Abstract
The human cytochrome b561 (hCytb561) family consists of electron transfer transmembrane proteins characterized by six conserved α-helical transmembrane domains and two β-type heme cofactors. These proteins contribute to the regulation of iron metabolism and numerous different physiological and pathological processes by recycling ascorbic acid and maintaining iron reductase activity. Key members of this family include cytochrome b561 (CYB561), duodenal CYB561 (Dcytb), lysosomal CYB561 (LCytb), stromal cell-derived receptor 2 (SDR2) and 101F6, which are widely expressed in human tissues and participate in the pathogenesis of several diseases and tumors. They are associated with the promotion or inhibition of tumor growth and progression in various malignancies and are potential therapeutic targets for malignant tumors. The present review summarizes the existing literature regarding the structure of the Cytb561 family, the basic functional characteristics of hCytb561 family members, and the roles of the CYB561, Dcytb, LCytb, SDR2 and 101F6 in various diseases and tumors.
Keywords: cytochrome b561, CYB561, iron metabolism, Dcytb/CYBRD1, tumor
1. Introduction
The B-type cytochrome family comprises electron transport proteins containing heme. The redox-active center of these proteins is iron protoporphyrin IX, which can be non-covalently bound to other protein matrices. Cytochrome b561 (Cytb561) is a member of the B-type cytochrome family, and is a transmembrane protein containing two heme-b subunits embedded within the membrane. It has a maximum absorbance wavelength in the redox absorption spectrum of ~561 nm, which is reflected in its name. The Cytb561 protein consists of 200–300 amino acids, approximately half of which are embedded within the membrane bilayer. Cytb561 can transmit electrons across chromaffin granule membranes (1,2) and facilitate transmembrane electron transfer (3).
The Cytb561 family is named after the specific cytochrome b561 (CYB561), which was identified and named in bovine chromaffin granule membranes in 1971 by Flatmark et al (4). Based on an analysis of the CYB561 gene sequence from chromaffin granules of the bovine adrenal gland, it was discovered that this protein family exists in various organs and cells across multiple species, of animals, including humans (5), mice (6), Drosophila melanogaster (7), Anopheles gambiae (8), Caenorhabditis elegans (9) and planarian species (10), and plants such as Arabidopsis thalania (11) and cultivated rice (12). Cytb561, unique to eukaryotes, exhibits a high degree of conservation, implying similar structures and functions across different species (13). Mammalian Cytb561 proteins and predicted plant Cytb561 proteins are highly hydrophobic and can transfer electrons from the cytoplasmic side of the cell membrane to the extracellular space or intracellular vesicles. They have important roles in various physiological processes, including iron absorption, cellular defense, nitrate reduction and signal transduction. Through multiple sequence alignment, Cytb561 family members from various sources have been categorized into seven categories (14): Animals/neuroendocrine, plants, insects, fungi, animals/tumor suppression factor (TSF), plants containing a DoH domain, and stromal cell-derived receptor 2 (SDR2). In 1974, Silsand and Flatmark (15) purified Cyb561 from bovine chromaffin granules; however, other natural Cytb561 proteins have not been purified due to their low abundance in natural sources. Using the Basic Local Alignment Search Tool, six members of the human Cytb561 (hCytb561) family (16) have been identified: CYB561 encoded by CYB561A1; duodenal CYB561 (Dcytb) encoded by CYB561A2 (17); lysosomal CYB561 (LCytb) encoded by CYB561A3; SDR2 encoded by ferric chelate reductase 1 (FRRS1); 101F6/human TSF encoded by CYB561D2 (14); and 101F6 analogs (16). To date, the most extensively studied members of this family, which are associated with tumors, are CYB561, Dcytb, LCytb, SDR2 and 101F6.
In the present article, the structures of the Cytb561 family, basic functional characteristics of hCytb561 family members, and the roles of the five key family members in various diseases and tumors are reviewed.
2. Iron metabolism, ASC and tumors
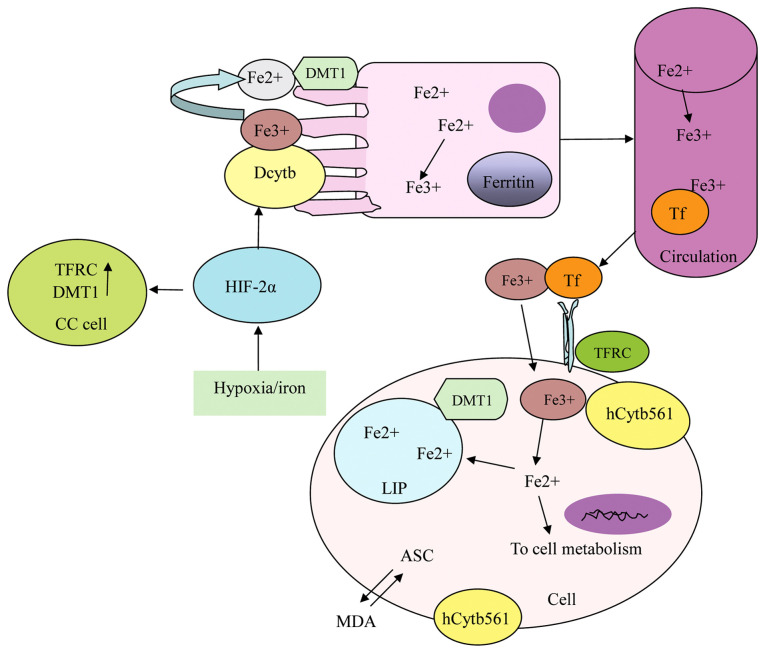
Iron metabolism in the human body is a tightly regulated system, involving iron absorption, transport and distribution, the formation of various important functional ferritin molecules, storage and excretion (18). Iron is absorbed into the bloodstream via active transport by mucosal cells in the duodenum and upper segment of the jejunum. In the bloodstream, ferric ions (Fe3+) are bound and transported by transferrin. After entering cells through transferrin receptor 1 (TFRC), these ions are reduced and released into the cytoplasmic labile iron pool (LIP), and excess iron is stored in ferritin (19). TFRC is an iron import protein that is post-transcriptionally regulated by iron levels; it contributes to iron absorption and is the main means of iron uptake in proliferating cells (20). Ferritin is an iron-binding protein that primarily functions to store and stabilize iron, and also acts as an iron oxidase, converting ferrous ions (Fe2+) to Fe3+ during iron ion internalization and storage (21). Divalent metal transporter 1 (DMT1) is a proton-coupled transmembrane metal ion transporter in mammals, which mediates iron absorption in the small intestine and facilitates iron transport from the endosome (22). Fe2+ transported from the endosome by DMT1 contributes to the LIP, which is involved in cellular metabolism (23). Iron absorption is regulated by multifactorial feedback, and the synthesis of ferritin, transferrin and its receptor TFRC is regulated by iron levels in the body.
Ascorbic acid (ASC) acts as a cofactor for numerous important enzymes involved in mammalian metabolism (24), including dopamine β-hydroxylase in the vesicles of catecholamine storage granules (25) and peptide-amide monooxygenase in neuropeptide storage granules (24). The regeneration of ASC in neuropeptide storage vesicles is necessary to sustain amidated peptide biosynthesis. The cyclical regeneration of ASC contributes to cell physiology and serves as a key regulator of iron metabolism (26). This vitamin affects iron metabolism by promoting the intestinal absorption of non-heme iron (27), increasing transferrin-dependent iron uptake, and promoting the ASC/dehydroascorbic acid (DHA) cycle at the plasma membrane to increase the uptake of non-transferrin-bound iron (28–30). It also promotes ferritin synthesis by increasing or maintaining the level of iron regulatory protein 1 in its non-iron-bound form to promote cytoplasmic aconitase reactivity (31,32). Additionally, ASC regulates iron metabolism by inhibiting the autophagy of ferritin (33), inhibiting lysosomal ferritin degradation (33,34) and regulating cell iron efflux (30,35).
Iron is one of the most important trace elements in mammals due to its important role in cell replication, metabolism and growth (36). Iron also contributes to the generation of free radicals via oxidation and reduction, such as in the Fenton reaction (37), the destruction of lipids and proteins, DNA base modification, DNA strand breakage and other mutations associated with oxidative DNA damage (38), as well as the occurrence and development of cancer (39). To sustain high intracellular iron levels and promote the function of iron-dependent proteins, malignant tumor cells modulate the expression or activity of various iron-related proteins. For example, elevated plasma ferritin levels are associated with advanced clinical stages and poor prognosis in patients with hepatocellular carcinoma (40) and hepatobiliary cancer (41). DMT1 is highly expressed in colorectal cancer, and inhibiting its activity can restrict tumor growth by inhibiting the Janus kinase-signal transducer and activator of transcription 3 signaling pathway (42). Increased serum concentrations of heavy-chain ferritin (FTH) in patients with melanoma are associated with increased numbers of circulating CD4+ CD25+ regulatory T cells, which contribute to the suppression of antitumor immune function (43). Moreover, the proliferation of CD8+ T cells, critical in the antitumor immune response, requires iron stored in FTH (44). As highlighted in a previous review, disrupted iron homeostasis can be observed at all stages of cancer development (45). Various iron metabolism-related proteins have been demonstrated to be involved in the initiation, proliferation and metastasis of malignant tumors (46). The expression levels of certain genes associated with iron metabolism have been indicated to be powerful indicators of tumor prognosis. For example, upregulated TFRC expression in breast cancer (20,47) and downregulated ferritin (48) or ferriportin expression (49) are associated with a poor prognosis. The regulation of iron homeostasis, by iron depletion and the targeting of iron metabolism, has shown strong and extensive antitumor effects, suggesting that iron metabolism is a potential target for cancer therapy (50). As the hCytb561 family plays an important role in iron metabolism and cancer, it is expected to become a key target for the treatment of malignant tumors.
3. Basic characteristics of the Cytb561 family
Basic structure
The Cytb561 family of transmembrane proteins typically contain six transmembrane α-helix domains (51–53). Sequence alignment indicates that all retrieved sequences exhibit four completely conserved sequences located in the second to fifth transmembrane segments (14). Four consecutive central helices form the core domain of CYB561 (Fig. 1). The core of the four helices comprises two pairs of histidine (His) residues (14,51); the first and third residues are located on the cytoplasmic side, while the second and fourth are located at the boundary region of the outer (or intravesicular) hydrophilic ring and transmembrane helix (54–56). The CYB561 core domain, via its four His residues, coordinates to heme-b groups on both sides of the membrane (57–59). The core domain facilitates intramolecular electron transfer by accepting electrons from ASC (60). It is structurally similar to other redox domains, such as dopamine β-monoxygenase redox domains and the dopamine β-monooxygenase N-terminal (DOMON) domain (24), and can be a component of other proteins. The basic characteristics of hCytb561 family members, including their genomic locations and sequence sizes, are shown in Table I. They are divided into different categories according to their specific motifs (Fig. 1). CYB561, Dcytb and LCytb have been assigned to the animals/neuroendocrine class due to presence of motif 1 [FN(X)HP(X)2M(X)2G(X)5G(X)ALLVYR] and motif 2 [YSLHSW(X)G] in their core structures (15). 101F6 and its analogs have been classified in the animals/TSF class due to the presence of modified motif 1 [LFSWHP(X)2M(X)3F(X)3M(X)EAIL(X)SP(X)2SS] in their structures. SDR2 belongs to the SDR2 class due to the presence of three DOH domains or a DOMON domain (14), which is homologous to a domain found in dopamine-β-hydroxylase (61). All family members have transmembrane electron transfer activity or contain at least two heme groups that may contribute to the integrity or binding of the transmembrane protein structure (56). The double-electrode voltage-clamp technique has demonstrated the ability of SDR2 to conduct electric current (62).
Figure 1.
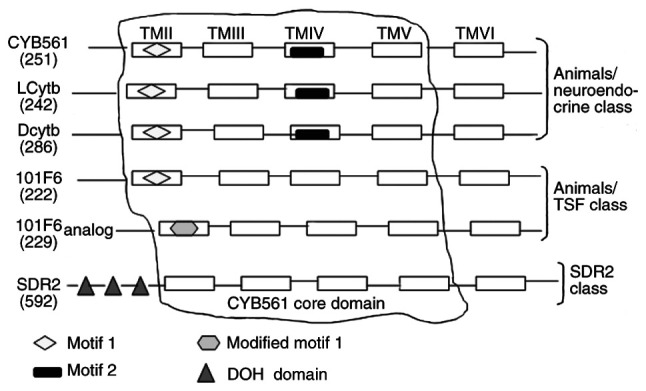
Location of the protein domain for human cytochrome b561 family members and their classifications. The number in parentheses under the name of each family member is the number of amino acids. TMII–VI, transmembrane domain II–VI; CYB561, cytochrome b561; Dcytb, duodenal CYB561; LCytb, lysosomal CYB561; SDR2, stromal cell-derived receptor 2; TSF, tumor suppression factor.
Table I.
Basic characteristics of the hCytb561 family.
| Characteristics | CYB561 | Dcytb | LCytb | SDR2 | 101F6 |
|---|---|---|---|---|---|
| Gene name | CYB561A1/FRRS2 | CYBRD1/CYB561A2 | CYB561A3 | FRRS1 | CYB561D2 |
| Genomic location | 17q23.3 | 2q31.1 | 11q12.2 | 1p21.2 | 3p21.31 |
| Sequence size, bp | 14,336 | 35,897 | 13,539 | 62,666 | 17,353 |
| Amino acids, n | 251 | 286/174 | 242 | 592 | 222 |
| Domain | 19-220 CYB561 | 15-220/1-174 (isomorph) CYB561 | 12-219 CYB561 | 13-179 Reelin; 216-331 DOMON; 335-534 CYB561 | 14-217 CYB561 |
| Subcellular location | CV, SV, CGM | CM, ACM | LEM, LM | CM | CV, ERM |
| Highest tissue expression | Brain, adrenal gland | Brush border membrane of duodenum | Adrenal gland | Liver, kidney | Pancreas, brain |
hCytb561, human Cytb561; Cytb561/CYB561, cytochrome b561; Dcytb, duodenal CYB561; LCytb, lysosomal CYB561; SDR2, stromal cell-derived receptor 2; FRRS1/2, ferric chelate reductase 1/2; CYBRD1, cytochrome b reductase 1; DOMON, dopamine b-monooxygenase N-terminal; CV, cytoplasmic vesicle; SV, secretory vesicle; CGM, chromaffin granule membrane; CM, cell membrane; ACM, apical cell membrane; LEM, late endosome membrane; LM, lysosome membrane; ERM, endoplasmic reticulum membrane.
Human CYB561 is estimated to lack the first 22 amino acids at the N-terminal of the cytoplasmic side predicted for the bovine cell sequence. Srivastava et al (63) hypothesized that the gene product of human CYB561 contains five transmembrane helical structures. The crystal structure of Arabidopsis thaliana Cytb561 suggests that this protein may function as a dimer (13). Additionally, two highly conserved amino acids, Lys-81 and His-106, have been demonstrated to be important in substrate recognition and catalysis (64). To illustrate the structures of hCytb561 family members, data on their 3-dimensional structures have been obtained from the UniProt (Universal Protein Resource) website (https://www.uniprot.org/uniprotkb) and are presented in Fig. 2.
Figure 2.
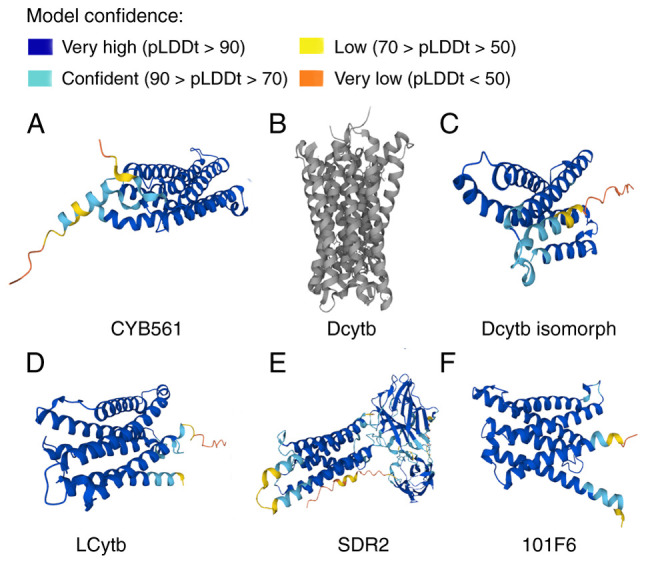
Structure of human Cytb561 family members. Protein structures of (A) CYB561, (B) Dcytb, (C) Dcytb isomorph, (D) LCytb, (E) SDR2 and (F) 101F6. The structures are from the UniProt website (https://www.uniprot.org/uniprotkb). The pLDDT is a per-residue confidence score generated by the AlphaFold prediction tool, which ranges between 0 and 100. The higher the value, the more confident the model is in predicting that residue. Some regions with low pLDDT may be unstructured in isolation, Due to the lack of stable structural features in these regions, the accuracy of model predictions is low. Cytb561/CYB561, cytochrome b561; Dcytb, duodenal CYB561; LCytb, lysosomal CYB561; SDR2, stromal cell-derived receptor 2; UniProt, Universal Protein Resource; pLDDT, Predicted Local Distance Difference Test.
Basic function
The Cytb561 family uses ASC, an extracapsular electron donor, on one side of the membrane to provide electrons, catalyze transmembrane electron transport, and reduce monodehydroascorbate (MDA), a partially oxidized form of ASC that acts as an intracapsular electron receptor, thereby facilitating the regeneration of ASC (64,65). Motif 2 was originally thought to be an MDA free-radical-binding sequence (60). The reduced expression of the CYB561 homologue in Drosophila memory mutant (nemy) has been shown to lead to defective memory retention, confirming the role of Cytb561 in memory retention and its influence on neural function (66). In addition, the role of Cytb561 in ASC regeneration has been confirmed by the fact that ASC in erythrocytes can reduce extracellular MDA (67,68). ASC has been confirmed to function as an electron donor for mouse DCytb (67). Furthermore, B-type cytochromes with biophysical properties similar to those of Cytb561 from bovine chromaffin granules have been reported to reduce ASC in the cell membranes of certain plants (69,70).
In vitro, all Cytb561 members exhibit Fe3+ reductase activity (71). DCytb in the cell membranes of mouse duodenal epithelial cells has been shown to reduce intestinal Fe3+ and thereby facilitate the uptake of iron, which ameliorates hypoxia and iron deficiency (17). In addition, the increased expression of DCytb in mice with total iron overload (hemochromatosis) promotes iron uptake (72). The transmembrane Fe (3+) reductase activity of CYB561 has been demonstrated in ASC-loaded protein liposomes (57). Furthermore, DCytb uses cytoplasmic ASC as an electron donor to reduce extracellular Fe3+ to Fe2+ in rabbit peritoneal neutrophils (73). Three members of the Cytb561 family, namely mouse Cytb561 and mouse and Drosophila SDR2, have been found to have iron reductase activity (71). These findings indicate that when free Fe3+ and ASC are in the same membrane compartment, Fe3+ easily obtains electrons from ASC and is reduced to Fe2+, which can generate free radicals via the Fenton reaction. However, when Fe3+, ASC and Cytb561 are separated, the ASC-mediated reduction of Fe3+ may be moderated. Whether Cytb561 family members reduce Fe3+or MDA in vivo is likely to depend on the availability of the substrate and the biological environment.
4. Expression and function of the hCytb561 family
CYB561
First identified in chromaffin vesicles that synthesize catecholamines, CYB561 was later confirmed to be present in neuroendocrine tissues (74). RNA sequencing has shown that CYB561 is widely expressed in eight systems (locomotion, digestive, respiratory, circulatory, urinary, reproductive, endocrine and nervous systems) of the normal human body. According to gene expression data from the Bgee database (https://bgee.org), the CYB561 gene is expressed in the adrenal gland and 223 other tissues. Pathologically, CYB561 is expressed in HeLa cervical cancer cells and melanoma cells (74). In addition, its mRNA levels are significantly upregulated in SW480 colon cancer, HL-60 T-cell lymphoma and K-562 chronic myeloid leukemia cells but not in Burkitt's lymphoma (75). Under physiological conditions, CYB561 in natural chromaffin granules, vesicles and recombinant membrane systems exhibits ‘electron shuttle’ activity between ASC and membrane permeable ferricyanide or MDA (57,76), which is associated with cell metabolism and mitochondrial activation (77) (Fig. 3), and affects heart rate and blood pressure through regulation of the adrenaline pathway (78).
Figure 3.
Function of the hCytb561 family in iron metabolism and the possible mechanism of Dcytb. All HCytb561 family members have iron reductase activity and participate in the regeneration cycle of ASC and the Fe redox reaction. Dcytb participates in and regulates the absorption of Fe in duodenal mucosal epithelial cells and is regulated by hypoxia or iron content through HIF-2a. Absorbed iron (Fe3+) combines with Tf in the circulation for transport, combines with TFRC to enter tissue cells and participates in cell metabolism. hCytb561, human cytochrome b561; Dcytb, duodenal CYB561; ASC, ascorbic acid; HIF-2a, hypoxia-inducible factor-2α; Fe, iron; Tf, transferrin; TFRC, transferrin receptor 1; DMT1, divalent metal transporter 1; CC, colon cancer; LIP, labile iron pool; MDA, monodehydroascorbate.
Pathogenic homozygous mutations in the CYB561 gene have been reported to lead to ASC deficiency in catecholamine-secreting vesicles and functional dopamine b-hydroxylase deficiency, resulting in orthostatic hypotension syndrome (78). In CYB561(−/-) mice, it was found that the concentration of norepinephrine in whole brain homogenate and adrenal glands was reduced compared with that in wild-type mice (78). CYB561 is also involved in the progression of certain tumors and influences their prognosis. For example, a low expression level of CYB561 mRNA was found to be associated with a poor prognosis in patients with ovarian cancer, suggesting that CYB561 may be a single-gene prognostic biomarker (79). In addition, in another study CYB561 was demonstrated to promote the growth and metastatic potential of castration-resistant neuroendocrine prostate cancer (80). Our previous study found that CYB561 expression was upregulated in breast cancer; associated with human epidermal growth factor receptor 2 (HER2), immune cells, histological grade and molecular subtypes; and associated with the poor prognosis of patients (81), which is consistent with other results reported in the literature (82). Moreover, our previous study demonstrated that CYB561 promotes the proliferation, migration and invasion of breast cancer cells and inhibits apoptosis (83). The knockdown of CYB561 changes the Fe2+ and total iron content of cancer cells and the expression of the iron absorption and transport-related proteins TFRC and DMT1, indicating that it is involved in the iron metabolism of breast cancer cells (83). In addition, recent literature shows that CYB561 plays a role in promoting the proliferation of HER2-positive breast cancer cells by inhibiting the degradation of H2A histone family member Y (84).
Dcytb/cytochrome b reductase 1 (CYBRD1)/CYB561A2
Dcytb is a plasma membrane protein with 40–50% homology to CYB561 (17). The predicted ASC- and DHA-binding motifs in the Cytb561 family are highly conserved in Dcytb (17) (Fig. 3). The gene encoding Dcytb is widely expressed in human tissues and highly expressed in the duodenal brush border membrane. Dcytb serves as the plasma membrane iron reductase of mammals, facilitating the absorption of dietary iron at the brush edge of duodenal cells by reducing Fe3+ to Fe2+ and cooperating with DMT1 to transport Fe2+ to cells in the small intestine (21) (Fig. 3). It also participates in the regeneration of extracellular ASC (85) by reducing extracellular MDA (86), and may also be a transmembrane reductase of copper (87). The inactivation of the L-gulono-g-lactone oxidase (GULO) gene under physiological conditions prevents the synthesis of the protein GULO enzyme and therefore ASC synthesis. Dcytb can reduce and regenerate oxidized ASC while reducing Fe(3+) to Fe(2+), thereby promoting iron absorption. Therefore, it is speculated that the demand for this reductase increases when the demand for iron absorption is strong (88).
Under normoxic conditions, hemoglobin in the reticulocytes of CYBRD1(−/-) mice is impaired, and under hypoxic conditions, these mice exhibit reduced duodenal reductase activity and spleen iron reserves, leading to abnormal erythropoiesis (89). As no evidence of hypoxia-induced iron reductase activity has been observed in the CYBRD1(−/-) mice, this suggests that Dcytb may be the only hypoxia-induced iron reductase in the duodenum. Hypoxia has been demonstrated to strongly upregulate Dcytb expression through a hypoxia-inducible factor-2α (HIF-2α)-dependent mechanism. This activation of HIF-2α also upregulates DMT1 expression, which disrupts local iron homeostasis and promotes cell proliferation, which contributes to the development of colon cancer (90) (Fig. 3). In addition, the upregulation of the iron-import-related proteins Dcytb, DMT1 and TFRC in colorectal cancer cells is associated with increased intracellular iron content (91). An analysis of data from The Cancer Genome Atlas (TCGA) database revealed that the expression of CYBRD1 is upregulated in ovarian cancer (92), and its increased expression is associated with a reduction in overall survival times (92). In another study it was shown that CYBRD1 is upregulated in glioma cell lines, and its silencing reduces the aggressiveness of glioma cells, indicating its potential as a biomarker for glioma recurrence (93) (Fig. 3). Boult et al (94) reported that the expression of CYBRD1 is upregulated during the progression of Barrett's esophagus to adenocarcinoma, suggesting its involvement in the development of esophageal adenocarcinoma through increased iron absorption. In addition, tumor-initiating cells from the breast and prostate origin have been found to exhibit the altered expression of various genes associated with iron metabolism, including CYBRD1, endothelial PAS domain-containing protein 1/HIF-2α and TFRC (95). By contrast, CYBRD1 has also been reported to exhibit tumor-inhibiting effects. Analysis of gene expression in a Gene Expression Omnibus dataset showed that CYBRD1 expression is downregulated in bladder cancer compared with normal bladder mucosa, suggesting that it may have a tumor-inhibitory role (96). Dcytb has been shown to be an important predictor of prognosis in patients with breast cancer, with high Dcytb expression serving as an overall predictor of distant metastasis-free survival and exhibiting a significant association with increased relapse-free survival (23). However, the role of Dcytb in breast cancer cells was indicated to be unrelated to iron metabolism; instead it was demonstrated to delay tumor progression by inhibiting the activation of focal adhesion kinase, a kinase that plays a central role in tumor cell adhesion and metastasis (23). In addition, analysis of TCGA data has shown that the CYBRD1 protein level decreases with increasing lung adenocarcinoma stage and grade, indicating a negative correlation between CYBRD1 expression and the progression of lung adenocarcinoma. Moreover, the low expression of CYBRD1 is associated with poor overall and disease-specific survival (97).
LCytb/CYB561A3
This reductase is present in the late endolysosome-lysosome membrane; it is expressed at high levels in the adrenal gland, lymph nodes, B lymphocytes and monocytes, but is downregulated in sepsis (98) and skin warts caused by human papillomavirus infection (99).
LCytb demonstrates iron reductase activity in yeast (67,75). It also participates in iron homeostasis in endosomes and lysosomes. In late endosomes and lysosomes, LCytb uses ASC as an electron donor to reduce Fe3+ to Fe2+, which is then transferred by DMT1 into the cytoplasm. In addition, LCytb regulates the iron cycle in macrophages by reducing the expression of lysosomal reductase, which is part of the innate immune response that prevents the proliferation of pathogens and sepsis (100). The knockout of LCytb/CYB561A3 can cause catastrophic damage to lysosomes and mitochondria and damage mitochondrial respiratory function (101). A study evaluated the relationship between LCytb/CYB561A3 and tumors, including Burkitt's lymphoma caused by EBV infection (101). CYB561A3 was demonstrated to be critical for the proliferation of Burkitt's lymphoma cells, but to have no effect on the proliferation of lymphoblastic lymphoma cells or other cancer cells subjected to EBV transformation. Although the autophagic degradation of ferritin and plasma membrane transferrin were upregulated, CYB561A3 knockout induced severe iron starvation in Burkitt's cells (101).
SDR2/FRRS1
SDR2, encoded by the gene FRRS1, is a homolog of CYB561 and Dcytb, which is localized in the cell membrane and expressed at high levels in the liver (62), kidney (62), esophageal mucosa, oral epithelium, gallbladder, testicles and heart, as well as in lesions associated with asthma, dermatitis, and rhinitis (102). A recent study found that FRRS1 is upregulated in cervical squamous cell carcinoma (103). SDR2 exhibits iron chelate reductase activity and functions as an active iron reductase that regulates catecholamines in the brain. It reduces Fe3+ to Fe2+ prior to transportation from the endosome to the cytoplasm (67,104) (Fig. 3).
FRRS1 has been predicted by machine learning methods to be upregulated in the brain tissue of patients with Alzheimer's disease and suggested to be a potential risk gene for Alzheimer's disease (105). Linton et al (106) found that the expression of FRRS1 is downregulated in primary soft tissue sarcoma of the extremities and is negatively correlated with metastatic recurrence. In addition, they tested the combined prognostic effects of FRRS1, helicase 4, complement Factor H and mesenchymal-epithelial transition factor using a simple equal-weight scoring system, and found that they had a greater prognostic effect than tumor grading. However, by contrast, other researchers found that FRRS1 is upregulated in SiHa and HeLa cervical squamous cell carcinoma cells, and transfection with short hairpin-FRRS1 inhibited the growth of these cells and promoted their apoptosis (103).
101F6/CYB561D2
Encoded by CYB561D2, 101F6 is highly expressed in the pancreas, nervous system, granulocytes and human glioma (107). It is a transmembrane reductase with iron reductase activity (108), which uses ASC in the cytoplasm as an electron donor to transfer electrons through the endoplasmic reticulum for reduction of lumenal MDA and Fe3+ (109–111) (Fig. 3).
CYB561D2 is a putative tumor suppressor gene that is located in the 3p21.3 region of the human chromosome, where allelic deletions and genomic changes are frequently found in lung cancer and numerous other cancers (112–114). The overlap of heterozygosity deletion and homozygosity deletion in this region occurs frequently in lung and breast cancer, suggesting that one or more genes in this region play an important role in the pathogenesis of these cancers (112–115). Recombinant adenovirus-mediated transfection of 101F6 demonstrated that 101F6 inhibits cell growth alters the cell cycle and induces apoptosis in human lung cancer cells (116). In addition, the exogenous expression of 101F6 enhances the uptake of ASC by lung cancer cells, leading to the accumulation of cytotoxic H2O2. This cooperatively kills tumor cells through apoptosis and autophagy pathways, independently of caspase activation (116). By contrast, the upregulated expression of CYB561D2 has been found to be associated with a higher clinical grade and shorter survival time in patients with glioma. In vitro experiments revealed that the overexpression of CYB561D2 in glioma increased the expression of the immunosuppressive genes PD-L1, chemokine (C-C motif) ligand 2 and tryptophan 2,3-dioxygenase in co-cultured T cells (117), while CYB561D2 knockout inhibited the growth, colony formation and migration of glioma cells and promoted cell apoptosis (117).
5. Summary
The Cytb561 family is a class of transmembrane proteins characterized by six transmembrane helices and two heme groups, which exhibit electron transfer and ferric reductase activities. The main members of this family, namely CYB561, Dcytb, LCytb, SDR2 and 101F6, are involved in ASC recycling and iron metabolism. They are widely expressed in human tissues, upregulated or downregulated in different tumors, and involved in the pathogenesis of a variety of diseases and tumors. In addition, they are potential prognostic indicator for certain cancers. Although all members of this family theoretically exhibit iron reductase activity, they are not necessarily involved in iron metabolism in tumors, and Dcytb is a notable example. Members of the Cytb561 family play crucial roles in the promotion or suppression of numerous types of malignant tumors; for example, CYB561 promotes breast cancer growth, Dcytb facilitates glioma invasion and inhibits the growth of breast cancer and bladder cancer, CYB561A3 is crucial in the proliferation of Burkitt's lymphoma cells, and SDR2 accelerates cervical squamous cell carcinoma growth. In addition, 101F6 represses the growth of human lung cancer cells but contributes to the growth of glioma cells. These transmembrane proteins also have the potential to serve as therapeutic targets in various tumors. The reasons are as follows: i) Their expression levels are up- or downregulated in various tumor tissues compared with corresponding normal tissues, and this differential expression is associated with the prognosis of patients; for example, the upregulated expression of CYB561 in breast cancer is a poor prognostic factor. ii) At the cellular level, altering their expression levels can alter the functional status of tumor cells, such as by promoting or inhibiting cell proliferation, migration, invasion and apoptosis. iii) At the animal level, modulating their expression levels in tumor cells can affect the tumor size in nude mice; for example, the knockdown of CYB561 expression level in HER2-positive breast cancer cells reduced tumorigenicity compared with that in the control (84). The seemingly contradictory roles and tumor specificity of the members of the Cytb561 family suggest that their intricate mechanisms of action in tumors merit further study. At present, the role of this family in tumors and the associated mechanisms are still under investigation. To develop precise targeted treatments for patients with tumors, it is essential to conduct further research to elucidate the role of these iron metabolism-related proteins.
Acknowledgements
Not applicable.
Funding Statement
This review was supported by grants from the Department of Science and Technology of Qinghai Province in China (project no. 2024-SF-L01) and the Thousand Talents of Program of High-end Innovation of Qinghai Province in China.
Availability of data and materials
Not applicable.
Authors' contributions
XZ and ZA conceived of the review and acted as mentors and guarantors of the work. XZ wrote the manuscript. HLe was involved in drafting the manuscript. HLi modified the manuscript. XG reviewed and revised the text. Data authentication is not applicable. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kelley PM, Njus D. Cytochrome b561 spectral changes associated with electron transfer in chromaffin-vesicle ghosts. J Biol Chem. 1986;261:6429–6432. doi: 10.1016/S0021-9258(19)84579-9. [DOI] [PubMed] [Google Scholar]
- 2.Srivastava M. Xenopus cytochrome b561: Molecular confirmation of a general five transmembrane structure and developmental regulation at the gastrula stage. DNA Cell Biol. 1986;15:1075–1080. doi: 10.1089/dna.1996.15.1075. [DOI] [PubMed] [Google Scholar]
- 3.Asard H, Horemans N, Caubergs RJ. Transmembrane electron transport in ascorbate-loaded plasma membrane vesicles from higher plants involves a b-type cytochrome. FEBS Lett. 1992;306:143–146. doi: 10.1016/0014-5793(92)80986-Q. [DOI] [PubMed] [Google Scholar]
- 4.Flatmark T, Terland O, Helle KB. Electron carriers of the bovine adrenal chromaffin granules. Biochim. Biophys. Acta. 1971;226:9–19. doi: 10.1016/0005-2728(71)90173-3. [DOI] [PubMed] [Google Scholar]
- 5.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 6.Mouse Genome Sequencing Consortium, corp-author. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 7.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 8.Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 9.C. elegans Sequencing Consortium, corp-author. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 10.Asada A, Kusakawa T, Orii H, Agata K, Watanabe K, Tsubaki M. Planarian cytochrome b561: Conservation of a six transmembrane structure and localization along the central and peripheral nervous system. J Biochem. 2002;131:175–182. doi: 10.1093/oxfordjournals.jbchem.a003085. [DOI] [PubMed] [Google Scholar]
- 11.Arabidopsis Genome Initiative, corp-author. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2004;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 12.Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 13.Lu P, Ma D, Yan C, Gong X, Du M, Shi Y. Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase. Proc Natl Acad Sci USA. 2014;111:1813–1818. doi: 10.1073/pnas.1323931111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsubaki M, Takeuchi F, Nakanishi N. Cytochrome b561 protein family: Expanding roles and versatile transmembrane electron transfer abilities as predicted by a new classification system and protein sequence motif analyses. Biochim Biophys Acta. 2005;1753:174–190. doi: 10.1016/j.bbapap.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 15.Silsand T, Flatmark T. Purification of cytochrome b-561: An integral heme protein of the adrenal chromaffin granule membrane. Biochim Biophys Acta. 1974;359:257–266. doi: 10.1016/0005-2795(74)90223-2. [DOI] [PubMed] [Google Scholar]
- 16.Bérczi A, Zimányi L. The trans-membrane cytochrome b561 proteins: Structural information and biological function. Curr Protein Pept Sci. 2014;15:745–760. doi: 10.2174/1389203715666140828100351. [DOI] [PubMed] [Google Scholar]
- 17.McKie AT, Barrow D, Latunde-Dada GO, Rolfs A, Sager G, Mudaly E, Mudaly M, Richardson C, Barlow D, Bomford A, et al. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science. 2001;291:1755–1759. doi: 10.1126/science.1057206. [DOI] [PubMed] [Google Scholar]
- 18.Abbate V, Hider R. Iron in biology. Metallomics. 2017;9:1467–1469. doi: 10.1039/C7MT90039B. [DOI] [PubMed] [Google Scholar]
- 19.Galy B, Conrad M, Muckenthaler M. Mechanisms controlling cellular and systemic iron homeostasis. Nat Rev Mol Cell Biol. 2024;25:133–155. doi: 10.1038/s41580-023-00648-1. [DOI] [PubMed] [Google Scholar]
- 20.Kawabata H. Transferrin and transferrin receptors update. Free Radic Biol Med. 2019;133:46–54. doi: 10.1016/j.freeradbiomed.2018.06.037. [DOI] [PubMed] [Google Scholar]
- 21.Srai SK, Sharp P. Proteins of Iron Homeostasis. In: Anderson GJ, McLaren GD, editors. Iron Physiology and Pathophysiology in Humans. Humana Press; Totowa NJ, USA: 2012. pp. pp3–25. ISBN 978-1-60327-484-5. [DOI] [Google Scholar]
- 22.Hubert N, Hentze MW. Previously uncharacterized isoforms of divalent metal transporter (DMT)-1: Implications for regulation and cellular function. Proc Natl Acad Sci USA. 2002;99:12345–12350. doi: 10.1073/pnas.192423399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemler DJ, Lynch ML, Tesfay L, Deng Z, Paul BT, Wang X, Hegde P, Manz DH, Torti SV, Torti FM. DCYTB is a predictor of outcome in breast cancer that functions via iron-independent mechanisms. Breast Cancer Res. 2017;19:25. doi: 10.1186/s13058-017-0814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menniti FS, Knoth J, Diliberto EJ., Jr Role of ascorbic acid in dopamine beta-hydroxylation. The endogenous enzyme cofactor and putative electron donor for cofactor regeneration. J Biol Chem. 1986;261:16901–16908. doi: 10.1016/S0021-9258(19)75974-2. [DOI] [PubMed] [Google Scholar]
- 25.Kent UM, Fleming PJ. Purified cytochrome b561 catalyzes transmembrane electron transfer for dopamine beta-hydroxylase and peptidyl glycine alpha-amidating monooxygenase activities in reconstituted systems. J Biol Chem. 1987;262:8174–8178. doi: 10.1016/S0021-9258(18)47545-X. [DOI] [PubMed] [Google Scholar]
- 26.Lane DJ, Richardson DR. The active role of vitamin C in mammalian iron metabolism: Much more than just enhanced iron absorption! Free Radic Biol Med. 2014;75:69–83. doi: 10.1016/j.freeradbiomed.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Atanassova BD, Tzatchev KN. Ascorbic acid-important for iron metabolism. Folia Med (Plovdiv) 2008;50:11–16. [PubMed] [Google Scholar]
- 28.Lane DJR, Lawen A. Non-transferrin iron reduction and uptake are regulated by transmembrane ascorbate cycling in K562 cells. J Biol Chem. 2008;283:12701–12708. doi: 10.1074/jbc.M800713200. [DOI] [PubMed] [Google Scholar]
- 29.Lane DJ, Robinson SR, Czerwinska H, Bishop GM, Lawen A. Two routes of iron accumulation in astrocytes: Ascorbate-dependent ferrous iron uptake via the divalent metal transporter (DMT1) plus an independent route for ferric iron. Biochem J. 2010;432:123–132. doi: 10.1042/BJ20101317. [DOI] [PubMed] [Google Scholar]
- 30.Lane DJ, Chikhani S, Richardson V, Richardson DR. Transferrin iron uptake is stimulated by ascorbate via an intracellular reductive mechanism. Biochim Biophys Acta. 2013;1833:1527–1541. doi: 10.1016/j.bbamcr.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Toth I, Rogers JT, McPhee JA, Elliott SM, Abramson SL, Bridges KR. Ascorbic acid enhances iron-induced ferritin translation in human leukemia and hepatoma cells. J Biol Chem. 1995;270:2846–2852. doi: 10.1074/jbc.270.6.2846. [DOI] [PubMed] [Google Scholar]
- 32.Toth I, Bridges KR. Ascorbic acid enhances ferritin mRNA translation by an IRP/aconitase switch. J Biol Chem. 1995;270:19540–19544. doi: 10.1074/jbc.270.33.19540. [DOI] [PubMed] [Google Scholar]
- 33.Bridges KR. Ascorbic acid inhibits lysosomal autophagy of ferritin. J Biol Chem. 1987;262:14773–1478. doi: 10.1016/S0021-9258(18)47862-3. [DOI] [PubMed] [Google Scholar]
- 34.Hoffman KE, Yanelli K, Bridges KR. Ascorbic acid and iron metabolism: Alterations in lysosomal function. Am J Clin Nutr. 1991;54((6 Suppl)):S1188S–S1192S. doi: 10.1093/ajcn/54.6.1188s. [DOI] [PubMed] [Google Scholar]
- 35.Richardson DR. Role of ceruloplasmin and ascorbate in cellular iron release. J Lab Clin Med. 1999;134:454–465. doi: 10.1016/S0022-2143(99)90166-X. [DOI] [PubMed] [Google Scholar]
- 36.Crichton R. John Wiley and Sons; 2009. In Iron Metabolism: From Molecular Mechanisms to Cinical Consequences; pp. 17–58. [Google Scholar]
- 37.Sun H, Zhang C, Cao S, Sheng T, Dong N, Xu Y. Fenton reactions drive nucleotide and ATP syntheses in cancer. J Mol Cell Biol. 2018;10:448–459. doi: 10.1093/jmcb/mjy039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akatsuka S, Yamashita Y, Ohara H, Liu YT, Izumiya M, Abe K, Ochiai M, Jiang L, Nagai H, Okazaki Y, et al. Fenton reaction induced cancer in wild type rats recapitulates genomic alterations observed in human cancer. PLoS One. 2012;7:e43403. doi: 10.1371/journal.pone.0043403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torti SV, Torti FM. Iron and cancer: 2020 vision. Cancer Res. 2020;80:5435–5448. doi: 10.1158/0008-5472.CAN-20-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bian Z, Hann HW, Ye Z, Yin C, Wang Y, Fang W, Wan S, Wang C, Tao K. Ferritin level prospectively predicts hepatocarcinogenesis in patients with chronic hepatitis B virus infection. Oncol Lett. 2018;16:3499–3508. doi: 10.3892/ol.2018.9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song A, Eo W, Kim S, Shim B, Lee S. Significance of serum ferritin as a prognostic factor in advanced hepatobiliary cancer patients treated with Korean medicine: A retrospective cohort study. BMC Complement Altern Med. 2018;18:176. doi: 10.1186/s12906-018-2240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue X, Ramakrishnan SK, Weisz K, Triner D, Xie L, Attili D, Pant A, Győrffy B, Zhan M, Carter-Su C, et al. Iron uptake via DMT1 integrates cell cycle with JAK-STAT3 signaling to promote colorectal tumorigenesis. Cell Metab. 2016;24:447–461. doi: 10.1016/j.cmet.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gray CP, Arosio P, Hersey P. Association of increased levels of heavy-chain ferritin with increased CD4+ CD25+ regulatory T-cell levels in patients with melanoma. Clin Cancer Res. 2003;9:2551–2559. [PubMed] [Google Scholar]
- 44.Liu NQ, De Marchi T, Timmermans AM, Beekhof R, Trapman-Jansen AM, Foekens R, Look MP, van Deurzen CH, Span PN, Sweep FC, et al. Ferritin heavy chain in triple negative breast cancer: A favorable prognostic marker that relates to a cluster of differentiation 8 positive (CD8+) effector T-cell response. Mol Cell Proteomics. 2014;13:1814–1827. doi: 10.1074/mcp.M113.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lelièvre P, Sancey L, Coll JL, Deniaud A, Busser B. Iron dysregulation in human cancer: Altered metabolism, biomarkers for diagnosis, prognosis, monitoring and rationale for therapy. Cancers (Basel) 2020;12:3524. doi: 10.3390/cancers12123524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Yu L, Ding J, Chen Y. Iron metabolism in cancer. Int J Mol Sci. 2018;20:95. doi: 10.3390/ijms20010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Habashy HO, Powe DG, Staka CM, Rakha EA, Ball G, Green AR, Aleskandarany M, Paish EC, Douglas Macmillan R, Nicholson RI, et al. Transferrin receptor (CD71) is a marker of poor prognosis in breast cancer and can predict response to tamoxifen. Breast Cancer Res Treat. 2010;119:283–293. doi: 10.1007/s10549-009-0345-x. [DOI] [PubMed] [Google Scholar]
- 48.Alkhateeb AA, Han B, Connor JR. Ferritin stimulates breast cancer cells through an iron-independent mechanism and is localized within tumor-associated macrophages. Breast Cancer Res Treat. 2013;137:733–744. doi: 10.1007/s10549-012-2405-x. [DOI] [PubMed] [Google Scholar]
- 49.Pinnix ZK, Miller LD, Wang W, D'Agostino R, Jr, Kute T, Willingham MC, Farris M, Petty WJ, de Hoyos A, Weaver KE, Wentworth S. Ferroportin and iron regulation in breast cancer progression and prognosis. Sci Transl Med. 2010;2:43ra56. doi: 10.1126/scitranslmed.3001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morales M, Xue X. Targeting iron metabolism in cancer therapy. Theranostics. 2021;11:8412–8429. doi: 10.7150/thno.59092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bashtovyy D, Bérczi A, Asard H, Páli T. Structure prediction for the di-heme cytochrome b561 protein family. Protoplasma. 2003;221:31–40. doi: 10.1007/s00709-002-0065-0. [DOI] [PubMed] [Google Scholar]
- 52.Perin MS, Fried VA, Slaughter CA, Südhof TC. The structure of cytochrome b561, a secretory vesicle-specific electron transport protein. EMBO J. 1988;7:2697–2703. doi: 10.1002/j.1460-2075.1988.tb03123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asard H, Kapila J, Verelst W, Bérczi A. Higher-plant plasma membrane cytochrome b561: A protein in search of a function. Protoplasma. 2001;217:77–93. doi: 10.1007/BF01289417. [DOI] [PubMed] [Google Scholar]
- 54.Degli Esposti M, Kamensky YuA, Arutjunjan AM, Konstantinov AA. A model for the molecular organization of cytochrome beta-561 in chromaffin granule membranes. FEBS Lett. 1989;254:74–78. doi: 10.1016/0014-5793(89)81012-9. [DOI] [PubMed] [Google Scholar]
- 55.Tsubaki M, Nakayama M, Okuyama E, Ichikawa Y, Hori H. Existence of two heme B centers in cytochrome b561 from bovine adrenal chromaffin vesicles as revealed by a new purification procedure and EPR spectroscopy. J Biol Chem. 1997;272:23206–23210. doi: 10.1074/jbc.272.37.23206. [DOI] [PubMed] [Google Scholar]
- 56.Oakhill JS, Marritt SJ, Gareta EG, Cammack R, McKie AT. Functional characterization of human duodenal cytochrome b (Cybrd1): Redox properties in relation to iron and ascorbate metabolism. Biochim Biophys Acta. 2008;1777:260–268. doi: 10.1016/j.bbabio.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 57.Bérczi A, Su D, Lakshminarasimhan M, Vargas A, Asard H. Heterologous expression and site-directed mutagenesis of an ascorbate-reducible cytochrome b561. Arch Biochem Biophys. 2005;443:82–92. doi: 10.1016/j.abb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 58.Kamensky Y, Liu W, Tsai AL, Kulmacz RJ, Palmer G. Axial ligation and stoichiometry of heme centers in adrenal cytochrome b561. Biochemistry. 2007;46:8647–8658. doi: 10.1021/bi700054g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okuyama E, Yamamoto R, Ichikawa Y, Tsubaki M. Structural basis for the electron transfer across the chromaffin vesicle membranes catalyzed by cytochrome b561: Analyses of cDNA nucleotide sequences and visible absorption spectra. Biochim. Biophys. Acta. 1998;1383:269–278. doi: 10.1016/s0167-4838(97)00216-1. [DOI] [PubMed] [Google Scholar]
- 60.Takeuchi F, Kobayashi K, Tagawa S, Tsubaki M. Ascorbate inhibits the carbethoxylation of two histidyl and one tyrosyl residues indispensable for the transmembrane electron transfer reaction of cytochrome b561. Biochemistry. 2001;40:4067–4076. doi: 10.1021/bi002240x. [DOI] [PubMed] [Google Scholar]
- 61.Aravind L. DOMON: An ancient extracellular domain in dopamine beta-monooxygenase and other proteins. Trends Biochem Sci. 2001;26:524–526. doi: 10.1016/S0968-0004(01)01924-7. [DOI] [PubMed] [Google Scholar]
- 62.Picco C, Scholz-Starke J, Naso A, Preger V, Sparla F, Trost P, Carpaneto A. How are cytochrome b561 electron currents controlled by membrane voltage and substrate availability? Antioxid Redox Signal. 2014;21:384–391. doi: 10.1089/ars.2013.5809. [DOI] [PubMed] [Google Scholar]
- 63.Srivastava M, Gibson KR, Pollard HB, Fleming PJ. Human cytochrome b561: A revised hypothesis for conformation in membranes which reconciles sequence and functional information. Biochem J. 1994;303:915–921. doi: 10.1042/bj3030915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakanishi N, Takeuchi F, Tsubaki M. Histidine cycle mechanism for the concerted proton/electron transfer from ascorbate to the cytosolic haem b centre of cytochrome b561: A unique machinery for the biological transmembrane electron transfer. J Biochem. 2007;142:553–560. doi: 10.1093/jb/mvm181. [DOI] [PubMed] [Google Scholar]
- 65.Kipp BH, Kelley PM, Njus D. Evidence for an essential histidine residue in the ascorbate-binding site of cytochrome b561. Biochemistry. 2001;40:3931–3937. doi: 10.1021/bi002214z. [DOI] [PubMed] [Google Scholar]
- 66.Iliadi KG, Avivi A, Iliadi NN, Knight D, Korol AB, Nevo E, Taylor P, Moran MF, Kamyshev NG, Boulianne GL. Nemy encodes a cytochrome b561 that is required for Drosophila learning and memory. Proc Natl Acad Sci USA. 2008;105:19986–19991. doi: 10.1073/pnas.0810698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Su D, Asard H. Three mammalian cytochromes b561 are ascorbate-dependent ferrireductases. FEBS J. 2006;273:3722–3734. doi: 10.1111/j.1742-4658.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- 68.VanDuijn MM, Tijssen K, VanSteveninck J, Van Den Broek PJ, Van Der Zee J. Erythrocytes reduce extracellular ascorbate free radicals using intracellular ascorbate as an electron donor. J Biol Chem. 2000;275:27720–27725. doi: 10.1074/jbc.M910281199. [DOI] [PubMed] [Google Scholar]
- 69.Asard H, Venken M, Caubergs R, Reijnders W, Oltmann FL, De Greef JA. b-Type cytochromes in higher plant plasma membranes. Plant Physiol. 1989;90:1077–1083. doi: 10.1104/pp.90.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Askerlund P, Larsson C, Widell S. Cytochromes of plant plasma membranes. Characterization by absorbance difference spectroscopy and redox titration. Physiol Plant. 1989;76:123–134. doi: 10.1111/j.1399-3054.1989.tb05621.x. [DOI] [Google Scholar]
- 71.Vargas JD, Herpers B, McKie AT, Gledhill S, McDonnell J, van den Heuvel M, Davies KE, Ponting CP. Stromal cell-derived receptor 2 and cytochrome b561 are functional ferric reductases. Biochim Biophys Acta. 2003;1651:116–123. doi: 10.1016/S1570-9639(03)00242-5. [DOI] [PubMed] [Google Scholar]
- 72.Herrmann T, Muckenthaler M, van der Hoeven F, Brennan K, Gehrke SG, Hubert N, Sergi C, Gröne HJ, Kaiser I, Gosch I, et al. Iron overload in adult Hfe-deficient mice independent of changes in the steady-state expression of the duodenal iron transporters DMT1 and Ireg1/ferroportin. J Mol Med. 2004;82:39–48. doi: 10.1007/s00109-003-0508-x. [DOI] [PubMed] [Google Scholar]
- 73.Escriou V, Laporte F, Garin J, Brandolin G, Vignais PV. Purification and physical properties of a novel type of cytochrome b from rabbit peritoneal neutrophils. J Biol Chem. 1994;269:14007–14014. doi: 10.1016/S0021-9258(17)36747-9. [DOI] [PubMed] [Google Scholar]
- 74.Pruss RM, Shepard EA. Cytochrome b561 can be detected in many neuroendocrine tissues using a specific monoclonal antibody. Neuroscience. 1987;22:149–157. doi: 10.1016/0306-4522(87)90205-3. [DOI] [PubMed] [Google Scholar]
- 75.Srivastava M. Genomic structure and expression of the human gene encoding cytochrome b561, an integral protein of the chromaffin granule membrane. J Biol Chem. 1995;270:22714–22720. doi: 10.1074/jbc.270.39.22714. [DOI] [PubMed] [Google Scholar]
- 76.Njus D, Kelley PM. The secretory-vesicle ascorbate-regenerating system: A chain of concerted H+/e(−)-transfer reactions. Biochim Biophys Acta. 1993;1144:235–248. doi: 10.1016/0005-2728(93)90108-R. [DOI] [PubMed] [Google Scholar]
- 77.Olak ME, Thirdborough SM, Ung CY, Elliott T, Healy E, Freeman TC, Ardern-Jones MR. Distinct molecular signature of human skin langerhans cells denotes critical differences in cutaneous dendritic cell immune regulation. J Invest Dermatol. 2014;134:695–703. doi: 10.1038/jid.2013.375. [DOI] [PubMed] [Google Scholar]
- 78.Van den Berg MP, Almomani R, Biaggioni I, van Faassen M, van der Harst P, Silljé HHW, Mateo Leach I, Hemmelder MH, Navis G, Luijckx GJ, et al. Mutations in CYB561 causing a novel orthostatic hypotension syndrome. Circ Res. 2018;122:846–854. doi: 10.1161/CIRCRESAHA.117.311949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Willis S, Villalobos VM, Gevaert O, Abramovitz M, Williams C, Sikic BI, Leyland-Jones B. Single gene prognostic biomarkers in ovarian cancer: A meta-analysis. PLoS One. 2016;11:e0149183. doi: 10.1371/journal.pone.0149183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olarte CK, Bagamasbad DP. SAT-132 the secretory vesicle membrane protein, CYB561, promotes the growth and metastatic potential of castration-resistant neuroendocrine prostate cancer. J Endocr Soc. 2020;4((Suppl 1)):SAT–132. doi: 10.1210/jendso/bvaa046.1194. [DOI] [Google Scholar]
- 81.Zhou X, Shen G, Ren D, Guo X, Han J, Guo Q, Zhao F, Wang M, Dong Q, Li Z, Zhao J. Expression and clinical prognostic value of CYB561 in breast cancer. J Cancer Res Clin Oncol. 2022;148:1879–1892. doi: 10.1007/s00432-022-03928-z. [DOI] [PubMed] [Google Scholar]
- 82.Yang X, Zhao Y, Shao Q, Jiang G. Cytochrome b561 serves as a potential prognostic biomarker and target for breast cancer. Int J Gen Med. 2021;14:10447–10464. doi: 10.2147/IJGM.S338878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou X, Guo X, Han J, Wang M, Liu Z, Ren D, Zhao J, Li Z. Cytochrome b561 regulates iron metabolism by activating the Akt/mTOR pathway to promote Breast Cancer Cells proliferation. Exp Cell Res. 2023;431:113760. doi: 10.1016/j.yexcr.2023.113760. [DOI] [PubMed] [Google Scholar]
- 84.Zhao T, Wang C, Zhao N, Qiao G, Hua J, Meng D, Liu L, Zhong B, Liu M, Wang Y, et al. CYB561 promotes HER2+ breast cancer proliferation by inhibiting H2AFY degradation. Cell Death Discov. 2024;10:38. doi: 10.1038/s41420-024-01804-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ganasen M, Togashi H, Takeda H, Asakura H, Tosha T, Yamashita K, Hirata K, Nariai Y, Urano T, Yuan X, et al. Structural basis for promotion of duodenal iron absorption by enteric ferric reductase with ascorbate. Commun Biol. 2018;1:120. doi: 10.1038/s42003-018-0121-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Su D, May JM, Koury MJ, Asard H. Human erythrocyte membranes contain a cytochrome b561 that may be involved in extracellular ascorbate recycling. J Biol Chem. 2006;281:39852–39859. doi: 10.1074/jbc.M606543200. [DOI] [PubMed] [Google Scholar]
- 87.Wyman S, Simpson RJ, McKie AT, Sharp PA. Dcytb (Cybrd1) functions as both a ferric and a cupric reductase in vitro. FEBS Lett. 2008;582:1901–1906. doi: 10.1016/j.febslet.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 88.Asard H, Barbaro R, Trost P, Bérczi A. Cytochromes b561: Ascorbate-mediated trans-membrane electron transport. Antioxid Redox Signal. 2013;19:1026–1035. doi: 10.1089/ars.2012.5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choi J, Masaratana P, Latunde-Dada GO, Arno M, Simpson RJ, McKie AT. Duodenal reductase activity and spleen iron stores are reduced and erythropoiesis is abnormal in Dcytb knockout mice exposed to hypoxic conditions. J Nutr. 2012;142:1929–1934. doi: 10.3945/jn.112.160358. [DOI] [PubMed] [Google Scholar]
- 90.Xue X, Taylor M, Anderson E, Hao C, Qu A, Greenson JK, Zimmermann EM, Gonzalez FJ, Shah YM. Hypoxia-inducible factor-2α activation promotes colorectal cancer progression by dysregulating iron homeostasis. Cancer Res. 2012;72:2285–2293. doi: 10.1158/0008-5472.CAN-11-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brookes MJ, Hughes S, Turner FE, Reynolds G, Sharma N, Ismail T, Berx G, McKie AT, Hotchin N, Anderson GJ, et al. Modulation of iron transport proteins in human colorectal carcinogenesis. Gut. 2006;55:1449–1460. doi: 10.1136/gut.2006.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen R, Cao J, Jiang W, Wang S, Cheng J. Upregulated expression of CYBRD1 predicts poor prognosis of patients with ovarian cancer. J Oncol. 2021;2021:7548406. doi: 10.1155/2021/7548406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qing M, Zhou J, Chen W, Cheng L. Highly expressed CYBRD1 associated with glioma recurrence regulates the immune response of glioma cells to interferon. Evid Based Complement Alternat Med. 2021;2021:2793222. doi: 10.1155/2021/2793222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boult J, Roberts K, Brookes MJ, Hughes S, Bury JP, Cross SS, Anderson GJ, Spychal R, Iqbal T, Tselepis C. Overexpression of cellular iron import proteins is associated with malignant progression of esophageal adenocarcinoma. Clin Cancer Res. 2008;14:379–387. doi: 10.1158/1078-0432.CCR-07-1054. [DOI] [PubMed] [Google Scholar]
- 95.Rychtarcikova Z, Lettlova S, Tomkova V, Korenkova V, Langerova L, Simonova E, Zjablovskaja P, Alberich-Jorda M, Neuzil J, Truksa J. Tumor-initiating cells of breast and prostate origin show alterations in the expression of genes related to iron metabolism. Oncotarget. 2017;8:6376–6398. doi: 10.18632/oncotarget.14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee HY, Li CC, Li WM, Hsu YL, Yeh HC, Ke HL, Yeh BW, Huang CN, Li CF, Kuo PL, Wu WJ. Identification of potential genes in upper tract urothelial carcinoma using next-generation sequencing with bioinformatics and in vitro analyses. PeerJ. 2021;9:e11343. doi: 10.7717/peerj.11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma J, Huang W, Zhu C, Sun X, Zhang Q, Zhang L, Qi Q, Bai X, Feng Y, Wang C. miR-423-3p activates FAK signaling pathway to drive EMT process and tumor growth in lung adenocarcinoma through targeting CYBRD1. J Clin Lab Anal. 2021;35:e24044. doi: 10.1002/jcla.24044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang J, Cheng Y, Duan M, Qi N, Liu J. Unveiling differentially expressed genes upon regulation of transcription factors in sepsis. Biotech. 2017;7:46. doi: 10.1007/s13205-017-0713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Al-Eitan LN, Tarkhan AH, Alghamdi MA, Al-Qarqaz FA, Al-Kofahi HS. Transcriptome analysis of HPV-induced warts and healthy skin in humans. BMC Med Genomics. 2020;13:35. doi: 10.1186/s12920-020-0700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Meng F, Fleming BA, Jia X, Rousek AA, Mulvey MA, Ward DM. Lysosomal iron recycling in mouse macrophages is dependent upon both LcytB and Steap3 reductases. Blood Adv. 2022;6:1692–1707. doi: 10.1182/bloodadvances.2021005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Z, Guo R, Trudeau SJ, Wolinsky E, Ast T, Liang JH, Jiang C, Ma Y, Teng M, Mootha VK, Gewurz BE. CYB561A3 is the key lysosomal iron reductase required for Burkitt B-cell growth and survival. Blood. 2021;138:2216–2230. doi: 10.1182/blood.2021011079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lemonnier N, Melén E, Jiang Y, Joly S, Ménard C, Aguilar D, Acosta-Perez E, Bergström A, Boutaoui N, Bustamante M, et al. A novel whole blood gene expression signature for asthma, dermatitis, and rhinitis multimorbidity in children and adolescents. Allergy. 2020;75:3248–3260. doi: 10.1111/all.14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu H, Liu L, Liu Q, He F, Zhu H. LncRNA HOXD-AS1 affects proliferation and apoptosis of cervical cancer cells by promoting FRRS1 expression via transcription factor ELF1. Cell Cycle. 2022;21:416–426. doi: 10.1080/15384101.2021.2020962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ponting CP. Domain homologues of dopamine b-hydroxylase and ferric reductase: Roles for iron metabolism in neurodegenerative disorders? Hum Mol Genet. 2001;10:1853–1858. doi: 10.1093/hmg/10.17.1853. [DOI] [PubMed] [Google Scholar]
- 105.Binder J, Ursu O, Bologa C, Jiang S, Maphis N, Dadras S, Chisholm D, Weick J, Myers O, Kumar P, et al. Machine learning prediction and tau-based screening identifies potential Alzheimer's disease genes relevant to immunity. Commun Biol. 2022;5:125. doi: 10.1038/s42003-022-03068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Linton KM, Hey Y, Saunders E, Jeziorska M, Denton J, Wilson CL, Swindell R, Dibben S, Miller CJ, Pepper SD, et al. Acquisition of biologically relevant gene expression data by Affymetrix microarray analysis of archival formalin-fixed paraffin-embedded tumours. Br J Cancer. 2008;98:1403–1414. doi: 10.1038/sj.bjc.6604316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li S, Shi J, Gao H, Yuan Y, Chen Q, Zhao Z, Wang X, Li B, Ming L, Zhong J, et al. Identification of a gene signature associated with radiotherapy and prognosis in gliomas. Oncotarget. 2017;8:88974–88987. doi: 10.18632/oncotarget.21634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.El Behery M, Fujimura M, Kimura T, Tsubaki M. Direct measurements of ferric reductase activity of human 101F6 and its enhancement upon reconstitution into phospholipid bilayer nanodisc. Biochem Biophys Rep. 2020;21:100730. doi: 10.1016/j.bbrep.2020.100730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mizutani A, Sanuki R, Kakimoto K, Kojo S, Taketani S. Involvement of 101F6, a homologue of cytochrome b561, in the reduction of ferric ions. J Biochem. 2007;142:699–705. doi: 10.1093/jb/mvm185. [DOI] [PubMed] [Google Scholar]
- 110.Recuenco MC, Fujito M, Rahman MM, Sakamoto Y, Takeuchi F, Tsubaki M. Functional expression and characterization of human 101F6 protein, a homologue of cytochrome b561 and a candidate tumor suppressor gene product. Biofactors. 2008;34:219–230. doi: 10.1002/biof.5520340306. [DOI] [PubMed] [Google Scholar]
- 111.Recuenco MC, Rahman MM, Takeuchi F, Kobayashi K, Tsubaki M. Electron transfer reactions of candidate tumor suppressor 101F6 protein, a cytochrome b561 homologue, with ascorbate and monodehydroascorbate radical. Biochemistry. 2013;52:3660–3668. doi: 10.1021/bi301607s. [DOI] [PubMed] [Google Scholar]
- 112.Ji L, Nishizaki M, Gao B, Burbee D, Kondo M, Kamibayashi C, Xu K, Yen N, Atkinson EN, Fang B, et al. Expression of several genes in the human chromosome 3p21.3 homozygous deletion region by an adenovirus vector results in tumor suppressor activities in vitro and in vivo. Cancer Res. 2002;62:2715–2720. [PMC free article] [PubMed] [Google Scholar]
- 113.Ji L, Minna JD, Roth JA. 3p21.3 tumor suppressor cluster: Prospects for translational applications. Future Oncol. 2005;1:79–92. doi: 10.1517/14796694.1.1.79. [DOI] [PubMed] [Google Scholar]
- 114.Lerman MI, Minna JD. The international lung cancer chromosome 3p21.3 tumor suppressor gene consortium. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: Identification and evaluation of the resident candidate tumor suppressor genes. Cancer Res. 2000;60:6116–6133. [PubMed] [Google Scholar]
- 115.Zabarovsky ER, Lerman MI, Minna JD. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene. 2002;21:6915–6935. doi: 10.1038/sj.onc.1205835. [DOI] [PubMed] [Google Scholar]
- 116.Ohtani S, Iwamaru A, Deng W, Ueda K, Wu G, Jayachandran G, Kondo S, Atkinson EN, Minna JD, Roth JA, Ji L. Tumor suppressor 101F6 and ascorbate synergistically and selectively inhibit non-small cell lung cancer growth by caspase-independent apoptosis and autophagy. Cancer Res. 2007;67:6293–6303. doi: 10.1158/0008-5472.CAN-06-3884. [DOI] [PubMed] [Google Scholar]
- 117.Tao B, Shi J, Shuai S, Zhou H, Zhang H, Li B, Wang X, Li G, He H, Zhong J. CYB561D2 up-regulation activates STAT3 to induce immunosuppression and aggression in gliomas. J Transl Med. 2021;19:338. doi: 10.1186/s12967-021-02987-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.