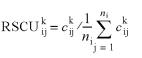Abstract
Genomics projects have resulted in a flood of sequence data. Functional annotation currently relies almost exclusively on inter-species sequence comparison and is restricted in cases of limited data from related species and widely divergent sequences with no known homologs. Here, we demonstrate that codon composition, a fusion of codon usage bias and amino acid composition signals, can accurately discriminate, in the absence of sequence homology information, cytoplasmic ribosomal protein genes from all other genes of known function in Saccharomyces cerevisiae, Escherichia coli and Mycobacterium tuberculosis using an implementation of support vector machines, SVMlight. Analysis of these codon composition signals is instructive in determining features that confer individuality to ribosomal protein genes. Each of the sets of positively charged, negatively charged and small hydrophobic residues, as well as codon bias, contribute to their distinctive codon composition profile. The representation of all these signals is sensitively detected, combined and augmented by the SVMs to perform an accurate classification. Of special mention is an obvious outlier, yeast gene RPL22B, highly homologous to RPL22A but employing very different codon usage, perhaps indicating a non-ribosomal function. Finally, we propose that codon composition be used in combination with other attributes in gene/protein classification by supervised machine learning algorithms.
INTRODUCTION
Our understanding of biology has been greatly influenced by the numerous whole genome sequencing projects, beginning with microbial genomes, continuing with the eukaryotic species Saccharomyces cerevisiae, Caenorhabditis elegans (worm), Drosophila melanogaster (fruit fly) and Arabidopsis thaliana (mustard weed) and culminating most recently with the human and mouse genomes. Others, either on the way or being contemplated, include the genomes of rat, zebrafish, puffer fish and non-human primates. Meanwhile, the widespread use of electronic literature, introduction of high throughput assays for gene expression and other large-scale projects (for example, mutagenesis screens and phenotyping projects for the mouse; 1,2) are also vastly increasing the amount of digital information that is available. Although researchers still need to practise critical thought, they are now able to perform data-driven experiments by devising new ways to handle and isolate appropriate subsets of complex observational data derived from nature (3).
Genomic data of all types (e.g. sequence information) are of relatively low value without the incorporation of data obtained from classical ad hoc experimental approaches. As stand-alone data, they do not immediately address questions concerning function, mechanism and regulation, issues of the greatest interest to biologists. Furthermore, all forms of genomic data are prone to error, for example, the annotated information of the function of a gene product inferred by sequence homology. However, with the advent of both computational capacity and underlying mathematical logic used to make inferences, statistical learning theory (4), typically the support vector machine (SVM) (4,5), is now in a phase of success characteristic of an observational stage of science and is now capable of providing additional insight into, for instance, gene expression and function (6). The SVM is a type of supervised machine learning algorithm that can be integrated with a priori knowledge based on investigation and knowledge accumulated in each domain of science, such as the Gene Ontology (GO) system (7). Here, all genes of known function have been organized into a directed acyclic graph (DAG) according to molecular function, localization and biological processes their products are involved in. By using a dynamic, controlled vocabulary applicable to all eukaryotes, the GO system is rapidly gaining popularity and has been applied to the S.cerevisiae, fruit fly, mouse and worm genomes to build a knowledge database of the roles of genes and proteins in cells. Such a priori knowledge can be easily exploited for the careful choice of genomic datasets; SVMs trained on these datasets usually yield a robust classification in practice.
In this paper, we have investigated the ability of SVMs to discriminate ribosomal protein coding genes (rp genes) from all other genes of known function based on their codon composition in Escherichia coli, Mycobacterium tuberculosis and S.cerevisiae. Codon composition is inherently the fusion of both codon usage bias and amino acid composition signals. It is well recognized that there is a high correlation between codon bias and gene expression levels, which in turn is related to function and/or similarity in regulation. Amino acid composition is related to the physico-chemical properties of the protein and, hence, perhaps ultimately to its function. Here we demonstrate that rp genes exhibit markedly different conserved codon composition patterns from other genes in E.coli, M.tuberculosis and S.cerevisiae. We also show that a careful analysis of the classification by the SVMs can provide valuable insights into the specific features that confer individuality to this set of genes. Finally, based on our results, we propose that codon composition is a potentially efficacious attribute that can be used in combination with other attributes in the classifying of genes/proteins by supervised machine learning algorithms.
MATERIALS AND METHODS
Sequence data
The whole genomes of E.coli K-12 (8) and M.tuberculosis CDC1551 and full-length sequences of the 16 S.cerevisiae chromosomes along with gene annotations were retrieved from the Genome division of GenBank. All coding sequences (CDS) and their translated sequences were checked locally with respect to the corresponding translation tables (translation table 11 for E.coli and M.tuberculosis and the standard table for S.cerevisiae) to avoid annotation errors present in the original datasets. There were 4289 putative protein coding genes in E.coli, 4187 in M.tuberculosis and 6312 in S.cerevisiae.
Codon composition and codon usage
Each protein coding gene sequence (excluding initiation and stop codons) was represented by a 61-dimensional vector with respect to the 61 sense codons,
ck = (ckij), k = 1, 2, …, K; i = 1, 2, …, 20; j = 1, …, ni
where, ck is the vector representing the kth protein coding gene (out of a total of K genes) and ni is the number of synonymous codons (j represents the jth synonymous codon) of the ith amino acid (of the possible 20). In our study, K = 4289 for E.coli, 4187 for M.tuberculosis and 6312 for S.cerevisiae (see above). Based on the dataset of 61-dimensional sense codon vectors, the codon composition of each gene was calculated as the frequency of each codon of the gene. Codon bias (of the kth gene), measured by its relative synonymous codon usage (RSCU; 9), was calculated thus:
Training and test datasets for E.coli, M.tuberculosis and S.cerevisiae
Ribosomal protein genes were extracted from the 4289 genes of E.coli and the 4187 genes of M.tuberculosis by a keyword search of the annotation field of the CDS in the feature table of the complete genome sequences. In E.coli, the 55 rp genes obtained (Table 1) were taken as positive training examples for the SVMs. We felt that 55 genes were too few to be split into training and testing datasets, so we used this same set for training and testing in E.coli. The rest of the genes were split into two groups. One group consisted of 1432 genes of unknown function whose products were annotated as ‘hypothetical’, ‘unclassified’, ‘putative’ or ‘similar to…’. The other group comprised 2802 genes whose functions are well known. This group was further subdivided randomly into two groups that were used as the negative training dataset (1408 genes) and the negative test dataset (1394 genes), respectively. Similarly, in M.tuberculosis, the 56 genes annotated as rp genes (Table 1) were used as the positive training and testing datasets. Of the remaining protein coding genes the 2146 with known function were randomly and equally divided into the negative training and testing datasets. The trained model was also applied on the set of 1905 genes of unknown function.
Table 1. The list of ribosomal protein coding genes in E.coli and S.cerevisiae.
| Organism | Subunit | Length | Genes |
|---|---|---|---|
| E.coli | Large subunit | 33a (34) | rpmF, rplT, rpmI, rplY, rplS, rpmA, rplU, rplM, rplQ, rpmJ, rplO, rpmD, rplR, rplF, rplE, rplX, rplN, rpmC, rplP, rplV, rplB, rplW, rplD, rplC, rpmG, rpmB, rpmH, rpmE, rplK, rplA, rplJ, rplL, rplI |
| Small subunit | 22a (21) | rpsT, rpsB, rpsA, rpsV, rpsP, rpsU, rpsO, rpsI, rpsD, rpsK, rpsM, rpsE, rpsH, rpsN, rpsQ, rpsC, rpsS, rpsJ, rpsG, rpsL, rpsF, rpsR | |
| M.tuberculosis (CDC1551) | Large subunit | 34 | MT0669.1, MT0680, MT0669, MT3548, MT0741, MT0748, MT0735, MT3563, MT0745, MT2972, MT0731, MT1681, MT2518, MT0733, MT0730, MT0741.1, MT2517, MT3052.2, MT2118, MT0114, MT0736, MT0728, MT0747, MT1337, MT2117.1, MT0663, MT4041.1, MT1680, MT3567.1, MT0729, MT0742, MT0744, MT0681, MT0062 |
| Small subunit | 22 | MT1666, MT0727, MT3566, MT0710, MT3567, MT2117, MT0742.1, MT2855, MT2977, MT0737, MT2116, MT0061, MT0732, MT2958, MT2485, MT0734, MT3565, MT0746, MT0059, MT0711, MT0743, MT3547 | |
| S.cerevisiae | Large subunit | 46 | RPP0, RPP1A, RPP2A, RPL1A, RPL2A, RPL3, RPL4A, RPL5, RPL6A, RPL7A, RPL8A, RPL9A, RPL10, RPL11A, RPL12A, RPL13A, RPL14A, RPL15A, RPL16A, RPL17A, RPL18A, RPL19A, RPL20A, RPL21A, RPL22A, RPL23A, RPL24A, RPL25, RPL26A, RPL27A, RPL28, RPL29, RPL30, RPL31A, RPL32, RPL33A, RPL34A, RPL35A, RPL36A, RPL37A, RPL38, RPL39, RPL40A, RPL41A, RPL42A, RPL43A |
| 35 duplicates | RPP1B, RPP2B, RPL1B, RPL2B, RPL4B, RPL6B, RPL7B, RPL8B, RPL9B, RPL11B, RPL12B, RPL13B, RPL14B, RPL15B, RPL16B, RPL17B, RPL18B, RPL19B, RPL20B, RPL21B, RPL22B, RPL23B, RPL24B, RPL26B, RPL27B, RPL31B, RPL33B, RPL34B, RPL35B, RPL36B, RPL37B, RPL40B, RPL41B, RPL42B, RPL43B | ||
| Small subunit | 32 | RPS0A, RPS1A, RPS2, RPS3, RPS4A, RPS5, RPS6A, RPS7A, RPS8A, RPS9A, RPS10A, RPS11A, RPS12, RPS13, RPS14A, RPS15, RPS16A, RPS17A, RPS18A, RPS19A, RPS20, RPS21A, RPS22A, RPS23A, RPS24A, RPS25A, RPS26A, RPS27A, RPS28A, RPS29A, RPS30A, RPS31 | |
| 24 duplicates | RPS0B, RPS1B, RPS4B, RPS6B, RPS7B, RPS8B, RPS9B, RPS10B, RPS11B, RPS14B, RPS16B, RPS17B, RPS18B, RPS19B, RPS21B, RPS22B, RPS23B, RPS24B, RPS25B, RPS26B, RPS27B, RPS28B, RPS29B, RPS30B |
aMost authors state that there are 34 r proteins in the large subunit and 21 in the small subunit of E.coli. However, according to annotation information extracted from GenBank records there are 33 and 22 r proteins in the large and small subunits, respectively.
Similarly, using the GO counterpart of the classification of molecular function of genes of the Saccharomyces Genome Database (SGD) (http://www.geneontology.org/gene_association. sgd), the 6312 genes of S.cerevisiae were classed as genes of unknown function (3039 genes). Among the genes of known function, the 137 cytoplasmic rp genes (10,11) were divided into two groups. One of them included 78 non-duplicated genes and was used as the positive training dataset; the other 59 duplicated genes were used as the positive test dataset. Also, genes coding for histones (9) and enzymes (1041) were chosen as the negative training dataset; the rest of the genes of known function (2086) were taken as the negative test dataset.
Support vector machines
In theory, a simple and intuitive way to build a binary classifier is to construct a hyperplane, which separates class members from non-members. Unfortunately, most real world problems are not linearly separable based on the collected data. One solution is to map the data into higher dimensional space (feature space) and define a separating hyperplane there. However, this usually invokes both computational and learning algorithmic costs, which SVMs elegantly bypass (4,5). SVMs avoid over-fitting by choosing the maximum (soft) margin separating hyperplane in the feature space and reduce computational complexity by using kernel functions which connect input space and feature space directly for similarity comparison computing. Kernel functions allow one to work in feature space without explicitly computing all elements. Though an SVM is essentially a binary classifier, it can also deal with multi-class classification problems (4,12). Success with SVMs requires careful attention to two key aspects: the kernel function and the magnitude of the trade-off between accuracy and generalization.
In this study, we used SVMlight v.3.5 (13; http://ais.gmd.de/~thorsten/svm_light/) for SVM data training and classifying. SVMlight is an implementation of SVMs in C. Its main features include a fast optimization algorithm, efficient computation of leave-one-out estimates and the capacity to handle many thousands of support vectors and several tens of thousands of training examples, as well as the sparse vector representation of input objects that are either trained or classified. Different kernel functions were applied in our experiments, including linear function, polynomial function and radial basis function (RBF). We found that the RBF along with well-chosen parameters (100 ≤ γ ≤ 120; we usually chose 110) performed best compared to the other two types of kernel function, implying that our classification problem was highly non-linear.
Measurements of SVM performance
Performance of the SVMs was measured using the indices: cost, cost savings, error rate, recall and precision. Cost is defined as C = FP + (2 × FN), where FP is the number of false positives for a SVM classifier and FN is the number of false negatives. We weighted false negatives more heavily than false positives because, in our datasets, the number of positive examples is much smaller than the number of negative examples. Cost savings is defined as S = C – C′, where C′ is the cost of the null learning procedure that classifies all test examples as negatives. Error rate, recall and precision are determined thus:
error rate = (FP + FN)/(FP + FN + TP + TN)
recall = TP/(TP + FN)
precision = TP/(TP + FP)
where TP and TN are the number of true positives and true negatives, respectively.
RESULTS
Functional classification based on codon composition
The codon composition of each gene in E.coli, M.tuberculosis and S.cerevisiae was represented as a vector in 61-dimensional space (considering only sense codons). SVMs were trained on the positive and negative training datasets from the three organisms (see Materials and Methods) by using the leave-one-out cross-validation method. To evaluate the performances of the trained models, each of them was applied to training and test datasets. Evaluation on the training dataset is important as the model can find ‘outliers’, elements that may have been wrongly assigned to the dataset in the first place. The results (Table 2) indicate that the SVM learning technique was able to accurately recognize rp genes, indicating that this set of genes has a unique codon composition profile compared with all other functional classes of genes. The classification was quite accurate in E.coli and M.tuberculosis: none of the false positives and negatives obtained had significant decision values, except for MT1666 (training dataset, –1.17; testing dataset, –1.13) and MT2958 (training dataset, –0.8; testing dataset, –0.76) (Table 2). (Throughout this analysis, false positives and false negatives were considered significant if they had decision values >0.2 and <–0.2, respectively.) Even in S.cerevisiae, the only significant false negatives obtained were RPP0 (training dataset; –0.35; discussed below), RPL22B (test dataset; –1.37; discussed below) and RPS22B (test dataset; –0.28); the significant false positives were TEF1 (0.27) and TEF2 (0.26) (Table 2). It is interesting to note that TEF1 and TEF2 encode an identical protein, the translation elongation factor eEF1α A chain, which, like ribosomal proteins, is part of the translation machinery of the cell. The performances of the SVM on E.coli, M.tuberculosis and S.cerevisiae were 98.1, 95.5 and 88.4% (on the training datasets) and 91.3, 94.5 and 87.5% (on the testing datasets), respectively (Table 2). As mentioned earlier, the information contained in codon composition is representative of both codon bias and amino acid composition. It is therefore implicit in our finding that ribosomal proteins, as a class, have very similar amino acid compositions. To test if this was a result of homology among ribosomal proteins, we performed a ClustalW (14) multiple sequence alignment of all the ribosomal proteins and examined the output for homology. We found no significant homology across the ribosomal proteins (data not shown); it is fascinating that despite this, ribosomal proteins have similar amino acid compositions. We were therefore interested to understand the physiological implications of this finding and also to determine the relative contribution of amino acid composition and codon bias to the uniqueness of the codon composition of rp genes.
Table 2. The performance of support vector machines.
| Dataset | FP | FN | TP | TN | Savings | Error (%) | Recall (%) | Precision (%) | Performance | Significant FPs (>0.2) | Significant FNs (<–0.2) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E.coli training | 0 | 5 | 50 | 1408 | 100 | 0.3 | 90.9 | 100 | 95.5 | ||
| E.coli test | 1 | 5 | 50 | 1393 | 99 | 0.4 | 90.9 | 98 | 94.5 | ||
| M.tuberculosis training | 0 | 13 | 43 | 1073 | 82 | 1.6 | 76.8 | 100 | 88.4 | MT1666 (–1.1650) MT2958 (–0.8045) | |
| M.tuberculosis test | 0 | 14 | 42 | 1059 | 84 | 1.3 | 75 | 100 | 87.5 | MT1666 (–1.1333) MT2958 (–0.7572) | |
| S.cerevisiae training | 2 | 1 | 77 | 1048 | 152 | 0.3 | 98.7 | 97.5 | 98.1 | RPP0 (–0.3510) | |
| S.cerevisiae test | 3 | 7 | 52 | 2083 | 101 | 0.5 | 88.1 | 94.5 | 91.3 | TEF1 (0.2676) TEF2 (0.2628) | RPL22B (–1.3718) RPS22B (–0.2846) |
SVMs were able to recognize cytoplasmic rp genes in both S.cerevisiae and E.coli with high precision and recall. Further, although false positives are scored in the S.cerevisiae and E.coli datasets, the decision values are very low. FP is the number of false positives predicted by the SVM, FN is the number of false negatives, TP is the number of true positives and TN is the number of true negatives, respectively. Savings is a method to measure the performance of SVMs. The exact definition and meaning of each of these indices in the table is defined in Materials and Methods.
The contribution of amino acid composition
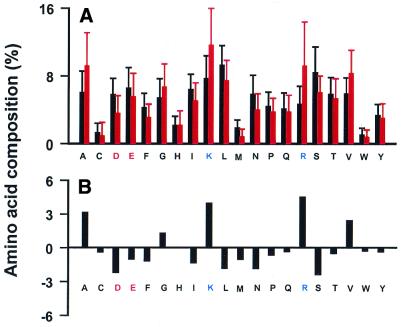
At the outset we compared the amino acid composition of ribosomal (cytoplasmic ribosomal in S.cerevisiae) and non-ribosomal proteins from the three organisms. There is a marked enrichment in basic amino acids (Lys and Arg) and small, hydrophobic amino acids (Ala, Val and Gly), as well as significant depletion in the negatively charged amino acids Glu and Asp (in S.cerevisiae), in ribosomal proteins (Fig. 1). To test the contribution of this skewed amino acid composition on the SVM classification we trained models in which the input vectors of the examples were only based on the protein’s amino acid composition. The accuracies of these models, understandably, were not as good as those trained based on codon usage composition (Table 3). Furthermore, in the three organisms, the performances of all the models decreased due to more false negatives with moderately high values of the decision function. For instance, in E.coli, rpsB (decision value –1.18), rpsA (–0.61), rpsF (–0.49), rpsO (–0.43), rpsJ (–0.36), rpmC (–0.32), rplK (–0.30) and rplD (–0.21) were confidently classified as non-rp genes (Table 3A). In S.cerevisiae, HHT1 and HHT2 (both of which code for the identical protein histone H3) were misclassified as rp genes (both with decision value 0.33) and rp genes RPP0 (–0.87), RPL1A (–0.49), RPL5 (–0.49), RPL30 (–0.43), RPL27A (–0.31), RPL22A (–0.27) and RPS0A (–0.22) were not recognized, with decision values <–0.2 (Table 3B). Hence, while amino acid composition does contribute to the segregation of ribosomal proteins by SVMs, it is not the sole discriminator.
Figure 1.
A comparison of amino acid composition between cytoplasmic ribosomal proteins (red) and all known function proteins (black) in S.cerevisiae. (A) Average compositions with their standard deviations. (B) The difference in composition between cytoplasmic ribosomal proteins and all known function proteins. Ribosomal proteins have a much higher frequency of Arg, Lys, Ala, Val and Gly residues, but fewer Asp and Ser. Similar trends were observed in E.coli and M.tuberculosis.
Table 3. A comparison of SVM performance using codon composition, amino acid composition alone and codon composition excluding {K, R}, {A, V, G} and {D, E} on the training datasets.
| Dataset | FP | FN | TP | TN | Savings | Error (%) | Recall (%) | Precision (%) | Performance | Significant FPs (>0.2) | Significant FNs (<–0.2) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (A) | ||||||||||||
| Codon composition | 0 | 5 | 50 | 1408 | 100 | 0.3 | 90.9 | 100 | 95.455 | |||
| Amino acid composition | 0 | 15 | 40 | 1408 | 80 | 1 | 72.7 | 100 | 86.364 | rpsB (–1.1785) rpsA (–0.6130) | ||
| Codon composition excluding | KR | 1 | 10 | 45 | 1407 | 89 | 0.8 | 81.8 | 97.8 | 89.822 | rpsH (–0.6287) | |
| AVG | 0 | 11 | 44 | 1408 | 88 | 0.8 | 80 | 100 | 90 | rpsF (–0.4867) rpsA (–0.4414) | ||
| DE | 0 | 2 | 53 | 1408 | 106 | 0.1 | 96.4 | 100 | 98.182 | |||
| (B) | ||||||||||||
| Codon composition | 2 | 1 | 77 | 1048 | 152 | 0.3 | 98.7 | 97.5 | 98.093 | RPP0 (–0.3510) | ||
| Amino acid composition | 3 | 12 | 66 | 1047 | 129 | 1.3 | 84.6 | 95.7 | 90.134 | HHT1 (0.3352) HHT2 (0.3352) | RPP0 (–0.8666) RPL1A (–0.4939) | |
| Codon composition excluding | KR | 1 | 5 | 73 | 1049 | 145 | 0.5 | 93.6 | 98.6 | 96.119 | RPS9A (–0.6761) RPL11A (–0.4220) | |
| AVG | 5 | 3 | 75 | 1045 | 145 | 0.7 | 96.2 | 93.8 | 94.952 | HHT1 (0.7299) HHF1 (0.2936) | RPP0 (–0.8073) RPS0A (–0.2215) | |
| DE | 2 | 2 | 76 | 1048 | 150 | 0.4 | 97.4 | 97.4 | 97.436 | GPM1 (0.2917) | RPP0 (–0.5199) RPP1A (–0.3181) |
The upper half of the table represents E.coli training datasets (A), the lower half the S.cerevisiae training datasets (B). Training using amino acid composition alone resulted in the prediction of more false negatives and false positives with higher decision values. Each of the sets of amino acids (that were excluded) contributed to performance in classification. Results here are similar to those obtained with M.tuberculosis (data not shown). Only top two values for the last two columns are listed due to space limitation.
We then determined if the uniqueness of ribosomal proteins was due to their abundance of basic or small hydrophobic amino acids. The mapping of conserved amino acid residues onto the structure of the ribosome has revealed that these exposed charged residues frequently form surface patches that reflect RNA-binding sites (15). It is therefore probable that these residues (at least Arg and Lys) may be present more frequently in ribosomal proteins than in most of the other functional groups. Accordingly, we omitted Arg and Lys and retrained the SVMs on codon composition. On the training sets one significant false negative was obtained for E.coli (rpsH, –0.63; Table 3A); in S.cerevisiae, RPS9A (–0.68), RPL11A (–0.42) and RPL12A (–0.27) were significant false negatives (Table 3B). On the E.coli test dataset, elongation factor Ts gene tsf (0.23) and triosephosphate isomerase gene tpiA (0.22) were significant false positives; rpsH (–0.63) was a confident false negative. Interestingly, the elongation factor Ts is also part of the translation machinery of the cell and is found just 257 nt downstream (3′) of rpsB. On the S.cerevisiae test dataset, SNU13 (0.31), a component of the U4/U6.U5 snRNP which is involved in pre-mRNA splicing, was a significant false positive; RPL22B (–1.10), RPS22B (–0.56) and RPL11B (–0.44) were significant false negatives. Overall, the number of false negatives in E.coli, M.tuberculosis (data not shown) and S.cerevisiae was greater than that obtained with SVMs trained on the complete codon composition.
Similar experiments were conducted, training SVMs on codon composition excluding the amino acid sets {Ala, Val, Gly} and {Asp, Glu} (Tables 3 and 4). It is worth noting that in all instances the SVMs failed to recall gene RPP0 (coding for an acidic ribosomal protein containing an unusually low number of basic residues) except when trained excluding {Lys, Arg} (Table 3). Interestingly, in S.cerevisiae, SVMs trained excluding {Ala, Val, Gly} confidently predicted genes HHT1 (histone H3) and HHF1 (histone H4) as cytoplasmic rp genes on the training dataset (Table 3B). This indicates that the {Ala, Val, Gly} content in histones and rp genes differs sufficiently to help the SVMs discriminate between them. On the S.cerevisiae test dataset, NOP10, a nucleolar rRNA processing protein, and TEF2 were significant false positives when {Ala, Val, Gly} were excluded, and STM1 (whose product has affinity for quadruplex nucleic acids) and TEF1 when {Asp, Glu} were omitted (Table 4). Similarly, on the E.coli test dataset, when {Asp, Glu} were omitted hupB (encoding the β subunit of the DNA-binding histone-like protein HU) was recognized (Table 4). It is important to note that all the above falsely predicted genes are involved in binding to nucleic acids, similar to ribosomal proteins. Hence, what sets cytoplasmic ribosomal proteins apart from them is the significant contribution to their distinctive codon composition profile from the positively charged, negatively charged and small hydrophobic residues. This strongly suggests the importance of these sets of residues in the unique functionality of the ribosome.
Table 4. A comparison of SVM performance using codon composition excluding {K, R}, {A, V, G} and {D, E} on the test datasets in S.cerevisiae and E.coli.
| Dataset | FP | FN | TP | TN | Savings | Error (%) | Recall (%) | Precision (%) | Performance | Significant FPs (>0.2) | Significant FNs (<–0.2) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E.coli codon composition excluding | KR | 3 | 10 | 45 | 1391 | 87 | 0.9 | 81.8 | 93.8 | 87.784 | tsf (0.2253) tpiA (0.2232) | rpsH (–0.6287) |
| AVG | 1 | 11 | 44 | 1393 | 87 | 0.8 | 80 | 97.8 | 88.889 | rpsF (–0.4867) rpsA (–0.4414) | ||
| DE | 3 | 2 | 53 | 1391 | 103 | 0.3 | 96.4 | 94.6 | 95.503 | hupB (0.2070) | ||
| S.cerevisiae codon composition excluding | KR | 6 | 9 | 50 | 2080 | 94 | 0.7 | 84.7 | 89.3 | 87.016 | SNU13 (0.3056) | RPL22B (–1.1006) RPS22B (–0.5589) |
| AVG | 7 | 8 | 51 | 2079 | 95 | 0.7 | 86.4 | 87.9 | 87.186 | NOP10 (0.3352) TEF2 (0.2264) | RPL22B (–1.7212) RPS22B (–0.7575) | |
| DE | 4 | 4 | 55 | 2082 | 106 | 0.4 | 93.2 | 93.2 | 93.22 | STM1 (0.4306) TEF1 (0.2901) | RPL22B (–1.3539) RPS22B (–0.4144) |
Results here were similar to those obtained with the training datasets (Table 3). The overall performance was similar to M.tuberculosis (data not shown).
The contribution of codon bias
To determine the contribution of the codon bias signal to the ability of SVMs to distinguish cytoplasmic ribosomal proteins from all others, we used RCSU data (see Materials and Methods for details) alone as the learning attributes for the SVMs. However, none of the SVMs were able to discriminate ribosomal protein genes, regardless of the parameters that were specified (data not shown).
It is well known that highly expressed genes have a highly biased codon usage in order to maximize efficiency and accuracy of translation in bacteria and unicellular eukaryotes (16). Although the level of expression is unknown for all genes in the genome, genes such as ribosomal proteins, elongation factors and RNA polymerase subunits are known to be highly expressed in all bacterial species analyzed so far. For example, in S.cerevisiae, at least 40 ribosomes must be made every second with a 90 min generation time (17). Further, it has also been shown earlier that ribosomal protein genes have a highly biased codon usage (10,18). Taken together, we conclude that with our representation in the SVMs, the mixture of both amino acid composition and codon bias signals were detected well, combined and the fused signals augmented to perform accurate classification.
DISCUSSION
The uniqueness of ribosomal proteins
Ribosomal proteins have been shown to be unique among cellular proteins in E.coli, M.tuberculosis and S.cerevisiae in terms of their codon composition (see Results; 10,18). As already emphasized, this implies that they have a unique amino acid composition as well as codon bias. It is interesting to speculate as to what the physiological implications of this may be.
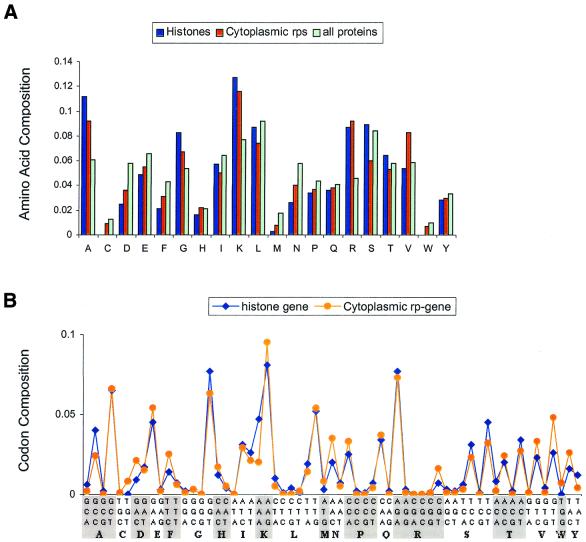
The ribosome is a pivotal molecular machine in the cell because it synthesizes all proteins by the execution of two main functions: decoding the genetic message and the formation of peptide bonds. During the past year, significant insight has been gained into structural, functional and mechanistic aspects of the ribosome, based to a large extent on the availability of 3-dimensional structures of the ribosome and parts thereof (for reviews see 19–21). However, according to our current understanding, the primary function of ribosomal proteins seems only to be as a stabilizer of the highly compact rRNA structure to guarantee the peptidyltransferase activity based on catalysis by RNA (22), despite extra-ribosomal functions of ribosomal proteins having been described (23). From the 3-dimensional structure of the ribosome it is obvious that the protein–rRNA contacts are far greater than expected from earlier in vitro studies with isolated proteins and RNAs (20). This kind of intimacy between negatively charged nucleic acids and proteins dictates that ribosomal proteins possess a high proportion of basic residues, a unique requirement they perhaps share only with proteins that stabilize chromatin, such as histones. Hence it is likely that the high proportion of positively charged amino acids (Lys and Arg) and the relative depletion of negatively charged amino acids (Glu and Asp) are sensed by the SVMs to be significant discriminators of ribosomal proteins from the rest of the cellular proteins. The amino acid composition profile of histones is very similar to that of ribosomal proteins: there is a high proportion of Lys, Arg, Ala and Gly residues and significantly fewer Asp residues than other cellular proteins (Fig. 2A). While the presence of a high proportion of positively charged amino acids has a physiological explanation, what about the large number of small hydrophobic amino acids, notably alanine? It has recently been shown that highly expressed proteins in S.cerevisiae are enriched in alanine (24). Hence, the enrichment of alanine is likely linked to the high expression of ribosomal genes. Therefore, the skewed amino acid composition of ribosomal proteins, despite the lack of sequence homology among themselves, is probably vital to the structural and functional integrity of the ribosome. It would hence be highly conserved throughout all life forms, as the near universality of the genetic code implies.
Figure 2.
(A) A comparison of the amino acid composition of histone coding genes, cytoplasmic rp genes and all known function genes in S.cerevisiae. The amino acids that contribute to the discrimination of cytoplasmic ribosomal proteins from histones are mainly Ser and Val. (B) A comparison of the average codon composition of histone genes with cytoplasmic ribosomal protein genes. There is a significant difference in use of the codons GCC, GGT, GTT, TCT, etc., indicating that, in addition to amino acid composition, codon usage in histones also differs from that of cytoplasmic ribosomal proteins.
The high expression level of ribosomal proteins has been alluded to. It is a well known fact that synonymous codon usage in various genomes is non-random. The occurrence of codons in a gene strongly correlates with the relative abundance of their respective tRNA pools in many species. Furthermore, there is a clear positive correlation between codon usage and gene expression level in E.coli and S.cerevisiae (25–27). Consistent with this trend, most of the ribosomal protein coding genes we investigated here use significantly biased synonymous codons to code corresponding amino acids. For example, these genes have a high codon adaptation index (CAI) with values usually from 0.6 to 0.9 in S.cerevisiae (10). The preference for a major codon, i.e. the binding of a relatively abundant tRNA species to the ribosome, has a direct positive impact on translational efficacy and accuracy for highly expressed genes (16,28,29). Hence the strong codon bias, in addition to the high proportion of alanine residues, is likely linked to high expression levels of ribosomal genes, although perhaps the former attribute is what discriminates ribosomal genes from histone genes (Fig. 2). In addition, the frequency of occurrence of Ser, Val, Ile and Leu in histones is significantly different from that in cytoplasmic ribosomal proteins and matches more closely the average frequency in all genes of known function in S.cerevisiae (Fig. 2A).
Lessons learnt from machine learning
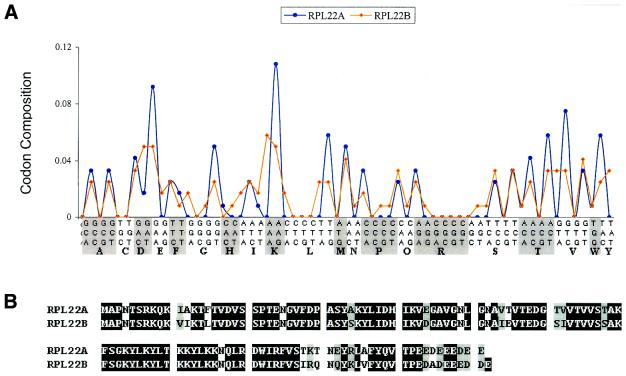
As mentioned earlier, the rationale behind choosing codon composition as a classifying attribute was because it is linked to the physico-chemical properties of the protein and could perhaps have functional implications (see Introduction). The SVMs were able to accurately separate ribosomal genes from non-ribosomal genes. Analysis of the signals that the SVMs perceived provided data on this class of proteins that permitted biochemical interpretation, with the benefit of a priori information (see above). Further, as a result of our experiments some interesting data emerged. The S.cerevisiae gene RPL22B (a duplicate of the RPL22A gene), encoding ribosomal protein RPL22B, was misclassified by the SVMs as a non-ribosomal protein coding gene with a very high decision value (–1.4; Table 2). We determined the codon compositions of these two genes (Fig. 3A). Interestingly, though these two genes share high identity (84.4%) at the amino acid sequence level (Fig. 3B), they employ a very different codon usage pattern. The codon usage bias of RPL22B is quite low (CAI = 0.29) while that of RPL22A is much higher (CAI = 0.86) (10). Since codon bias is strongly linked to expression level in S.cerevisiae (25–27), this would indicate that these two genes probably have very different expression profiles. Microarray expression data determined at 17 different time points of the yeast mitotic cycle in synchronized cells (30) reveals that RPL22A is expressed on average 4.32 ± 0.70-fold higher than RPL22B. This leads us to hypothesize that RPL22B may not be a bona fide ribosomal protein coding gene, but might have a very different function from RPL22A.
Figure 3.
(A) The codon composition patterns of cytoplasmic ribosomal genes RPL22A and RPL22B in yeast. (B) The protein sequence alignment of Rpl22A and Rpl22B. Although these two proteins have high identity (84.4%) at the amino acid sequence level, their codon usage is significantly different (CAI of RPL22A = 0.86; CAI of RPL22B = 0.29). We hence hypothesize that Rpl22B may have a non-ribosomal function.
The multiplicity of function of genes (ribosomal protein or otherwise) may pose a wider problem in the functional classification of genes. Although they are species-related, we argue that the high conservation of structure and function of ribosomal proteins would enable us to accurately identify them based on codon composition in multicellular organisms such as D.melanogaster, rat and human. However, preliminary results indicate that though there are conserved patterns of codon composition in D.melanogaster (K.Lin, Y.Kuang, J.S.Joseph and P.R.Kolatkar, unpublished results), the accuracy of discrimination of ribosomal genes is lower. We speculate that cytoplasmic ribosomal proteins in these organisms may be involved in multiple functions (23) and may hence possess less obvious similarities as a class at the codon composition level. There is some precedence for this: we found that the MT1666 gene in M.tuberculosis and the rpsA gene in E.coli, both encoding their respective 30S ribosomal subunit protein S1, were more likely to be classified as non-ribosomal protein coding genes, with decision values of –1.17 (Table 2) and –0.12 (data not shown), respectively. It is known that, at least in E.coli, S1 is also involved in extra-ribosomal functions (21).
Codon composition as a classifying attribute
The proof of the efficacy of a classifying attribute is its ability to accurately predict the role of genes of unknown function. Therefore, we attempted to identify ribosomal genes in the datasets consisting of genes of unknown function from E.coli, M.tuberculosis and S.cerevisiae. We were aware, of course, that due to their crucial roles, most (if not all) ribosomal proteins and their genes in these organisms would already have been investigated at the genetic, biochemical, genomic and structural levels. It would hence be unlikely that we would identify a hitherto unknown ribosomal gene. Not surprisingly, we did not find any new candidates among them. However, the method that we have described in this paper may have the potential to be used to predict ribosomal genes in divergent genomes where sequence homology alone may not be sufficient to identify them all.
More importantly, we have demonstrated here that codon composition has strong potential to be used as an attribute in the functional classification of genes. Codon usage alone has been earlier used in the prediction (using factorial component analysis) of the location of ribosomal proteins and aminoacyl-tRNA synthetases in eukaryotic cells (31). However, the segregation of genes into functional classes by purely computational means will realistically necessitate the employment of an array of complementary attributes rather than a single one. For example, the promoters of ribosomal protein genes have a characteristic architecture (for a review see 32) that has been exploited for their classification. As better supervised learning methods in analyzing gene expression profiles, especially methods derived from statistical learning theories (4), emerge, these can be combined with the knowledge obtained from better structured gene functional taxonomies (for example the GO system; 7) and various other data sources, including the published literature, DNA and protein sequence databases, gene expression data, 3-dimensional structural data, metabolic pathways and localization information of gene products. These methods, especially the application of SVMs to the increasing a priori knowledge, will become more and more important (33,34; and references therein) in the post-genome era. The data presented in this paper strongly suggests that machine learning theories, especially supervised methods, could provide the best initial approaches to characterizing and assigning gene function in functional genomics.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Phil Long, Prospero Naval and Amirul Islam for helpful discussions. We also wish to thank the Economic Development Board and the National Science and Technology Board of Singapore for financial support of the project.
REFERENCES
- 1.Nadeau J.H. (2000) Muta-genetics or muta-genomics: the feasibility of large-scale mutagenesis and phenotyping programs. Mamm. Genome., 11, 603–607. [DOI] [PubMed] [Google Scholar]
- 2.Paigen K. and Eppig,J.T. (2000) A mouse phenome project. Mamm. Genome., 11, 715–717. [DOI] [PubMed] [Google Scholar]
- 3.Brent R. (2000) Genomic biology. Cell, 100, 169–183. [DOI] [PubMed] [Google Scholar]
- 4.Vapnik V.N. (1998) Statistical Learning Theory. John Wiley & Sons, New York.
- 5.Evgenious T., Pontil,M. and Poggio,T. (2000) Statistical learning theory: a primer. Int. J. Comput. Vis., 38, 9–13. [Google Scholar]
- 6.Brown M.P., Grundy,W.N., Lin,D., Cristianini,N., Sugnet,C.W., Furey,T.S.,Jr, Ares,M. and Haussler,D. (2000) Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl Acad. Sci. USA., 97, 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (The Gene Ontology Consortium) (2000) Gene ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner F.R., Plunkett,G., Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F., Gregor,J. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 9.Sharp P.M., Tuohy,T.M. and Mosurski,K.R. (1986) Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res., 14, 5125–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Planta R.J. and Mager,W.H. (1998) The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast, 14, 471–477. [DOI] [PubMed] [Google Scholar]
- 11.Mager W.H., Planta,R.J., Ballesta,J.G., Lee,J.C., Mizuta,K., Suzuki,K., Warner,J.R. and Woolford,J. (1997) A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res., 25, 4872–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allwein E.L., Schapire,R.E. and Singer,Y. (2000) Reducing multiclass to binary: a unifying approach for margin classifiers. J Machine Learning Res., 1, 113–141. [Google Scholar]
- 13.Joachims T. (1999) Making large-scale SVM learning practical. In Schölkopf,B., Burges,C. and Smola,A. (eds), Advances in Kernel Methods—Support Vector Learning. MIT Press, Cambridge, MA.
- 14.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramakrishnan V. and White,S.W. (1998) Ribosomal protein structures: insights into the architecture, machinery and evolution of the ribosome. Trends Biochem. Sci., 23, 208–212. [DOI] [PubMed] [Google Scholar]
- 16.Akashi H. and Eyre-Walker,A. (1998) Translational selection and molecular evolution. Curr. Opin. Genet. Dev., 8, 688–693. [DOI] [PubMed] [Google Scholar]
- 17.Tollervey D., Lehtonen,H., Carmo-Fonseca,M. and Hurt,E.C. (1991) The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in S. cerevisiae. EMBO J., 10, 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlin S., Campbell,A.M. and Mrazek,J. (1998) Comparative DNA analysis across diverse genomes. Annu. Rev. Genet., 32, 185–225. [DOI] [PubMed] [Google Scholar]
- 19.Maguire B.A. and Zimmermann,R.A. (2001) The ribosome in focus. Cell, 104, 813–816. [DOI] [PubMed] [Google Scholar]
- 20.Moore P.B. (2001) The ribosome at atomic resolution. Biochemistry, 40, 3243–3250. [DOI] [PubMed] [Google Scholar]
- 21.Ramakrishnan V. and Moore,P.B. (2001) Atomic structures at last: the ribosome in 2000. Curr. Opin. Struct. Biol., 11, 144–154. [DOI] [PubMed] [Google Scholar]
- 22.Nissen P., Hansen,J., Ban,N., Moore,P.B. and Steitz,T.A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science, 289, 920–930. [DOI] [PubMed] [Google Scholar]
- 23.Wool I.G. (1996) Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci., 21, 164–165. [PubMed] [Google Scholar]
- 24.Jansen R. and Gerstein,M. (2000) Analysis of the S. cerevisiae transcriptome with structural and functional categories. Nucleic Acids Res., 28, 1481–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikemura T. (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol., 2, 13–34. [DOI] [PubMed] [Google Scholar]
- 26.Gouy M. and Gautier,C. (1982) Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res., 10, 7055–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharp P.M. and Cowe,E. (1991) Synonymous codon usage in Saccharomyces cerevisiae. Yeast, 7, 657–678. [DOI] [PubMed] [Google Scholar]
- 28.Bulmer M. (1991) The selection-mutation-drift theory of synonymous codon usage. Genetics, 129, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell J.R. and Moriyama,E.N. (1997) Evolution of codon usage bias in Drosophila. Proc. Natl Acad. Sci. USA, 94, 7784–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho R.J., Campbell,M.J., Winzeler,E.A., Steinmetz,L., Conway,A., Wodicka,L., Wolfsberg,T.G., Gabrielian,A.E., Landsman,D., Lockhart,D.J. and Davis,R.W. (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell, 2, 65–73. [DOI] [PubMed] [Google Scholar]
- 31.Chiapello H., Ollivier,E., Landes-Devauchelle,C., Nitschke,P. and Risler,J.L. (1999) Codon usage as a tool to predict the cellular location of eukaryotic ribosomal proteins and aminoacyl-tRNA synthetases. Nucleic Acids Res., 15, 2848–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mager W.H. and Planta,R.J. (1990) Multifunctional DNA-binding proteins mediate concerted transcription activation of S. cerevisiae ribosomal protein genes. Biochim. Biophys. Acta, 1050, 351–355. [DOI] [PubMed] [Google Scholar]
- 33.Kell D.B. and King,R.D. (2000) On the optimization of classes for the assignment of unidentified reading frames in functional genomics programmes: the need for machine learning. Trends Biotechnol., 18, 93–98. [DOI] [PubMed] [Google Scholar]
- 34.Altman R.B. and Raychaudhuri,S. (2001) Whole-genome expression analysis: challenges beyond clustering. Curr. Opin. Struct. Biol., 11, 340–347. [DOI] [PubMed] [Google Scholar]