Abstract
Recurrence of metabolic dysfunction-associated steatotic liver disease (MASLD) after liver transplantation (LT) is a continuing concern. The role of gut microbiome dysbiosis in MASLD initiation and progression has been well established. However, there is a lack of comprehensive gut microbiome shotgun sequence data for patients experiencing MASLD recurrence after LT. In this data descriptor, we describe a dataset of deep metagenomic sequences of a well-defined LT recipient population. Community-based analysis revealed a high abundance of Akkermansia muciniphila, consistently observed in most patient samples with a low (0–2) MASLD Activity Score (NAS). We constructed 357 metagenome-assembled genomes (MAGs), including 220 high-quality MAGs (>90% completion). The abundance of different species of Bacteroides MAGs dominated in patient samples with NAS > 5 (“definite MASH”). In contrast, the MAGs of A. muciniphila, Akkermansia sp., and Blutia sp. dominated in samples from patients without MASH (NAS = 0–2). In addition, the phylogenetic analysis of A. muciniphila and Akkermansia sp. MAGs identified two new phylogroups of Akkermansia that are distinct from the previously reported three phylogroups.
Subject terms: Non-alcoholic fatty liver disease, Metagenomics
Background & Summary
Metabolic dysfunction-associated steatotic liver disease (MASLD), formerly known as nonalcoholic fatty liver disease (NAFLD), encompasses a wide range of liver disorders, and its escalating prevalence has become a global concern1. Estimates indicate that the prevalence of MASLD was approximately 25.5% in 2005, which increased to 38.7% in 20162. The disruption of the gut-liver axis due to an imbalance in the gut microbial community can have a negative impact on energy homeostasis, leading to the development of various metabolic syndromes such as obesity and MASLD3,4. Intestinal health is a crucial aspect of MASLD, and consequently, various studies have assessed the makeup of the gut microbial community and its abundance using sequence-based metagenomic approaches3–6. For microbiome analysis, deep shotgun sequencing (with more than 10 million reads/sample) provides several advantages over shallow sequencing (<10 million reads/sample) and 16S rRNA amplicon sequencing-based approaches, such as identifying rare microbial taxa (at species levels), classification of uncultivated bacteria, metabolic profiling, host-microbe interactions, novel gene discovery, identification of gene clusters responsible for secondary metabolite production, and for constructing metagenome-assembled genomes (MAGs)7–9. The significance of gut microbiota composition and its functions in MASLD warrants the use of deep shotgun metagenomic sequencing. To the best of our knowledge, as of the current time, there is no publicly available ultra-deep shotgun sequencing dataset (with sequencing depth >20 million reads/sample) from patients who have undergone liver transplantation (LT) and subsequently developed MASLD recurrence.
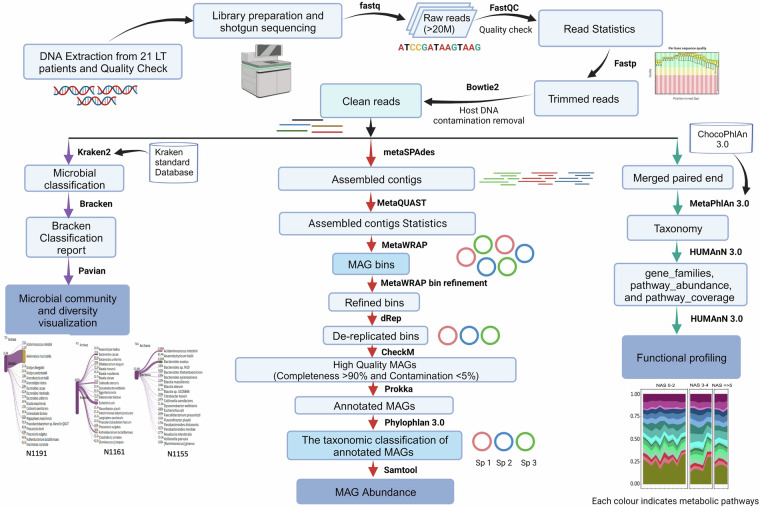
In our previous prospective observational study, the gut microbial community status of LT patients with varying pathologies of metabolic dysfunction-associated steatohepatitis (MASH), formerly known as nonalcoholic steatohepatitis (NASH), recurrence was reported by utilizing the 16S rRNA amplicon sequencing-based approach4. As mentioned before, data generated from deep shotgun sequencing of the metagenomic samples is essential for comprehensive community-based functional analyses and constructing draft MAGs, enabling a deeper understanding of the disease outcomes10. In this study, we employed a deep shotgun sequencing approach (that generated over 20 million reads per sample) to investigate the gut microbial flora and construct MAGs from liver transplant (LT) patients manifesting varying degrees of MASH recurrence, as illustrated in Fig. 1.
Fig. 1.
The detail workflow from DNA extraction to bioinformatics analysis. Every step and their associated software packages are given for better understanding of the analysis.
Based on the NAS score, all samples have been categorized into three groups according to conventional clinical practices11,12: “no MASH” (NAS 0–2), “borderline MASH” (NAS 3-4), and “definite MASH” (NAS ≥ 5) samples. Patient-level demographic and clinical data at the time of stool sample collection are provided in Table S1. At the phylum level, we observed variations in the abundance of three phyla — Fusobacteria, Euryarchaeota, and Verrucomicrobiota — across these three sample groups. Remarkably, our findings align with our previous research involving 16S rRNA sequencing of the same samples. In this study, we observed a substantial increase in the abundance of A. muciniphila and Akkermansia sp. in the majority of samples from patients with low NAS (0–2), reaffirming our earlier observations4. The species-level functional profiling indicated that the elevated abundance of three amino acid biosynthesis pathways positively correlates with samples from patients with no MASH outcomes [NAS (0–2)]13,14. Additionally, we constructed and taxonomically classified the MAGs from all these samples (Fig. 2) and estimated their abundance using a mapping-based approach. The abundance of MAGs of A. muciniphila and Blautia sp. were very high in most patient samples with low or no MASH activities [NAS (0–2)]. We also have analyzed and compared the MAGs of A. muciniphila and Akkermansia sp. to explore the phylogenetic groups. This exploration led to identifying two potentially new phylogenetic clusters within A. muciniphila and Akkermansia sp. The information regarding pathway abundance, in conjunction with the MAGs, may contribute significantly to enhancing our understanding of the underlying disease mechanisms during the progression of MASLD.
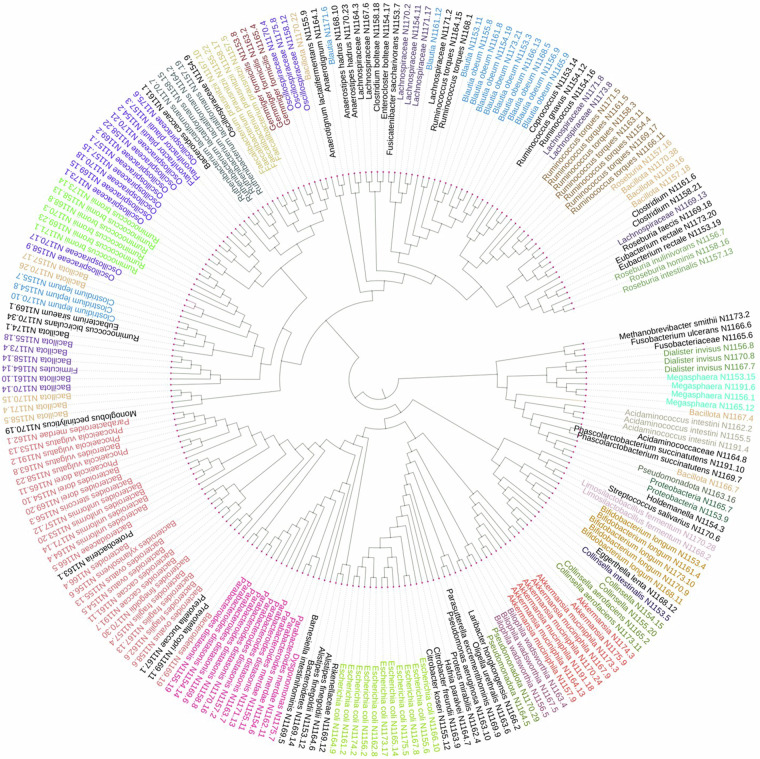
Fig. 2.
The phylogenetic tree of high-quality MAGs constructed using maximum likelihood method (completeness >90% and contamination <5%) prepared by PhyloPhlAn 3.0 and visualized by iTOL. The different colors in the tree represented a separate cluster of MAGs and their close relatives. The red colored cluster shows the A. muciniphila group.
Methods
Study location, ethical clearance, and sample collection
The study was conducted at James D. Eason Transplant Institute of Methodist University Hospital, affiliated with the University of Tennessee Health Sciences Center, Memphis, TN. Adult LT recipients (age >18) with MASH as an indication for LT, who had a liver biopsy one-year post-transplant, were recruited for this study4. The signed consent of all the participants had been taken prior to enrollment in the study. The protocol and the study design were approved by the University of Tennessee Institutional Review Board (Study Protocol # 15-03891-XP UM). The stool samples from each participant were collected in accordance with the specified methodology described previously4.
Fecal microbiota DNA extraction and quality check
The genomic DNA was extracted following the protocol described in the PowerFecal DNA extraction kit (MO BIO Laboratories, Carlsbad, CA). Initially, the quality and the quantity of the extracted DNA were checked on 0.8% agarose gel and NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE), respectively.
Shotgun library preparation and sequencing
The library preparation for shotgun sequencing was done following the protocol of the Kapa Hyper Stranded kit (Roche). Subsequently, the quality of the prepared libraries was assessed using the 5200 Fragment Analyzer (Agilent Technologies, USA). The libraries were then pooled; quantitated by qPCR and subjected to sequencing (NovaSeq. 6000) on one SP lane for 151 cycles from both ends of the fragments. The sequencing was done using paired-end reads (2 × 150 bp), yielding more than 25 million reads per sample on average, ensuring a robust coverage for subsequent analysis and interpretation of the genomic data.
Host contamination removal
To enhance the accuracy of the downstream analysis, it is crucial to remove host DNA contamination from metagenomic reads. Here, we employed a mapping based method using bowtie2 v2.5.015 to remove human DNA contamination. In brief, bowtie2 index command was used to index the human reference genome (GRCh38) obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/data-hub/genome/GCF_000001405.26/), followed by an alignment step. Any reads that were successfully mapped to the human genome were identified as host DNA and subsequently removed from the dataset.
Microbial diversity and community analysis
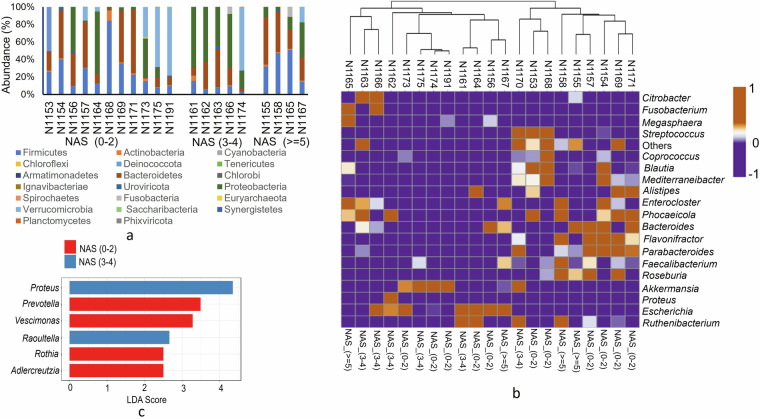
The clean microbial reads obtained from bowtie2 were subjected to assign taxonomic composition using Kraken2 package v2.0.816. The mapping of clean reads was done against Kraken2 standard database (https://benlangmead.github.io/aws-indexes/k2) (accessed on 11/02/2022). The output file (Kraken.report) generated in Kraken2 was further used as input in Bracken v2.817 to produce accurate phylum and species level abundance (Fig. 3). Results indicated that Firmicutes (also known as Bacillota) and Bacteroidetes were the most abundant phyla across all samples (Fig. 3a). Interestingly, our analysis revealed notable differences in the presence of the Euryarchaeota and Verrucomicrobia phyla among the sample groups. Specifically, these phyla were detected in the majority of “no MASH” (NAS 0–2) samples but were largely absent in the “borderline MASH” (NAS 3-4), and “definite MASH” (NAS ≥ 5) samples (Fig. S1). Furthermore, the relative abundance at the genus level was calculated, and the top 20 genera are presented in Fig. 3b. The results indicated that the genus Akkermansia was highly abundant in samples from the NAS 0–2 and NAS 3-4 groups, with abundances ranging from 0.43 to 0.79 (Fig. 3b). However, in patients with NAS ≥ 5, the abundance of Akkermansia was either low or absent in most samples. A. muciniphila is generally considered a next-generation probiotic, and its high abundance in the gut is associated with various positive health outcomes, including MASLD18. To identify the differential abundance of the key genera in the MASLD sample groups, we performed Linear Discriminant Analysis (LDA) using LEfSe19,20 with a threshold of p < 0.05 and an LDA score of 2.0. Although the abundance of Akkermansia was high in samples from the NAS 0–2 and NAS 3-4 groups, it was not significantly enriched in these groups., and thus its abundance cannot be correlated with MAFLD outcome. The results indicated that the genera Proteus (LDA 4.35) and Raoultella (LDA 2.66) were enriched in the NAS 3-4 group (Fig. 3c). In contrast, the genera Prevotella (LDA 3.49) and Vescimonas (LDA 3.28) were enriched in the NAS 0–2 group. Interestingly, no significant differential effects were observed in the NAS ≥ 5 group.
Fig. 3.
Bracken report based microbial diversity and community in each sample. (a) The distribution and abundance of top 20 phyla based on sample groups (b) heatmap representing the relative abundance of top 20 genera in different MASLD samples. (c) LDA enrichment of significant genera in different MASLD groups calculated using LEfSe considering g p < 0.05 and LDA score 2.0.
Species level diversity was further analyzed and visualized using the Pavian platform through Sankey flow diagrams (Fig. 4)21. The high abundance of A. muciniphila was consistently observed in samples from patients without MASH outcomes [NAS (0–2)] (Fig. 4r,t,u).
Fig. 4.
The snakey diagram of the top 20 species generated from the Pavian platform and their absolute abundance in terms of read counts.
Furthermore, the alpha diversity indices (Table S2) strongly support our diversity findings. For example, sample N1191 (Fig. 4u) is highly dominated by a single species A. muciniphila, and thus its overall species diversity is low which is indicated by high Berger Parker’s dominance value (0.78571) and lower Simpson’s index value (0.3798).
Functional profiling of gut microbial community
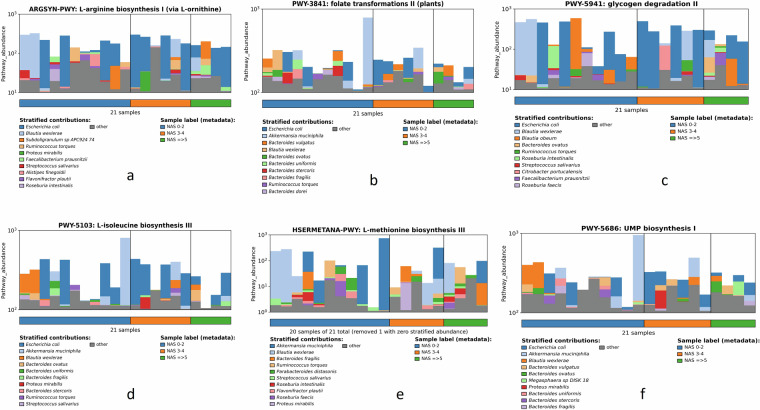
Functional profiling was done to gain a deeper understanding of the functional potential of the microbial communities and how it may relate to the observed pathologies associated with MASLD progression. In brief, the paired-end clean reads of each sample (r1 and r2) were first merged with Cat command in the Linux platform. The merged file was then used in MetaPhlAn v4.022 to generate the taxonomy file. The merged clean read file and the corresponding taxonomy file were used as input in HUMAnN v3.023 to generate three files: gene_families, pathway_abundance, and pathway_coverage using ChocoPhlAn database v201901b (accessed on 04/23/2023). All pathway_abundance files obtained from 21 samples were merged, normalized in HUMAnN v3.0 using humann_renorm_table command, and plotted using humann_barplot command (Fig. 5). Several pathways like the folate transformations (Fig. 5b), L-isoleucine biosynthesis (Fig. 5d), and L-methionine biosynthesis (Fig. 5e) exhibited higher abundance in “no MASH” (NAS 0–2) samples compared to “definite MASH” (NAS ≥ 5) samples, and interestingly, all these pathways were dominated by A. muciniphila. Similarly, the abundance of the other two microbial pathways: L-arginine biosynthesis (Fig. 5a) and glycogen degradation (Fig. 5c) were also high in low NAS (NAS 0–2) samples compared simples with NAS ≥ 5 (definite MASH), and dominated by the probiotic bacteria Blautia sp14. Previous research highlighted that an abundance of these pathways are associated with normal liver function13,14,24–26.
Fig. 5.
The functional profile of the top 10 species in each sample. (a-f) Here we have demonstrated six important pathways that might directly correlate with MASLD outcome. The important pathways were either dominated by A. muciniphila or Blautia sp.
Metagenomic assembly, contig generation, and quality check
Metagenome assembly is the first step in metagenome assembled genome (MAG) construction27. Here, we used SPAdes v3.15.528 to construct long contigs from the clean reads using a de novo approach using –meta option. The quality and length of these assembled contigs were assessed by MetaQUAST29.
Binning and refinement of MAGs
Binning is the most critical step in the construction of MAG and here we used MetaWRAP30 for binning the contigs obtained from metaSPAdes. MetaWRAP is a wrapper of three binning packages: MaxBin2, MetaBAT2, and CONCOCT. The bins obtained from MetaWRAP were often fragmented due to uneven coverage and inter-species overlapping; thus, bin refinement is also recommended. The metawrap bin_refinement command was used to refine the bins generated from MaxBin2, MetaBAT2, and CONCOCT using -c 50 -x 5 option, which has generated a total of 357 draft genomes or MAGs.
Completeness, contamination, and taxonomy of MAGs
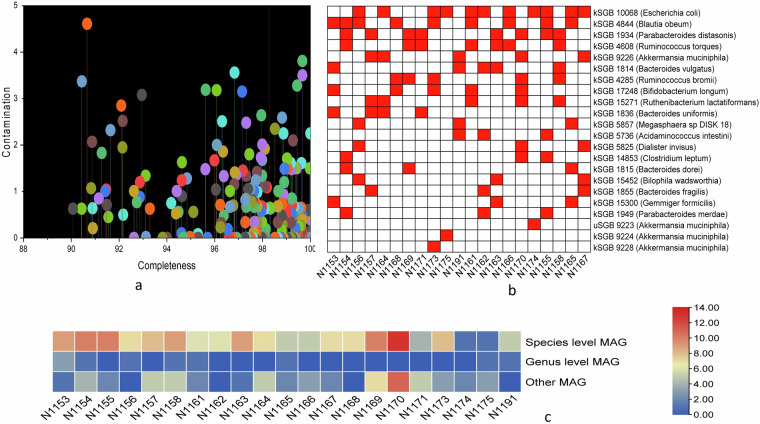
The completeness and contamination of MAGs were further assessed by using CheckM v1.1.331, and the result of high-quality MAGs (>90% completeness and <5% contamination) was tabulated accordingly (Table S3, Fig. 6a). We have documented the most abundant species-level MAGs, providing insights into the specific microbial species that are abundant within the samples (Fig. 6b). The overall distribution of high-quality MAGs, categorizing them at different taxonomic levels, including species, genus, and other taxonomic ranks was illustrated in Fig. 6c. The high-quality MAGs were annotated with Prokka v1.14.632 using a default command. The taxonomic classification of annotated MAGs was performed in Phylophlan 3.033, and the generated tree file was visualized in iTOL online platform (https://itol.embl.de/) (Fig. 2).
Fig. 6.
The distribution of meta genome assembled genome (MAGs) in each sample. (a) Indicated the completeness and contamination of 220 high-quality MAGs. Each color represented a single MAG, and the attached line indicated the percentage of contamination. (b) Represent the top known species level genome bins or MAGs in different samples. Here, red and white blocks mean the presence and absence, respectively. (c) Along with species-level MAGs, several genus-level and other lineage MAGs were also constructed and presented here. The color density bar on the right side indicated the number of specific level MAGs in different samples.
Abundance of MAG
The abundance of each MAG in respective samples was calculated following the method described by Zorrilla et al.34. In brief, fasta files of each MAGs generated in Prokka were merged (each sample separately) using Cat command, followed by mapping in bwa v0.7.1735 to generate sam files. The Samtool_view and samtools_ sort commands36 were used to convert the sam file to the sorted bam file. The samtools flagstat command was finally employed to calculate the mapping reads, and the relative abundance of each MAG was calculated as the total number of mapped reads divided by the total number of reads in the corresponding sample (Table S4). Interestingly, in all “definite MASH” samples (NAS ≥ 5) except one sample (N1167), the dominant species level MAGs belong to Bacteroides ovatus (N1155, 36.58%), and Bacteroides vulgatus (N1158, 15.11%), and Bacteroides dorei (N1165, 10.8%) (Table S4). However, in most of the samples from patients without MASLD or MASH (NAS 0–2) the abundant MAGs belong to the species Blautia obeum (ranges between 12.41% to 41.46%)), A. muciniphila (ranges between 25.31% to 72.49%) (and Akkermansia sp. (54.38%) (Table S4).
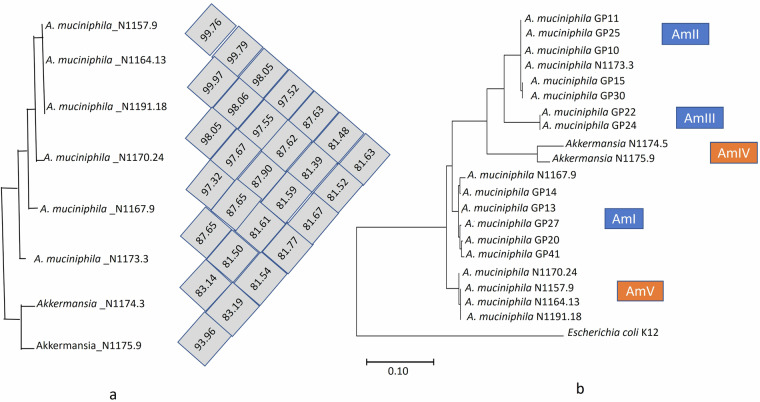
Average nucleotide identity and phylogenetic analysis of Akkermansia MAGs
Most of the “no MASH” (NAS 0–2) samples have a high abundance of A. muciniphila, which indicates a positive correlation with MASLD status. Therefore, we calculated the average nucleotide identity (ANI) among these A. muciniphila and Akkermansia sp. MAGs using OrthoANI tool v0.93.137 (Fig. 7a). Interestingly, A. muciniphila MAGs obtained from samples with low NAS (0–2) such as AK_N1157.9, AK_N1164.13, AK_N1191.18 clustered together which indicates that the A. muciniphila strains from different samples with favorable NAS scores share a high degree of genomic similarity. In order to check the phylogenetic group of the constructed A. muciniphila MAGs, we selected a few A. muciniphila strains randomly representing different phylogenetic clusters (AmI, AmII, and AmIII)38. The fasta files of these strains were downloaded from the NCBI genome database, followed by annotation in Prokka using the default command. The 16S rRNA gene sequences were then extracted and aligned in MEGA 6 software39 to generate the phylogenetic tree to compare the phylogenetic position of A. muciniphila MAGs and Akkermansia sp. MAGs (Fig. 7b). A. muciniphila MAG obtained from the N1173 sample (NAS 0) and N1167 (NAS 6) clustered with AmII and AmI, respectively. Furthermore, A. muciniphila MAGs constructed from “borderline MASH” (NAS 3–4) sample (N1157) and “no MASH” (NAS 0–2) samples (N1164, N1170, and N1191) clustered together and formed a new phylogroup AMV (Fig. 6B). However, genus level Akkermansia MAGs obtained from N1174 and N1175 exhibited an new phylogroup AMIV, along with previously described phylogenetic groups (AmI, AmII, AmIII)38. This phylogenetic analysis can shed light on the genetic diversity and adaptation strategies of A. muciniphila within the context of MASLD and its correlation with disease outcomes.
Fig. 7.
The comparison of A. muciniphila MAGs constructed from different samples. (a) Represented the average nucleotide identity among different MAGs. (b) demonstrate the phylogenetic tree of A. muciniphila showing different phylogenetic groups. Escherichia coli was taken as an out-group.
Data Records
The Illumina NovaSeq sequencing reads are available in the NCBI Sequence Read Archive (SRA) under BioProject identifier PRJNA97082040, with accession number SRP43822141. High quality MAGs (n = 220) are available at SAMN36703611- SAMN36703829 and SAMN36726309 under the same BioProject identifier40. The information regarding patient fat percentage and NAS score (Supplementary Table 1), microbial alpha diversity (Supplementary Table 2), MAG quality assessment (Supplementary Table 3), MAG abundance in each sample set (Supplementary Table 4), and differential abundance of three phyla in all sample sets (Supplementary Figure 1) were deposited to figshare42 with 10.6084/m9.figshare.27730911.
Technical Validation
Here, we have explored the microbial diversity and abundance of MAGs in stool samples of LT patients using deep shotgun Illumina sequencing. Microbial community assessment and construction of MAGs underwent a series of quality control processes, including removing host contamination (Fig. 1, Table 1). The sequencing platform generated a total of 538.6 million reads. Following quality filtering with a threshold of q < 25, 535.1 million reads were retained (Table 1). This stringent quality filtering process ensures that only high-quality reads are included in downstream analyses, enhancing the reliability and accuracy of the results obtained from the sequencing data. The mapping percentage of classified clean reads of most of the sample is above 60% (Table 2), confirming the reading quality and depth.
Table 1.
Sequencing reads processing statistics of each data set based on FastQC result.
| Sample ID | Raw reads count | Trimmed reads (q > 25) | % removal after initial trimming | Human DNA removed (clean reads) | % removal of host contamination |
|---|---|---|---|---|---|
| N1153 | 25424028 | 25281010 | 0.562530847 | 25026660 | 1.006091 |
| N1154 | 34717232 | 34525330 | 0.552757201 | 34431878 | 0.270677 |
| N1155 | 41353794 | 41044262 | 0.748497224 | 40816658 | 0.554533 |
| N1156 | 25341390 | 25187138 | 0.608695892 | 25184750 | 0.009481 |
| N1157 | 20468100 | 20339394 | 0.62881264 | 20334194 | 0.025566 |
| N1158 | 23389134 | 23251812 | 0.587118788 | 23212412 | 0.169449 |
| N1161 | 21739642 | 21568308 | 0.788117854 | 21543746 | 0.11388 |
| N1162 | 23113510 | 22965722 | 0.639400939 | 22962290 | 0.014944 |
| N1163 | 20194568 | 20065904 | 0.637121824 | 20061580 | 0.021549 |
| N1164 | 29329826 | 29179320 | 0.513149993 | 29171336 | 0.027362 |
| N1165 | 25934688 | 25766786 | 0.6474032 | 24149980 | 6.274768 |
| N1166 | 20228664 | 20118430 | 0.544939597 | 20103872 | 0.072362 |
| N1167 | 21321082 | 21191406 | 0.608205531 | 21171620 | 0.093368 |
| N1168 | 19026784 | 18924362 | 0.538304319 | 18840866 | 0.441209 |
| N1169 | 18455136 | 18339154 | 0.628453781 | 18334706 | 0.024254 |
| N1170 | 34380902 | 34123660 | 0.7482119 | 34102748 | 0.061283 |
| N1171 | 22904742 | 22745032 | 0.697279192 | 22723724 | 0.093682 |
| N1173 | 30135136 | 29913668 | 0.734916212 | 29790838 | 0.410615 |
| N1174 | 25592112 | 25417010 | 0.684203008 | 25414844 | 0.008522 |
| N1175 | 20839126 | 20723470 | 0.554994485 | 20681918 | 0.200507 |
| N1191 | 34750680 | 34520478 | 0.662438836 | 34492538 | 0.080937 |
Table 2.
Mapping percentage of clean reads using Kraken 2 standard database.
| Sample ID | Classified reads (%) | Microbial reads (%) | Bacterial reads (%) |
|---|---|---|---|
| N1153 | 61.3 | 61.3 | 60.8 |
| N1154 | 56.8 | 56.8 | 54.8 |
| N1155 | 77.6 | 77.4 | 77.4 |
| N1156 | 81.6 | 81.5 | 80.9 |
| N1157 | 71.6 | 71.6 | 71.5 |
| N1158 | 51.9 | 51.8 | 50.5 |
| N1161 | 82.7 | 82.4 | 82.3 |
| N1162 | 92.6 | 92.5 | 91.8 |
| N1163 | 86.6 | 86.5 | 82.5 |
| N1164 | 80.5 | 79.9 | 78.8 |
| N1165 | 63 | 62.6 | 62.6 |
| N1166 | 86.7 | 86.3 | 86.3 |
| N1167 | 57.7 | 57.5 | 56.9 |
| N1168 | 39.6 | 39.4 | 39.4 |
| N1169 | 48.8 | 48.8 | 48.7 |
| N1170 | 52.3 | 52.2 | 52.2 |
| N1171 | 54.8 | 54.7 | 54.7 |
| N1173 | 70.9 | 70.7 | 69.7 |
| N1174 | 86.1 | 85.8 | 85.8 |
| N1175 | 82 | 81.9 | 81.9 |
| N1191 | 90.3 | 90.3 | 89.5 |
A total number of 968898 contigs were prepared from clean reads during the MAGs construction process (Table 3), which varies from 95554 (highest) to 16091 (lowest). The number of long contigs ≥5000 bp and very long contigs ≥50000 bp varies from 4621 (highest) to 747 (lowest) and 316 (highest) to 51 (lowest), respectively, which indicates the high quality and depth of the sequencing reads, as well as the effectiveness of the assembler. To increase the accuracy of binning and construction of MAGs, we have excluded the contigs ≲2,500 bp to avoid high contamination and low completeness. The MAGs were validated following the standards defined by the Minimum Information about a Metagenome-Assembled Genome (MIMAG) of bacteria and archaea consortium43. In brief, CheckM v1.1.331 was used to calculate the completeness and contamination of each MAG using CheckM marker gene list. Only the high-quality MAGs (completeness >90% and contamination <5%) showing single-copy genes within a phylogenetic lineage31 were considered and deposited in the NCBI genome database (BioProject number PRJNA970820). Furthermore, the quality of Akkermansia MAGs was assessed considering the type strain of A. muciniphila (ATCC BAA-835) (Table 4). The lower number of contigs (varies from 39 to 14), along with scaffold-gap at extensive misassemblies (0) and a number of uncalled bases or N’s (0), confirmed the accuracy of assembly and draft genome quality.
Table 3.
Assembly statistics of each data set assessed through MetaQuast.
| Sample ID | N50 | Contigs | Contigs (≥1000 bp) | Contigs (≥5000 bp) | Contigs (≥50000 bp) | Largest contigs | Total length |
|---|---|---|---|---|---|---|---|
| N1153 | 6227 | 48041 | 18566 | 2489 | 258 | 746854 | 105729105 |
| N1154 | 9909 | 45466 | 15502 | 2647 | 254 | 564156 | 103691738 |
| N1155 | 8925 | 47210 | 21537 | 3309 | 316 | 430385 | 124325910 |
| N1156 | 5041 | 33399 | 10654 | 1195 | 140 | 494532 | 62771082 |
| N1157 | 6482 | 49291 | 16145 | 2586 | 203 | 658334 | 101152500 |
| N1158 | 6622 | 60260 | 26331 | 4621 | 254 | 344279 | 144792077 |
| N1161 | 3570 | 51603 | 16760 | 2121 | 158 | 426442 | 91504015 |
| N1162 | 3324 | 38157 | 12526 | 1298 | 124 | 622898 | 67759276 |
| N1163 | 10804 | 36485 | 17629 | 2633 | 268 | 551962 | 101588829 |
| N1164 | 2291 | 69590 | 21598 | 1771 | 209 | 432752 | 111061798 |
| N1165 | 7099 | 32105 | 13663 | 2319 | 142 | 539170 | 76610458 |
| N1166 | 11262 | 23907 | 10892 | 1786 | 169 | 916721 | 67927401 |
| N1167 | 13388 | 28346 | 8822 | 957 | 179 | 1001980 | 60863485 |
| N1168 | 5012 | 31015 | 12414 | 2033 | 90 | 305683 | 64411737 |
| N1169 | 2728 | 95554 | 32177 | 3148 | 256 | 518380 | 162005710 |
| N1170 | 7415 | 73343 | 26590 | 4400 | 353 | 690106 | 163873475 |
| N1171 | 2931 | 71052 | 27683 | 2504 | 230 | 416246 | 129883700 |
| N1173 | 3562 | 48745 | 18083 | 2057 | 146 | 444211 | 91532050 |
| N1174 | 3557 | 16091 | 6867 | 747 | 51 | 368322 | 31960148 |
| N1175 | 9015 | 20302 | 9222 | 1596 | 136 | 383053 | 54938244 |
| N1191 | 2642 | 48936 | 18965 | 2083 | 99 | 503892 | 83493951 |
Table 4.
Genome statistics of Akkermansia MAGs taking A. muciniphila ATCC BAA-835 as reference.
| Samples bin ID | N50 | Contigs | Contigs (≥5000 bp) | scaffold gap ext. mis. | GC (%) | N’s |
|---|---|---|---|---|---|---|
| N1157.9 | 124433 | 37 | 31 | 0 | 55.41 | 0 |
| N1164.13 | 113582 | 36 | 36 | 0 | 55.59 | 0 |
| N1167.9 | 177866 | 39 | 30 | 0 | 55.84 | 0 |
| N1170.24 | 177913 | 26 | 23 | 0 | 55.34 | 0 |
| N1173.3 | 227844 | 33 | 24 | 0 | 58.13 | 0 |
| N1174.3 | 179887 | 39 | 33 | 0 | 55.96 | 0 |
| N1175.9 | 110625 | 41 | 41 | 0 | 56.8 | 0 |
| N1191.18 | 113537 | 14 | 13 | 0 | 55.6 | 0 |
Supplementary information
Acknowledgements
Partial support was received from University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA, and the Division of Hepatology, Sandra Atlas Bass Center for Liver Diseases & Transplantation, Donald and Barbara Zucker School of Medicine at Hofstra/Northwell Health, Manhasset, NY, USA.
Author contributions
Sampling: P.B. and S.K.S.; Study design: P.B., G.B.; Methodology: G.B., S.R.P.; Writing – original draft: G.B., P.B., S.R.P.; Writing – review & editing: P.B., G.B., H.H., S.K.S.
Code availability
We used all open-source software or packages to analyze our data and did not use any custom codes. The version of each package was provided with non-default parameters when required.
Competing interests
We declare that none of the authors except Pratik Banerjee have any conflict of interest. Pratik Banerjee is a member of the Editorial Board at Scientific Data.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sanjaya Kumar Satapathy, Email: ssatapat@northwell.edu.
Pratik Banerjee, Email: pratik@illinois.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-024-04153-8.
References
- 1.Gadiparthi, C. et al. NAFLD epidemiology, emerging pharmacotherapy, liver transplantation implications and the trends in the United States. Journal of Clinical and Translational Hepatology8, 215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riazi, K. et al. The prevalence and incidence of NAFLD worldwide: a systematic review and meta-analysis. The Lancet Gastroenterology & Hepatology (2022). [DOI] [PubMed]
- 3.Albhaisi, S. A. & Bajaj, J. S. The Influence of the Microbiome on NAFLD and NASH. Clinical liver disease17, 15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satapathy, S. K. et al. Characterization of Gut Microbiome in Liver Transplant Recipients With Nonalcoholic Steatohepatitis. Transplantation direct6 (2020). [DOI] [PMC free article] [PubMed]
- 5.Pirola, C. J. et al. Cross talk between the liver microbiome and epigenome in patients with metabolic dysfunction-associated steatotic liver disease. EBioMedicine101, 104996 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornejo-Pareja, I. et al. Non-alcoholic fatty liver disease in patients with morbid obesity: the gut microbiota axis as a potential pathophysiology mechanism. J Gastroenterol (2024). [DOI] [PMC free article] [PubMed]
- 7.Stothart, M. R., McLoughlin, P. D. & Poissant, J. Shallow shotgun sequencing of the microbiome recapitulates 16S amplicon results and provides functional insights. Mol Ecol Resour23, 549–564 (2023). [DOI] [PubMed] [Google Scholar]
- 8.La Reau, A. J. et al. Shallow shotgun sequencing reduces technical variation in microbiome analysis. Sci Rep13, 7668 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh, N. K. et al. Characterization of metagenome-assembled genomes from the International Space Station. Microbiome11, 1–27 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerjee, G. et al. Application of advanced genomic tools in food safety rapid diagnostics: challenges and opportunities. Current Opinion in Food Science47, 100886 (2022). [Google Scholar]
- 11.Juluri, R. et al. Generalizability of the nonalcoholic steatohepatitis Clinical Research Network histologic scoring system for nonalcoholic fatty liver disease. J Clin Gastroenterol45, 55–58 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanyal, A. J. et al. Diagnostic performance of circulating biomarkers for non-alcoholic steatohepatitis. Nat Med29, 2656–2664 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voloshin, I., Hahn-Obercyger, M., Anavi, S. & Tirosh, O. L-arginine conjugates of bile acids-a possible treatment for non-alcoholic fatty liver disease. Lipids in Health and Disease13, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, Y. et al. Uridine attenuates obesity, ameliorates hepatic lipid accumulation and modifies the gut microbiota composition in mice fed with a high-fat diet. Food & function12, 1829–1840 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nature methods9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. Genome biology20, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, J., Breitwieser, F. P., Thielen, P. & Salzberg, S. L. Bracken: estimating species abundance in metagenomics data. PeerJ Computer Science3, e104 (2017). [Google Scholar]
- 18.Banerjee, G., Papri, S. R., Satapathy, S. K. & Banerjee, P. Akkermansia muciniphila-a potential next-generation probiotic for non-alcoholic fatty liver disease. Curr. Pharm. Biotechnol.25, 426–433 (2024). [DOI] [PubMed] [Google Scholar]
- 19.Chang, F., He, S. & Dang, C. Assisted selection of biomarkers by linear discriminant analysis effect size (LEfSe) in microbiome data. J. Vis. Exp., e61715 (2022). [DOI] [PubMed]
- 20.Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol.12, R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breitwieser, F. P. & Salzberg, S. L. Pavian: interactive analysis of metagenomics data for microbiome studies and pathogen identification. Bioinformatics36, 1303–1304 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanco-Míguez, A. et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat. Biotechnol., 1-12 (2023). [DOI] [PMC free article] [PubMed]
- 23.Beghini, F. et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. elife10, e65088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karmin, O., Woo, C. W., Sid, V. & Siow, Y. L. in Molecular Nutrition 323-343 (Elsevier, 2020).
- 25.Macotela, Y. et al. Dietary leucine-an environmental modifier of insulin resistance acting on multiple levels of metabolism. PloS one6, e21187 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rinella, M. E. et al. Mechanisms of hepatic steatosis in mice fed a lipogenic methionine choline-deficient diet. J. Lipid Res.49, 1068–1076 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banerjee, G., Papri, S. R. & Banerjee, P. Protocol for the construction and functional profiling of metagenome-assembled genomes for microbiome analyses. STAR Protoc.5, 103167 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res.27, 824–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikheenko, A., Saveliev, V. & Gurevich, A. MetaQUAST: evaluation of metagenome assemblies. Bioinformatics32, 1088–1090 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Uritskiy, G. V., DiRuggiero, J. & Taylor, J. MetaWRAP — a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome6, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res.25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Asnicar, F. et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun.11, 2500 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zorrilla, F., Buric, F., Patil, K. R. & Zelezniak, A. metaGEM: reconstruction of genome scale metabolic models directly from metagenomes. Nucleic Acids Res.49, e126–e126 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv1303.3997 (2013).
- 36.Li, H. et al. The sequence alignment/map format and SAMtools. bioinformatics25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, I., Ouk Kim, Y., Park, S.-C. & Chun, J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol.66, 1100–1103 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Guo, X. et al. Genome sequencing of 39 Akkermansia muciniphila isolates reveals its population structure, genomic and functional diverisity, and global distribution in mammalian gut microbiotas. BMC genomics18, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol.30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.NCBI BioProjecthttps://identifiers.org/ncbi/bioproject:PRJNA970820 (2023).
- 41.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRP438221 (2023).
- 42.Banerjee, G., Papri, S. R., Huang, H., Satapathy, S. K. & Banerjee, P. Deep sequencing-derived Metagenome Assembled Genomes from the gut microbiome of liver transplant patients. Figshare10.6084/m9.figshare.27730911 (2023). [DOI] [PMC free article] [PubMed]
- 43.Bowers, R. M. et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol.35, 725–731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRP438221 (2023).
Supplementary Materials
Data Availability Statement
We used all open-source software or packages to analyze our data and did not use any custom codes. The version of each package was provided with non-default parameters when required.