score and Immune Score for each sample, and combine them to generate the ESTIMATE Score. A higher score indicates a greater proportion of these components in the tumor microenvironment. Additionally, the Tumor Immune Dysfunction and Exclusion (TIDE, http://tide.dfci.harvard.edu/login/) score and Immunophenoscore (IPS) were leveraged to predict the response of UCEC patients to immune checkpoint blockade (ICB) therapy. The TIDE algorithm assesses tumor immune escape potential by analyzing gene expression profiles, incorporating T cell dysfunction and exclusion mechanisms to predict responses to ICB28. The IPS evaluates a patient’s likely responsiveness to immunotherapy based on the expression of specific immune-related genes29. The IPS of each UCEC sample was obtained from The Cancer Immunome Atlas (TCIA, https://tcia.at/home) database.
Mutation analysis
Somatic mutation data were sourced from the TCGA database. Utilizing the “maftools” package, we generated waterfall plots to depict the mutational landscape in high-risk and low-risk patient groups, and computed the TMB score for each sample. Additionally, we employed the " DEPTH” package to determine the intratumor heterogeneity (ITH) score for each sample. Furthermore, the microsatellite instability (MSI) status of each UCEC sample was obtained from TCIA database. We then analyzed the variations in TMB, ITH, and MSI between high-risk and low-risk groups, as well as the correlations between the risk score and TMB, ITH, and MSI.
Drug sensitivity analysis
We employed the R package “pRRophetic” to predict the 50% inhibitory concentration (IC50) values for common chemotherapeutic and small molecule drugs in each UCEC sample. Comprehensive information on the drugs was sourced from the Genome of Drug Sensitivity in Cancer (GDSC, https://www.cancerrxgene.org/)30. Subsequently, we examined the differences in drug sensitivity between the high-risk and low-risk groups.
Cell culture
The human endometriosis cell line (hEM15A) and UCEC cell lines (Ishikawa, HEC-1-A, and RL95-2) were obtained from the American Type Culture Collection (ATCC). All cell lines were cultured in DMEM/F12 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. The cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2.
Tissue collection
We obtained UCEC tissue samples, along with paired adjacent non-cancerous tissues, from 16 patients treated at the Cancer Hospital of Shantou University Medical College. All patients had not received prior radiotherapy or chemotherapy. The study involving human participants was conducted in accordance with the Declaration of Helsinki and relevant institutional and national guidelines and regulations. Ethical approval for this study was granted by the Ethics Review Committee of the Cancer Hospital of Shantou University Medical College. Written informed consent was obtained from all patients prior to their participation in the study.
Quantitative real-time PCR (qRT-PCR)
Total cellular RNA was isolated with TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. The extracted RNA was then reverse transcribed into cDNA using HiScript RT Mix (Vazyme, Nanjing, China). Quantitative real-time PCR (qRT-PCR) was performed using SYBR Green Master Mix (Vazyme, Nanjing, China), and the relative expression levels were calculated using the 2-∆∆CT method, with GAPDH as the internal reference gene. All primers are listed in Supplementary Table 2.
Statistical analysis
All statistical analyses were conducted using R software (version 4.2.1). The Wilcoxon test was used to compare differences in variables between two groups, while the Kruskal-Wallis test was employed for comparisons among more than two groups. Correlation analysis was performed using Spearman’s rank correlation. Differences in clinicopathological characteristics between the training and testing cohorts were assessed using the chi-square test. In this study, a P-value of less than 0.05 was considered statistically significant.
Results
Differential gene expression and function analyses between UCEC and normal groups
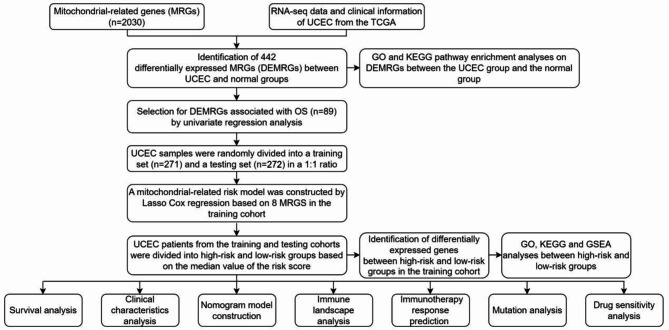
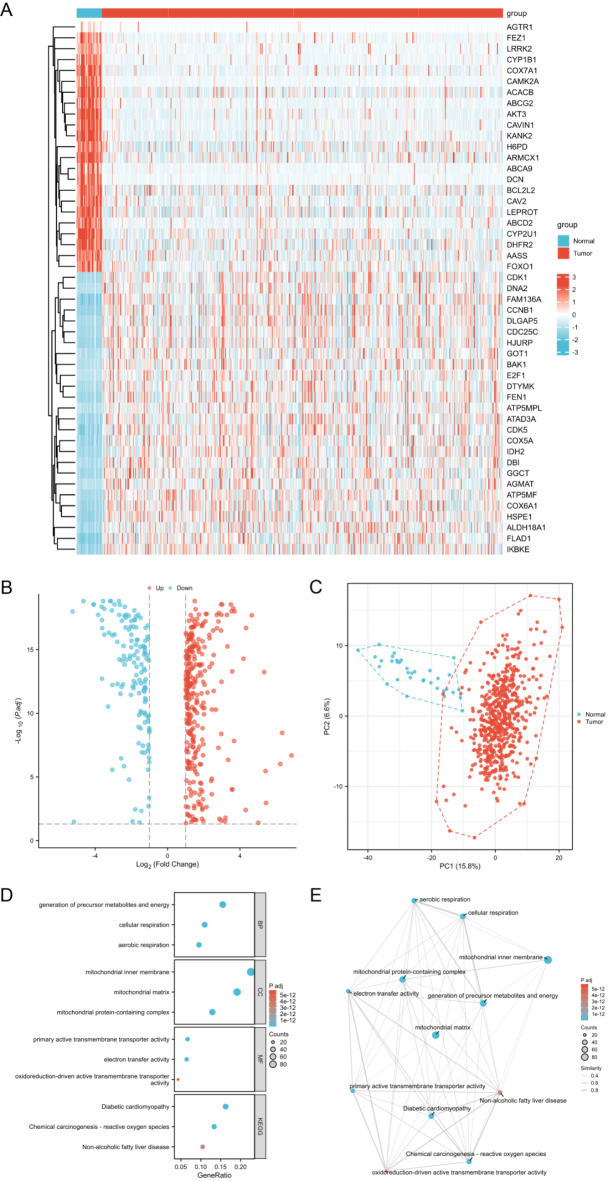
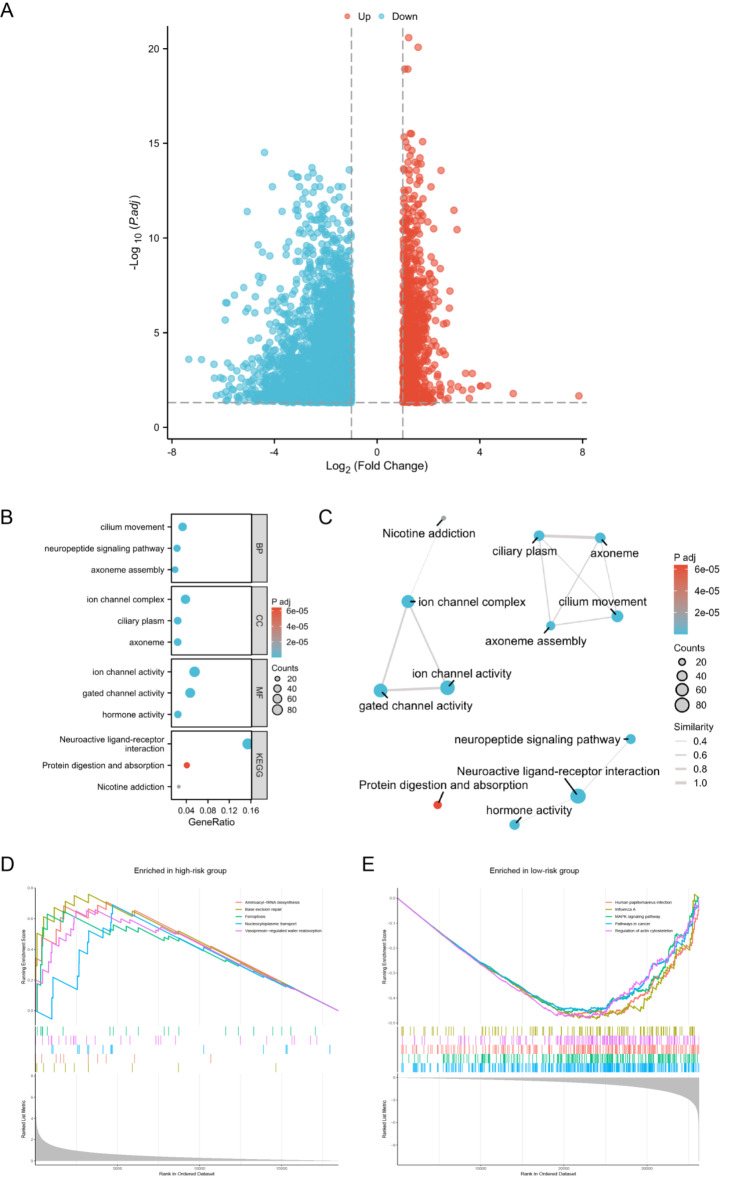
The analytical process methodology employed in this study is illustrated in Fig. 1. We analyzed the differential expression of 2030 MRGs between UCEC and normal groups, ultimately identifying 442 DEMRGs (Supplementary Table S3). The heatmap illustrates the expression patterns of some of the most significantly DEMRGs (Fig. 2A). The results of the differential analysis are presented using a volcano plot (Fig. 2B). A total of 303 DEMRGs were up-regulated and 149 were down-regulated in UCEC samples compared with normal samples. Principal component analysis (PCA) demonstrates that these MRGs can effectively distinguish between tumor and normal samples (Fig. 2C). Additionally, we performed GO and KEGG pathway enrichment analyses based on the DEMRGs (Supplementary Table S4). A bubble chart displays the primary results of the functional enrichment analysis (Fig. 2D), and an enrichment map shows the interrelationships among the enrichment results (Fig. 2E). GO analysis indicates that these MRGs are mainly involved in energy production and metabolism, particularly ATP production through aerobic respiration and the electron transport chain, which are essential for the survival of cancer cells. KEGG pathway enrichment analysis underscores the carcinogenic effects triggered by ROS accumulation resulting from mitochondrial dysfunction.
Fig. 1.
The flowchart graph of this study.
Fig. 2.
Differential gene expression and function analyses between UCEC and normal groups. (A) Heatmap illustrates the expression patterns of some of the most significantly DEMRGs between UCEC and normal groups. (B) Volcano plot of DEMRGs between UCEC and normal groups. (C) PCA demonstrates that these DEMRGs can effectively distinguish between tumor and normal samples. (D) Bubble chart displays the primary results of the GO and KEGG pathway enrichment analyses based on the DEMRGs between normal and UCEC groups. (E) Enrichment map shows the interrelationships among the GO and KEGG pathway enrichment analyses results.
Mitochondria-related risk model construction
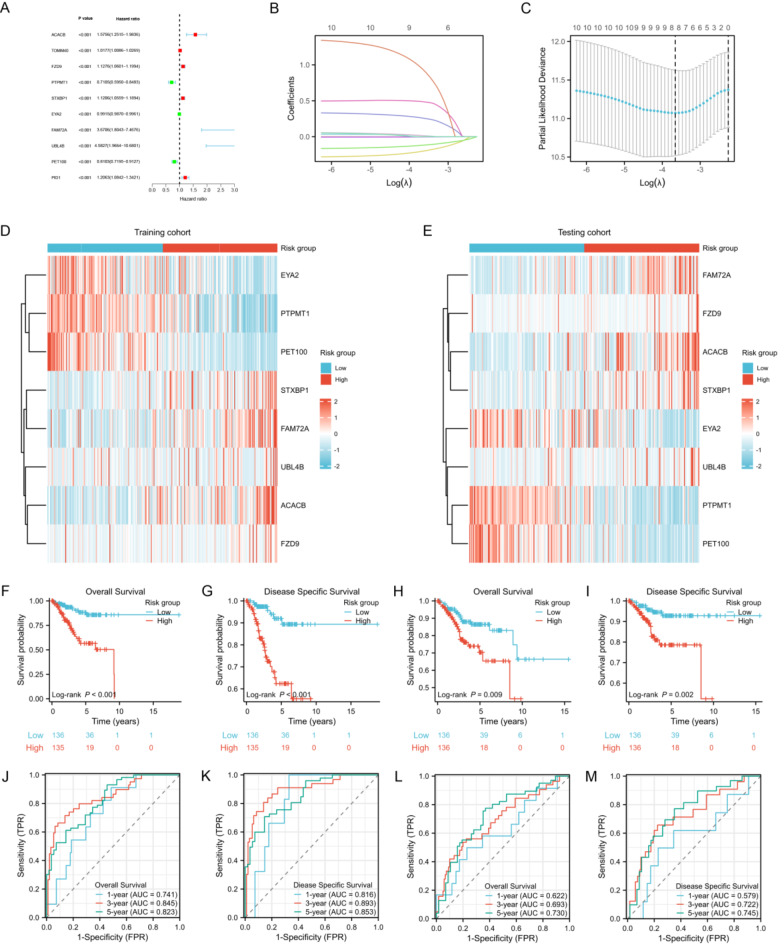
Through univariate regression analysis, we identified 89 DEMRGs associated with OS (Supplementary Table S5). From these, we selected the ten DEMRGs most closely related to prognosis and visualized the results of the univariate regression analysis using a forest plot (Fig. 3A). Using these ten MRGs, we conducted Lasso Cox regression analysis in the training set to construct a risk model. The distribution of LASSO coefficients and the regression penalty plot are illustrated in Fig. 3B and C, respectively. Ultimately, we developed a risk model based on eight MRGs, with the risk score calculation formula for each UCEC sample as follows: Risk score = (0.267 * ACACB) + (0.017 * FZD9) + (− 0.210 * PTPMT1) + (0.018 * STXBP1) + (− 0.003 * EYA2) + (0.492 * FAM72A) + (0.986 * UBL4B) + (− 0.123 * PET100). Based on the median value of the risk score, we categorized the samples from both the training and test sets into high-risk and low-risk groups. We employed heat maps to illustrate the expression levels of these eight MRGs in the high-risk and low-risk groups within both the training and validation sets (Fig. 3D,E).
Fig. 3.
Risk model construction and survival analysis. (A) Univariate Cox regression analysis of the ten DEMRGs most closely related to OS. (B) The LASSO coefficient profiles of the eight MRGs used to construct the risk model. (C) Penalty plot for the LASSO regression model. (D,E) Heatmaps illustrate the expression patterns of the eight MRGs between low-risk and high-risk groups in the training (D) and testing (E) cohorts. (F,G) KM survival analysis for the OS (F) and DSS (G) between two risk groups in the training cohort. (H,I) KM survival analysis for the OS (H) and DSS (I) analysis between two risk groups in the testing cohort. (J,K) Time-dependent ROC analysis for predicting 1-year, 3-year, and 5-year OS (J) and DSS (K) in the training cohort. (L,M) Time-dependent ROC analysis for predicting 1-year, 3-year, and 5-year OS (L) and DSS (M) in the testing cohort.
Differential expression analysis of the eight MRGs between UCEC and normal groups
We conducted an analysis of the differential expression of the eight MRGs between the UCEC group and the normal group (Supplementary Figure S1A,B). The results indicated that FZD9, PTPMT1, EYA2, FAM72A, and PET100 were significantly up-regulated in the UCEC group compared to the normal group, whereas ACACB, STXBP1, and UBL4B were down-regulated (all p < 0.01). Furthermore, we validated the differential expression of these eight MRGs in cell lines (Supplementary Figure S1C). Compared to the human endometriosis cell line hEM15A, FZD9, PTPMT1, EYA2, FAM72A, and PET100 were up-regulated in UCEC cell lines, whereas ACACB, STXBP1, and UBL4B were down-regulated (all p < 0.05). Additionally, we validated the differential expression of the eight MRGs in paired UCEC tumor tissues and corresponding adjacent non-cancerous tissues (Supplementary Figure S1D). Furthermore, the correlation of the expression of the eight MRGs in UCEC is depicted in Supplementary Figure S2.
Survival analysis between risk groups
In the training cohort, the survival rate was 71.9% for high-risk patients and 91.9% for low-risk patients. In the testing cohort, the survival rates for high- and low-risk patients were 81.6% and 87.5%, respectively. Through KM survival analysis, we discovered that both OS and DSS were markedly shorter in high-risk patients compared to low-risk patients in both the training and testing cohorts (Fig. 3F-I, all p < 0.01). Moreover, time-dependent ROC analysis demonstrated the model’s prognostic predictive capability. In the training cohort, the risk score predicted 1-year, 3-year, and 5-year OS with AUCs of 0.741, 0.845, and 0.823, respectively (Fig. 3J). For DSS, the AUCs for 1-year, 3-year, and 5-year predictions were 0.816, 0.893, and 0.853, respectively (Fig. 3K). These results were validated in the testing cohort (Fig. 3L and M). Furthermore, we collected UCEC tissues along with matched adjacent non-cancerous tissues from 16 patients who underwent surgery. Among these, the survival rate in the high-risk group was 50%, compared to 87.5% in the low-risk group. KM survival analysis revealed significantly poorer prognosis in the high-risk group compared to the low-risk group (Supplementary Figure S3).
Survival analysis of the eight MRGs
Kaplan-Meier survival analysis was performed on the eight MRGs incorporated into the risk model. As demonstrated in Supplementary Figure S4, higher expression levels of ACACB, STXBP1, and FAM72A were associated with reduced overall survival (OS) and disease-specific survival (DSS), whereas PTPMT1, EYA2, and PET100 showed a positive correlation with these outcomes (all p < 0.05).
Clinical characteristics analysis
As demonstrated in Table 1, the Chi-square test results reveal no statistically significant differences in the clinicopathological characteristics of UCEC patients between the training and testing cohorts. Heatmaps were employed to distinctly illustrate the clinicopathological attributes of patients within the training and testing cohorts (Fig. 4A,B). In the training cohort, it was observed that patients over the age of 60 exhibited higher risk score compared to those aged 60 or younger (Fig. 4C, p < 0.05). Patients with endometrioid UCEC presented lower risk score than those with serous or mixed UCEC (Fig. 4D, p < 0.001). Additionally, higher risk score correlated with advanced stage (Fig. 4E, p < 0.001), while no association was found between risk score and grade (Fig. 4F). These findings were corroborated in the testing cohort (Fig. 4G–J).
Table 1.
Clinical characteristics of UCEC patients in training and testing cohorts.
| Clinical Characteristics | Training cohort | Testing cohort | P value |
|---|---|---|---|
| No. | 271 | 272 | |
| Age | |||
| ≤ 60 | 103 (38.0%) | 104 (38.2%) | 0.956 |
| > 60 | 168 (62.0%) | 168 (61.8%) | |
| Histological type | |||
| Endometrioid | 200 (73.8%) | 207 (76.1%) | 0.644 |
| Serous | 58 (21.4%) | 56 (20.6%) | |
| Mixed | 13 (4.8%) | 9 (3.3%) | |
| Stage | |||
| I | 160 (59.0%) | 179 (65.8%) | 0.255 |
| II | 30 (11.1%) | 22 (8.1%) | |
| III | 63 (23.2%) | 60 (22.1%) | |
| IV | 18 (6.6%) | 11 (4.0%) | |
| Grade | |||
| G1 | 45 (16.6%) | 54 (19.9%) | 0.778 |
| G2 | 61 (22.5%) | 59 (21.7%) | |
| G3 | 160 (59.0%) | 153 (56.3%) | |
| G4 | 5 (1.8%) | 6 (2.2%) | |
Fig. 4.
Clinical characteristics analysis of the risk score. (A,B) Heatmaps illustrate the clinical characteristics attributes of patients in the training (A) and testing (B) cohorts. (C–F) Correlation between risk score and clinical characteristics in the training cohort. (G–J) Correlation between risk score and clinical characteristics in the testing cohort. NS indicates no statistical difference, *P < 0.05, **P < 0.01, ***P < 0.001.
Nomogram model construction
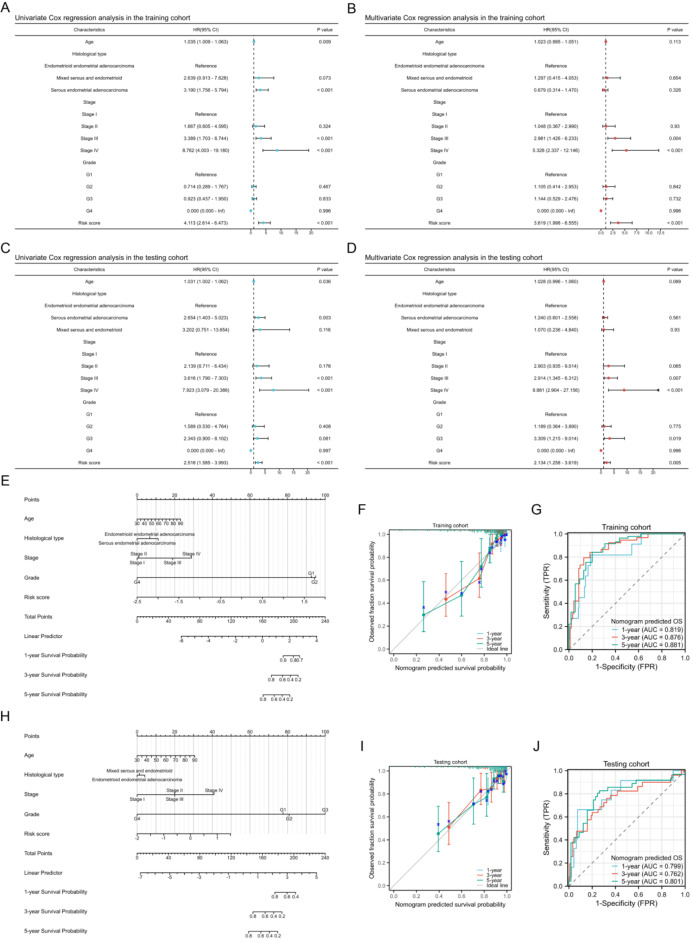
To discern potential prognostic factors, we conducted univariate and multivariate COX regression analyses, incorporating variables such as age, histological type, stage, grade, and risk score. The results from both univariate and multivariate COX regression analyses indicated that the risk score was a significant prognostic factor for predicting OS in the training cohort (Fig. 5A,B, both p < 0.001). Similarly, the risk score was identified as a prognostic factor in the testing cohort (Fig. 5C,D, both p < 0.01). Subsequently, by integrating the risk score with clinical characteristics, we constructed a nomogram model in the training cohort to predict the 1-year, 3-year, and 5-year OS of UCEC patients (Fig. 5E). The calibration curves demonstrated satisfactory consistency between the predicted and observed values (Fig. 5F). The time-dependent ROC curves revealed that the AUC values for predicting 1-year, 3-year, and 5-year OS were 0.819, 0.876, and 0.881, respectively (Fig. 5G), highlighting the nomogram model’s exceptional prognostic performance. We also constructed a nomogram model in the testing cohort and validated these findings (Fig. 5H–J).
Fig. 5.
Nomogram model construction. (A,B) Univariate (A) and multivariate (B) Cox regression analysis of the risk score and clinical characteristics in the training cohort. (C,D) Univariate (C) and multivariate (D) Cox regression analysis of the risk score and clinical characteristics in the testing cohort. (E) Nomogram model construction for predicting 1-year, 3-year, and 5-year OS in the training cohort. (F,G) Calibration curve (F) and time-dependent ROC curves (G) of the nomogram model in the training cohort. (H) Nomogram model construction for predicting 1-year, 3-year, and 5-year OS in the testing cohort. (I,J) Calibration curve (I) and time-dependent ROC curves (J) of the nomogram model in the testing cohort.
DEGs identification between risk groups and functional enrichment analysis
In the training cohort, we identified 4,594 differentially expressed genes (DEGs) between high-risk and low-risk groups (Supplementary Table S6), with 3830 upregulated in the low-risk group and 764 in the high-risk group (Fig. 6A). Utilizing these DEGs, we conducted GO and KEGG pathway enrichment analyses (Supplementary Table S7). The principal findings were depicted through a bubble chart (Fig. 6B) and their correlations were illustrated via an enrichment map (Fig. 6C). These functional enrichment analyses unveiled significant differences in various biological processes between the high-risk and low-risk groups, notably in pathways related to cell movement, signal transduction, metabolism, and ion channel activity. These insights provide a deeper understanding of UCEC’s biological mechanisms. We also performed GSEA to analyze differential pathway expression between the high-risk and low-risk groups (Supplementary Tables S8 and S9). The GSEA results revealed that pathways enriched in the high-risk group were predominantly associated with the regulation of cell death, DNA repair, protein synthesis, molecular transport, and body fluid regulation (Fig. 6D), whereas the low-risk group’s critical signaling pathways and pathogenic infections might collectively disrupt normal cell functions, thereby facilitating cancer development and progression (Fig. 6E).
Fig. 6.
DEGs identification between risk groups and functional enrichment analysis. (A) Volcano plot of DEGs between high-risk and low-risk groups. (B) Bubble chart displays the primary results of the GO and KEGG pathway enrichment analyses based on the DEGs between high-risk and low-risk groups. (C) Enrichment map shows the interrelationships among the GO and KEGG pathway enrichment analyses results. (D,E) GSEA enriched pathways in high-risk (D) and low-risk (E) groups.
Immune landscape analysis and immunotherapy response prediction
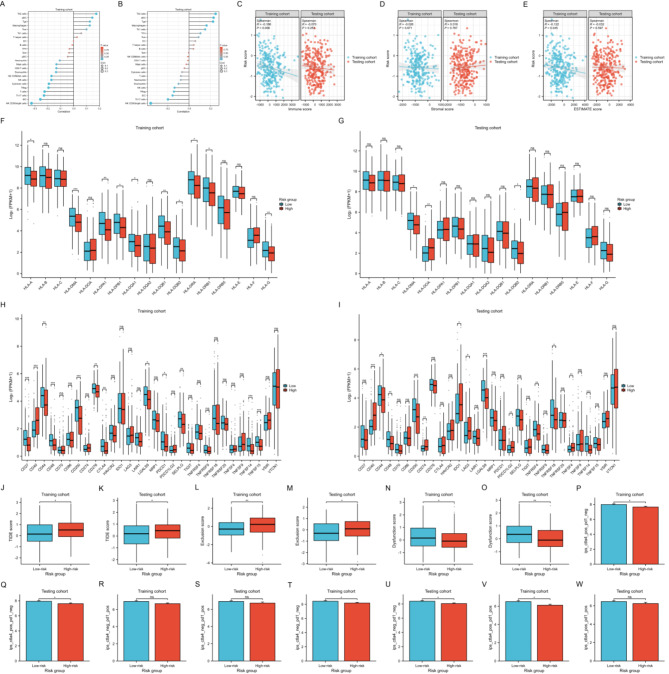
Considering the intricate relationship between the immune microenvironment and tumor progression, we conducted an analysis of the correlation between the risk score and the infiltration levels of 24 common immune cells. Our findings revealed that, in both the training and testing cohorts, the risk score was predominantly negatively correlated with the majority of immune cells, particularly NK CD56bright cells, iDC, Th17, Treg, and NK cells (Fig. 7A,B). In contrast, the risk score exhibited a positive correlation with Th2, aDC, Tgd, and macrophages. Additionally, we observed a negative correlation between the risk score and both the immune score and ESTIMATE score in the training cohort, with a similar trend observed in the testing cohort, albeit without statistical significance (Fig. 7C,E). No significant correlation was detected between the risk score and the stromal score (Fig. 7D). These results suggest that a higher risk score is associated with tumor immune suppression and immune evasion within the tumor microenvironment. A further examination of the differential expression of HLA family members and ICGs between high-risk and low-risk groups revealed that, in both the training and testing cohorts, the majority of HLA family members and ICGs were expressed at lower levels in high-risk patients compared to low-risk patients (Fig. 7F,I). These findings underscore significant differences in the immune landscape between high-risk and low-risk patients, implying potential variations in their responses to immunotherapy. Subsequently, we utilized the TIDE algorithm to predict the response of UCEC samples to ICB therapy. In both the training and testing cohorts, the high-risk group displayed higher TIDE and Exclusion scores, but lower Dysfunction score compared to the low-risk group (Fig. 7J,O). Moreover, we evaluated tumor immunogenicity using the IPS, where higher scores indicate a greater potential response to ICB therapy. The results demonstrated that, regardless of statistical significance, the low-risk group exhibited higher IPS in all states of CTLA4 and PD1 compared to the high-risk group (Fig. 7P,W). These findings indicate that our mitochondrial risk model can effectively predict the immunotherapy response in UCEC patients, with those having a lower risk score being more likely to benefit from immunotherapy.
Fig. 7.
Immune landscape analysis and immunotherapy response prediction (A,B) Correlation between risk score and infiltration Levels of 24 immune cells in the training (A) and testing (B) cohorts. (C–E) Correlation between risk score and immune score (C), stromal score (D) and ESTIMATE score (E). (F,G) HLA family members expression between high-risk and low-risk groups in the training (F) and testing (G) cohorts. (H,I) ICGs expression between high-risk and low-risk groups in the training (H) and testing (I) cohorts. (J–O) Differences of TIDE, Exclusion and Dysfunction scores in the training and testing cohorts. (P–W) Differences of IPS in the training and testing cohorts. NS indicates no statistical difference, *P < 0.05, **P < 0.01, ***P < 0.001.
Mutation analysis
We delineated the mutational landscape of the top 20 most frequently mutated genes in high-risk and low-risk UCEC patients using a mutation map. Our analysis revealed that the frequency of PTEN mutations was markedly elevated in the low-risk group, surpassing 80% in both the training and validation cohorts (Fig. 8A and C). Notably, the most significantly mutated gene in the high-risk group was TP53 (Fig. 8B,D), which did not show a significant mutation rate in the low-risk group. Furthermore, we discovered that the TMB in the low-risk group was higher than that in the high-risk group in the training cohort (Fig. 8E), and there was a negative correlation between risk score and TMB (Fig. 8G), although this was not corroborated in the test cohort (Fig. 8F). We also examined the differences in ITH between the high-risk and low-risk groups and investigated the correlation between risk score and ITH score, but no significant differences or correlations were identified (Fig. 8H,J). Additionally, the distribution of MSI status between the two risk groups in the training and testing cohorts is illustrated in Fig. 8K,L, respectively. We observed that the risk score of microsatellite stable (MSS) patients was higher than that of microsatellite instability-high (MSI-H) patients in both the training and test cohorts (Fig. 8M,N).
Fig. 8.
Mutation analysis in two risk groups. (A–D) Mutational landscape of the top 20 most frequently mutated genes in high-risk and low-risk groups in the training and testing cohort. (E,F) Diffecences of TMB between high-risk and low risk groups in the training (E) and testing (F) cohorts. (G) Correlation between risk score and TMB. (H,I) Diffecences of ITH score between high-risk and low risk groups in the training (H) and testing (I) cohorts. (J) Correlation between risk score and ITH score. (K,L) Distribution of MSI status between the two risk groups in the training (K) and testing (L) cohorts. (M,N) Differences in risk score among groups with different MSI statuses in the training (M) and testing (N) cohorts. NS indicates no statistical difference, *P < 0.05, **P < 0.01.
Drug sensitivity analysis
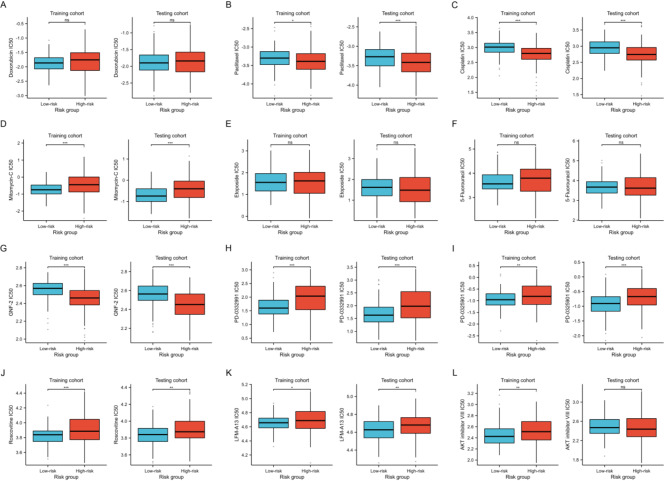
Chemotherapy and small molecule drugs are prevalent treatments for UCEC cancer. By leveraging the GDSC database, we assessed the IC50 differences of frequently used chemotherapeutic and small molecule drugs between two risk groups. Our findings indicated that in both the training and testing cohorts, the IC50 values for Paclitaxel, Cisplatin, and GNF-2 were significantly lower in the high-risk group. Conversely, the IC50 values for Mitomycin-C, PD-0332991, PD-0325901, Roscovitine, and LFM-A13 were markedly lower in the low-risk group (Fig. 9, all p < 0.05).
Fig. 9.
Analysis of the IC50 differences of frequently used chemotherapeutic and small molecule drugs between two risk groups in the training and testing cohorts. NS indicates no statistical difference, *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Mitochondria, fundamental organelles within cells, are pivotal not only in energy metabolism but also in the onset and advancement of cancer. The significance of mitochondria in cancer has been increasingly recognized, with research demonstrating that MRGs are crucial for prognostic evaluation and predicting therapeutic responses across diverse cancer types31–33. In the past few decades, research has elucidated the pivotal role of MRGs in the pathogenesis and progression of UCEC. For instance, mutations in the FBXO7 gene lead to excessive mitochondrial fission, thereby inducing UCEC, which underscores the critical impact of the disequilibrium between mitochondrial fission and fusion on the onset and advancement of UCEC13. Additionally, aberrantly expressed MRGs in UCEC present promising avenues for novel therapeutic targets. For example, IMT1, a newly identified POLRMT-specific inhibitor, has demonstrated efficacy in curbing UCEC progression by impairing mitochondrial function34. At present, research investigating the correlation between MRGs and both the prognosis and immunotherapy of UCEC remains in its exploratory phase. Consequently, the development of prognostic models based on MRGs possesses substantial clinical application potential.
In this study, we leveraged transcriptional data of MRGs along with clinical information from the TCGA-UCEC dataset to pinpoint MRGs significantly associated with prognosis. Consequently, we developed a prognostic risk model for UCEC patients based on eight MRGs, which were ACACB, FZD9, PTPMT1, STXBP1, EYA2, FAM72A, UBL4B, PET100. Although these eight MRGs are pivotal in the development and progression of various cancers, their mechanistic roles in UCEC remain undocumented. The ACACB gene encodes acetyl-CoA carboxylase beta, a crucial enzyme in fatty acid synthesis, and its expression in colorectal cancer promotes tumor growth and drug resistance35. FZD9, a member of the Frizzled protein family, is significantly upregulated in numerous malignancies, regulating cell proliferation and invasion through the Wnt pathway activation36,37. PTPMT1, belonging to the PTEN family, primarily catalyzes phosphatidylation modifications and is highly expressed in various cancers38. STXBP1, essential for neurotransmitter release, has seldom been studied in tumor progression, but studies show its high expression in lung adenocarcinoma is linked to poor prognosis39. EYA2, a member of the eyes absent protein family, promotes cancer progression by inducing TGF-β, epithelial-mesenchymal transition in cancer cells, or downregulating PTEN40,41. FAM72 is a protein-coding gene specific to neural stem cells, implicated in cancer cell division, proliferation, and differentiation. Additionally, elevated levels of FAM72 are associated with hypomethylation and influence the prognosis of various cancers42,43. As a member of the ubiquitin-like protein family, UBL4B plays a significant role in protein metabolism and the maintenance of cellular homeostasis44. However, current research on the mechanisms of UBL4B in cancer remains limited. Further investigations are warranted to elucidate the potential role of UBL4B in tumor biology. Additionally, the PET100 gene PET100 gene is crucial for the maturation of cytochrome coxidase and the overall functionality of mitochondria45. Although research on the specific role of PET100 in cancer remains limited, mutations in this gene may be linked to cancer development. Based on the outcomes of the risk model construction, the prognosis for patients in the high-risk group was markedly worse than that for those in the low-risk group. Utilizing both univariate and multivariate COX regression analyses, we identified the risk score as an independent prognostic indicator for predicting OS in UCEC patients. By integrating the risk score with the clinical features of the patients, we subsequently developed a nomogram model. Time-dependent ROC analysis revealed that this model exhibits outstanding predictive accuracy for 1-year, 3-year, and 5-year OS. The development of the mitochondria-related risk model provides a novel approach to assessing the prognosis of UCEC patients. This model not only elucidates the potential mechanistic roles of these genes in UCEC but also establishes a scientific foundation for crafting personalized treatment strategies.
To elucidate the potential mechanisms underlying the prognostic disparities between high-risk and low-risk groups, we identified DEGs and conducted a functional enrichment analysis between the two cohorts. Our analysis revealed significant distinctions in the expression of DEGs associated with cellular biological functions and signaling pathways. In the high-risk group, pathways related to DNA repair and protein synthesis were enriched, suggesting that these genes may play a pivotal role in tumor cell survival and genomic stability maintenance46,47. Furthermore, the enrichment of cancer-related pathways such as MAPK in the low-risk group is closely linked to the progression of UCEC48. Based on our extensive analysis of the immunological landscape, we identified a significant association between elevated risk score and the presence of immunosuppressive and immune-evading mechanisms within the tumor microenvironment. A clear negative correlation was identified between the risk score and the infiltration levels of various antitumor immune cells, particularly NK CD56bright cells and immature dendritic cells (iDCs). Under normal conditions, CD56bright NK cells are primarily engaged in immunoregulatory activities. However, studies have shown that upon specific activation, these cells can exhibit potent antitumor effects49. Tumor cells frequently employ multiple mechanisms to inhibit the maturation and function of dendritic cells. IDCs are essential for continuously capturing and processing tumor antigens, thereby maintaining a level of immune surveillance to prevent tumor immune evasion50. Conversely, we observed that risk scores positively correlate with the infiltration of pro-tumor immune cells. For instance, Th2 cells facilitate tumor growth and progression by secreting cytokines such as IL-451. Macrophage infiltration promotes angiogenesis through various mechanisms, thereby supporting tumor growth and progression52. In high-risk patients, the expression levels of HLA family members and ICGs are generally diminished. HLA molecules are pivotal in antigen presentation and immune recognition, while ICGs are crucial in modulating immune responses53,54. Therefore, this reduced expression implies that high-risk patients may exhibit compromised immune function, thereby diminishing the efficacy of tumor immune surveillance.
The differences in the immune landscape between the two groups suggested a variance in the potential sensitivity of UCEC patients to immunotherapy. The extensive implementation of molecular subtyping in UCEC has greatly advanced the application of immune checkpoint inhibitors55. Research has demonstrated that PD-1 inhibitors possess significant efficacy in the treatment of advanced or recurrent endometrial cancer. Nevertheless, prior studies reveal that most cancer patients exhibit insensitivity to ICB therapy56. Accurately predicting sensitivity to ICB treatment is essential for identifying patient populations suitable for precision therapy. In this study, the high-risk group exhibited elevated TIDE and Exclusion scores, but a lower Dysfunction score compared to the low-risk group. These findings suggested that UCEC samples in the high-risk group are more likely to evade immune system attacks through T cell exclusion rather than T cell dysfunction, potentially resulting in a poorer response to ICB therapy. IPS analysis revealed that the low-risk group score higher, signifying that low-risk patients have greater immunogenicity. Moreover, high TMB levels are often linked to increased tumor immunogenicity, indicating a higher presence of mutated antigens within the tumor that can be identified and targeted by the immune system, thus enhancing immunotherapy effectiveness57. Our subsequent analysis demonstrated that the low-risk group in the training cohort exhibited higher TMB levels than the high-risk group. In summary, our risk model not only reflects the immune microenvironment status of UCEC patients but also effectively predicts their responsiveness to immunotherapy. Patients classified as low-risk are more likely to benefit from ICB therapy, offering valuable guidance for personalized immunotherapy approaches in UCEC. The further application of this model could aid in identifying patients who are particularly well-suited for immunotherapy, thereby enhancing treatment outcomes. This work not only deepens our understanding of tumor immune mechanisms in UCEC but also serves as a crucial reference for developing immunotherapy strategies in precision medicine. Moreover, through an analysis of drug sensitivity to commonly utilized chemotherapy agents and potential small molecule drugs in UCEC, it was discovered that the high-risk group demonstrates increased sensitivity to Paclitaxel, Cisplatin, and GNF-2, while the low-risk group exhibits greater sensitivity to Mitomycin-C, PD-0332991, PD-0325901, among others. These insights provide a foundational basis for the personalized and precision treatment of UCEC patients.
Nonetheless, our study is subject to several limitations. Primarily, it represents a retrospective analysis based on a single-source dataset, the TCGA database, which predominantly comprises patients from Western regions. The relatively limited sample size and regional bias may affect the model’s generalizability across diverse populations. Consequently, further validation of the model’s applicability requires in East Asian populations and other geographic regions is necessary through independent datasets. Furthermore, the biological mechanisms underlying the identified MRGs require additional elucidation through cellular and animal experiments. To address these limitations, our future research will concentrate on multi-center, large-scale prospective studies to validate the predictive capabilities of this model for prognosis and immunotherapy response in UCEC patients. Additionally, further investigation into the underlying mechanisms of MRGs in UCEC development will be essential to identify potential therapeutic targets.
Conclusion
In conclusion, we have developed a risk model based on eight MRGs, which demonstrates substantial efficacy in predicting the prognosis of UCEC patients and their responsiveness to ICB therapy. This model provides a scientific basis for personalized and precise treatment strategies in clinical practice, suggesting that low-risk patients may derive greater benefits from immunotherapy. By differentiating patients with varying risk profiles, this model offers clinicians a tool to make more informed decisions on therapy options, potentially improving patient outcomes. Future research should focus on prospectively validating these findings across diverse populations and exploring the underlying mechanisms associated with the identified MRGs in UCEC progression.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
R.G.L, J.H.W and Y.T.F participated in the data analysis, organized the article writing, and critically modified the manuscript. L.Z and F.Z were responsible for the experimental section. Y.Q.Z designed research and modified the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This work was supported by the Guangdong Basic and Applied Basic Research Foundation (Grant No.2024A1515011271), Youth Science Foundation of the Cancer Hospital of Shantou University Medical College (Grant No. 2023A002).
Data availability
The data used in this study were obtained from The Cancer Genome Atlas (TCGA) database, which is publicly available at https://www.cancer.gov/tcga.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Ethical approval for this study was granted by the Ethics Review Committee of the Cancer Hospital of Shantou University Medical College.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors contributed equally: Ru-Gen Liao and Jin-Hong Wang.
Contributor Information
Yu-Tong Fang, Email: fyt1996@126.com.
Li Zhou, Email: zlyyzl@126.com.
Yong-Qu Zhang, Email: zhangyq@stu.edu.cn.
References
- 1.Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.74 (3), 229–263 (2024). [DOI] [PubMed] [Google Scholar]
- 2.Ma, S., Chen, Q., Li, X., Fu, J. & Zhao, L. UBE2C serves as a prognosis biomarker of uterine corpus endometrial carcinoma via promoting tumor migration and invasion. Sci. Rep.13 (1), 16899 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller, K. D. et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J. Clin.72 (5), 409–436 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Dhani, N. C. et al. Phase II trial of Cabozantinib in Recurrent/Metastatic endometrial Cancer: a study of the Princess Margaret, Chicago, and California Consortia (NCI9322/PHL86). Clin. Cancer Res.26 (11), 2477–2486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raglan, O. et al. Risk factors for endometrial cancer: an umbrella review of the literature. Int. J. Cancer. 145 (7), 1719–1730 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. C., Carson, T. L., Thompson, H. J. & Agurs-Collins, T. The Triple Health Threat of Diabetes, obesity, and Cancer-Epidemiology, disparities, mechanisms, and interventions. Obes. (Silver Spring). 29 (6), 954–959 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spencer, N. Y. & Stanton, R. C. The Warburg Effect, Lactate, and nearly a century of trying to Cure Cancer. Semin Nephrol.39 (4), 380–393 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Ward, P. S. & Thompson, C. B. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell.21 (3), 297–308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren, T. et al. MCUR1-Mediated Mitochondrial Calcium Signaling Facilitates Cell Survival of Hepatocellular Carcinoma via reactive oxygen species-dependent P53 degradation. Antioxid. Redox Signal.28 (12), 1120–1136 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Kirtonia, A., Sethi, G. & Garg, M. The multifaceted role of reactive oxygen species in tumorigenesis. Cell. Mol. Life Sci.77 (22), 4459–4483 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang, S. F., Tseng, L. M. & Lee, H. C. Role of mitochondrial alterations in human cancer progression and cancer immunity. J. Biomed. Sci.30 (1), 61 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao, H. et al. Mitochondrial calcium Uniporter (MCU) that modulates mitochondrial calcium uptake and facilitates endometrial Cancer Progression through Interaction with VDAC1. Curr. Cancer Drug Targets. 24 (3), 354–367 (2024). [DOI] [PubMed] [Google Scholar]
- 13.Zhang, H. et al. FBXO7, a tumor suppressor in endometrial carcinoma, suppresses INF2-associated mitochondrial division. Cell. Death Dis.14 (6), 368 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bokhman, J. V. Two pathogenetic types of endometrial carcinoma. Gynecol. Oncol.15 (1), 10–17 (1983). [DOI] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research Network et al. Integrated genomic characterization of endometrial carcinoma. Nature497 (7447), 67–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 102 (43), 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med.16 (4), 385–395 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Zhao, F. et al. Elucidating the role of tumor-associated ALOX5 + mast cells with transformative function in cervical cancer progression via single-cell RNA sequencing. Front. Immunol.15, 1434450 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao, W. et al. Single-cell RNA sequencing reveals that MYBL2 in malignant epithelial cells is involved in the development and progression of ovarian cancer. Front. Immunol.15, 1438198 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao, J. et al. Construction of N-7 methylguanine-related mRNA prognostic model in uterine corpus endometrial carcinoma based on multi-omics data and immune-related analysis. Sci. Rep.12 (1), 18813 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, X. et al. Unveiling the cellular landscape: insights from single-cell RNA sequencing in multiple myeloma. Front. Immunol.15, 1458638 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res.28 (1), 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci.28 (11), 1947–1951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res.51 (D1), D587–D592 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun, L. et al. Single-cell RNA sequencing explored potential therapeutic targets by revealing the tumor microenvironment of neuroblastoma and its expression in cell death. Discov Oncol.15 (1), 409 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature462 (7269), 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun.4, 2612 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med.24 (10), 1550–1558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charoentong, P. et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell. Rep.18 (1), 248–262 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Yang, W. et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res.41 (Database issue), D955–D961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang, Y. et al. Mitochondrial-related genes as prognostic and metastatic markers in breast cancer: insights from comprehensive analysis and clinical models. Front. Immunol.15, 1461489 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi, Y. et al. Deciphering a mitochondria-related signature to supervise prognosis and immunotherapy in hepatocellular carcinoma. Front. Immunol.13, 1070593 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang, Y. et al. Overexpressed VDAC1 in breast cancer as a novel prognostic biomarker and correlates with immune infiltrates. World J. Surg. Oncol.20 (1), 211 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, S. P., Ou, L., Zhang, Y., Shen, F. R. & Chen, Y. G. A first-in-class POLRMT specific inhibitor IMT1 suppresses endometrial carcinoma cell growth. Cell. Death Dis.14 (2), 152 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong, H. J. et al. ACACB is a novel metabolism-related biomarker in the prediction of response to cetuximab therapy inmetastatic colorectal cancer. Acta Biochim. Biophys. Sin (Shanghai). 54 (11), 1671–1683 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, G. et al. Frizzled7 promotes epithelial-to-mesenchymal transition and Stemness Via activating Canonical Wnt/β-catenin pathway in gastric Cancer. Int. J. Biol. Sci.14 (3), 280–293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King, T. D., Zhang, W., Suto, M. J. & Li, Y. Frizzled7 as an emerging target for cancer therapy. Cell. Signal.24 (4), 846–851 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding, P. P. et al. PTPMT1 regulates mitochondrial death through the SLC25A6-NDUFS2 axis in pancreatic cancer cells. Am. J. Cancer Res.13 (3), 992–1003 (2023). [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, X. et al. Membrane location of syntaxin-binding protein 1 is correlated with poor prognosis of Lung Adenocarcinoma. Tohoku J. Exp. Med.250 (4), 263–270 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Farabaugh, S. M., Micalizzi, D. S., Jedlicka, P., Zhao, R. & Ford, H. L. Eya2 is required to mediate the pro-metastatic functions of Six1 via the induction of TGF-β signaling, epithelial-mesenchymal transition, and cancer stem cell properties. Oncogene31 (5), 552–562 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, Z., Qiu, R., Qiu, X. & Tian, T. EYA2 promotes lung cancer cell proliferation by downregulating the expression of PTEN. Oncotarget8 (67), 110837–110848 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rahane, C. S., Kutzner, A. & Heese, K. A cancer tissue-specific FAM72 expression profile defines a novel glioblastoma multiform (GBM) gene-mutation signature. J. Neurooncol. 141 (1), 57–70 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Chatonnet, F. et al. The hydroxymethylome of multiple myeloma identifies FAM72D as a 1q21 marker linked to proliferation. [DOI] [PMC free article] [PubMed]
- 44.Yu, C. et al. The dispensable roles of X-Linked Ubl4a and its autosomal counterpart Ubl4b in spermatogenesis represent a New Evolutionary type of X-Derived Retrogenes. Front. Genet.12, 689902 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oláhová, M. et al. A truncating PET100 variant causing fatal infantile lactic acidosis and isolated cytochrome c oxidase deficiency. Eur. J. Hum. Genet.23 (7), 935–939 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang, R. & Zhou, P. K. DNA damage repair: historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal. Transduct. Target. Ther.6 (1), 254 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovalski, J. R., Kuzuoglu-Ozturk, D. & Ruggero, D. Protein synthesis control in cancer: selectivity and therapeutic targeting. EMBO J.41 (8), e109823 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, J., Sun, X., Zhang, H., Wang, Y. & Li, Y. MPA influences tumor cell proliferation, migration, and invasion induced by RANKL through PRB involving the MAPK pathway in endometrial cancer. Oncol. Rep.33 (2), 799–809 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Poznanski, S. M. & Ashkar, A. A. Shining light on the significance of NK cell CD56 brightness. Cell. Mol. Immunol.15 (12), 1071–1073 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dudek, A. M., Martin, S., Garg, A. D., Agostinis, P. & Immature Semi-mature, and fully mature dendritic cells: toward a DC-Cancer cells interface that augments anticancer immunity. Front. Immunol.4, 438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiang, W. et al. Monoacylglycerol lipase regulates cannabinoid receptor 2-dependent macrophage activation and cancer progression. Nat. Commun.9 (1), 2574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kitamura, T., Qian, B. Z. & Pollard, J. W. Immune cell promotion of metastasis. Nat. Rev. Immunol.15 (2), 73–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, H. & Wang, Q. Zhongguo Fei Ai Za Zhi ;13(2):149–153. (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu, F. F., Liu, C. J., Liu, L. L., Zhang, Q. & Guo, A. Y. Expression profile of immune checkpoint genes and their roles in predicting immunotherapy response. Brief. Bioinform.22 (3), bbaa176 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Ott, P. A. et al. Safety and Antitumor Activity of Pembrolizumab in Advanced programmed death ligand 1-Positive endometrial Cancer: results from the KEYNOTE-028 study. J. Clin. Oncol.35 (22), 2535–2541 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Haslam, A. & Prasad, V. Estimation of the percentage of US patients with Cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw. Open.2 (5), e192535 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anagnostou, V., Bardelli, A., Chan, T. A. & Turajlic, S. The status of tumor mutational burden and immunotherapy. Nat. Cancer. 3 (6), 652–656 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this study were obtained from The Cancer Genome Atlas (TCGA) database, which is publicly available at https://www.cancer.gov/tcga.