Abstract
Previous studies have shown that the guanine plus cytosine (G+C) content of ribosomal RNAs (rRNAs) is highly correlated with bacterial growth temperatures. This correlation is strongest in the double-stranded stem regions of the rRNA, a fact that can be explained by selection for increased structural stability at high growth temperatures. In this study, we examined the single-stranded regions of 16S rRNAs. We reasoned that, since these regions of the molecule are subject to less structural constraint than the stem regions, their nucleotide content might simply reflect the overall nucleotide content of the genome. Contrary to this expectation, however, we found that all of the single-stranded regions are characterized by very high adenine (A) and relatively low cytosine (C) contents. Moreover, the nucleotide content of these single-stranded regions is surprisingly constant between species, despite dramatic differences in optimal growth temperatures, and despite large differences in the overall genomic G+C content. This provides compelling evidence for strong stabilizing selection acting on 16S rRNA single-stranded regions. We found that selection favors purines (A+G), and especially adenine (A), in the single-stranded regions of these rRNAs.
INTRODUCTION
The thermal stability of double-stranded DNA is dependent on the nucleotide content of the molecule. Specifically, DNA molecules that are rich in guanine (G) and cytosine (C) are more thermostable than those with an excess of adenine (A) and thymine (T). This is because G:C base pairs have an additional hydrogen bond compared with A:T pairs; consequently, it has been proposed that the presence of extra G:C pairs should help stabilize DNA and RNA secondary structures at elevated temperatures (1). This has led to the further prediction that there should be a correlation between the G+C content of genomes and environmental temperature. Recent studies of prokaryotic genomes, however, failed to demonstrate any correlation between the overall G+C content of the genome and optimal growth temperature (2–4). Nevertheless, there is a very strong relationship between the G+C content of structural RNAs (including small subunit ribosomal RNA, large subunit ribosomal RNA, transfer RNA and 5S RNA) and bacterial growth temperature (2,3). In the case of the ribosomal RNAs (rRNAs), the elevated G+C content was concentrated primarily in the double-stranded stem regions of the molecule, and it was largely independent of the average G+C content of the bacterial genome (3,4). These results indicate that, while thermal adaptation does not affect the overall nucleotide content of the genome, it has a very significant effect on the composition of the double-stranded regions of structural RNAs.
In this study, we focused on the single-stranded regions of the 16S rRNA genes. We calculated the nucleotide compositions of these regions in both mesophilic and thermophilic eubacteria and archaea. We were especially interested in the covariation with both optimal growth temperature and the average nucleotide content of the genome. Our expectation was that, in contrast to the double-stranded stems, these unpaired regions of the rRNA molecule would be affected by the same mutational pressures that determine the overall nucleotide content of the genome. As a control, we also scored the nucleotide content of the genes encoding ribosomal proteins in these genomes. This study was made possible by the recent availability of complete genomic sequences for many bacterial species.
MATERIALS AND METHODS
We assembled a database that contained the nucleotide contents and optimal growth temperatures for 44 prokaryotic species. Nucleotide contents of the total 16S rRNAs, along with their component secondary structures (stems, loops, bulge loops and internal loops), were computed from the European Small Subunit RNA Database [http://rrna.uia.ac.be (5)]. Ambiguous nucleotides, which account for ∼4.8% of an average sequence length (1520 nucleotides), were ignored in the calculation. An RNA stem is defined as a right-handed double helix of base pairs; a hairpin loop is a loop of unpaired nucleotides at the termini of stems; a bulge loop is formed by unpaired nucleotides in one strand of a double-stranded region, where the other strand has contiguous base pairing; and an internal loop contains several unpaired nucleotides in both strands of a double-stranded region (6). Stems, bulge loops and internal loops are each identified in this database. The remainder of the molecule consists of hairpin loops, multiple branched loops, pseudoknot loops and dangling ends (5′ terminal and 3′ termini). These latter regions, together with the bulge loops and internal loops, are collectively called single-stranded regions in this study.
In all, we have data from 28 mesophilic species (optimal growth temperature <45°C) and 16 thermophilic species (growth temperature ≥45°C). The entire genomes of 31 of these species (21 eubacteria and 10 archaea) have been completely sequenced and these genomic sequences were retrieved from GenBank. For these species, in addition to obtaining data on the 16S rRNA sequences, we computed nucleotide compositions for the entire genome and for the genes encoding ribosomal proteins. Growth temperature data on the thermophiles having complete genomic sequences available were taken from the original papers describing the genomic sequence. Growth temperature data for the remaining species were mainly obtained from the data sets described in Galtier and Lobry (3), Hurst and Merchant (4) and DSMZ German Collection of Microorganisms and Cell Cultures (http://www.dsmz.de/species/strains.htm). Additional data may be viewed at http://www.bact.wisc.edu/microtextbook/NutritionGrowth/Temperature.html.
Statistical analyses were performed using the statistics package SYSTAT version 10 (SPSS Science, 2000). For the correlation analyses, the Pearson correlation coefficient r was used to evaluate the strength of correlation. A ‘strong’ correlation is defined as an absolute value of r >0.9; a value between 0.5 and 0.9 represents a ‘moderate’ correlation; and a value <0.5 is defined as a ‘weak’ or no correlation (7). A negative value of r means that the correlation is negative.
RESULTS
Average nucleotide composition in mesophiles and thermophiles
We first compared the average nucleotide composition of the entire genome with the optimal growth temperature for each species. Specifically, we wished to test for a correlation between the G+C content of the entire genome and optimal growth temperature. The results are shown in Table 1. As can be seen in Table 1, although there is a wide variation in G+C content within both the mesophiles and the thermophiles, there is no obvious difference between the two groups. In fact, we found no significant difference in the average genomic G+C content between these two sets of species (Mann–Whitney U-test, P = 0.811).
Table 1. GC content and optimal growth temperature (Topt, °C) of completely sequenced genomes used in this study.
| Organism | G+C (%) | Topt | References |
|---|---|---|---|
| (A) Mesophile |
|
|
|
|
Bacillus subtilis |
43.5 |
38.8 |
(3) |
|
Borrelia burgdorferi |
28.6 |
37.0 |
DSMZa |
|
Campylobacter jejuni |
30.5 |
37.0 |
DSMZ |
|
Caulobacter crescentus |
67.2 |
22.5 |
(3) |
|
Chlamydophila pneumoniae |
40.6 |
37.0 |
b |
|
Chlamydia trachomatis |
41.3 |
37.0 |
b |
|
Deinococcus radiodurans |
66.6 |
37.0 |
(3) |
|
Escherichia coli |
50.8 |
37.0 |
(3) |
|
Haemophilus influenzae |
38.1 |
36.0 |
(3) |
|
Halobacterium sp. |
67.9 |
37.0 |
DSMZ |
|
Helicobacter pylori |
38.9 |
37.0 |
DSMZ |
|
Mycoplasma genitalium |
31.7 |
37.0 |
b |
|
Mycoplasma pneumoniae |
40.0 |
37.0 |
(3) |
|
Mycobacterium tuberculosis |
65.6 |
37.0 |
(3) |
|
Neisseria meningitides |
51.5 |
36.0 |
(3) |
|
Pasteurella multocida |
40.4 |
37.0 |
(3) |
|
Pseudomonas aeruginosa |
66.6 |
37.0 |
(3) |
|
Rickettsia prowazekii |
29.0 |
37.0 |
b |
|
Streptococcus pyogenes |
38.5 |
37.0 |
(3) |
|
Treponema pallidum |
52.8 |
37.0 |
c |
| Vibrio cholerae | 47.5 | 37.0 | c |
| Ureaplasma urealyticum | 25.5 | 37.0 | (3) |
| Mean ± SE | 45.6 ± 2.9 | 36.3 ± 0.7 | |
| |
|
|
|
| (B) Thermophile |
|
|
|
|
Aeropyrum pernix |
56.3 |
90.0 |
(16) |
|
Aquifex aeolicus |
43.5 |
95.0 |
(17) |
|
Archaeoglobus fulgidus |
48.6 |
83.0 |
(18) |
|
Methanobacterium thermoautotrophicum |
49.5 |
65.0 |
(19) |
|
Methanococcus jannaschii |
31.4 |
85.0 |
(20) |
|
Pyrococcus horikoshii |
41.9 |
95.0 |
(21) |
|
Sulfolobus solfataricus |
35.8 |
80.0 |
(22) |
|
Thermoplasma acidophilum |
46.0 |
59.0 |
(23) |
|
Thermotoga maritima |
46.2 |
80.0 |
(24) |
| Mean ± SE | 44.4 ± 2.5 | 81.3 ± 4.1 |
aDSMZ, German Collection of Microorganisms and Cell Cultures (http://www.dsmz.de/species/strains.htm).
bThe optimal growth temperature of this species was not available from the temperature data we collected. However, since it is an obligate intracellular pathogen of humans or mammals, its growth temperature was assumed to be 37°C in this study.
chttp://www.bact.wisc.edu/microtextbook/NutritionGrowth/Temperature.html.
Next, we compared the average nucleotide content of the entire genome with that of the 16S rRNA sequences (Table 2). In this case, we calculated the frequency of each of the four nucleotides separately. Again, we found that mesophiles and thermophiles have very similar base compositions when we score the entire genome (Table 2A). For example, the mean total genomic adenine content of the 22 mesophiles is 27.2 ± 1.5%; while the mean genomic adenine content of the nine thermophiles is very similar at 27.8 ± 1.3%, and the values are not significantly different (Mann–Whitney U-test, P = 0.794). Likewise, there is no significant difference in the average frequency of the other three bases between the genomes of the mesophiles and the thermophiles. The result is quite different, however, when we compare the nucleotide compositions of 16S rRNA genes. In this case, there is a clear and highly significant difference between mesophiles and thermophiles (Table 2B). For example, the average adenine content of the 16S rRNA sequences among the mesophiles is 26.1 ± 0.4%, while the mean adenine content among the thermophiles is only 20.3 ± 0.4%, and this difference is statistically significant (Mann–Whitney U-test, P < 0.001). A similar difference between mesophiles and thermophiles is seen for the other three bases. Overall, the 16S rRNA sequences of the thermophiles are relatively G+C rich, as expected, and we note that, in mesophiles, the purines (G and A) are the two most abundant bases.
Table 2. Average nucleotide composition (mean% ± SE) of whole genomes and 16S rRNA genes of mesophiles and thermophiles.
| Nucleotide | Mesophile | Thermophile | Significancea | |||
|---|---|---|---|---|---|---|
| (A) Nucleotide composition of entire genome, for the 31 genomes listed in Table 1 | ||||||
| A |
27.2 ± 1.47 |
27.8 ± 1.26 |
NSb |
|||
| C |
22.8 ± 1.48 |
22.1 ± 1.26 |
NS |
|||
| G |
22.8 ± 1.46 |
22.2 ± 1.22 |
NS |
|||
| T |
27.2 ± 1.47 |
27.8 ± 1.22 |
NS |
|||
| (B) Nucleotide composition of 16S rRNA genes |
|
|
|
|||
| A |
26.1 ± 0.38 |
20.3 ± 0.41 |
P < 0.001 |
|||
| C |
21.6 ± 0.31 |
28.8 ± 0.49 |
P < 0.001 |
|||
| G |
30.5 ± 0.38 |
36.0 ± 0.47 |
P < 0.001 |
|||
| T | 21.8 ± 0.31 | 14.9 ± 0.55 | P < 0.001 | |||
aProbability of Mann–Whitney U-test of respective nucleotide composition in the two sets of mesophiles and thermophiles.
bNS, not significant (P > 0.5).
Since our main focus was on the unpaired regions of the 16S rRNA sequence, we subdivided the rRNA molecule according to its secondary structure, and the base compositions of these structural components are summarized in Table 3. In the double-stranded stem regions (Table 3A), although both mesophiles and thermophiles are rich in G and C, the frequency of these two bases is significantly higher in thermophiles (Mann–Whitney U-test, P < 0.001). These results are consistent with the hypothesis that G:C base pairs are selected because of their role in stabilizing the stem regions, and that this selection is especially strong among the thermophiles. When we look at the unpaired regions of the 16S rRNA, i.e. the single-stranded loops, bulges and dangling ends, a very different picture emerges (Table 3B and C). We note two trends: first, these segments of the rRNA sequence are not particularly rich in G+C, and secondly, there is not a large difference in any of the four base frequencies between the thermophiles and the mesophiles. For instance, whereas there is a 2-fold difference in adenine content of the stem regions between the mesophiles and thermophiles (Table 3A), there is no significant difference between mesophiles and thermophiles as regards their adenine content in the loops and dangling ends (Mann–Whitney U-test, P = 0.893; Table 3B). Likewise, although there are differences for the other three bases, the magnitude of these differences is relatively small. What is most striking about the data on the single-stranded regions are the high levels of adenine (A) and of purines in general (A and G) in both the mesophiles and the thermophiles.
Table 3. Average nucleotide composition (mean% ± SE) of structural components of 16S rRNA genes of 44 mesophiles and thermophiles.
| Nucleotide | Mesophile | Thermophile | Significancea |
|---|---|---|---|
| (A) Nucleotide composition of the stems; the average length of stems in a 16S rRNA is 856 nucleotidesb | |||
| A |
15.7 ± 0.46 |
7.5 ± 0.59 |
P < 0.001 |
| C |
26.7 ± 0.42 |
37.0 ± 0.74 |
P < 0.001 |
| G |
34.9 ± 0.45 |
42.8 ± 0.58 |
P < 0.001 |
| T |
22.7 ± 0.41 |
12.7 ± 0.73 |
P < 0.001 |
| (B) Nucleotide composition of the loops (including hairpin loops, multiple branched loops, psudoknot loops and dangling ends); the average length of loops in a 16S rRNA is 427 nucleotidesb | |||
| A |
37.6 ± 0.35 |
37.8 ± 0.34 |
NSc |
| C |
16.5 ± 0.24 |
17.8 ± 0.21 |
P = 0.001 |
| G |
23.9 ± 0.28 |
25.1 ± 0.37 |
P = 0.012 |
| T |
21.9 ± 0.26 |
19.2 ± 0.43 |
P < 0.001 |
| (C) Nucleotide composition of the internal loops and bulge loops; the average length of internal/bulge loops in a 16S rRNA is 163 nucleotidesb | |||
| A |
48.2 ± 0.46 |
45.5 ± 0.42 |
P = 0.001 |
| C |
9.2 ± 0.30 |
11.3 ± 0.46 |
P = 0.001 |
| G |
25.5 ± 0.59 |
27.1 ± 0.55 |
NS |
| T | 17.0 ± 0.56 | 16.0 ± 0.40 | NS |
aProbability of Mann-Whitney U-test of respective nucleotide composition in the two sets of mesophiles and thermophiles.
bAmbiguous nucleotides were excluded from the calculation.
cNS, not significant (P > 0.05).
In order to explore these patterns further, we examined the distribution of nucleotide frequencies in the 16S rRNA sequences from the individual species. We were especially interested in the correlations between these nucleotide frequencies and (i) the optimal growth temperature of the species and (ii) the average nucleotide content of the entire genome.
16S rRNA stems and loops are affected very differently by growth temperature
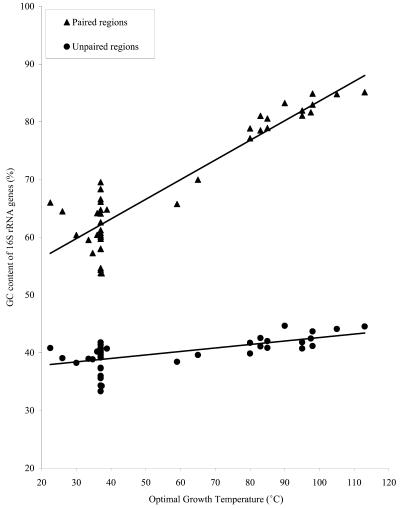
We plotted the relationship between the G+C content of the 16S rRNA genes and the optimal growth temperatures of the 44 species. For this plot, we separated the data into two sets: one set for the double-stranded stem regions, and the other for the single-stranded regions. The results are shown in Figure 1. As can be seen in Figure 1, the G+C content of the stem regions rises rapidly with increasing temperature (slope of regression line = 0.34, P < 0.001). A much weaker trend, however, is seen for the dependence of the G+C content of the single-stranded regions on growth temperature (slope of regression line = 0.06). From this result, it is clear that the selection for an increased G+C content of 16S rRNA sequences at higher temperatures is operating almost entirely within the stem regions. It is interesting to note that the two species with moderate thermophilicity (Thermoplasma acidophilum, with an optimum growth temperature of 57°C and Methanobacterium thermoautotrophicum, with an optimal temperature of 65°C) are also intermediate with respect to the nucleotide content of their rRNA stem regions.
Figure 1.
G+C content of 16S rRNA paired regions (stems) and unpaired regions (single-strand regions) plotted against optimal growth temperature (°C). The slope of the regression line for G+Cpaired regions versus temperature is 0.34. The slope of the line for G+Cunpaired regions versus temperature is only 0.06.
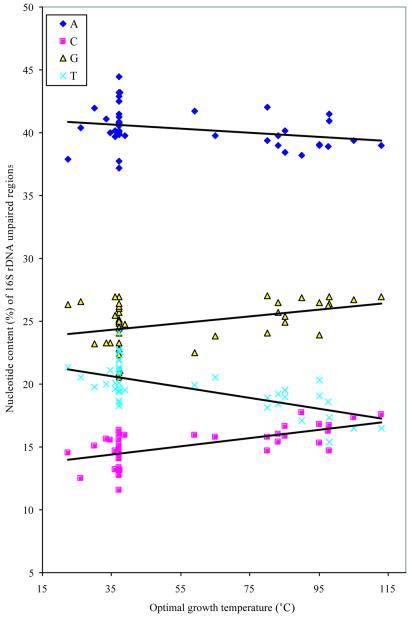
In order to get more detailed information on the patterns of nucleotide distribution in the single-stranded regions, we plotted the frequencies for each of the four bases separately (Fig. 2). In Figure 2, we can observe the slight increase in the frequencies of G and C with increasing growth temperature, and the concomitant decreases in the frequencies of A and T. What is much more marked than these slight changes related to growth temperature, however, are the very large and relatively constant differences in the frequencies of the four bases. Adenine is at a uniformly high frequency, followed by guanine, whereas the two pyrimidines, cytosine and thymine, are at low frequencies at all growth temperatures. In summary, these data show that nucleotide contents in single-stranded regions of the 16S rRNA are relatively constant between the mesophiles and thermophiles, and they show that adenine is the most abundant nucleotide in both groups. In contrast to this weak relationship between the nucleotide content of the single-stranded regions and optimal growth temperature, there is a very strong temperature dependence in the double-stranded stem regions (compare Table 4A with B). In the case of the paired regions, the G and C contents are strongly, and positively, correlated with growth temperature, whereas both A and T show a strong negative correlation with growth temperature in these regions (Table 4A).
Figure 2.
Individual nucleotide composition of unpaired regions of 16S rRNA genes plotted against optimal growth temperature (°C).
Table 4. Correlation and regression analysis of nucleotide composition of 16S rRNA and optimal growth temperature (Topt).
| ra | Slopeb | P-value on slopec | |
|---|---|---|---|
| (A) Paired regions | |||
| A |
–0.891 |
–0.153 |
<0.001 |
| C |
0.931 |
0.192 |
<0.001 |
| G |
0.889 |
0.147 |
<0.001 |
| T |
–0.928 |
–0.187 |
<0.001 |
| (B) Unpaired regions | |||
| A |
–0.297 |
–0.017 |
0.051 |
| C |
0.643 |
0.033 |
<0.001 |
| G |
0.412 |
0.027 |
0.005 |
| T | –0.657 | –0.043 | <0.001 |
aPearson correlation coefficient.
bThe slope of the linear regression line.
cThe associated probability associated with the null hypothesis (regression line slope = 0).
The relationship between the nucleotide content of the 16S rRNA and the nucleotide content of the whole genome
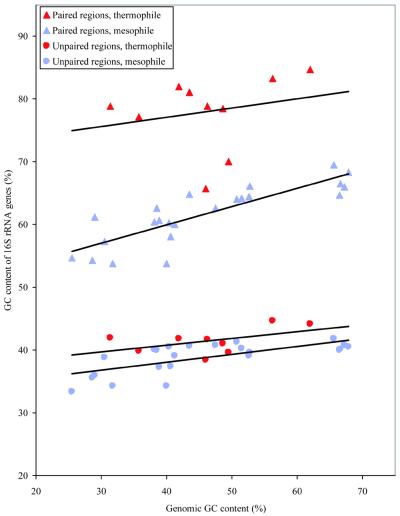
It is already known that the nucleotide content of the stem regions of the rRNAs are distinct from the background genomic levels (3). Our working hypothesis was that the loop regions, which are not subject to selection for elevated G+C levels, might show a pattern of nucleotide composition that resembled the background genomic levels. Since we now have complete genomic sequences for many species, we were able to test this hypothesis directly. In Figure 3, we show the relationship between the G+C content of the 16S rRNA and the average G+C content of the corresponding whole genome. For this analysis the data were separated into stem and loop regions, and into thermophiles and mesophiles. Although there is a significant positive relationship between G+C content of the stem regions and total genomic G+C content among the mesophiles (slope of the regression line = 0.29, P < 0.001), there is no such relationship for the stem regions of the thermophiles (slope of the regression line = 0.149, P = 0.532). To our surprise, we found that the relationship between the nucleotide content of the single-stranded regions and that of the whole genome was extremely weak.
Figure 3.
G+C content of 16S rRNA paired and unpaired regions plotted against the average G+C content of the whole genome. Data for mesophiles and thermophiles are shown separately.
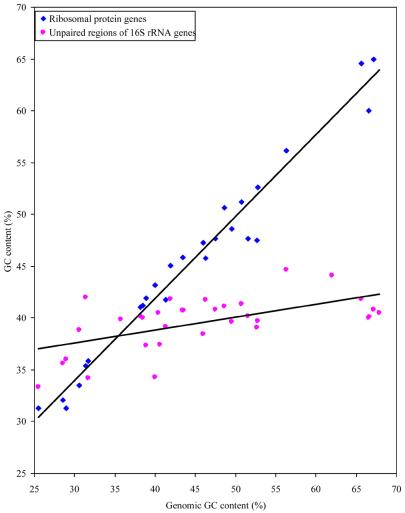
Whereas Figure 2 shows that the nucleotide composition of the single-stranded regions is not very sensitive to changes in optimal growth temperatures, Figure 3 shows that these regions are equally immune to the effects of large changes in the nucleotide content of the whole genome. To highlight this relative constancy of nucleotide content in very different genomic backgrounds, we compared the patterns of nucleotide content of the single-stranded regions with that of the ribosomal protein genes from the same species. The results are shown in Figure 4 and in Table 5. Unlike the single-stranded regions of the rRNA, the ribosomal protein coding genes show a very strong effect of the genomic background among both the mesophiles and the thermophiles (the slopes of the regression lines are 0.787 and 0.805, respectively). This difference between the rRNA and the ribosomal protein genes reinforces the impression that there is a positive selection maintaining the relatively constant nucleotide levels in the single-stranded regions of the rRNAs.
Figure 4.
The relationship between the average genomic G+C content and the G+C content of (i) ribosomal protein genes and (ii) 16S rRNA unpaired regions.
Table 5. The relationship between the overall G+C content of the genome and the G+C content of (A) 16S rRNA unpaired regions and (B) the ribosomal protein gene coding sequences.
| ra | Slopeb | P-value on slopec | |
|---|---|---|---|
| (A) 16S rRNA unpaired regions | |||
| Mesophile |
0.689 |
0.126 |
<0.001 |
| Thermophile |
0.493 |
0.107 |
0.147 |
| (B) Ribosomal protein coding sequences | |||
| Mesophile |
0.985 |
0.787 |
<0.001 |
| Thermophile | 0.981 | 0.805 | <0.001 |
aPearson correlation coefficient.
bThe slope of the linear regression line.
cThe associated probability associated with the null hypothesis (regression line slope = 0).
DISCUSSION
The main finding of this study is that the nucleotide content of the single-stranded regions of 16S rRNA (hairpin loops, multiple branched loops, psudoknot loops, internal loops, bulge loops and dangling ends) are remarkably constant between thermophiles and mesophiles, and also among genomes that have very different average nucleotide frequencies. Essentially, these regions are characterized by a uniformly low G+C content in all of the species studied. At first glance, this seems to contradict the previous studies, which showed that the G+C contents of rRNA genes are positively correlated with bacterial growth temperature (2,3). This seeming paradox can easily be resolved, however, by the realization that the increased G+C content of the thermophiles is concentrated almost entirely within the double-stranded stem regions of the molecule.
Since the sequences within the loop regions do not contribute to the formation of the secondary structure of the rRNA molecule, it is not surprising that they are not affected by temperature-based selection in the same way as the stem regions. One might expect, however, that they would reflect the nucleotide biases that affect the genome as a whole. It has now been well established that such biases in nucleotide composition can have a major effect on the coding capacity of most genes within the genome, causing predicable changes in the amino acid composition of the encoded proteins (8,9). Given these findings, we reasoned that those regions of the rRNA, which were not critical for forming the secondary structure, might respond to nucleotide pressures that affect the genome as a whole. Based on our results presented here, this is clearly not the case. As shown in Figure 4, even the highly conserved ribosomal protein-coding sequences are affected by the overall nucleotide content of the genome, but this is not the case for the single-stranded regions of the rRNA. This indicates that there are very strong selective constraints acting on these single-stranded regions to maintain their constant nucleotide frequencies.
An examination of the frequencies of the individual nucleotides gives some clues about the nature of these selective forces. In single-stranded regions, we found that adenine is the most abundant nucleotide base followed by guanine, thymine and cytosine (A>> G ≥ T ≥ C). On the other hand the frequency of adenine is lowest in the stem regions (Table 3A). One possible explanation might be that selection for high G and C levels in the stem regions has resulted in a local depletion of these two nucleotides in the free nucleotide pool, thus leaving an excess of A and T. We can exclude this possibility, however, by noting that the free nucleotide pool for each species varies over a wide range, as indicated by the variation in the nucleotide content of the whole genome. A more probable explanation of high level of A (and also G) in the unpaired regions of rRNA in all species is that it plays an essential role in maintaining the secondary and tertiary structure of the molecule.
Ribosomal RNA may contain many non-canonical base pairs, other than Watson–Crick pairs (http://prion.bchs.uh.edu/bp_type/bp_structure.html). Purines are especially good at forming base triples and non-canonical pairs such as sheared GA, GA imino, Hoogsteen, reverse Hoogsteen and wobble pairs (10). The high amount of A and G in the single-stranded regions may be involved in tertiary interactions between these regions. The detailed structures of 16S rRNA of Thermus thermophilus and 23S rRNA of Haloarcula marismortui are now known (11,12). It will be interesting to examine these structures and to make an inventory of tertiary interactions between nucleotides of single-stranded regions.
We wondered if the characteristically high G+C contents of rRNA genes among the thermophiles could be used as a diagnostic test of thermophily. To test this idea, we calculated and sorted G+C contents of all sequence entries deposited in the small subunit rRNA database of Ribosomal Database Project (http://rdp.cme.msu.edu/). Species with the high rRNA G+C contents are indeed thermophiles. The two species having the highest G+C contents in their rRNAs (68.99% in Pyrolobus fumarius and 68.81% in Pyrodictium occultum) were found to be hyperthermophiles (P.fumarius can live at temperatures up to 113°C; P.occultum grows at 85–105°C). A number of recent reports (13,14) have proposed that structural RNAs could be identified as high G+C islands in a low G+C genomic background. We have confirmed that the 16S rRNA sequences correspond to such islands in the genomes of Methanococcus jannaschii, Mycoplasma genitalium and Borrelia burgdorferi (all of which have A+T-rich genomes).
Our finding of very stable nucleotide contents among 16S rRNA sequences, despite large variations in the nucleotide content of the entire genome, is intriguing in light of an earlier report (15) that the small subunit rRNA sequences are subject to compositional biases that can affect phylogenetic inference based on these sequences. A re-analysis of three protist sequences (Giardia lamblia, Vairimorpha necatrix and Entamoeba histolytica) that were used in that earlier study showed that there are, indeed, very large differences in G+C content among these rRNAs. For instance, the rRNA of G.lamblia contains >70% G+C, which is equivalent to that seen among the bacterial thermophiles. In contrast, the G+C content of the 16S rRNA in E.histolytica is only 38%. This suggests that the strong constraint on nucleotide content that is seen among the prokaryotes may not apply to eukaryotes.
In summary, we have used the recently available large genomic datasets to confirm earlier reports that the double-stranded regions of rRNA are subject to intense selection for increased G+C content, especially in organisms with high optimal growth temperatures, and even in an A+T-rich genomic background. More important, we have found that the single-stranded regions of the rRNA are subject to equally intense selection to maintain a very constant nucleotide composition, in the face of large variations in both optimal growth temperature and nucleotide composition of the whole genome. The fact that this pattern in seen in both eubacteria and archaea provides further evidence for the action of selection, as opposed to an accident of phylogenetic history. This selective force strongly favors purines, and especially adenine, in the unpaired regions of the rRNA. The biochemical basis for this selective preference remains to be elucidated, although it is tempting to speculate that these nucleotides are critical for the maintenance of higher order rRNA structure.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the two referees for their helpful comments. This work was supported by a Research Grant from NSERC Canada (D.A.H.) and a graduate scholarship from the University of Ottawa (H.-C.W.).
REFERENCES
- 1.Wada A. and Suyama,A. (1986) Local stability of DNA and RNA secondary structure and its relation to biological functions. Prog. Biophys. Mol. Biol., 47, 113–157. [DOI] [PubMed] [Google Scholar]
- 2.Dalgaard J.Z. and Garrett,R.A. (1993) Archaeal hyperthermophile genes. In Kates,M., Kushner,D.J. and Matheson,A.T. (eds), The Biochemistry of Archaea (Archaebacteria). Elsevier, Amsterdam, pp. 535–563.
- 3.Galtier N. and Lobry,J.R. (1997) Relationships between genomic G+C content, RNA secondary structures and optimal growth temperature in prokaryotes. J. Mol. Evol., 44, 632–636. [DOI] [PubMed] [Google Scholar]
- 4.Hurst L.D. and Merchant,A.R. (2001) High guanine–cytosine content is not an adaptation to high temperature: a comparative analysis amongst prokaryotes. Proc. R. Soc. Lond. B, 268, 493–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van de Peer Y., De Rijk,P., Wuyts,J., Winkelmans,T. and De Wachter,R. (2000) The European small subunit ribosomal RNA database. Nucleic Acids Res., 28, 175–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Rijk P. (1995) Optimisation of a database for ribosomal RNA structure and application in structural and evolutionary research. PhD Thesis, University of Antwerp, Belgium.
- 7.Milton J.S. (1992) Statistical Methods in the Biological and Health Sciences. McGraw-Hill, New York, p. 368.
- 8.Singer G.A.C. and Hickey,D.A. (2000) Nucleotide bias causes genomewide bias in the amino acid composition of proteins. Mol. Biol. Evol., 17, 1581–1588. [DOI] [PubMed] [Google Scholar]
- 9.Kreil D.P. and Ouzounis,C.A. (2001) Identification of thermophilic species by the amino acid compositions deduced from their genomes. Nucleic Acids Res., 29, 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagaswamy U., Larios-Sanz,M., Hury,J., Collins,S., Zhang,Z., Zhao,Q. and Fox,G.E. (2002) NCIR: a database of non-canonical interactions in known RNA structures. Nucleic Acids Res., 30, 395–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wimberly B.T., Brodersen,D.E., Clemons,W.M.,Jr, Morgan-Warren,R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- 12.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 13.Rivas E. and Eddy,S.R. (2000) Secondary structure alone is generally not statistically significant for the detection of noncoding RNAs. Bioinformatics, 16, 583–605. [DOI] [PubMed] [Google Scholar]
- 14.Carter R.J., Dubchak,I. and Holbrook,S.R. (2001) A computational approach to identify genes for functional RNAs in genomic sequences. Nucleic Acids Res., 29, 3928–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasegawa M. and Hashimoto,T. (1993) Ribosomal RNA trees misleading? Nature, 361, 23. [DOI] [PubMed] [Google Scholar]
- 16.Kawarabayasi Y., Hino,Y., Horikawa,H., Yamazaki,S., Haikawa,Y., Jin-no,K., Takahashi,M., Sekine,M., Baba,S., Ankai,A. et al. (1999) Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res., 6, 83–101. [DOI] [PubMed] [Google Scholar]
- 17.Deckert G., Warren,P.V., Gaasterland,T., Young,W.G., Lenox,A.L., Graham,D.E., Overbeek,R., Snead,M.A., Keller,M., Aujay,M. et al. (1998) The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature, 392, 353–358. [DOI] [PubMed] [Google Scholar]
- 18.Klenk H.P., Clayton,R.A., Tomb,J., White,O., Nelson,K.E., Ketchum,K.A., Dodson,R.J., Gwinn,M., Hickey,E.K., Peterson,J.D. et al. (1997) The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature, 390, 364–370. [DOI] [PubMed] [Google Scholar]
- 19.Smith D.R., Doucette-Stamm,L.A., Deloughery,C., Lee,H.-M., Dubois,J., Aldredge,T., Bashirzadeh,R., Blakely,D., Cook,R., Gilbert,K. et al. (1997) Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol., 179, 7135–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bult C.J., White,O., Olsen,G.J., Zhou,L., Fleischmann,R.D., Sutton,G.G., Blake,J.A., FitzGerald,L.M., Clayton,R.A., Gocayne,J.D. et al. (1996) Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science, 273, 1058–1073. [DOI] [PubMed] [Google Scholar]
- 21.Kawarabayasi Y., Sawada,M., Horikawa,H., Haikawa,Y., Hino,Y., Yamamoto,S., Sekine,M., Baba,S., Kosugi,H., Hosoyama,A. et al. (1998) Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res., 5, 55–76. [DOI] [PubMed] [Google Scholar]
- 22.She Q., Singh,R.K., Confalonieri,F., Zivanovic,Y., Allard,G., Awayez,M.J., Chan-Weiher,C.C., Clausen,I.G., Curtis,B.A., De Moors,A. et al. (2001) The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl Acad. Sci. USA, 98, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruepp A., Graml,W., Santos-Martinez,M.L., Koretke,K.K., Volker,C., Mewes,H.W., Frishman,D., Stocker,S., Lupas,A.N. and Baumeister,W. (2000) The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature, 407, 508–513. [DOI] [PubMed] [Google Scholar]
- 24.Nelson K.E., Clayton,R.A., Gill,S.R., Gwinn,M.L., Dodson,R.J., Haft,D.H., Hickey,E.K., Peterson,J.D., Nelson,W.C., Ketchum,K.A. et al. (1999) Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima. Nature, 399, 323–329. [DOI] [PubMed] [Google Scholar]