Abstract
This research aimed to identify novel indicators for sepsis by analyzing RNA sequencing data from peripheral blood samples obtained from sepsis patients (n = 23) and healthy controls (n = 10). 5148 differentially expressed genes were identified using the DESeq2 technique and 5636 differentially expressed genes were identified by the limma method(|Log2 Fold Change|≥2, FDR < 0.05). A total of 1793 immune-related genes were identified from the ImmPort database, with 358 genes identified in both groups. Next, a Biological association network was constructed, and five key hub genes (CD4, HLA-DOB, HLA-DRB1, HLA-DRA, AHNAK) were identified using a combination of three topological analysis algorithms (MCC, Closeness, and MNC) and four machine learning algorithms (Random Forest, LASSO regression, SVM, and XGBoost). immune cell distribution showed that the key genes correlated with multiple immune cell infiltrations. Gene Set Enrichment Analysis (GSEA) revealed that the key genes involved multiple immune response and inflammation-related signaling pathways. Subsequently, diagnostic models were constructed using four machine learning algorithms (Logistic regression, AdaBoost, KNN, and XGBoost) based on the identified key genes. Models with the highest performance were then selected. Ultimately, single-cell sequencing data revealed that the identified key genes were expressed in various immune cells, while Quantitative PCR (qPCR) tests confirmed their reduced expression in the sepsis group.
Keywords: Sepsis, RNA sequencing, Biomarkers, Differentially expressed genes, Immune response
Subject terms: RNA, Gene regulation
Introduction
Sepsis, a systemic inflammatory response triggered by infection, is associated with substantial morbidity and mortality1. Despite advancements in sepsis diagnosis and treatment over the past few years, current diagnostic markers and therapeutic strategies remain limited due to the complex and variable nature of its underlying pathophysiology2. Identifying novel biomarkers to enhance early diagnosis and therapeutic efficacy in sepsis has become a focal point of contemporary research.
Recent advancements in high-throughput sequencing technology have provided novel opportunities for identifying sepsis-related biomarkers through the differential analysis of gene expression profiles. RNA sequencing technology enables comprehensive and precise quantification of gene expression across diverse conditions, serving as a valuable tool for elucidating the molecular underpinnings of sepsis3.
Disruptions of the immune system are known to significantly contribute to the initiation and progression of sepsis4. Key pathological hallmarks of sepsis include cytokine storm, hyperactivation of immune cells, and a resulting imbalance in immune function5. Consequently, investigations focused on immune-related genes can offer deeper insights into the immunopathogenic mechanisms underlying sepsis and potentially lead to novel avenues for clinical intervention. Herein, we leveraged an immune gene database to identify sepsis-associated immune genes. Subsequently, we performed functional annotation and pathway analysis to elucidate potential diagnostic markers and therapeutic targets.
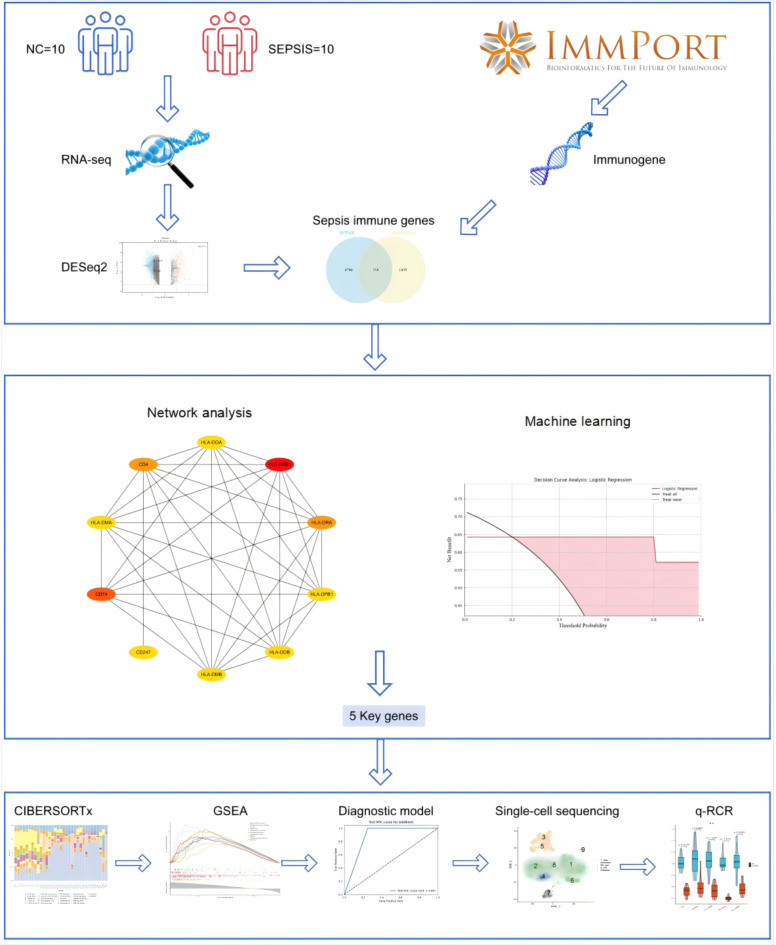
Analysis of Biological association network and the application of machine learning techniques are critical steps in biomarker identification6. Biological association network elucidate interactions among proteins, facilitating the pinpointing of key genes involved in disease mechanisms. Machine learning algorithms, trained and validated on large datasets of gene expression data, can efficiently identify feature genes that exhibit strong correlations with disease states. In summary, this study employs a comprehensive and systematic approach to screen and validate novel sepsis biomarkers. This approach integrates various techniques, including high-throughput RNA sequencing, Biological association network construction, machine learning-based screening, single-cell sequencing, and immune cell distribution. These methods offer valuable insights and pave the way for developing novel strategies for early diagnosis and personalized treatment of sepsis. Figure 1 provides a visual representation of the research workflow.
Fig. 1.
Workflow of the study. RNA sequencing was performed on peripheral blood samples from sepsis patients and healthy controls. Differentially expressed genes (DEGs) were identified using DESeq2. Immune-related genes were retrieved from the ImmPort database. Overlapping genes between DEGs and immune genes were used for downstream analyses. Biological association network was constructed and analyzed using MCC, Closeness, and MNC algorithms. Four machine learning techniques (Random Forest, LASSO regression, SVM, and XGBoost) were used to identify key genes. Immune infiltration and GSEA enrichment analyses were conducted on these key genes. A diagnostic model was developed to predict sepsis. Single-cell sequencing was used to determine the cellular localization of the key genes. Finally, qPCR validated the expression levels of these genes.
Methods
Clinical sample collection
This study utilized data from the Emergency Intensive Care Unit (ICU) of the Affiliated Hospital of Southwest Medical University. Peripheral blood samples were collected from 23 sepsis patients and 10 healthy volunteers between February 2019 and December 2020. Sepsis diagnosis was based on the most recent Sepsis 3.0 guidelines, and blood samples were obtained within the first 24 h of hospital admission. Control samples were collected from healthy volunteers during routine physical examinations conducted during the same period. Participants with severe organ failure, immune or hematological disorders, or those who were pregnant or lactating were excluded. Informed written consent was obtained from all participants, and the study was approved by the Hospital Ethics Committee (Ethics Approval No. ky2018029, Clinical Trial No. ChiCTR1900021261). All procedures adhered to the Declaration of Helsinki.
RNA-seq
Total RNA was extracted from blood samples using TRIzol reagent, and its quality and quantity were assessed using an Agilent 2100 Bioanalyzer (Thermo Fisher Scientific, MA, USA). Ribosomal RNA was depleted using the Enzyme H reagent, specifically targeting oligonucleotide and nucleoside sequences. The purified RNA was fragmented into smaller segments through incubation with SPRI beads and divalent cations at elevated temperatures. First-strand cDNA synthesis was performed using reverse transcriptase and random primers. Second-strand cDNA was subsequently generated with DNA polymerase I and RNase H. The size distribution of cDNA fragments was evaluated using an Agilent 2100 Bioanalyzer. Library quantification was performed using Quantitative PCR (qPCR). Qualified libraries, as determined by manufacturer’s guidelines, were sequenced on a BGISEQ-500/MGISEQ-2000 platform (China Huada Genetics Shenzhen). Raw sequence data underwent quality control to remove adapter sequences, low-quality reads (defined as quality value < 10 and > 20% bases with a quality score below 10), and reads containing more than 5% undetermined bases (N). The processed data, in FASTQ format, was aligned to the reference genome using HISAT and Bowtie2 software.
Differential gene screening and immune gene acquisition
Preprocessing of raw RNA sequencing data was performed using the online iDEP96 tool (http://bioinformatics.sdstate.edu/idep96/)7. This process involved an initial quality control step, followed by data normalization to ensure consistency and comparability between samples. Principal component analysis (PCA) was conducted on the cleaned data to assess sample differentiation and data integrity. Differential expression analysis was performed on the refined dataset using the DESeq2 technique, applying a threshold of |Log2 Fold Change| ≥ 2 and FDR < 0.05 to identify significantly differentially expressed genes (DEGs). In addition, we also used the limma method for differential expression analysis, which is applicable to RNA sequencing data through voom transformation and is particularly suitable for Fold Change estimation of small sample data(|Log2 Fold Change| ≥ 2 and FDR < 0.05). Volcano plots were generated using R version 4.4.1 to visualize the distribution of DEGs. Furthermore, hierarchical clustering analysis was conducted on the identified DEGs using the pheatmap package in R 4.4.1. This analysis resulted in heatmaps that depicted the expression profiles of these genes, highlighting differences in expression between sepsis patients and healthy individuals. To identify immune-related genes with significant expression changes, we utilized the ImmPort database, a public resource containing a comprehensive collection of immune-related genes and datasets8. We performed an intersection analysis between the identified DEGs and the list of immune-related genes from ImmPort. The resulting overlap of significantly altered immune genes was visualized using Venn diagrams.
Network analysis
To construct a Biological association network, the identified differentially expressed genes were uploaded to the STRING database (https://string-db.org/)9. The search criteria were set to include only interactions within the same species (“Homo sapiens”) and a minimum interaction score of 0.9. Unconnected nodes were excluded to maintain network simplicity. The resulting Biological association network was then imported into Cytoscape software (version 3.7.1) for further analysis. Three topological analysis algorithms, namely Maximal Clique Centrality (MCC), Closeness Centrality, and Maximal Neighborhood Component (MNC), were applied via the cytoHubba plugin to identify key hub genes within the network. Each algorithm identified the top 10 most central genes. These sets of genes were subsequently compared using a Venn diagram to identify the genes identified as central by all three algorithms.
Machine learning screening of core genes
In order to find the key immune genes that are closely related to sepsis, we try to use machine learning methods to screen from gene expression data. This approach helps us to process high-dimensional data and identify the gene features that are most important for the disease state. This study employed four machine learning algorithms—random forest, support vector machine (SVM), Lasso regression, and XGBoost—to identify key genes associated with sepsis. Random forest, an ensemble learning method, constructs multiple decision trees and aggregates their predictions to enhance model accuracy and robustness10. SVM, a supervised learning algorithm, seeks the optimal hyperplane to maximize the margin between classes. Feature selection is performed through cross-validation to optimize model performance11. LASSO regression is a linear regression model incorporating L1 regularization to induce sparsity and feature selection12. XGBoost is a gradient-boosting-based ensemble method known for its efficiency and performance13. In this study, we used screened differentially expressed immune-related genes as input to the machine learning algorithm. The screened gene expression data were normalized to ensure comparable data. Next, we constructed a feature matrix, where each row represents a sample, each column represents the expression value of a differential gene, and each sample is assigned a corresponding label (0 for healthy controls and 1 for septic patients) for the training of the machine learning model. Subsequently, the aforementioned machine learning algorithms were implemented using R language (version 4.4.1) to identify potential sepsis-associated core genes. The core genes identified by each algorithm were subjected to intersection analysis, and the overlapping genes were visualized using Venn diagrams.
Immune cell distribution
This study utilized CIBERSORTx, a deconvolution algorithm based on linear support vector regression, to estimate the relative abundance of various immune cell types within the mixed cell populations of sepsis patients14. RNA sequencing data was uploaded to the CIBERSORTx platform (https://cibersortx.stanford.edu/) to evaluate the presence and proportions of different immune cell types. The analysis employed the official LM22 signature matrix, encompassing the reference gene expression profiles for 22 human immune cell types. To enhance the reliability of the results, “Absolute Mode” was selected with 1000 permutations. To further investigate the relationship between the identified key genes and immune cell populations, the CIBERSORTx analysis results were imported into the R environment (version 4.4.1) for additional analysis. Spearman’s rank correlation coefficients were calculated using the cor.test function to assess the strength and direction of the correlations between each core gene and each immune cell type. The significance of these correlations was evaluated using p-values. Finally, the CIBERSORTx analysis results were visualized using the ggplot2 and pheatmap libraries within R version 4.4.1.
Expression of key genes and GSEA enrichment analysis
To explore the potential functions and associated signaling pathways of key genes identified in sepsis, we performed single-gene GSEA and gene expression analysis.Gene Set Enrichment Analysis (GSEA) is a computational method for determining whether a predefined set of genes is significantly enriched at the extremes of a list of sequenced genes, and thus for hypothesizing that the set of genes may be be involved in a biological process or signaling pathway. Specifically, single-gene GSEA is based on the principle of grouping samples according to the expression of a target gene (high and low expression groups), and then sorting the expression data of all genes to determine the expression patterns of other genes in samples with high or low expression of the target gene. Next, we performed enrichment analysis on predefined sets of functional genes (e.g., the KEGG gene set in the MSigDB database) to see whether these gene sets were significantly enriched at the top or bottom of the sequencing to hypothesize the biological pathways and functions that may be associated with this target gene15;16.
First, we entered the RNA sequencing data mentioned above as input data into R language 4.4.1, and then we targeted each key gene (CD4, HLA-DOB, HLA-DRB1, HLA-DRA, AHNAK) using the ‘Signal2Noise’ in R 4.4.1 ' mode for GSEA analysis using 1,000 gene alignments and setting FDR q-value < 0.25 and p-value < 0.05 as significance criteria to identify enriched signaling pathways, and finally generating enrichment Bubble Diagram using the enrichplot package to visualize the enrichment in different signaling pathways. In addition, we analyzed the expression levels of key genes using the DESeq2 package, while violin plots were drawn using the ggplot2 package to demonstrate the differential expression of key genes between the normal control (NC) and sepsis groups (SEPSIS).
Construction and screening of diagnostic models
This study employed four machine learning algorithms implemented in Python to construct and evaluate a sepsis detection model: logistic regression, AdaBoost, K-Nearest Neighbors (KNN), and XGBoost. The RNA sequencing data were first loaded and transposed. Labels were then added to each sample based on group assignment, with “0” representing the NC group and “1” representing the SEPSIS group. To ensure balanced and randomized training and testing sets, the data was split in a 6:4 ratio. Four algorithms were used to build and train the model on the training set: logistic regression, AdaBoost, KNN, and XGBoost. Logistic regression, a linear model for binary classification, learns the relationship between features and labels to classify samples17. AdaBoost, an ensemble learning method, combines multiple weak learners (e.g., decision trees) to enhance model accuracy18. KNN, a nonparametric classification algorithm, assigns class labels based on distance metrics between samples. In this case, the K nearest neighbors determine the classification19. XGBoost, a distributed gradient boosting algorithm, builds and combines multiple weak learners to improve overall model performance and accuracy. Specific hyperparameters were defined for each algorithm: L2 regularization was applied in logistic regression, AdaBoost used a decision tree base learner with 100 iterations, KNN employed 5 neighbors for classification, and XGBoost utilized a learning rate of 0.3 with a maximum tree depth of 6. Following training, each model’s performance on the test set was evaluated using precision-recall curves, ROC curves, AUC values, and calibration curves. Calibration curves assess whether predicted probabilities correspond to actual outcomes. To further evaluate the robustness and generalization ability of the constructed machine learning models on external datasets, we selected the public dataset GSE65682 for external validation. Training and internal validation of the models were done on a previous training set, while independent testing was performed on an external dataset. Additionally, decision curve analysis evaluated the net benefit of each model across different classification thresholds. The DeLong test was used to compare the AUC values of all four models to determine the best-performing model. Finally, SHAP (SHapley Additive exPlanations) values were employed for the top model to interpret its decision-making process. SHAP values quantify the influence of each feature on the model’s output, allowing the identification of genes with the most significant impact on model predictions.
Single-cell sequencing
To further explore the specific immune cell types and their expression patterns of the identified key genes in sepsis, we performed single-cell RNA sequencing analysis. This approach allowed us to understand the cellular context and heterogeneity of these key genes at the single-cell level, thereby revealing the potential role of each immune cell subtype in the immune response to sepsis. 10× Genomics single-cell RNA sequencing was performed on each sample. Cell Ranger, the proprietary software from 10× Genomics, was utilized to assess the quality of each sample. Quality control metrics, such as the number of high-quality cells, detected genes, and genome alignment rate, were generated by comparing raw data reads to the reference genome. Following this initial quality assessment, further filtering steps were implemented to remove multicellular, bicellular, and non-cellular events from the experimental data. Single-cell transcriptome sequencing was then performed. This technique utilizes unique molecular identifiers (UMIs) and cell barcodes alongside identified transcript sequences to determine the precise count of each transcript molecule within an individual cell. To visualize the dimensionality reduction results derived from the mutual nearest neighbors (MNN) method for clustering single-cell populations, the t-SNE algorithm was employed. This analysis ultimately identified the optimal cell clusters for further investigation. Marker genes are characterized by high expression within a specific cell type and minimal expression in other cell clusters. They are essentially up-regulated in a particular cell population compared to others. The bimod test was used to identify marker genes specific to each cell population by comparing the expression profiles of these populations to all remaining populations. Cell type annotation was performed using the SingleR package and the HPCA reference dataset20. This technique assigns cell types based on the strongest correlation with a reference dataset. It accomplishes this by calculating the correlation between the single-cell reference expression profiles and the expression profiles of the target cells. Following the gene expression analysis at the single-cell level within peripheral blood cells, a two-dimensional t-SNE map was generated to visualize the cellular distribution. These cell populations were initially classified based on established cell type markers, including monocytes, natural killer (NK) cells, T cells, and B cells.
LPS inflammatory cell modeling and q-PCR experiments
To validate the expression of key genes in a simulated sepsis model, we performed in vitro experiments using LPS-stimulated THP-1 human monocytic leukemia cells. THP-1 cells were seeded into 6-well plates (3.0 × 105 cells/well) and cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. To induce macrophage differentiation, cells were treated with 50 ng/mL PMA for 24 h. Afterward, the medium was replaced with a fresh RPMI-1640 medium for maintenance. Transfection assays involved adding 125 µL Opti-MEM® medium without antibiotics or serum, followed by 100 pmol siRNA and 4 µL Lipo8000™ Transfection Reagent. The mixture was incubated at room temperature for 20 min before being added to the wells. After 24 h, cells were stimulated with 100 ng/mL LPS for 24 h to mimic a sepsis model. Following cell processing, total RNA was extracted using RNAiso Plus reagent and quantified using a Nanodrop 2000 spectrophotometer. cDNA synthesis was performed by reverse transcription using the PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) following the manufacturer’s protocol (15 min at 37 °C, 5 sec at 85 °C, hold at 4 °C). PCR was performed using the PerfectStart® Green qPCR SuperMix kit and the qTOWER 3G Real-Time PCR System. The qPCR reaction mixture contained 3.6 µL water, 5 µL 2× SuperMix, 0.2 µL each of forward and reverse primers, and 1 µL template cDNA. The PCR program consisted of an initial denaturation step (94 °C for 30 s) followed by 40 cycles of denaturation (94 °C for 5 sec) and annealing/extension (60 °C for 30 s). Gene expression levels were assessed in triplicate biological replicates. Relative gene expression was calculated using the 2-ΔΔCt method. Data analysis and visualization were performed using R version 4.4.1. Violin plots were generated to depict the expression levels of key genes in both the NC and SEPSIS groups, with statistical significance determined by t-tests.
Results
Differential gene screening and immune gene acquisition
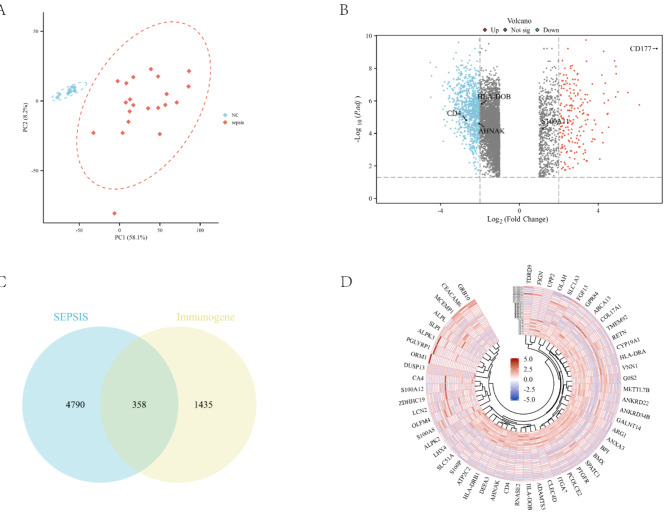
To explore the differences in gene expression between sepsis patients and healthy populations, we performed a differential gene screen to identify gene expression patterns that were significantly different between the two groups, which in turn identified immune genes associated with sepsis. PCA of gene expression data revealed a clear separation between the NC and SEPSIS groups along principal components 1 (PC1) and 2 (PC2), accounting for 58.1% and 8.2% of the variance, respectively (Fig. 2A). This indicates distinct gene expression patterns between the two groups. Querying the ImmPort database identified 1,793 immune-related genes. Differential expression analysis identified 5,148 genes with significant changes in expression levels, including 822 upregulated and 4,326 downregulated genes (Fig. 2B). In addition, limma differential analysis identified a total of 5636 differentially expressed genes (890 up-regulated and 4746 down-regulated). The volcano plot (Fig. 2B) visualized the significance and fold change of differentially expressed genes. Notably, genes such as CD4, HLA-DOB, HLA-DRB1, HLA-DRA, and AHNAK exhibited significant downregulation, while CD177 showed significant upregulation in the sepsis group. To identify immune genes associated with sepsis, a Venn plot (Fig. 2C) was constructed. This analysis revealed 358 genes potentially crucial for the immune response to sepsis and linked to sepsis-related immune functions. Finally, a heatmap (Fig. 2D) depicted the expression patterns of a subset of differentially expressed genes across different samples, providing a visual representation of their expression variation. In the analysis of key genes such as AHNAK, HLA-DOB, HLA-DRB1, HLA-DRA and CD4, we applied DESeq2 and limma methods respectively and compared the Fold Change (FC) and adjusted p-values of the two. The results showed that DESeq2 and limma were basically consistent in terms of p-value, and both could significantly differentiate these key genes between the sepsis and control groups. However, on FC estimation, limma had slightly higher FC values for these genes(Table 1).
Fig. 2.
Identification of overlapping genes. (A) Principal component analysis (PCA) demonstrating distinct clustering of normal control (NC) and sepsis samples along PC1 and PC2 axes. (B) Volcano plot illustrating differentially expressed genes, with upregulated genes in red, downregulated genes in blue, and non-significant genes in grey. (C) Venn diagram depicting the intersection of differentially expressed sepsis genes and immune-related genes from the ImmPort database. (D) Heatmap visualizing the expression patterns of differentially expressed genes across samples, with color intensity representing gene expression levels.
Table 1.
Key genes in DESeq2 and limma analyses.
| Gene | DESeq2 | limma | ||
|---|---|---|---|---|
| log2 FC | Adj.Pval | log2 FC | Adj.Pval | |
| AHNAK | -2.03910555 | 2.52E-05 | -2.039382943 | 3.444E-05 |
| HLA-DOB | -2.214074852 | 7.96E-08 | -2.144355931 | 3.43514E-05 |
| HLA-DRB1 | -2.86628613 | 2.79E-04 | -2.881323693 | 0.000410655 |
| HLA-DRA | -2.203148241 | 3.12E-02 | -2.207214146 | 3.24509E-05 |
| CD4 | -2.70726075 | 1.55E-05 | -2.716740548 | 2.33809E-05 |
Network analysis
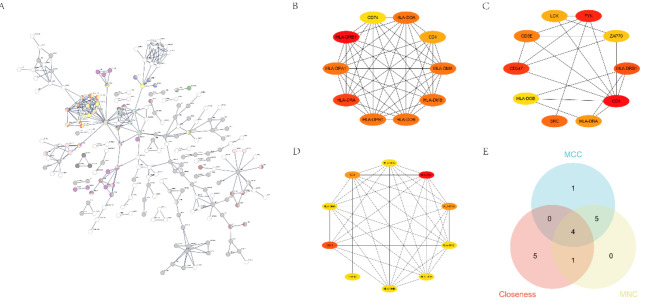
To understand the interactions between sepsis-related genes, we constructed a biological association network to identify core genes that may play key roles in the pathophysiological mechanisms of sepsis. After removing isolated nodes, the Biological association network consisted of 354 nodes and 339 edges, with CD4, HLA-DOB, HLA-DRB, HLA-DRB1, and AHNAK occupying central positions (Fig. 3A). To characterize the Biological association network topology in greater detail, three centrality algorithms—MCC, Closeness Centrality, and MNC—were employed to identify key network hubs (Fig. 3B-D). By integrating the results of these algorithms using Venn plots (Fig. 3E), CD4, HLA-DOB, HLA-DRB, and HLA-DRB1 were identified as shared core genes, suggesting their consistent importance within the Biological association network. These core genes may play critical roles in the initiation and progression of sepsis, providing a strong foundation for further investigations into the underlying mechanisms. By identifying these core genes, we expect to be able to provide valuable gene candidates for future targeted therapy studies.
Fig. 3.
Biological association network and key gene identification. (A) Biological association network visualization, with nodes representing proteins and edges representing interactions. Isolated nodes are excluded. (B) Top 10 gene sub-networks identified by the Maximal Clique Centrality (MCC) algorithm. Node color indicates gene significance (red: high, yellow: low). (C) Top 10 gene sub-networks identified by Closeness Centrality. Node color indicates gene significance (red: high, yellow: low). (D) Top 10 gene sub-networks identified by the Maximal Neighborhood Component (MNC) algorithm. Node color indicates gene significance (red: high, yellow: low). (E) Venn diagram illustrating the overlap of core genes identified by the MCC, Closeness, and MNC algorithms. Four genes are shared by all three methods.
Machine learning screening of core genes
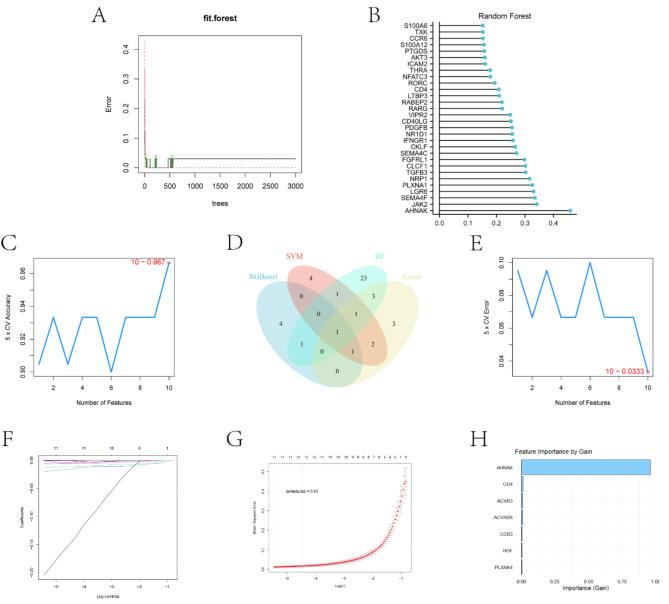
To further screen for key genes significantly associated with sepsis, we used four machine learning algorithms. By machine learning modeling of data from these differentially expressed immune genes, we expect to identify important genes that are significant in sepsis prediction or diagnosis. Each of the four machine learning algorithms identified genes with distinct biological characteristics (Table 2). The Random Forest model exhibited a gradual decrease in error rate during iterations, eventually stabilizing at a low level (Fig. 4A). Gene importance analysis revealed multiple key genes within this model (Fig. 4B). To optimize the SVM model, cross-validation was employed to assess the impact of varying feature numbers on model performance. Results indicated a significant increase in accuracy up to 96.7% with an increasing number of features (Fig. 4C). Conversely, the error rate assessment identified an optimal feature number of 10, corresponding to a minimum error rate of 0.0333 (Fig. 4E). In the Lasso regression model, most of the regression coefficients gradually approach zero as the Log Lambda value increases, showing the effect of regularization(Fig. 4F).LASSO regression analysis determined optimal model parameters by adjusting the lambda value (Fig. 4G) and identified key genes. XGBoost analysis highlighted AHNAK as the most critical gene in sepsis (Fig. 4H). A Venn plot (Fig. 4D) was generated, illustrating the overlap in feature genes identified by the four algorithms. Notably, AHNAK was consistently identified by all models, suggesting its potential significance in sepsis pathogenesis. Through the screening of these machine learning algorithms, we hope to find key genes that can significantly differentiate between sepsis and healthy states, thus enhancing the possibility of early diagnosis and precise treatment of sepsis.
Table 2.
Top10 genes screened by four algorithms.
| Random Forest | SVM | Lasso | XGBoost |
|---|---|---|---|
|
AHNAK JAK2 SEMA4F LGR6 PLXNA1 NRP1 TGFB3 CLCF1 FGFRL1 SEMA4C |
HTR3A CR2 AKT1 PDGFB AHNAK IL1R1 CD1C TGFB3 ACKR2 CCR3 |
CCR3 CD1C CD22 DES AHNAK HTR3A INSL6 NR1D1 NRP1 PDGFB |
AHNAK CD4 ACKR3 ACVR2B CCR3 HGF PLXNA4 |
Fig. 4.
Machine learning analysis and feature selection. (A) Error rate of the Random Forest model across increasing numbers of trees. (B) Gene importance scores determined by the Random Forest model. (C) Accuracy of the Support Vector Machine (SVM) model using 5-fold cross-validation with varying feature numbers. (D) Venn diagram illustrating the overlap of key genes identified by Random Forest, SVM, Lasso regression, and XGBoost. (E) Error rate of the SVM model using 5-fold cross-validation with varying feature numbers. (F) Changes in regression coefficients for different Log Lambda values in the Lasso regression model. (G) LASSO regression model performance with varying lambda values. (H) Gene importance scores determined by the XGBoost model.
Immune cell distribution
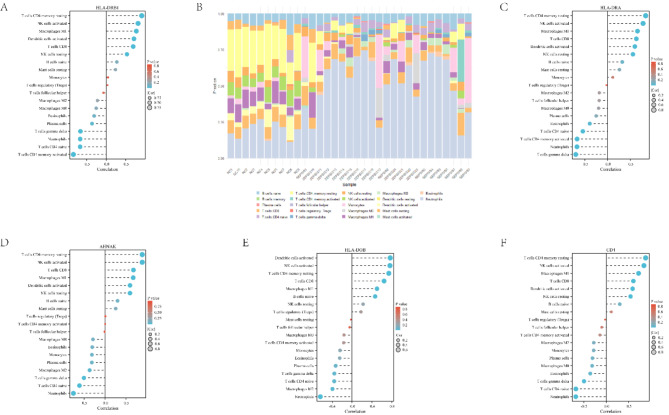
In order to explore the composition of different immune cells and their association with key genes in blood samples from sepsis patients, we performed an immune cell distribution analysis. By this method, we hope to reveal the interactions between immune cells and key genes involved in the pathological process of sepsis. Immune cell distribution revealed distinct differences in immune cell composition between sepsis and control groups (Fig. 5B). Figure 5A illustrates a positive correlation between the HLA-DRB1 gene and immune cell types, including T-cell CD4 memory quiescence, NK cell activation, and M1 macrophages. Figure 5C and D depict the relationships between HLA-DRA, AHNAK, and immune cell subsets. HLA-DRA exhibited positive associations with resting memory CD4 T cells, activated NK cells, and M1 macrophages, and negative associations with γδ T cells. AHNAK primarily positively correlated with resting memory CD4 T cells and activated NK cells. The associations between HLA-DOB, CD4, and immune cell types are presented in Fig. 5E and F, respectively. HLA-DOB exhibited strong positive correlations with dendritic cell and NK cell activation, while CD4 demonstrated positive associations with resting memory T cells and NK cell activation. These findings suggest that CD4, HLA-DOB, HLA-DRB, HLA-DRB1, and AHNAK are significantly associated with immune cell distribution and may play crucial roles in the immunopathogenesis of sepsis, warranting further investigation into the underlying immune mechanisms.
Fig. 5.
Immune cell distribution. (A) Bubble plot showing the correlation between HLA-DRB1 gene expression and immune cell type abundance. (B) Stacked bar plot illustrating the distribution of immune cell types in sepsis and control groups. (C) Scatter plot depicting the relationship between HLA-DRA gene expression and immune cell type abundance. (D) Bubble plot showing the correlation between AHNAK gene expression and immune cell type abundance. (E) Bubble plot showing the correlation between HLA-DOB gene expression and immune cell type abundance. (F) Bubble plot showing the correlation between CD4 gene expression and immune cell type abundance.
Expression of key genes and GSEA enrichment analysis
Fig. 6.
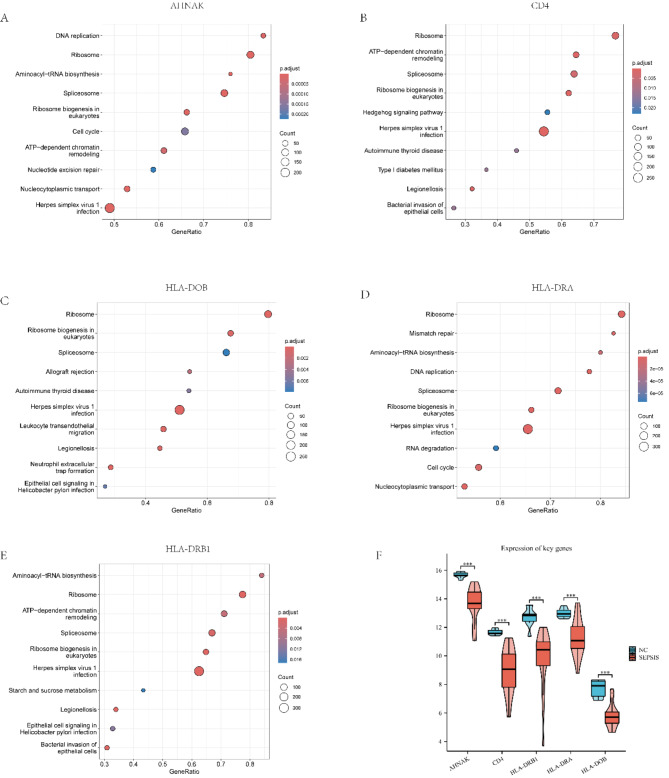
Gene Set Enrichment Analysis (GSEA) and gene expression. Figures A-E demonstrate the enrichment of key genes (AHNAK, CD4, HLA-DOB, HLA-DRA, HLA-DRB1) in different KEGG pathways. The horizontal coordinate is the GeneRatio, which indicates the proportion of target genes included in each pathway to the total number of genes, and a larger value indicates that the gene is more enriched in that pathway. The vertical coordinate is the name of the KEGG pathway enriched. The size of the point indicates the number of genes in the pathway (Count), the larger the point indicates that the pathway contains more relevant genes; the color shade indicates the corrected p-value (p.adjust), the darker the color indicates the higher the enrichment significance.(F) Violin plots comparing gene expression levels of key genes (AHNAK, CD4, HLA-DRA, HLA-DRB1, HLA-DOB) between normal control (NC) and sepsis (SEPSIS) groups. Red: sepsis group; blue: normal control group. *** indicates p-value < 0.001.
Construction and screening of diagnostic models
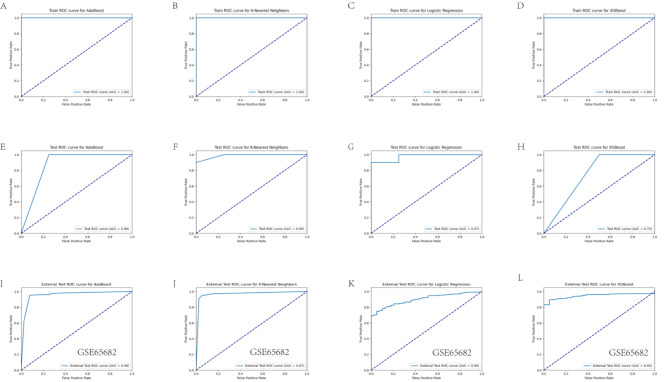
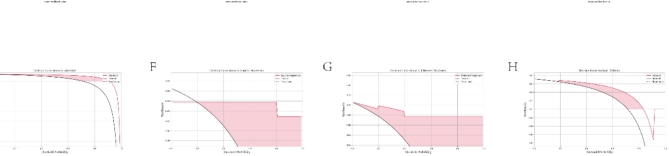
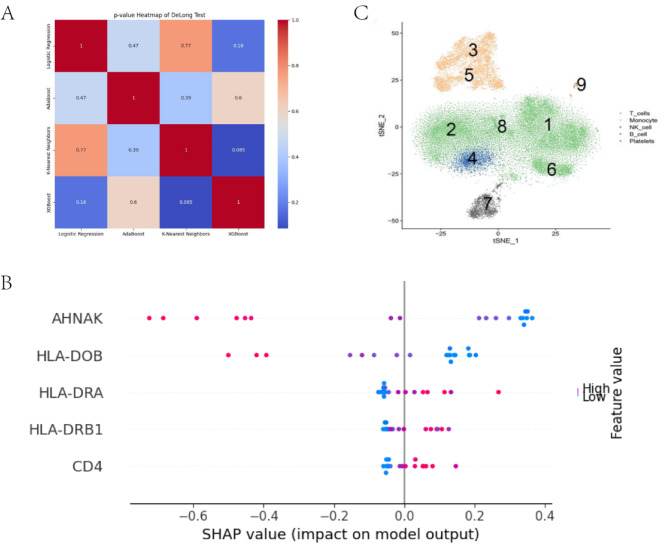
In order to achieve early diagnosis of sepsis, we constructed diagnostic models based on four machine learning algorithms (AdaBoost, KNN, Logistic regression and XGBoost). By training and screening these models, we expect to find the best model that can predict sepsis with high accuracy. We employed four machine learning algorithms (AdaBoost, KNN, Logistic Regression, and XGBoost) to identify the optimal diagnostic model for sepsis. Performance metrics for each model are summarized in Table 3. Model performance was evaluated using ROC curves and AUC values for training and test data. Figure 7A-D depict the ROC curves for each model on the training set, with all models achieving an AUC of 1.00, suggesting perfect performance on training data. Figure 7E-H show the ROC curves for each model on the test set. Here, the KNN model achieved the highest AUC of 0.99, while AdaBoost, Logistic Regression, and XGBoost achieved AUCs of 0.88, 0.97, and 0.75, respectively. The classification performance of four machine learning models was evaluated on the external validation set GSE65682. The results show that the ROC curves of these models on the external dataset perform differently, but generally exhibit high AUC values, indicating that the models have some differentiation ability. Models with AUC values close to 1 perform better, indicating that their predictions on the external validation set are better, thus validating the robustness and generalization ability of the models(Figs. 7I-L). To further assess model generalizability and reliability, calibration and decision curve analyses were performed (Fig. 8A-H). Calibration curves illustrate the agreement between predicted and actual probabilities. AdaBoost, KNN, and Logistic Regression models displayed closer agreement, while the XGBoost model showed slightly poorer calibration. Decision curve analysis evaluates the net benefit of using a model at different probability thresholds. AdaBoost, KNN, and Logistic Regression models displayed higher net benefit across most probability ranges than XGBoost. We further evaluated statistical differences in AUC values using the DeLong test (Fig. 9A). The results revealed significant differences (p < 0.05) between AdaBoost and KNN, KNN and Logistic Regression, as well as between Logistic Regression and XGBoost. Based on these analyses, the KNN model emerged as the optimal choice with the highest AUC (0.99) on the test set, superior calibration and decision curve characteristics, indicating strong predictive performance and generalizability. Therefore, the KNN model was identified as the most reliable tool for early sepsis diagnosis with high accuracy and stability. Figure 9B depicts the distribution of SHAP values for each key gene in the KNN model. SHAP values represent the importance and influence of each gene on the model’s output. The figure highlights AHNAK as the gene with the most significant influence on the model’s predictions. By constructing and screening diagnostic models, we aim to provide a reliable tool for the clinical diagnosis of sepsis, which will help to improve the efficiency of early diagnosis of sepsis and reduce the morbidity and mortality.
Table 3.
Bootstrap accuracy results.
| Model | Mean Accuracy | CI Lower Accuracy | CI Upper Accuracy |
|---|---|---|---|
| Logistic Regression | 0.885571429 | 0.642857143 | 1 |
| AdaBoost | 0.961642857 | 0.785714286 | 1 |
| KNN | 0.971214286 | 0.857142857 | 1 |
| XGBoost | 0.957857143 | 0.785714286 | 1 |
Fig. 7.
Receiver Operating Characteristic (ROC) curves. (A-D) ROC curves for Logistic Regression, AdaBoost, K-Nearest Neighbors (KNN), and XGBoost models on the training dataset. (E-H) ROC curves for the same models on the test dataset. Solid lines represent model performance, while dashed lines represent random classifier performance. Figures (I-L) show the ROC curves of the four machine learning models on the external validation set (GSE65682) for evaluating the classification performance of the models on the external dataset.
Fig. 8.
Model calibration and clinical utility. (A-D) Calibration curves for AdaBoost, Logistic Regression, K-Nearest Neighbors, and XGBoost models, assessing the agreement between predicted and observed probabilities. (E-H) Decision curves for AdaBoost, Logistic Regression, K-Nearest Neighbors, and XGBoost models, evaluating the net benefit of using each model at different probability thresholds.
Fig. 9.
Model evaluation and interpretation. (A) Heatmap illustrating p-values from the DeLong test comparing the AUC values of four machine learning models. (B) SHAP value plot visualizing the impact of significant genes on model predictions (red: high impact, blue: low impact). (C) Cell type identification: Cells 3 and 5 are macrophages, cell 4 is a natural killer cell, cells 1, 2, 6, and 8 are T cells, cell 7 is a B cell, and cell 9 is a platelet.
Single-cell sequencing
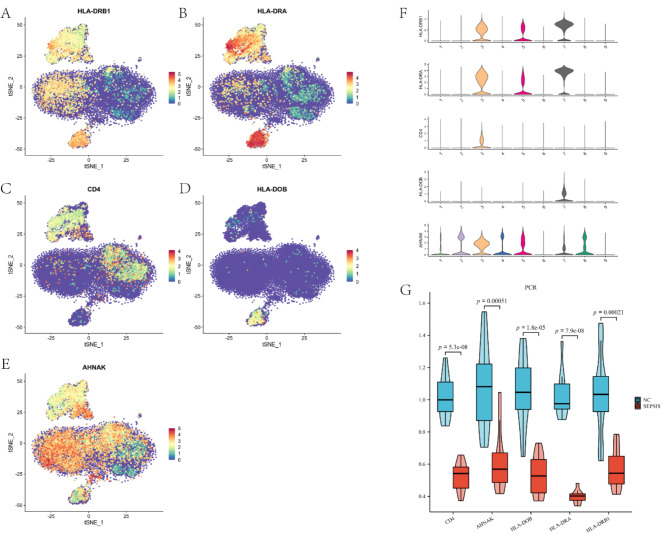
To understand the expression characteristics and cell type specificity of key genes in the peripheral blood of sepsis patients, we performed single-cell RNA sequencing analysis. With this analysis, we expect to determine the distribution of these key genes in different immune cells in order to reveal their specific roles in sepsis immunomodulation. We successfully processed five single-cell transcriptome sequencing samples. Following dimensionality reduction and clustering, nine cell groups were identified, comprising five distinct cell types: macrophages (clusters 3 and 5), natural killer cells (cluster 4), T cells (clusters 1, 2, 6, and 8), B cells (cluster 7), and platelets (cluster 9) (Fig. 9C). Single-cell sequencing data revealed a broad expression pattern for HLA-DRB1 and AHNAK across multiple immune cell types (Fig. 10A, E). In contrast, CD4 expression was predominantly observed in macrophages and T cells (Fig. 10C), while HLA-DOB and HLA-DRA were primarily expressed in macrophages and B cells (Fig. 10B, D). Violin plots illustrating the expression distribution of HLA-DRB1, HLA-DRA, CD4, HLA-DOB, and AHNAK across different immune cell types are presented in Fig. 10F. These findings indicate that the investigated key genes are actively expressed within various immune cell populations and may play critical roles in the pathophysiological mechanisms underlying sepsis. The results of the single-cell analysis complemented our overall RNA sequencing and machine learning results, revealing the distribution and expression levels of key genes in specific immune cell populations. This detailed cellular analysis helps validate the biomarkers we identified and provide a comprehensive understanding of the immune environment in sepsis, providing potential targets for future therapeutic interventions.
Fig. 10.
Single-cell gene expression and qPCR validation. (A-E) Single-cell gene expression patterns for key genes: HLA-DRB1 and AHNAK exhibit broad expression across multiple immune cell types, CD4 is primarily expressed in macrophages and T cells, while HLA-DOB and HLA-DRA are predominantly found in macrophages and B cells. (F) Violin plots illustrating gene expression distribution across different immune cell types. (G) Comparative gene expression analysis between sepsis and control groups, demonstrating significantly reduced expression of all key genes in the sepsis group (p < 0.001).
Validation of q-PCR experiments
To validate the expression patterns of key genes screened in sepsis patients, we performed qPCR experiments. With this experiment, we expect to confirm the down-regulation trend of the expression of these genes in sepsis patients to further support their potential diagnostic and therapeutic value in sepsis. Primer sequences are shown in Table 4. Gene expression was assessed in triplicate biological replicates, and statistical significance was determined using t-tests. qPCR analysis revealed significantly reduced mRNA levels of five key genes (CD4, AHNAK, HLA-DRB1, HLA-DRA, and HLA-DOB) in the sepsis group compared to the control group (p < 0.001 for all genes). These findings corroborate the previously observed gene expression patterns, further supporting the potential significance of these genes in sepsis pathogenesis(Fig. 10G).
Table 4.
Primer sequence.
| Primer Name | Sequence(5’-3’) | Product length |
|---|---|---|
| CD4-F | ATGCTGGCTCTGGAAACCTC | |
| CD4-R | CGAGACCTTTGCCTCCTTGT | 185 |
| HLA-DRB1-F | AGAGAAGACTGGGGTGGTGT | |
| HLA-DRB1-R | TCCGAGGAACTGTTTCCAGC | 89 |
| HLA-DOB-F | CCACTCCTGCACCAGCATAA | |
| HLA-DOB-R | CTGCCCATTCAGGAACCACT | 139 |
| AHNAK-F | GGGAGCGATGATGAGACAGG | |
| AHNAK-R | AAACTGACAGCTCCACCTCG | 90 |
| HLA-DRA-F | CCTGACCAATCAGGCGAGTT | |
| HLA-DRA-R | GTTGGCCAATGCACCTTGAG | 179 |
Discussion
This study aimed to elucidate the molecular underpinnings and critical genes implicated in sepsis through a comprehensive analysis of RNA sequencing, immune cell infiltration, machine learning, and single-cell sequencing data. Five key genes—CD4, HLA-DOB, HLA-DRB1, HLA-DRA, and AHNAK—were identified as significantly differentially expressed in sepsis patients. These genes were found to be integral components of signaling pathways associated with immune response and inflammation. Our findings suggest a strong association between the downregulation of these genes and immune dysfunction, as well as disease severity in sepsis. These results provide a foundation for further research exploring the potential of these genes as diagnostic and therapeutic targets for sepsis.
The CD4 gene exhibited significant downregulation in this study, particularly in qPCR validation, where its expression was markedly lower in the sepsis group compared to the normal control group (p < 0.001). CD4 protein is a critical marker for helper T cells, which play a pivotal role in the immune response. Through cytokine release and paracrine signaling, CD4+ T cells enhance antibody production by B cells and stimulate macrophage-mediated pathogen clearance21. immune cell distribution demonstrated a strong correlation between CD4 and resting memory CD4+ T cells. These findings suggest that CD4 downregulation may impair T helper cell function, leading to a weakened immune response and increased susceptibility to infection and organ dysfunction. Moreover, GSEA revealed the enrichment of CD4 within T-cell receptor signaling and antigen presentation pathways, emphasizing its crucial role in immune response. Single-cell sequencing confirmed the predominant expression of CD4 in macrophages and T cells, aligning with its established role in immune regulation.
HLA-DOB and HLA-DRB1 genes were also significantly downregulated in sepsis patients. Single-cell sequencing revealed predominant expression of HLA-DOB in macrophages and B cells, while HLA-DRB1 exhibited a broader distribution across multiple immune cell types. Both genes are integral to antigen presentation. HLA-DOB and HLA-DRB1 collaborate in the assembly and function of MHC class II molecules, facilitating the presentation of exogenous antigens to antigen-presenting cells and thereby initiating specific immune responses22;23. Downregulation of these genes may compromise antigen presentation, impairing the immune system’s ability to recognize and eliminate pathogens. immune cell distribution demonstrated a significant association between HLA-DRB1 and the inactivation of CD4 memory T cells, as well as the activation of NK cells and M1 macrophages, suggesting potential impacts on these cell types. Additionally, GSEA enrichment analysis confirmed the involvement of HLA-DOB and HLA-DRB1 in antigen processing, presentation, and immune system processes, reinforcing their critical role in immune regulation. qPCR analysis validated the significant downregulation of HLA-DOB and HLA-DRB1 in sepsis patients compared to healthy controls (p < 0.001), emphasizing their involvement in sepsis pathogenesis.
The HLA-DRA gene exhibited significant downregulation in sepsis patients, as confirmed by qPCR validation (p < 0.001). Single-cell sequencing localized HLA-DRA predominantly to macrophages and B cells. As a component of MHC class II molecules, HLA-DRA plays a critical role in antigen presentation. Research indicates that HLA-DRA enhances chemokine production (CCL5, CXCL9, CXCL10) within the tumor immune microenvironment, promoting an anti-tumor immune response by facilitating CD4+ and CD8+ T cell infiltration24. Furthermore, fluctuations in serum HLA-DRA levels can reflect the immune status in sepsis patients, with decreased levels potentially contributing to impaired T cell differentiation25. immune cell distribution revealed a strong association between HLA-DRA and the quiescence of CD4 memory T cells, the activation of NK cells, and the presence of M1 macrophages, suggesting its involvement in regulating these immune cell populations. GSEA confirmed the enrichment of HLA-DRA in immune system processes and antigen presentation pathways, emphasizing its critical role in immune function. These findings collectively suggest that HLA-DRA downregulation may profoundly impact immune function in sepsis patients.
The AHNAK gene exhibited significant downregulation in sepsis patients, as confirmed by qPCR validation (p < 0.001). Single-cell sequencing revealed a broad distribution of AHNAK expression across various immune cell types, with a particular abundance in macrophages and T cells. This gene encodes a large protein involved in crucial cellular processes such as cell signaling and cytoskeletal organization. Studies have demonstrated its critical role in calcium signaling and cytoskeletal remodeling26;27. Notably, the AHNAK gene was consistently identified by all four machine learning models employed, highlighting its potential as a diagnostic and prognostic marker for sepsis. immune cell distribution revealed a strong correlation between AHNAK expression and specific immune cell populations, including quiescent CD4 memory T cells and activated NK cells. These findings suggest that AHNAK plays a crucial role in modulating the function and structure of these immune cells. Furthermore, GSEA identified significant enrichment of AHNAK within pathways associated with cell signaling and cytoskeletal organization, suggesting a potential key role in cellular communication and maintaining structural integrity. The qPCR experiments confirmed the strong correlation between AHNAK expression and the quiescence of CD4 memory T cells and activation of NK cells observed in single-cell sequencing data. This qPCR validation further supports the downregulation of AHNAK in sepsis patients and suggests its potential involvement in disease progression.
This study investigated the expression and functional alterations of key genes in sepsis through a multi-faceted approach encompassing RNA sequencing, immune cell infiltration, machine learning, and single-cell sequencing. The downregulation of identified key genes was found to be associated with sepsis pathogenesis, suggesting their potential as diagnostic and prognostic biomarkers. In contrast to previous conventional studies, this study integrated multiple advanced techniques to provide a comprehensive analysis of gene expression and function. Previous studies have revealed significant genetic differences between sepsis patients and healthy controls through comparative gene expression analysis, and these genes have value as potential diagnostic and therapeutic targets. For example, one study identified the critical role of FYN and CD247 in sepsis through a bioinformatics approach and demonstrated an association between these genes and patient survival28. However, these analyses failed to completely elucidate the potential functions of these genes in the pathomechanism of sepsis and remain deficient in the in-depth functional exploration of gene interactions. Our findings not only corroborate existing knowledge regarding immunosuppression in sepsis but also elucidate the mechanistic roles of specific genes. The systematic integration of cutting-edge technologies employed in this study offers a novel perspective on the molecular underpinnings of sepsis and provides a robust foundation for future diagnostic and therapeutic strategies.
While this study employed a multi-faceted approach utilizing cutting-edge technologies to reveal the expression and functional changes of key genes in sepsis, some limitations warrant consideration. Firstly, the relatively small sample size employed may limit the generalizability of the findings and the statistical power to detect significant associations. In particular, it will bring a certain risk of bias to the training of machine learning models. In order to minimize the potential bias due to sample imbalance, we adopt data augmentation and stratified sampling strategies in machine learning model construction. Through these methods, we ensure that each machine learning algorithm can be adequately trained based on its own features, and maintain a reasonable ratio between the training and test set division. Secondly, the study primarily relied on RNA sequencing and immune infiltration analyses. These techniques, while valuable, do not fully elucidate the intricate mechanisms of gene regulation and protein expression in sepsis. To definitively establish the clinical significance of the identified key genes, comprehensive multi-omics analyses encompassing DNA methylation, protein-protein interactions, and metabolomics data, along with large-scale clinical trials, are necessary.DESeq2 and limma methods each have their own characteristics in terms of their applicability to differential expression analysis.DESeq2 has an advantage in correcting for between-sample variability, making it more reliable in calculating p-values for small sample data. However, as can be seen from the results of our comparisons, DESeq2 is slightly low in Fold Change (FC) estimation, e.g., the log2 FC of HLA-DRB1 is -2.87 in DESeq2, while it is -2.88 in limma. limma, by contrast, is more stable in FC estimation, which makes it particularly suitable for small-sample RNA sequencing data. To ensure the robustness of the results of this study, we retained the p-value calculations of DESeq2 and combined them with the FC estimation of limma for a more comprehensive assessment of the differential expression characteristics of key genes. Additionally, efforts to integrate these findings into the existing diagnostic framework for sepsis hold promise for improved patient outcomes.
Acknowledgements
We thank Home for Researchers editorial team (www.home-for-researchers.com) for language editing service.
Author contributions
The primary manuscript text was authored by H.W and L.L, while Figs were prepared by L.H and Y.H.The manuscript was reviewed by all authors. The study was approved for publication by all authors.
Funding
Key Clinical Specialty Construction Project of Sichuan Province. Sichuan Provincial Health and Health Committee Science and Technology Project (number 23LCYJ001).
Data availability
The datasets used in this study are stored in the CNGBdb database and can be accessed via the following link: https://db.cngb.org/search/project/CNP0002611/. The datasets are publicly available and do not require special permissions for access.
Declarations
Competing interests
The authors declare no competing interests.
Approval of human ethics and agreement to participate
Each patient and their family members voluntarily participated in this study and signed an informed consent form.Approval for the study was granted by the Ethics Committee at the Affiliated Hospital of Southwest Medical University (No.1. ky2018029), Clinical Trial No.:ChiCTR1900021261,Registration Date: February 4, 2019.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Huang, M., Cai, S. & Su, J. The pathogenesis of Sepsis and potential therapeutic targets. Int. J. Mol. Sci.20 (2019). [DOI] [PMC free article] [PubMed]
- 2.Arora, J., Mendelson, A. A., Fox-Robichaud, A. & Sepsis Network Pathophysiology and implications for early diagnosis. Am. J. Physiol. -Regul Integr. Comp. Physiol.324, R613–R624 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozsolak, F. & Milos, P. M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet.12, 87–98 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delano, M. J. & Ward, P. A. The immune system’s role in sepsis progression, resolution, and long-term outcome. Immunol. Rev.274, 330–353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conway-Morris, A. & Wilson, J. & Shankar-Hari, M. Immune activation in sepsis. Crit. Care Clin.34, 29–42 (2018). [DOI] [PubMed]
- 6.Greener, J. G., Kandathil, S. M., Moffat, L. & Jones, D. T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell. Biol.23, 40–55 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Ge, S. X., Son, E. W. & Yao, R. I. D. E. P. An Integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform.19, 524–534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharya, S. et al. Toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data. 5, 180015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu, W. et al. Announcing the launch of protein data bank China as an associate member of the Worldwide Protein Data Bank Partnership. Acta Crystallogr. Sect. D-Struct Biol.79, 792–795 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, J. & Szymczak, S. A. Review on longitudinal data analysis with random forest. Brief. Bioinform24 (2023). [DOI] [PMC free article] [PubMed]
- 11.Huang, S. et al. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteom.15, 41–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, C. & Hua, Z. C. Lasso peptides: Heterologous production and potential medical application. Front. Bioeng. Biotechnol.8, 571165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou, N. et al. Predicting 30-days mortality for MIMIC-III patients with Sepsis-3: A machine learning approach using XGboost. J. Transl. Med.18, 462 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steen, C. B., Liu, C. L., Alizadeh, A. A. & Newman, A. M. Profiling cell type abundance and expression in bulk tissues with CIBERSORTx. Methods Mol. Biol.2117, 135–157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu, Z. et al. Bioinformatic validation and machine learning-based exploration of purine metabolism-related gene signatures in the context of immunotherapeutic strategies for nonspecific orbital inflammation. Front. Immunol.15 (2024). [DOI] [PMC free article] [PubMed]
- 16.Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song, X., Liu, X., Liu, F. & Wang, C. Comparison of machine learning and logistic regression models in predicting acute kidney injury: A systematic review and meta-analysis. Int. J. Med. Inf.151, 104484 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Keshvari, S., Farizhendi, S. A., Ghiasi, M. M. & Mohammadi, A. H. AdaBoost metalearning methodology for modeling the incipient dissociation conditions of clathrate hydrates. ACS Omega. 6, 26919–26931 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ukey, N. et al. Survey on exact KNN queries over high-dimensional data space. Sensors23 (2023). [DOI] [PMC free article] [PubMed]
- 20.Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol.20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruterbusch, M., Pruner, K. B., Shehata, L. & Pepper, M. In vivo CD4(+) T cell differentiation and function: Revisiting the Th1/Th2 paradigm. Annu. Rev. Immunol.38, 705–725 (2020). [DOI] [PubMed]
- 22.Furukawa, H. et al. The role of common protective alleles HLA-DRB1*13 among systemic autoimmune diseases. Genes Immun.18, 1–7 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Nagarajan, U. M. et al. Class II transactivator is required for maximal expression of HLA-DOB in B cells. J. Immunol.168, 1780–1786 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Wang, B., Liu, Y., Xiong, F. & Wang, C. Improved immunotherapy outcomes via cuproptosis upregulation of HLA-DRA expression: Promoting the aggregation of CD4(+) and CD8(+)T lymphocytes in clear cell renal cell carcinoma. Pharmaceuticals17 (2024). [DOI] [PMC free article] [PubMed]
- 25.Xu, J. et al. Dynamic changes in human HLA-DRA gene expression and th cell subsets in sepsis: Indications of immunosuppression and associated outcomes. Scand. J. Immunol.91, e12813 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Haase, H. Ahnak, a new player in beta-adrenergic regulation of the cardiac L-type Ca2 + channel. Cardiovasc. Res.73, 19–25 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Sundararaj, S., Ravindran, A. & Casarotto, M. G. AHNAK: The quiet giant in calcium homeostasis. Cell. Calcium. 96, 102403 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Jiang, Y. et al. FYN and CD247: Key genes for septic shock based on bioinformatics and meta-analysis. Comb. Chem. High. Throughput Screen.25, 1722–1730 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used in this study are stored in the CNGBdb database and can be accessed via the following link: https://db.cngb.org/search/project/CNP0002611/. The datasets are publicly available and do not require special permissions for access.