Abstract
Using a bio-oligo pull-down DNA-binding assay we investigated the binding capacity of endogenous, DNA damage-induced p53 in human diploid fibroblasts to several p53-responsive elements (REs) present in p53-regulated genes. During the course of p53 accumulation, we observed a decrease in p53 binding to the GADD45 but not to the p21WAF1/CIP1 RE. Using mutated GADD45 sequences we show that this change is dependent on the presence of cytosines at position 3 in RE pentamers and on the p53 redox state. Site-directed mutagenesis experiments demonstrated that Cys277 (a residue directly contacting base 3 in a RE pentamer) is critical for differential regulation of GADD45 in DNA-damaged cells. These data represent a novel mechanism for differential affinity of p53 to distinct REs.
INTRODUCTION
p53 tumor suppressor protein has a crucial role in maintaining genomic integrity and cell cycle control (for a recent review see 1). Although several different activities of p53 have been reported (e.g. 3′→5′ exonuclease activity; 2), it mainly functions as a transcriptional activator of the cell cycle regulating genes p21WAF1/CIP1 (3) and 14-3-3σ (4), DNA repair-associated genes GADD45 (5) and p53R2, encoding a ribonucleotide reductase subunit (6) or apoptosis-associated genes BAX (7), NOXA (8) and p53AIP (9), as well as several p53-inducible genes (PIG) many of which control the redox balance of the cells (10). In addition to transcriptional activation, p53 represses transcription (1).
p53-mediated transcriptional activation is mainly dependent on its ability to specifically bind to a responsive element (RE) within the downstream gene promoters (or other regulatory regions), comprising two copies of a palindromic consensus sequence PuPuPuC(A/T)(T/A)GPyPyPy, separated by 0–13 bp (11). After binding to its RE, p53 stimulates the assembly of the transcription preinitiation complex TFIID–TFIIA at the TATA element (12). p53 also activates transcription by associating with other transcription factors such as Oct-1 and Sp1 (13,14). Most p53 mutations found in human tumors are located in the p53 sequence-specific DNA-binding domain and affect amino acid residues which contact DNA or are required for the correct folding of p53, thus abolishing RE binding and transcriptional activation (15). p53 has been shown to bind its RE exclusively as a tetramer (16). X-ray crystallography showed the precise arrangement of a p53 core domain monomer with the consensus DNA (15), which was later confirmed for the complete p21 RE in solution using footprinting techniques (17). Recent NMR studies of full-length versus C-terminally truncated p53 suggest that p53 binds its recognition site constitutively, in contrast to the previously proposed allosteric regulation by the C-terminus or by C-terminal modifications (18). This is supported by recent studies using DNA footprinting (19) and chromatin immunoprecipitation (20) showing that p53 binds chromatin-packed DNA constitutively. Thus, p53 present in unstressed cells seems to be capable of specific DNA binding (20).
Most of the natural p53 REs do not completely fit the consensus sequence (21). The fourth C and sixth G in the RE half-site are absolutely required for binding and are conserved in all known REs. Other bases, however, may differ from the canonical consensus sequence without major effects on binding, whereas some sequences sharing 100% homology with the consensus do not bind p53 (22). There are several known properties of REs influencing the strength of sequence-specific binding of p53, including the ability to form non-B-DNA structures (21) or the ability of RE DNA to bend upon p53 binding, which in turn depends on the composition of the central (A/T)(T/A) motif (17). In addition, the redox state of p53 influences its specific DNA binding. Reduction leads to enhanced binding, whereas oxidation abolishes it (23,24). The DNA-binding domain of human p53 contains 10 cysteine residues as potential targets for redox modification. Three of them, Cys176, 238 and 242, in the reduced but not oxidised state, coordinate a zinc atom (together with His179), thus creating a motif necessary for the binding of p53 to a RE (15). Mutation of each cysteine of this triad abolishes binding completely.
Several studies have reported the altered sequence specificity of p53 mutations in the central domain (25), but little is known about fine biochemical modifications of wild-type p53 leading to differential RE recognition. An example of such modifications, phosphorylation of Ser46 leading to enhanced recognition of a RE in an apoptosis-inducing gene, p53AIP, has recently been described (9). To analyse in detail the ability of endogenous p53 from DNA-damaged cells for sequence-specific binding we developed a bio-oligo pull-down assay. Utilising this method, we show that changes in the Cys277 redox state allow p53 to discriminate among individual REs according to their sequence, which thus represents another mechanism to regulate p53 sequence-specific DNA binding after DNA damage.
MATERIALS AND METHODS
Cell culture and transfections
WS-1 human fibroblasts were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% foetal calf serum, non-essential amino acids and penicillin/streptomycin antibiotic mixture. SaOS-2 p53-deficient human osteosarcoma cells were maintained in DMEM containing 15% foetal calf serum. Wild-type or mutant p53 expression vectors (1 µg) together with empty vector as carrier DNA (9 µg) were used to transiently transfect 5 × 106 SaOS-2 cells using calcium phosphate precipitation.
Bio-oligo pull-down assay
Cells were irradiated with UVC using a StrataLinker apparatus (Stratagene) or a 137Cs γ-ray source (BioBeam 8000; STS GmbH, Braunschweig, Germany) and nuclear extracts were prepared by a high salt buffer method as described previously (26), with minor modifications, and stored at –70°C. Biotinylated oligonucleotides (see Table 1) and unmodified complementary oligonucleotides were obtained either from Sigma-Genosys or from TAG Copenhagen. They were annealed into duplexes, purified and immobilised on streptavidin-coated magnetic beads (Dynal, Oslo, Norway) according to the manufacturer’s instructions (2 µg dsDNA/50 µl stock solution beads/reaction). For a typical binding reaction, 5–10 µl nuclear extract containing 2.5–5 µg protein was mixed with an equal volume of p53 binding buffer [40 mM HEPES–KOH pH 7.9, 50 mM KCl, 0.2 mM EDTA, 4 mM MgCl2, 1 mM dithiothreitol (DTT), 0.05% NP-40, 4 mM spermidine], thus adjusting the NaCl concentration to 0.2 M. Preincubation was performed for 15 min at room temperature with 5 µg competitor DNA [poly(dI:dC); Amersham Pharmacia Biotech]. In indicated experiments, 20 mM DTT (Sigma) or 0.5 mM azodicarboxylic acid bis(dimethylamide) (diamide; Sigma) was added and samples were incubated for 15 min at room temperature to oxidise/reduce the protein mixture. Double-stranded, immobilised oligonucleotides were added and the binding reaction was incubated for 30 min at room temperature with continuous mixing. DNA–bead complexes with the bound proteins were pelleted using a magnet and the supernatant was recovered. Beads were washed twice with 1× p53 binding buffer and bound proteins were released by addition of NaCl to a concentration of 0.5 M. Both fractions were then subjected to SDS–PAGE electrophoresis and western analysis using rabbit anti-p53 polyclonal antibody (FL-393) or mouse monoclonal anti-p53 antibody (DO-1) and detection by chemiluminescence (ECL; Amersham). Films were scanned and the intensity of the signals was determined using Multi-Analyst v.1.0.1 software (Bio-Rad). The ability of p53 to bind to the specific sequence at a given time point was expressed as the ratio of bound/unbound p53. To verify the amount of oligonucleotides in each reaction, DNA was released from the beads by incubation with 50% formamide for 5 min at +70°C, followed by ethidium bromide-containing agarose gel electrophoresis. Purified p53 core domain polypeptide containing amino acids 94–312 used in a control binding experiment was a gift from E. Palecek (Academy of Sciences of the Czech Republic, Brno, Czech Republic).
Table 1. p53 response elements and oligonucleotides used in the study.
| Name | Abbreviation | Sequencea | Remarks | Reference |
|---|---|---|---|---|
| Consensus | CON | 5′-bio-(N)20-AGGCATGTCT AGGCATGTCT-3′ | 100% consensus match | 11 |
| Mutant | MUT | 5′-bio-(N)20-AGGa c g tTCT AGGa c g tTCT-3′ | Non-binding mutant control | |
| p21 | p21 | 5′-bio-(N)20-GAACATGTCC cAACATGTT g-3′ | Cyclin-dependent kinase inhibitor | 3 |
| GADD45 | G45 | 5′-bio-(N)20-GAACATGTCT AAGCATGCTg-3′ | Cell cycle regulator, involved in DNA repair | 5 |
| GADD3T | G3T | 5′-bio-(N)20-GAACATGTCT AAGCATGTTg-3′ | Modified GADD45 | |
| GADD3C | G3C | 5′-bio-(N)20-GAACATGCCT AAGCATGCTg-3′ | Modified GADD45 | |
| GADD4T | G4T | 5′-bio-(N)20-GAACATGTCT AAACATGTTg-3′ | Modified GADD45 | |
| GADD4C | G4C | 5′-bio-(N)20-GAGCATGCCT AAGCATGCTg-3′ | Modified GADD45 | |
| BAX | BAX | 5′-bio-(N)20 -t cACAAGTTag AGACAAGCCTggg cgt GGGCTAtaTT-3′ | Apoptosis promoting | 7 |
aOnly the biotinylated strand is shown; consensus-fitting bases are in upper case and residues 3 and 8 are in bold.
Construction of p53 cysteine mutants
Human wild-type p53 cDNA cloned into the XbaI site of the pcDNA3.1 expression vector was provided by O. A. Jänne (University of Helsinki, Helsinki, Finland). Mutagenesis was performed using a QuickChange kit (Stratagene) with the following complementary primer pairs (only the upper strand primer shown, changed residues in bold, restriction enzyme sites underlined): Cys124Ser, AAG TCT GTG ACT AGT ACT TAC TCC CCT GCC spanning codons 120–129, newly introduced ScaI site; Cys135Ser, AAC AAG ATG TTT AGT CAA CTG GCC AAG ACC spanning codons 131–140, newly introduced HindII site; Cys275Ser, GAG GTG CGT GTT AGC GCC TGT CCT GGG AGA spanning codons 271–280, newly introduced HaeII site; Cys277Ser, G CGT GTT TGT GCT AGC CCT GGG AGA GAC CG spanning codons 273–281, newly introduced NheI site; Cys277Phe, G CGT GTT TGT GCC TTT CCC GGG AGA GAC CG spanning codons 273–281, newly introduced SmaI site. Arg273His human p53 cDNA in vector pBC12 was a gift from H.-W. Stürzbecher (University of Lübeck, Germany). Plasmid clones bearing the mutations were identified by restriction cleavage with the corresponding enzymes and base changes were confirmed by automated DNA sequencing.
RESULTS
Sequence-specific p53 DNA-binding assay
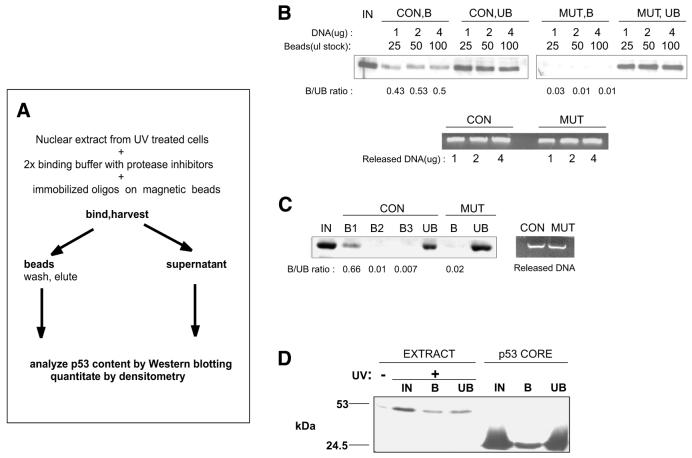
We and others have previously shown that UV-induced p53 can differentially activate its target genes depending on the extent of damage, leading to different biological responses of the cells (27,28). To address whether these differences are due to changes in recognition of DNA REs in its target genes by p53, we developed a bio-oligo pull-down assay based on magnetic isolation of beads with DNA–protein complexes (Fig. 1A). Nuclear extracts from UV-treated human diploid fibroblasts harbouring wild-type p53 were incubated with double-stranded oligonucleotides containing the consensus p53 RE (Table 1) and were immobilised on magnetic beads through biotin–streptavidin interaction. The DNA–protein complexes were then harvested using a magnet and p53 was released from the beads with 500 mM NaCl, which has been shown to disrupt tetrameric p53 and release it from DNA (29). First, we tested a consensus p53-binding element oligonucleotide (CON) (11) and its mutant variant where the central CATG motif was changed to ACGT (MUT) (Table 1 and Fig. 1B). A fraction of p53 present in the nuclear extract from UV-treated cells bound specifically to the CON but not the MUT oligonucleotide (Fig. 1B). To ascertain that the oligonucleotides used for the binding reaction were in molar excess and that there was no residual p53 with DNA-binding activity in the unbound fraction, we first increased the amount of DNA–bead complexes. The ratio of bound and unbound fractions, as measured by densitometry, did not change when the amount of immobilised DNA was elevated 2-fold (Fig. 1B). Secondly, we used a second and third round of binding reactions using the supernatant from the first binding reaction and fresh immobilised DNA. No additional binding of p53 to CON oligonucleotide was observed in the second and third reactions (Fig. 1C). As a third control experiment, we used the same amount of oligonucleotide and a large excess of purified bacterially expressed p53 core domain, possessing constitutive DNA-binding activity. The amount of DNA-bound p53 core domain was substantially higher than the amount of active p53 isolated from the nuclear extracts, indicating that the binding capacity was not saturated with endogenous p53 (Fig. 1D). These experiments thus indicated that, following UV treatment of human fibroblasts, the accumulated p53 consisted of two fractions, differing in their ability to specifically bind to DNA. To confirm that the unbound fraction was not inactivated due to a high NaCl concentration, we performed a binding experiment similar to Figure 1C using a NP-40 lysate of isolated nuclei instead of high salt buffer extraction (see Materials and Methods). The bound/unbound fraction ratio was similar to that in Figure 1C (not shown).
Figure 1.
Outline and optimisation of p53 DNA-binding assay. (A) Outline of experimental procedure. (B) Consensus (CON) or mutated (MUT) oligonucleotides were immobilised on beads and indicated amounts were incubated with 10 µg nuclear extract from WS-1 cells harvested 24 h after 35 J m–2 UVC irradiation. (C) Nuclear extract (20 µg) from UV-treated WS-1 cells was subjected to three consecutive binding reactions to CON oligonucleotide. As a negative control, a binding reaction with the MUT oligo is shown. The ratio of bound versus unbound amount of p53 is shown below the corresponding lanes. Oligonucleotides used for the binding were released from the beads as described in Materials and Methods and were run on an agarose gel containing ethidium bromide (Released DNA). (D) Immobilised CON oligonucleotide was used in a binding reaction with either 2.5 µg nuclear extract from UV-treated WS-1 cells (EXTRACT) or 600 ng purified, bacterially expressed p53 core domain (amino acids 92–312, p53 CORE). Nuclear extract from control cells was included to show the basal p53 levels. IN, input; B, bound fraction; UB, unbound fraction.
Time-dependent decrease in p53 sequence-specific binding to the GADD45 but not the CON or p21 RE
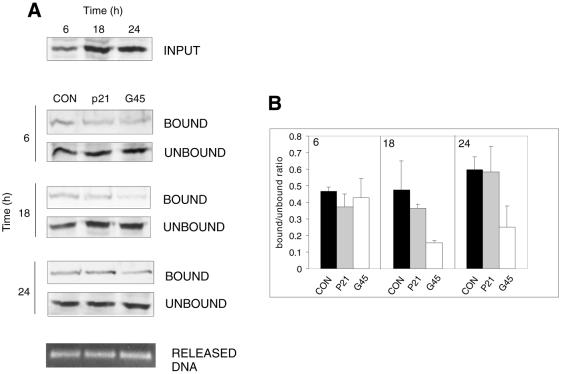
To analyse the sequence-specific DNA binding of p53 activated by UV irradiation to REs from several p53-responsive genes, we devised oligonucleotides with different, both synthetic and naturally occuring, RE sites (Table 1). Initially we used the synthetic CON sequence and its mutant (MUT) and sequences representing REs from three well-characterised p53 downstream genes: p21 from the human p21WAF1/CIP1 promoter, GADD45 from the GADD45 third intron and BAX from the BAX promoter. MUT failed to bind p53 as expected, as well as did the BAX oligonucleotide, probably because it fits the p53 consensus binding sequence less precisely than the other oligonucleotides used (results not shown). Additionally, the BAX sequence is specifically bound by other nuclear proteins, which may compete with p53 (30), and requires a coactivator binding to an Sp1-like responsive element immediately following the p53-binding site (31). Using the CON and p21 oligonucleotides, the amount of bound p53 was proportional to the total p53 level at all tested time points post-irradiation. On the other hand, binding of p53 to the GADD45 sequence was strongest in cell extracts 6 h after UV treatment but decreased afterwards (Fig. 2A and B). A similar result was obtained using endogenous murine p53 from UV-irradiated NIH 3T3 cells (not shown).
Figure 2.
Time-dependent decline in p53 binding activity to the GADD45 RE. (A) WS-1 cells were irradiated with 35 J m–2 UVC and harvested 6, 18 and 24 h thereafter. Equal amounts of nuclear extracts were subjected to a binding reaction with consensus (CON), p21 and GADD45 (G45) oligonucleotides. The amount of DNA in each reaction was verified as in Figure 1 (RELEASED DNA). (B) Bound and unbound p53 was quantitated by densitometry and the ratio of bound/unbound protein was calculated. Plots represent the average ± SE from at least two and mostly three independent experiments.
Thymine residues in positions contacting Cys277 stabilise p53 sequence-specific DNA binding upon DNA damage
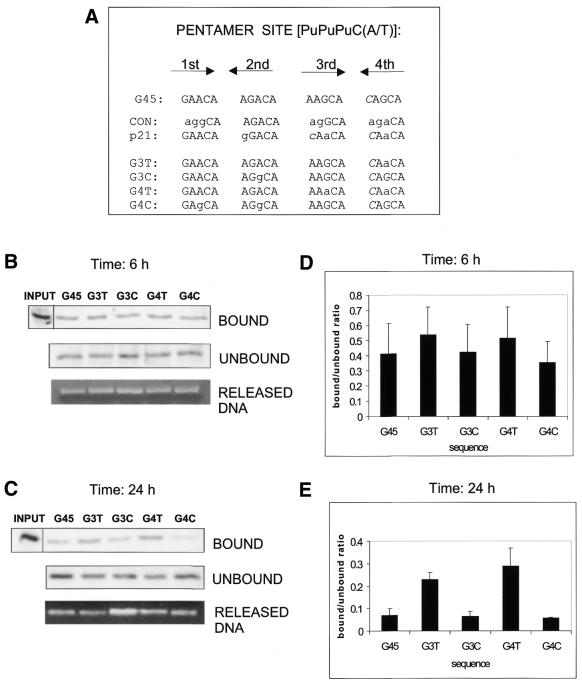
We compared all four p53-binding pentamers in GADD45 with the CON and p21 oligonucleotides (Fig. 3A). CON differed from GADD45 in 8 and p21 in only 4 of 20 bases (Fig. 3A, lower case letters). Two of these four bases (the first base in the second and third pentamers) are not contacted by p53, whereas the remaining two (the central bases in the third and fourth pentamers) are pyrimidines that form base pairs with Cys277 of the p53 core domain. In p21 the contacted residues are thymines, whereas in GADD45 these are cytosines. To test the possibility that different compositions of thymines/cytosines at the four positions contacted by Cys277 are the cause of differential p21 and GADD45 RE binding, we created GADD45 derivatives with increased (G3T and G4T) or decreased (G3C and G4C) numbers of thymines at positions contacted by Cys277 (Fig. 3A, note that the residues shown are the base-paired purines in the opposite strand) and used them in the following experiments. p53-binding assays using GADD45 and its derivatives were performed with nuclear extracts of WS-1 fibroblasts prepared 6 and 24 h after UV irradiation (Fig. 3). At the 6 h time point, binding of p53 to GADD45 and C-rich G3C and G4C was slightly decreased as compared with the T-rich oligonucleotides used (Fig. 3B and D). However, p53 from cells harvested 24 h after irradiation showed a strong preference for REs containing thymines but a very weak binding of REs that contained two or more cytosines at Cys277-contacted sites (Fig. 3C and E). Only one cytosine to thymine change at this position (G3T compared with GADD45) was able to markedly stabilise the DNA binding of late-accumulated p53.
Figure 3.
Thymine residues in position 3 of each RE half-site enhance the binding of late-accumulated p53 in UV-damaged cells. (A) Comparison of the p53-binding sequences in CON, p21 and GADD45 oligonucleotides. Shown are four pentamers contacted by four p53 subunits, in the 5′→3′ direction (first and third on one DNA strand, second and fourth on the other); lower case letters denote differences from the GADD45 sequence; differences from the consensus sequence are marked in italic. Note that Cys277-contacting pyrimidines are on the other strand of the pentamers. Nuclear extracts from UVC-irradiated (35 J m–2) WS-1 cells harvested at (B) 6 h and (C) 24 h were used in binding reactions with GADD45 oligonucleotide and its derivatives with increasing numbers of thymines/cytosines in position 3 (see Table 1). (D and E) Quantitations of the binding reactions in (B) and (C), respectively. Values were corrected according to the amount of DNA in each sample (RELEASED DNA).
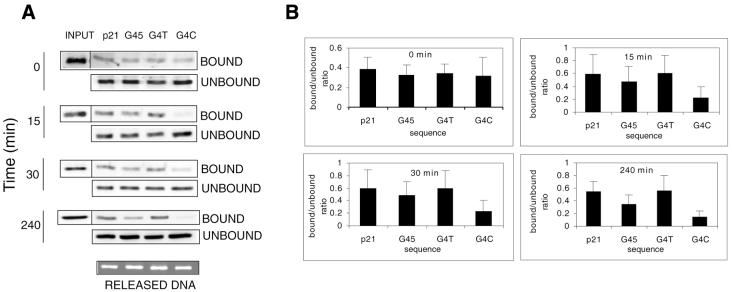
To compare p53 sequence-specific DNA binding with the P21/GADD45 REs after γ-radiation we irradiated WS-1 fibroblasts with 10 Gy and prepared nuclear extracts at various time points. Nuclear extracts were subjected to the binding assay using p21, G45, G4T and G4C oligonucleotides (Fig. 4). In control cells, p53 bound equally to all four oligonucleotides, despite the absence of genotoxic stress, with a bound/unbound ratio similar to that in cells after UV- or γ-radiation (Fig. 4A and B). However, diminished p53 binding to the G4C oligonucleotide was already apparent in extracts isolated 15 min after irradiation and to the G45 oligonucleotide by 4 h after irradiation (Fig. 4A and B). Thus, taking into account the previously demonstrated faster kinetics of p53 response after γ-radiation, the decrease in the binding capacity of p53 after γ-radiation reflected the UV-radiation response.
Figure 4.
Kinetics of p21, GADD45, G4T and G4C oligonucleotide binding by p53 from cells treated with ionising radiation. (A) WS-1 cells were either untreated (0) or treated with γ-radiation (10 Gy), nuclear extracts were harvested at the indicated time points and used in binding assays. (B) Quantitations of the binding reactions.
Reduction reactivates binding of late-accumulated p53 to the C-rich RE
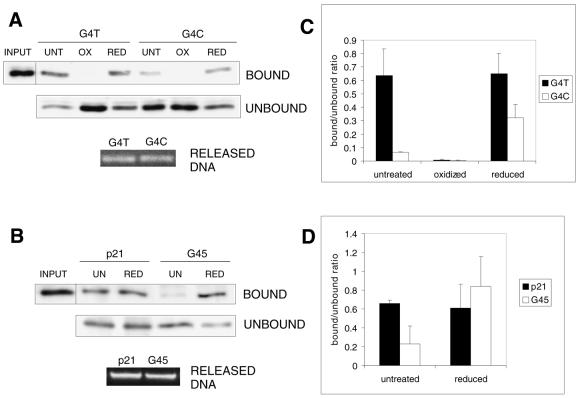
Cys277 directly contacts the first pyrimidine base in the consensus PyPyPy trinucleotide (15) and may be more accessible to redox modifications than the zinc-coordinating cysteines (32). Mutation of Cys277 to several other amino acids has been shown to abolish the binding or alter the sequence specificity of p53 (33). Since the DNA base residues, which regulated the affinity of p53 binding in our experiments, are contacted by Cys277, we next wanted to test whether the observed time-dependent changes in binding are influenced by changes in the overall redox state of the protein. We thus performed DNA-binding assays using the G4T and G4C oligonucleotides with nuclear extracts containing p53 from cells harvested 24 h post-UV irradiation. Nuclear extracts were left untreated or were treated with 0.5 mM diamide (oxidising agent) or 20 mM DTT (reducing agent). As expected, untreated nuclear extract showed markedly higher binding of p53 to G4T as compared with G4C (Fig. 5A and C). Oxidation by diamide completely hindered the binding of p53 to both oligonucleotides, presumably due to the abolition of zinc atom coordination by Cys176, 238 and 242 in the central domain. Reduction by DTT, on the other hand, enhanced the binding of p53 to G4C by ∼3-fold, whereas the same treatment had no effect on the binding of p53 to G4T (Fig. 5A and C). Similar results were obtained using p21 (equivalent to G4T) and GADD45 oligonucleotides in the same experimental arrangement (Fig. 5B and D) except that the oxidation reaction was omitted. These experiments suggested that in addition to zinc-coordinating Cys176, 238 and 242, Cys277 must be reduced to enable p53 to bind to a C-rich RE effectively.
Figure 5.
Reduction enhances the binding of late-accumulated p53 to C-rich but not to T-rich REs. (A) Nuclear extracts from UVC-irradiated (35 J m–2) WS-1 cells were harvested 24 h after treatment and were used in binding reactions with G4T and G4C oligonucleotides. Before the reactions, extracts were left untreated (UNT) or were treated with 5 mM diamide (oxidised, OX) or 20 mM DTT (reduced, RED). (B) The experimental arrangement was similar to that in (A) except that p21 and GADD45 oligonucleotides were used and the oxidation reaction was omitted. (C and D) Quantitations of the binding reactions in (A) and (B), respectively.
Specific DNA binding of Cys to Ser p53 mutants
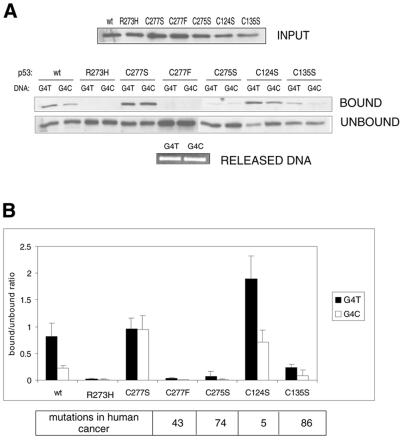
To address whether the above differences in p53 binding to C-rich RE are dependent on the redox changes of Cys277, and to determine the cysteine residue with which Cys277 possibly forms a disulphide bridge, we utilised site-directed mutagenesis of the relevant cysteines in p53. In addition to Cys277, the core domain of human p53 contains six other cysteine residues not involved in zinc coordination (Cys124, 135, 141, 182, 229 and 275). Of these, Cys275 is separated from Cys277 by one amino acid in the p53 polypeptide chain and Cys124 and Cys135 are in close vicinity to Cys277 within the folded p53 core domain (β-sheets 2 and 2′; 15). Thus, we generated a series of mutant p53 cDNAs with individual mutations of Cys277, 275, 124 and 135 to Ser, which structurally mimicks the thiol group of cysteine but cannot form a disulphide bridge under oxidising conditions. We also generated a p53 mutant of Cys277 to Phe, a missense mutation most frequently occurring at this site in human cancer (34). The constructs, together with a wild-type p53 construct and Arg273His mutant (completely lacking sequence-specific DNA-binding activity), were transiently transfected into SaOS-2 cells devoid of p53. Initially, we UVC-irradiated the transfected SaOS-2 cells. However, the high levels of ectopic wild-type p53 combined with UV-irradiation led to apoptosis of most of the cells, resulting in a very low yield of harvested proteins (not shown). Therefore, in the following experiments, the transfected cells were not subjected to any further treatments and the nuclear extracts were prepared 24 h after transfection and were tested for sequence-specific DNA-binding to G4T and G4C oligonucleotides (Fig. 6).
Figure 6.
Specific binding of different p53 cysteine mutants to G4T and G4C sequences. (A) Wild-type p53 (wt), a DNA-binding deficient p53 mutant (R273H, negative control) and a series of p53 cysteine mutants (C277S, C277F, C275S, C124S and C135S) were transfected into SaOS-2 cells, nuclear extracts were prepared 24 h after the end of the transfection, balanced for the amount of p53 and used for binding reactions with G4T and G4C oligonucleotides. (B) Quantitations of the binding reactions. Mutation frequency of the corresponding cysteine residue (to any amino acid) in human cancers (number of cases) according to the IARC database (34) is given below the graph.
Wild-type p53 had a strong affinity for the T-rich RE (G4T) and a lower affinity for the C-rich RE (G4C), similar to the endogenous p53 from WS-1 cells harvested 24 h after UV treatment, although the difference was not quite as prominent (compare for example Fig. 4A, untreated sample). The Arg273His contact mutant failed to bind both sequences completely. The binding capacity of Cys277Ser p53 towards G4T was comparable with wild-type p53, but the binding to G4C was enhanced, reaching a similar level to G4T (Fig. 6). Mutation of Cys277 to Phe completely abolished the specific binding regardless of the RE sequence, as did the Cys275Ser mutation. The Cys135Ser mutant retained low binding capacity, with G4T being bound stronger than G4C. Surprisingly, the Cys124Ser mutant showed an overall ∼2.5-fold increase in sequence-specific binding to both G4T and G4C as compared with wild-type p53 (Fig. 6). These results thus directly showed that REs that predominantly have cytosines at the Cys277 contact position require this residue in the reduced state for efficient binding to p53.
DISCUSSION
Sequence-specific DNA-binding activity of endogenous cellular p53 has previously been studied by electrophoretic mobility shift assay in vivo and chromatin footprinting techniques (19,35) and recently by chromatin immunoprecipitation (20,36). We developed a simple and sensitive DNA-binding assay that enabled us to follow the changes in sequence-specific DNA binding of endogenous p53 in unstressed and in DNA-damaged human diploid fibroblasts. We demonstrate that at any tested time point after DNA damage, the capacity of p53 for sequence-specific DNA binding varies depending on the DNA RE used. Importantly, the sensitivity of the assay permits the analysis of endogenous p53 and reveals its changes in DNA-damaged cells not observed in analyses utilising artificial expression of high levels of p53.
p53 was found to bind the REs from the P21 and GADD45 genes in both unstressed and in UV- or γ-radiation-damaged fibroblasts. The binding capacity of p53 towards the CON or p21 RE did not significantly increase after DNA damage, whereas the binding towards the GADD45 RE decreased. Approximately one-third of total p53 present in the cells possessed sequence-specific DNA-binding capacity and this fraction could not be increased by any experimental variation in the binding assay. The fraction of p53 lacking sequence-specific binding in the bio-oligo pull-down assay could represent the latent form proposed by the allosteric model of p53 regulation. However, the affinity could not be increased by, for example, antibody addition to the binding reaction (data not shown), suggesting that a latent fraction is not present. Thus, the p53 pool without binding capacity could reflect inability of p53 to form tetramers, p53 protein–protein interactions leading to steric hindrance or limitations in the bio-oligo pull-down assay. Recent chromatin immunoprecipitation and NMR studies have suggested that p53 has constitutive DNA-binding activity towards its recognition elements which is not dependent on a negative effect of the p53 C-terminus or C-terminal modifications (19,20,36). Whether all p53 molecules within the cells actually have binding capacity in the above studies cannot be ascertained. Using the bio-oligo pull-down assay, a significant increase in the binding of p53 derived from DNA-damaged cells to GADD45 and C-rich REs was observed only under reducing conditions, suggesting that redox conditions can determine the binding affinity of p53 towards certain of its REs.
Our results indicate that Cys277 in the C-terminal part of the p53 DNA-binding domain is essential for sequence-specific DNA binding and a target for redox regulation. Cys277 directly contacts the first pyrimidine base in the (A/T)GPyPyPy consensus pentamer (15). The presence of either Ser or reduced Cys at this position maintained strong binding of p53 to REs containing both C and T at the Cys277 contact site, whereas wild-type p53 harvested from stressed cells retained high affinity only to the T-rich REs, indicating the formation of an oxidised Cys277 side chain and a subsequent change in sequence specificity. A Cys to Ser change preserved the binding, presumably due to the equal ability of SH and OH side chains of these residues to make a hydrogen bond with a DNA base, as previously shown for murine p53 (24). Cho et al. (15) showed in their X-ray crystallographic study that the Cys277 side chain can either accept a hydrogen bond from N4 of the cytosine that it contacts or conversely donate a hydrogen bond to the O4 group of thymine. Chene (33) mutated Cys277 to several other amino acids (Phe, Ala, Val, Leu, Trp, Ser and Asp) and tested these for sequence specificity changes. The results indicate that the contact between thymine and Cys277 is based on hydrophobic interactions between SH and CH3 groups rather than on the hydrogen bonding proposed by Cho et al. (15). When a cysteine is present, however, hydrogen bonding between SH and NH2 would be necessary for protein–DNA interaction. Thus, Cys277 in the oxidised state may still form a hydrophobic interaction with a thymine, but a hydrogen bond with the NH2 group of cytosine probably cannot be formed, resulting in diminished binding to C-rich REs. On the other hand, mutating Cys277 to a longer side chain residue, Phe (the most common cancer-derived mutation found in this codon) caused a loss of overall binding, in agreement with other publications (33,37,38). The absence of binding can be explained by a steric hindrance which does not allow full contact of the p53 core domain with DNA.
Of the three cysteine residues that could be involved in disulphide bridge formation with Cys277, Cys275 would be a good candidate because of its close vicinity. Moreover, the CXC motif, corresponding to residues 275–277 in human p53, is conserved in all vertebrate p53 proteins, as well as in the Drosophila p53 homologue and the human p53 homologues p63 and p73 (SwissProt database retrieval; 39), which probably indicates its functional importance. However, mutating this residue to Ser caused a complete loss of p53 sequence-specific binding, possibly because of an unfavourable conformational change. Other tested residues, Cys124 and 135, lie within close vicinity of Cys277 in the folded p53 molecule. The Cys135Ser mutation led to a partial loss of p53 binding activity and Cys124Ser showed a gain of binding activity as compared with wild-type p53, but the difference between G4T and G4C binding was, in any case, similar to the wild-type protein. This suggests that neither of these two residues is involved in the redox control of Cys277 and that the observed binding differences are due to an altered p53 conformation, as in the case of the Cys275Ser mutation. Interestingly, Rainwater et al. (24) tested the DNA-binding properties of murine p53 by mutating all the Cys in the central domain to Ser and found little difference between wild-type p53 and mutants of sites corresponding to human residues 124, 135, 275 and 277. The discrepancy between these and our results is probably explained by the levels of p53 expression, by a difference between human and murine p53 or by different experimental approaches (electrophoretic mobility shift assay instead of oligonucleotide affinity isolation). However, a general enhancement of specific DNA binding in the Cys124Ser mutant corresponds to the low abundance of Cys124 mutations in human cancers (Fig. 6, table below the graph).
Redox regulation of p53 DNA-binding activity has been well documented (40,41), affecting zinc coordination and correct p53 folding. Thioredoxin activates p53 (42) and mutation of thioredoxin reductase (TRR1) overrides the growth inhibition caused by p53 when introduced into fission yeast (43), which indicates that redox activation of p53 is biologically relevant. On the other hand, intracellular p53 oxidation can be caused by elevated cellular levels of reactive oxygen intermediates (for a review see 32) or by an increased intracellular level of copper (44). The mechanism by which p53 residue 277 can be oxidised during the course of p53 accumulation is not clear. In any case, UV-radiation at the relatively high doses used here increases oxygen radicals and causes oxidative damage to the cells. It should be noted, however, that differential binding of p53 to the G4T and G4C sequences was also observed in p53 accumulated due to inhibition of the proteasome (not shown) and in wild-type p53 transiently expressed in SaOS-2 cells. The presence of oxidised cysteine residues in endogenous cellular p53 has been demonstrated by techniques that selectively label oxidised cysteine residues with maleimide derivatives (45,46), indicating that it is a common modification even under non-stress conditions. Nevertheless, the difference between G4T/G4C oligonucleotide binding was always less marked in ectopic p53 compared with DNA damage-induced endogenous protein, indicating that DNA damage contributes substantially to the Cys277 modification during p53 accumulation. In conclusion, the data presented suggest that p53 undergoes biochemical modification that allows it to discern among different promoters, according to the sequence composition of its REs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Olli A. Jänne and Horst-Werner Stürzbecher for providing us with plasmids, Dr Emil Palecek for a gift of p53 core domain and Ms Virpi Päivinen for excellent technical assistance. We also thank Dr Mirek Fojta (Institute of Biophysics, Academy of Sciences of the Czech Republic, Brno, Czech Republic) for helpful suggestions. This work was supported by Academy of Finland grants 32788 (M.L.) and 44885 (Finnish Centre of Excellence Program 2000–2005), the University of Helsinki, Biocentrum Helsinki, Sigrid Juselius Foundation, the Finnish Cancer Foundation and EU program grant QLG7-CT-1999-00851.
REFERENCES
- 1.Vogelstein B., Lane,D. and Levine,A.J. (2000) Surfing the p53 network. Nature, 408, 307–310. [DOI] [PubMed] [Google Scholar]
- 2.Mummenbrauer T., Janus,F., Müller,B., Wiesmüller,L., Deppert,W. and Grosse,F. (1996) p53 protein exhibits 3′-to-5′ exonuclease activity. Cell, 85, 1089–1099. [DOI] [PubMed] [Google Scholar]
- 3.el-Deiry W.S., Tokino,T., Velculescu,V.E., Levy,D.B., Parsons,R., Trent,J.M., Lin,D., Mercer,W.E., Kinzler,K.W. and Vogelstein,B. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell, 75, 817–825. [DOI] [PubMed] [Google Scholar]
- 4.Hermeking H., Lengauer,C., Polyak,K., He,T.C., Zhang,L., Thiagalingam,S., Kinzler,K.W. and Vogelstein,B. (1997) 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol. Cell, 1, 3–11. [DOI] [PubMed] [Google Scholar]
- 5.Kastan M.B., Zhan,Q., el-Deiry,W.S., Carrier,F., Jacks,T., Walsh,W.V., Plunkett,B.S., Vogelstein,B. and Fornace,A.J.,Jr (1992) A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell, 71, 587–597. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka H., Arakawa,H., Yamaguchi,T., Shiraishi,K., Fukuda,S., Matsui,K., Takei,Y. and Nakamura,Y. (2000) A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature, 404, 42–49. [DOI] [PubMed] [Google Scholar]
- 7.Miyashita T. and Reed,J.C. (1995) Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell, 80, 293–299. [DOI] [PubMed] [Google Scholar]
- 8.Oda E., Ohki,R., Murasawa,H., Nemoto,J., Shibue,T., Yamashita,T., Tokino,T., Taniguchi,T. and Tanaka,N. (2000) Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science, 288, 1053–1058. [DOI] [PubMed] [Google Scholar]
- 9.Oda K., Arakawa,H., Tanaka,T., Matsuda,K., Tanikawa,C., Mori,T., Nishimori,H., Tamai,K., Tokino,T., Nakamura,Y. and Taya,Y. (2000) p53AIP1, a potential mediator of p53-dependent apoptosis and its regulation by Ser-46-phosphorylated p53. Cell, 102, 849–862. [DOI] [PubMed] [Google Scholar]
- 10.Polyak K., Xia,Y., Zweier,J.L., Kinzler,K.W. and Vogelstein,B. (1997) A model for p53-induced apoptosis. Nature, 389, 300–305. [DOI] [PubMed] [Google Scholar]
- 11.el-Deiry W.S., Kern,S.E., Pietenpol,J.A., Kinzler,K.W. and Vogelstein,B. (1992) Definition of a consensus binding site for p53. Nature Genet., 1, 45–49. [DOI] [PubMed] [Google Scholar]
- 12.Xing J., Sheppard,H.M., Corneillie,S.I. and Liu,X. (2001) p53 stimulates TFIID–TFIIA–promoter complex assembly and p53–T antigen complex inhibits TATA binding protein–TATA interaction. Mol. Cell. Biol., 21, 3652–3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhan Q., Chen,I.T., Antinore,M.J. and Fornace,A.J.,Jr (1998) Tumor suppressor p53 can participate in transcriptional induction of the GADD45 promoter in the absence of direct DNA binding. Mol. Cell. Biol., 18, 2768–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koutsodontis G., Tentes,I., Papakosta,P., Moustakas,A. and Kardassis,D. (2001) Sp1 plays a critical role in the transcriptional activation of the human cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by the p53 tumor suppressor protein. J. Biol. Chem., 276, 29116–29125. [DOI] [PubMed] [Google Scholar]
- 15.Cho Y., Gorina,S., Jeffrey,P.D. and Pavletich,N.P. (1994) Crystal structure of a p53 tumor suppressor DNA complex—understanding tumorigenic mutations. Science, 265, 346–355. [DOI] [PubMed] [Google Scholar]
- 16.Friedman P.N., Chen,X., Bargonetti,J. and Prives,C. (1993) The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc. Natl Acad. Sci. USA, 90, 3319–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagaich A.K., Zhurkin,V.B., Sakamoto,H., Gorin,A.A., Clore,G.M., Gronenborn,A.M., Appella,E. and Harrington,R.E. (1997) Architectural accommodation in the complex of four p53 DNA binding domain peptides with the p21/waf1/cip1 DNA response element. J. Biol. Chem., 272, 14830–14841. [DOI] [PubMed] [Google Scholar]
- 18.Ayed A., Mulder,F.A., Yi,G.S., Lu,Y., Kay,L.E. and Arrowsmith,C.H. (2001) Latent and active p53 are identical in conformation. Nature Struct. Biol., 8, 756–760. [DOI] [PubMed] [Google Scholar]
- 19.Espinosa J.M. and Emerson,B.M. (2001) Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell, 8, 57–69. [DOI] [PubMed] [Google Scholar]
- 20.Kaeser M.D. and Iggo,R.D. (2002) Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc. Natl Acad. Sci. USA, 99, 95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim E., Albrechtsen,N. and Deppert,W. (1997) DNA-conformation is an important determinant of sequence-specific DNA binding by tumor suppressor p53. Oncogene, 15, 857–869. [DOI] [PubMed] [Google Scholar]
- 22.Halazonetis T.D., Davis,L.J. and Kandil,A.N. (1993) Wild-type p53 adopts a ‘mutant’-like conformation when bound to DNA. EMBO J., 12, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hainaut P. and Milner,J. (1993) Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res., 53, 4469–4473. [PubMed] [Google Scholar]
- 24.Rainwater R., Parks,D., Anderson,M.E., Tegtmeyer,P. and Mann,K. (1995) Role of cysteine residues in regulation of p53 function. Mol. Cell. Biol., 15, 3892–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freeman J., Schmidt,S., Scharer,E. and Iggo,R. (1994) Mutation of conserved domain II alters the sequence specificity of DNA binding by the p53 protein. EMBO J., 13, 5393–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haapajärvi T., Pitkänen,K., Tsubari,M. and Laiho,M. (1997) p53 transactivation and protein accumulation are independently regulated by UV light in different phases of the cell cycle. Mol. Cell. Biol., 17, 3074–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinke V. and Lozano,G. (1997) Differential activation of p53 targets in cells treated with ultraviolet radiation that undergo both apoptosis and growth arrest. Radiat. Res., 148, 115–122. [PubMed] [Google Scholar]
- 28.Latonen L., Taya,Y. and Laiho,M. (2001) UV-radiation induces dose-dependent regulation of p53 response and modulates p53–HDM2 interaction in human fibroblasts. Oncogene, 20, 6784–6793. [DOI] [PubMed] [Google Scholar]
- 29.Butcher S., Hainaut,P. and Milner,J. (1994) Increased salt concentration reversibly destabilizes p53 quarternary structure. Biochem. J., 298, 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thornborrow E.C. and Manfredi,J.J. (1999) One mechanism for cell type-specific regulation of the bax promoter by the tumor suppressor p53 is dictated by the p53 response element. J. Biol. Chem., 274, 33747–33756. [DOI] [PubMed] [Google Scholar]
- 31.Thornborrow E.C. and Manfredi,J.J. (2001) The tumor suppressor protein p53 requires a cofactor to activate transcriptionally the human BAX promoter. J. Biol. Chem., 276, 15598–15608. [DOI] [PubMed] [Google Scholar]
- 32.Meplan C., Richard,M.J. and Hainaut,P. (2000) Redox signalling and transition metals in the control of the p53 pathway. Biochem. Pharmacol., 59, 25–33. [DOI] [PubMed] [Google Scholar]
- 33.Chene P. (1999) Mutations at position 277 modify the DNA-binding specificity of human p53 in vitro. Biochem. Biophys. Res. Commun., 263, 1–5. [DOI] [PubMed] [Google Scholar]
- 34.Hainaut P., Hernandez,T., Robinson,A., Rodriguez-Tome,P., Flores,T., Hollstein,M., Harris,C.C. and Montesano,R. (1998) IARC database of p53 gene mutations in human tumors and cell lines: updated compilation, revised formats and new visualisation tools. Nucleic Acids Res., 26, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chin P.L., Momand,J. and Pfeifer,G.P. (1997) In vivo evidence for binding of p53 to consensus binding sites in the p21 and GADD45 genes in response to ionizing radiation. Oncogene, 15, 87–99. [DOI] [PubMed] [Google Scholar]
- 36.Szak S.T., Mays,D. and Pietenpol,J.A. (2001) Kinetics of p53 binding to promoter sites in vivo. Mol. Cell. Biol., 21, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thukral S.K., Lu,Y., Blain,G.C., Harvey,T.S. and Jacobsen,V.L. (1995) Discrimination of DNA binding sites by mutant p53 proteins. Mol. Cell. Biol., 15, 5196–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saller E., Tom,E., Brunori,M., Otter,M., Estreicher,A., Mack,D.H. and Iggo,R. (1999) Increased apoptosis induction by 121F mutant p53. EMBO J., 18, 4424–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brodsky M.H., Nordstrom,W., Tsang,G., Kwan,E., Rubin,G.M. and Abrams,J.M. (2000) Drosophila p53 binds a damage response element at the reaper locus. Cell, 101, 103–113. [DOI] [PubMed] [Google Scholar]
- 40.Parks D., Bolinger,R. and Mann,K. (1997) Redox state regulates binding of p53 to sequence-specific DNA, but not to non-specific or mismatched DNA. Nucleic Acids Res., 25, 1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fojta M., Kubicarova,T., Vojtesek,B. and Palecek,E. (1999) Effect of p53 protein redox states on binding to supercoiled and linear DNA. J. Biol. Chem., 274, 25749–25755. [DOI] [PubMed] [Google Scholar]
- 42.Ueno M., Masutani,H., Arai,R.J., Yamauchi,A., Hirota,K., Sakai,T., Inamoto,T., Yamaoka,Y., Yodoi,J. and Nikaido,T. (1999) Thioredoxin-dependent redox regulation of p53-mediated p21 activation. J. Biol. Chem., 274, 35809–35815. [DOI] [PubMed] [Google Scholar]
- 43.Casso D. and Beach,D. (1996) A mutation in a thioredoxin reductase homolog suppresses p53-induced growth inhibition in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 252, 518–529. [DOI] [PubMed] [Google Scholar]
- 44.Verhaegh G.W., Richard,M.J. and Hainaut,P. (1997) Regulation of p53 by metal ions and by antioxidants: dithiocarbamate down-regulates p53 DNA-binding activity by increasing the intracellular level of copper. Mol. Cell. Biol., 17, 5699–5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu H.H. and Momand,J. (1998) Pyrrolidine dithiocarbamate prevents p53 activation and promotes p53 cysteine residue oxidation. J. Biol. Chem., 273, 18898–18905. [DOI] [PubMed] [Google Scholar]
- 46.Wu H.H., Thomas,J.A. and Momand,J. (2000) p53 protein oxidation in cultured cells in response to pyrrolidine dithiocarbamate: a novel method for relating the amount of p53 oxidation in vivo to the regulation of p53-responsive genes. Biochem. J., 351, 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]