Abstract
Sequence analysis of bacterial genomes revealed a novel DNA-binding domain. This domain is found in several response regulators of the two-component signal transduction system, such as Pseudomonas aeruginosa AlgR, involved in the regulation of alginate biosynthesis and in the pathogenesis of cystic fibrosis; Clostridium perfringens VirR, a regulator of virulence factors, and in several regulators of bacteriocin biosynthesis, previously unified in the AgrA/ComE family. Most of the transcriptional regulators that contain this DNA-binding domain are involved in biosynthesis of extracellular polysaccharides, fimbriation, expression of exoproteins, including toxins, and quorum sensing. We refer to it as the LytTR (‘litter’) domain, after Bacillus subtilis LytT and Staphylococcus aureus LytR response regulators, involved in regulation of cell autolysis. In addition to response regulators, the LytTR domain is found in combination with MHYT, PAS and other sensor domains.
INTRODUCTION
Response regulators of the microbial two-component signal transduction systems consist of an N-terminal CheY-like receiver domain and a C-terminal output domain, typically a DNA-binding helix–turn–helix (HTH) or winged helix domain (1–3). In response to an environmental stimulus, a phosphoryl group is transferred from the sensor histidine kinase to an Asp residue in the CheY-like receiver domain of the cognate response regulator. Although the CheY-like domains do not bind DNA by themselves, phosphorylation of these domains induces conformational changes that affect the binding of the associated HTH domains to their recognition sites on the chromosomal DNA. Phosphorylation-induced conformational changes in the response regulator molecule have been demonstrated in direct structural studies (4,5).
Recent studies, including analyses of the response regulators encoded in complete bacterial genomes, illuminated a previously unseen diversity of their output domains. The HTH domains were found to fall into two main categories: NarL/FixJ family regulators contain a single HTH motif in the middle of a four-helix bundle (6), whereas regulators of the AraC/XylS family contain two HTH motifs separated by a linker helix and are usually flanked by two additional α-helices (7). A majority of known response regulators, including the well studied OmpR and PhoB proteins, were found to contain an alternative DNA-binding domain, referred to as ‘winged helix’ or ‘winged HTH’ (8). In addition, response regulators can contain other DNA-binding domains (1,9–11), as well as alternative output domains, such as AAA-type ATPase, methylesterase, GGDEF, EAL and HD-GYP domains, which do not bind DNA (1–3,12). Finally, alternative output domains can be combined with the DNA-binding domains, e.g. in the NtrC family of transcriptional regulators (2,4).
A detailed study of 13 response regulators encoded in the Streptococcus pneumoniae genome found a CheY-HTH domain organization in four of them; seven more had an OmpR-like winged helix DNA-binding domain (13). The two remaining response regulators, AgrA and BlpR, were found to have an unusual C-terminal DNA-binding domain, specific for the AgrA/ComE family (13). The importance of this domain was underscored by the observation that BlpR appeared to be the only response regulator essential for growth of S.pneumoniae cells (13,14).
Here, we present sequence analysis of this unusual DNA-binding domain, which shows no significant similarity to the HTH or winged-helix domains found in the previously characterized response regulators. Bacterial response regulators containing this novel DNA-binding domain comprise a well defined family of proteins that regulate production of important virulence factors, such as toxins, bacteriocins and extracellular polysaccharides.
MATERIALS AND METHODS
Initial similarity searches against the NCBI non-redundant protein database were performed using the PSI-BLAST program run to convergence with S.pneumoniae BlpS (NCBI protein database entry CAC03514.1) or the cytoplasmic portion of Oligotropha carboxidovorans CoxC (NCBI protein database entry CAB76400.1) as queries. C-terminal DNA-binding domains of various response regulators identified in these initial searches were used as queries for subsequent PSI-BLAST searches with varying parameters (because of the short size of the query proteins, to improve the sensitivity of the search, the expect value threshold was increased to 100 or more, the word size was decreased to 2, and the inclusion threshold was changed to 0.02) and for non-iterated BLAST searches against finished and unfinished microbial genome sequences (http://www.ncbi.nlm.nih.gov/BLAST). The multiple alignment of the representative sequences, retrieved in these searches, was generated by PSI-BLAST, followed by minimal manual editing. A major part of the LytTR domain (missing the first one or two β-strands) is listed in the ProDom database (http://prodes.toulouse.inra.fr/prodom/doc/prodom.html) as domain PD032235. Response regulators containing the LytTR domain comprise COG3279 in the COG database (http://www.ncbi.nlm.nih.gov/COG/).
Secondary structure predictions were performed using the full set of programs available at the PredictProtein (http://cubic.bioc.columbia.edu/predictprotein/), JPred (http://jpred.ebi.ac.uk/) and UCSC HMM (http://www.cse.ucsc.edu/research/compbio/) web servers. The pI values were calculated using the Compute pI/Mw tool at the ExPASy web server (http://www.expasy.org/).
The consensus sequence of the LytTR-binding site was deduced from a library of the upstream regulatory DNA sequences that have been experimentally demonstrated to bind response regulators with the LytTR domain. This library included AlgR-binding sites RB1, RB2 and RB3 for Pseudomonas aeruginosa algD gene (15,16) and the AlgR-binding site of the algC gene; sequences upstream of Lactobacillus plantarum plnABCD, plnEFI and plnJKLR operons that bind PlnC and PlnD (17); Lactobacillus sake sppA, sppIP, sppT_1 and sppT_3 probes that bind SppR, PlnC and PlnD (18); probes DR1, DR2 and DR3 with sequences upstream of S.pneumoniae comAB, comCDE and orf12 genes that bind ComE (19); and two probes covering the upstream region of Clostridium perfringens pfoA gene that binds VirR (20). With this library as an input, the SignalX program (21), a module of the GenomeExplorer program suite (22), was used to generate the profiles of monomeric and tandem LytTR-binding sites and to use these profiles to search for the likely LytTR-binding sites in selected DNA sequences.
RESULTS
LytTR, a novel conserved DNA-binding domain
PSI-BLAST searches of the NCBI non-redundant protein sequence database using the BlpS protein of S.pneumoniae as a query converged after five to eight iterations (depending on the search parameters used), retrieving a set of 94 proteins (as of January 1, 2002) with several characteristic sequence motifs (Fig. 1). Most of those hits comprised C-terminal portions of bacterial response regulators, including such well characterized ones as P.aeruginosa AlgR, Staphylococcus aureus AgrA and LytR, S.pneumoniae ComE and C.perfringens VirR (Table 1). Since these response regulators all contain N-terminal CheY-like domains, which do not interact directly with DNA, their C-terminal parts must be responsible for their well documented binding to specific DNA sequences in the upstream regions of target genes (9,10,15–20). Using the C-terminal domains of any of these response regulators as PSI-BLAST queries each time retrieved the same set of 94 proteins, indicating that these domains form a tight family, unrelated to any other DNA-binding domains. Because of a certain confusion with the nomenclature of individual proteins of this family, we refer to this conserved DNA-binding domain found in Bacillus subtilis protein LytT and S.aureus transcriptional regulator LytR as LytTR (‘litter’) domain, in line with the designation of the Staphylococcus lugdunensis synergistic hemolysin locus as slush (23). [In addition to the S.aureus LytR protein, the name LytR is also used for another group of transcriptional regulators (e.g. B.subtilis LytR), which are unrelated to members of the LytTR family (see COG1316 in the COG database, http://www.ncbi.nlm.nih.gov/COG/). Likewise, none of the proteins encoded in the comE operon of B.subtilis (ComEA, ComEB or ComEC) are related to the streptococcal ComE proteins or belongs to the LytTR family. Finally, the VirR protein from Agrobacterium tumefaciens is also unrelated to the LytTR family proteins with the same name. Instead, it belongs to a large family of uncharacterized ‘VirR-related’ proteins that comprise COG1720 in the COG database and UPF0066 in PROSITE, http://www.expasy.org/prosite/.] Accordingly, we refer to transcriptional regulators that contain the LytTR domain (Table 2) as the LytTR family proteins.
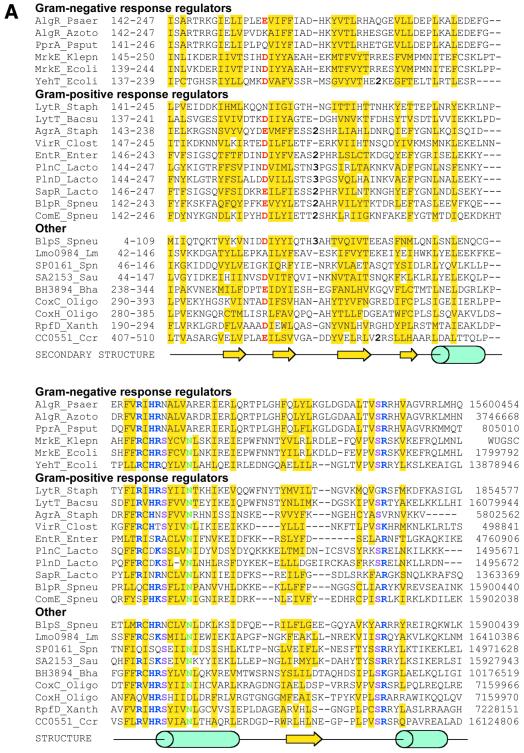
Figure 1.
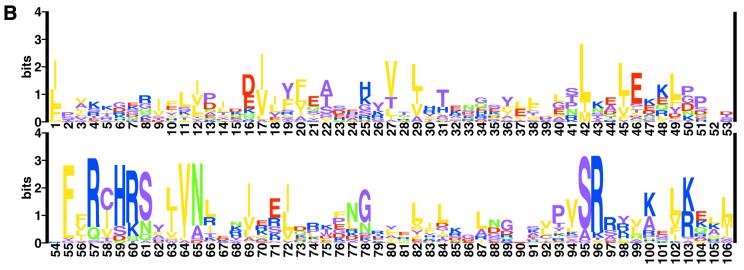
(Opposite) Sequence alignment of the LytTR-type DNA-binding domains. (A) LytTR-type domains were aligned on the basis of the PSI-BLAST outputs. The sequence of K.pneumoniae MrkE protein [SWISS-PROT accession no. P21649 (32)] has been modified according to the genomic data from the Washington University K. pneumoniae Genome Sequencing Project. Protein names, species abbreviations and the GenBank accession nos are as in Tables 1 and 2; the last column shows the protein ‘gi’ numbers in the NCBI protein database. Conserved amino acid residues that could be involved in DNA binding are shown in bold and colored as follows: blue, Arg, Lys, His; red, Asp, Glu; green, Asn and Gln; purple, other conserved residues. Yellow shading indicates conserved uncharged amino acid residues. The predicted secondary structure is a combination of the assignments produced by the PhD (33) and SAM-T99 (34) programs. (B) Consensus sequence of the LytTR domain, drawn from a multiple alignment of the 96 LytTR domains using the SeqLogo program (35) in the WWW-based implementation by S. E. Brenner (http://www.bio.cam.ac.uk/seqlogo). The first residue corresponds to Leu127 of the P.aeruginosa AlgR protein. The height of each letter indicates the degree of its conservation, the total height of each column reflects the statistical importance of the given position. The coloring is as in (A).
Table 1. Properties of the response regulators of the LytTR familya.
| Organism | Protein name | GenBank accession no. | Regulated processes (genes/operons) |
|---|---|---|---|
| Biosynthesis of extracellular polysaccharides | |||
| Pseudomonas aeruginosa | AlgR | AE004938 | Alginate biosynthesis (algC, algD), twitching motility |
| Azotobacter vinelandii | AlgR | AF077237 | Alginate biosynthesis, encystment |
| Pseudomonas syringae | AlgR1 | AF131199 | Alginate biosynthesis |
| Fimbrial expression | |||
| Klebsiella pneumoniae | MrkE | M55912 | Expression of type 3 fimbriae |
| Peptidoglycan turnover | |||
| Staphylococcus aureus | LytR | L42945 | Autolysis (lrgAB) |
| Virulence | |||
| Clostridium perfringens | VirR | D14877 | Production of perfringolysin O (pfoA), collagenase, hemagglutinin |
| Quorum-sensing systems involving autoinducing peptides | |||
| Virulence | |||
| Staphylococcus aureus | AgrA | M21854 | Expression of toxins, hemolysins, staphylo-kinase, other secreted proteins (vir factors) |
| Staphylococcus epidermidis | AgrA | Z49220 | Expression of exoproteins (vir factors) |
| Competence | |||
| Streptococcus pneumoniae | ComE | U76218 | Competence (comCDE, comAB, natAB)b |
| Streptococcus gordonii | ComE | X98109 | Competence |
| Streptococcus mitis | ComE | AJ000871 | Competence |
| Biosynthesis of bacteriocins | |||
| Carnobacterium piscicola | CbaR | AF207838 | Carnobacteriocin A production |
| Enterococcus faecium | EntR | AF099088 | Enterocin A production |
| Lactobacillus plantarum | PlnC | X94434 | Plantaricin A production (plnABCD, plnEFI, plnJKLR) |
| PlnD | X94434 | ||
| Lactobacillus plantarum | PlsR | Y15127 | Plantaricin S production |
| Lactobacillus sakei | SppR | Z48542 | Sakacin P production (sppA, sppIPKR, sppTE) |
| Lactobacillus sakei | SapR | Z46867 | Sakacin A production (sapA, sapK) |
| Staphylococcus warneri | ORF1 | AB034941 | Nukacin ISK-1 production |
| Streptococcus pneumoniae | BlpR (RR13) | AJ276410 | Bacteriocin-like peptide production, essential for survival |
aAn expanded version of this table, which includes additional proteins, accession numbers and references, is available as Supplementary Material.
bSequence analysis of the S.pneumoniae orf12 operon showed that these genes are homologs of B.subtilis natAB genes, encoding a Na+-transporting ATPase (data not shown).
Table 2. Domain organization of various LytTR-containing proteins.
| Domain organizationa | Total no. of proteinsb | Examples [organism, protein name (accession no.)]c |
|---|---|---|
| Cytoplasmic | ||
| CheY-LytTR | 76 | Escherichia coli MrkE (BAA16251), YehT (P33356), Bacillus subtilis LytT (CAB14852), those listed in Table 1 |
| LytTR only | 2 | Streptococcus pneumoniae BlpS (AAK74683), Pseudomonas phage D3 Orf50 (AAF80809) |
| LMO-LytTR | 5 | Lactococcus lactis L121252 (AAK04633), Listeria innocua Lin0983 (CAC96214), Listeria monocytogenes Lmo0984 (CAC99062), Staphylococcus aureus SA2153 (BAB43455), Streptococcus pneumoniae SP0161 (AAK74343), Streptococcus pneumoniae SP1915 (AAK75983) |
| ABC-LytTR | 1 | Bacillus halodurans BH3894 (AP001520) |
| PAS-LytTR | – | Burkholderia cepaciad, Geobacter sulfurreducensd |
| Membrane-bound | ||
| MHYT-LytTR | 2 | Oligotropha carboxidovorans CoxC (CAB76246), CoxH (CAB76400) |
| MHYE-LytTR | 3 | Xanthomonas campestris RpfD (CAB77034), Caulobacter crescentus CC1610 (AAK23589), Mesorhizobium loti mll0891 (BAB48381) |
| 3TM-LytTR | 1 | Caulobacter crescentus CC0295 (AAK22282) |
| 4TM-LytTR | 3 | Agrobacterium tumefaciens AGR_C_3954p (AAK87922), Caulobacter crescentus CC0330 (AAK22317), CC3036 (AAK24998) |
| 8TM-LytTR | 1 | Caulobacter crescentus CC0551 (AAK22538) |
aABC, the ATP-binding domain of ABC-type transporters (ATP-binding cassette); LMO, an uncharacterized 40 amino acid domain with a coiled-coil region (Fig. S2A in the Supplementary Material); MHYE, a conserved membrane-associated domain (Fig. S2B in the Supplementary Material); 3TM, 4TM and 8TM, uncharacterized domains with, respectively, three, four and eight predicted transmembrane segments. PAS and MHYT domains have been described earlier (25,36).
bThe total number of proteins with the given domain organization in the NCBI protein database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Protein) as of January 1, 2002. Many of those proteins are copies of the same protein sequenced from different species or even different strains of the same species.
cAccession numbers for the NCBI, EBI and DDBJ protein databases.
dUnfinished genome sequences from the DOE Joint Genome Institute (B.cepacia) and The Institute for Genomic Research (G.sulfurreducens).
Sequence analysis of the LytTR domain indicates that it is most likely composed of four β-strands, followed by two α-helices, a β-strand and one more α-helix (Fig. 1A). The most conserved sequence motif FhRhH[RK][SNQ]hhVN (where ‘h’ indicates a hydrophobic amino acid residue) is located at the beginning of the second α-helix in the region that could potentially comprise a HTH motif (Fig. 1A and B). However, the variable distance between the two α-helices in different proteins and a lack of sequence conservation in this spacer region (Fig. 1A), argue against its role in DNA binding. In addition, some of the programs used predict, in this region, a short β-strand, while others predict a single long α-helix, which makes the structural assignment in this region somewhat questionable. In any case, the predicted secondary structure of the LytTR domain is not similar to any protein with known three-dimensional structure, indicating that it comprises a novel type of a DNA-binding domain.
The C-terminus of the LytTR domain contains a cluster of Lys and Arg residues (Fig. 1B) that confer strong positive charge to the whole domain, bringing its pI into the 9.0–11.0 range. This positive charge has been previously proposed to play a role in the DNA binding by the response regulators of the AgrA/ComE family (10). However, the LytTR domain appears to bind DNA in a sequence-specific fashion, interacting with direct repeats of 10–12 bp with a relatively well defined consensus sequence (see below). In addition, there are several exceptions, including the B.subtilis LytT protein whose LytTR domain has a pI of 6.5. These observations argue against direct involvement of those Lys and Arg residues in DNA binding by the LytTR domain.
Alternative domain combinations involving LytTR domain
In addition to numerous response regulators with CheY-LytTR domain organization, the LytTR domain has been found in other contexts (Table 2). The deep-water marine bacterium Bacillus halodurans encodes a unique protein, combining an ATP-binding domain of ABC-type transporter family with a C-terminal LytTR domain. Such a domain combination has not been seen elsewhere and the regulatory signal transmitted by this protein remains enigmatic.
In CoxC and CoxH proteins from the CO-metabolizing bacterium Oligotropha carboxidovorans (24), the LytTR domain can be found in association with MHYT, a recently described sensor domain, which contains six transmembrane segments carrying conserved His and Met residues (25). The coxC and coxH genes flank the operon encoding the CO dehydrogenase and are involved in transcriptional regulaton of the autotrophic metabolism in O.carboxidovorans (24). A LytTR domain-containing protein is also encoded upstream of the CO dehydrogenase-encoding genes in Hydrogenophaga pseudoflava (data not shown).
Several Gram-positive bacteria combine the LytTR domain with a short (∼40 amino acids) poorly conserved N-terminal domain (see Table 2, and Fig. S2A in the Supplementary Material), dubbed LMO (‘elmo’). These proteins are likely to be transcriptional regulators whose binding to DNA is controlled by protein–protein interactions involving the LMO domain.
The RpfD protein from the plant pathogen Xanthomonas campestris (26) combines the LytTR domain with an N-terminal membrane-bound domain that consists of three predicted transmembrane segments with conserved Glu, Asp, His and Tyr residues (see Fig. S2B in the Supplementary Material). The membrane topology of this domain is somewhat similar to that of the MHYT domain (25), suggesting that it, too, might be membrane-bound and metal-binding. We refer to it as the MHYE domain. Although the rpfD gene is located in the rpf (regulation of pathogenicity factors) locus of X.campestris, mutants with transposon insertion in the rpfD gene did not exhibit any discernible phenotype (26), just as MHYT-less mutants of B.subtilis (25). While the function of this protein still remains unknown, it is worth noting that very similar proteins are encoded in the genomes of Caulobacter crescentus and Mesorhizobium loti (Table 2).
The binding site for the LytTR domain
Studies of the DNA binding by response regulators of the AgrA family established that these proteins bind to imperfect direct repeats of the sequence pattern [TA][AC][CA]GTTN[AG][TG] separated by a 12–13 bp spacer (10,14,17–19). Remarkably similar binding sites were also identified in front of AlgR- and VirR-regulated promoters (15,20). Using an alignment of the experimentally characterized LytTR-binding sites in a profile-based search with the SignalX program (21) identified potential LytTR-binding sites in the upstream region of the S.aureus lrgAB operon (27) and in front of coxC, coxD and coxH genes (24) of the O.carboxidovorans cox gene cluster (data not shown). One of the most interesting findings from these searches was the prediction of the fourth LytTR-binding site in the near-upstream region of the P.aeruginosa algD gene. This site (–71 TCCGTTTGA –79) is located next to the previously identified RB3 AlgR-binding site (16) and can actually be seen as the second AlgR-protected DNA region on figure 3 in Mohr et al. (16). The adjacency of these two AlgR-binding sites suggest that they might function together in binding an AlgR dimer in a manner similar to that of other LytTR-containing response regulators. A detailed analysis of the potential LytTR-binding sites in completely sequenced bacterial genomes will be reported elsewhere.
DISCUSSION
The LytTR family of response regulators
Involvement of the transcriptional regulators that contain the LytTR domain in the pathogenesis of cystic fibrosis (P.aeruginosa AlgR), toxic shock syndrome (S.aureus AgrA), gas gangrene (C.perfringens VirR) and other deadly diseases has ensured sustained interest in their structure and function. However, a detailed analysis of these proteins has been hampered by the remarkable sequence conservation of their N-terminal CheY domain, which largely masked (dis)similarities of the DNA-binding domains of various response regulators. When Shimizu et al. (28) sequenced C.perfringens virR gene, they noticed the similarity of its product to MrkE, AlgR and AgrA response regulators, but paid most attention to their common N-terminal CheY domains. Likewise, Brunskill and Bayles (29) aligned S.aureus LytR with YehT, AgrA, AlgR and MrkE, but limited that alignment to the CheY portion of the protein. Nevertheless, several previous studies reported the lack of sequence similarity of the LytTR domain to the HTH or winged-helix DNA-binding domains and succeeded in assigning LytTR-containing response regulators to two families. On one hand, Whitchurch et al. (30) linked AlgR with YehT and LytR in a new family of response regulators and Mizuno (11) identified Escherichia coli YehT and MrkE (o244) as response regulators with unique output domains. On the other hand, Pestova et al. (9) identified the S.pneumoniae ComE response regulator and demonstrated its similarity to the quorum-sensing regulators AgrA, CbnR, PlnC and PlnD and to the C.perfringens VirR. Likewise, Grebe and Stock (1) assigned AlgR, LytR, LytT and MrkE to the RC3 group of response regulators with the LytR-type output domain; in contrast, AgrA, CbnR, ComE, PlnC, PlnD, PlsR and SppR were assigned to the RD group of response regulators with the ComE-type output domain. Here, we have unified the AlgR/LytR and AgrA/ComE families into one and identified other proteins with the LytTR domain that are encoded in complete microbial genomes.
Phylogenetic distribution of the LytTR domain
Although transcriptional regulators with the LytTR domain are widely distributed among the bacteria of the Bacillus/Clostridium group (low G+C Gram-positive bacteria) and are found, albeit usually in just one copy, in many proteobacteria, they are conspicuously missing in other groups of Gram-negative bacteria, in archaea and in eukaryotes (see Table S3 in the Supplementary Material). Although the LytTR domain is missing in Mycobacterium tuberculosis and Mycobacterium leprae, response regulators with this domain are encoded in other actinobacteria (high G+C Gram-positive bacteria), such as Streptomyces coelicolor and Rhodococcus ruber, as well as in the unfinished genome of Mycobacterium smegmatis (data not shown). This skewed phylogenetic distribution generally correlates with the genome size, as only those proteobacteria with genomes >2 MB (i.e. encoding at least 2000 proteins) encode a LytTR domain. Thus, although LytTR-containing transcriptional regulators play important roles in regulating virulence factors, this domain appears to be more typical for free-living bacteria and non-obligate parasites that tend to have relatively large genomes. This correlates with the involvement of LytTR-containing transcriptional regulators in complex behavioral responses, such as quorum sensing (9,18).
It is worth noting that in α-proteobacteria, the LytTR domain is found in combinations with various membrane-bound domains, including all five instances of this domain in C.crescentus (Table 2). Transcriptional regulation mediated by these proteins should occur only when the corresponding target genes are positioned in close vicinity to the cell membrane and may account for the temporary expression of those genes, which could be particularly important for the complex cell cycle of C.crescentus.
Is LytTR a HTH domain?
As discussed above, the LytTR domain consists of three (predicted) α-helices and four (predicted) β-strands with a potential additional short β-strand between the first and second α-helices (Fig. 1A). Remarkably, some of the available structure prediction programs predict that the conserved F[FYVL][RQ][CIV] motif forms an additional β-strand, while others predict that these residues belong to very long α-helix, extending into the α-helix shown in Figure 1. Thus, although different programs disagree on the exact structure of this region, they all consider a loop (or turn) structure in that region unlikely. A possibility of helix-to-strand conversion has been reported in the DNA-binding region of the transcriptional regulator Fis (31). In any case, this region has no apparent similarity to the turn regions of the classical HTH motifs and can be confidently predicted to comprise a novel type of DNA-binding domain. Of course, all secondary structure assignments discussed above are just predictions that need to be verified by determining the three-dimensional structure of the LytTR domain (currently in progress). Knowledge of the LytTR structure should also help in refining its binding site, shedding some light on the range of cellular functions regulated by this remarkable system.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Eugene Koonin and Igor Rogozin for helpful suggestions, Mikhail Gelfand and Andrey Mironov for letting us use an updated version of their SignalX program, and Chester Price for critically reading the manuscript. We acknowledge the availability of unfinished genome sequences from the Institute for Genomic Research (Geobacter sulfurreducens), Washington University (Klebsiella pneumoniae) and the DOE Joint Genome Institute (Burkholderia cepacia).
REFERENCES
- 1.Grebe T.W. and Stock,J.B. (1999) The histidine protein kinase superfamily. Adv. Microb. Physiol., 41, 139–227. [DOI] [PubMed] [Google Scholar]
- 2.Stock A.M., Robinson,V.L. and Goudreau,P.N. (2000) Two-component signal transduction. Annu. Rev. Biochem., 69, 183–215. [DOI] [PubMed] [Google Scholar]
- 3.West A.H. and Stock,A.M. (2001) Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci., 26, 369–376. [DOI] [PubMed] [Google Scholar]
- 4.Lee J., Owens,J.T., Hwang,I., Meares,C. and Kustu,S. (2000) Phosphorylation-induced signal propagation in the response regulator NtrC. J. Bacteriol., 182, 5188–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis R.J., Scott,D.J., Brannigan,J.A., Ladds,J.C., Cervin,M.A., Spiegelman,G.B., Hoggett,J.G., Barak,I. and Wilkinson,A.J. (2002) Dimer formation and transcription activation in the sporulation response regulator Spo0A. J. Mol. Biol., 316, 235–245. [DOI] [PubMed] [Google Scholar]
- 6.Baikalov I., Schroder,I., Kaczor-Grzeskowiak,M., Grzeskowiak,K., Gunsalus,R.P. and Dickerson,R.E. (1996) Structure of the Escherichia coli response regulator NarL. Biochemistry, 35, 11053–110561. [DOI] [PubMed] [Google Scholar]
- 7.Gallegos M.T., Schleif,R., Bairoch,A., Hofmann,K. and Ramos,J.L. (1997) Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev., 61, 393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez-Hackert E. and Stock,A.M. (1997) Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol., 269, 301–312. [DOI] [PubMed] [Google Scholar]
- 9.Pestova E.V., Havarstein,L.S. and Morrison,D.A. (1996) Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol., 21, 853–862. [DOI] [PubMed] [Google Scholar]
- 10.Diep D.B., Havarstein,L.S. and Nes,I.F. (1996) Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol., 178, 4472–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuno T. (1997) Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res., 4, 161–168. [DOI] [PubMed] [Google Scholar]
- 12.Galperin M.Y., Nikolskaya,A.N. and Koonin,E.V. (2001) Novel domains of the prokaryotic two-component signal transduction system. FEMS Microbiol. Lett., 203, 11–21. [DOI] [PubMed] [Google Scholar]
- 13.Lange R., Wagner,C., de Saizieu,A., Flint,N., Molnos,J., Stieger,M., Caspers,P., Kamber,M., Keck,W. and Amrein,K.E. (1999) Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene, 237, 223–234. [DOI] [PubMed] [Google Scholar]
- 14.de Saizieu A., Gardes,C., Flint,N., Wagner,C., Kamber,M., Mitchell,T.J., Keck,W., Amrein,K.E. and Lange,R. (2000) Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol., 182, 4696–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohr C.D., Hibler,N.S. and Deretic,V. (1991) AlgR, a response regulator controlling mucoidy in Pseudomonas aeruginosa, binds to the FUS sites of the algD promoter located unusually far upstream from the mRNA start site. J. Bacteriol., 173, 5136–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohr C.D., Leveau,J.H., Krieg,D.P., Hibler,N.S. and Deretic,V. (1992) AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J. Bacteriol., 174, 6624–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Risoen P.A., Havarstein,L.S., Diep,D.B. and Nes,I.F. (1998) Identification of the DNA-binding sites for two response regulators involved in control of bacteriocin synthesis in Lactobacillus plantarum C11. Mol. Gen. Genet., 259, 224–232. [DOI] [PubMed] [Google Scholar]
- 18.Risoen P.A., Brurberg,M.B., Eijsink,V.G. and Nes,I.F. (2000) Functional analysis of promoters involved in quorum sensing-based regulation of bacteriocin production in Lactobacillus. Mol. Microbiol., 37, 619–628. [DOI] [PubMed] [Google Scholar]
- 19.Ween O., Gaustad,P. and Havarstein,L.S. (1999) Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol., 33, 817–827. [DOI] [PubMed] [Google Scholar]
- 20.Cheung J.K. and Rood,J.I. (2000) The VirR response regulator from Clostridium perfringens binds independently to two imperfect direct repeats located upstream of the pfoA promoter. J. Bacteriol., 182, 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelfand M.S., Koonin,E.V. and Mironov,A.A. (2000) Prediction of transcription regulatory sites in Archaea by a comparative genomic approach. Nucleic Acids Res., 28, 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mironov A.A., Vinokurova,N.P. and Gelfand,M.S. (2000) Software for analysis of bacterial genomes. Mol. Biol., 34, 222–231. [PubMed] [Google Scholar]
- 23.Donvito B., Etienne,J., Denoroy,L., Greenland,T., Benito,Y. and Vandenesch,F. (1997) Synergistic hemolytic activity of Staphylococcus lugdunensis is mediated by three peptides encoded by a non-agr genetic locus. Infect. Immun., 65, 95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santiago B., Schubel,U., Egelseer,C. and Meyer,O. (1999) Sequence analysis, characterization and CO-specific transcription of the cox gene cluster on the megaplasmid pHCG3 of Oligotropha carboxidovorans. Gene, 236, 115–124. [DOI] [PubMed] [Google Scholar]
- 25.Galperin M.Y., Gaidenko,T.A., Mulkidjanian,A.Y., Nakano,M. and Price,C.W. (2001) MHYT, a new integral membrane sensor domain. FEMS Microbiol. Lett., 205, 17–23. [DOI] [PubMed] [Google Scholar]
- 26.Dow J.M., Feng,J.X., Barber,C.E., Tang,J.L. and Daniels,M.J. (2000) Novel genes involved in the regulation of pathogenicity factor production within the rpf gene cluster of Xanthomonas campestris. Microbiology, 146, 885–891. [DOI] [PubMed] [Google Scholar]
- 27.Brunskill E.W. and Bayles,K.W. (1996) Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol., 178, 5810–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu T., Ba-Thein,W., Tamaki,M. and Hayashi,H. (1994) The virR gene, a member of a class of two-component response regulators, regulates the production of perfringolysin O, collagenase and hemagglutinin in Clostridium perfringens. J. Bacteriol., 176, 1616–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brunskill E.W. and Bayles,K.W. (1996) Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol., 178, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitchurch C.B., Alm,R.A. and Mattick,J.S. (1996) The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA, 93, 9839–9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W.Z., Ko,T.P., Corselli,L., Johnson,R.C. and Yuan,H.S. (1998) Conversion of a beta-strand to an alpha-helix induced by a single-site mutation observed in the crystal structure of Fis mutant Pro26Ala. Protein Sci., 7, 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen B.L., Gerlach,G.F. and Clegg,S. (1991) Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J. Bacteriol., 173, 916–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rost B. and Sander,C. (1993) Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol., 232, 584–599. [DOI] [PubMed] [Google Scholar]
- 34.Karplus K. and Hu,B. (2001) Evaluation of protein multiple alignments by SAM-T99 using the BAliBASE multiple alignment test set. Bioinformatics, 17, 713–720. [DOI] [PubMed] [Google Scholar]
- 35.Schneider T.D. and Stephens,R.M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res., 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor B.L. and Zhulin,I.B. (1999) PAS domains: internal sensors of oxygen, redox potential and light. Microbiol. Mol. Biol. Rev., 63, 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.