Abstract
OGG1 is the major DNA glycosylase in human cells for removal of 7,8 dihydro-8-oxoguanine (8-oxoG), one of the most frequent endogenous base lesions formed in the DNA of aerobic organisms. During replication, 8-oxoG will frequently mispair with adenine, thus forming G:C → T:A transversions, a common somatic mutation associated with human cancers. In the present study, we have constructed a stable transfectant cell line expressing hOGG1 fused at the C-terminal end to green fluorescent protein (GFP) and investigated the cellular distribution of the fusion protein by fluorescence analysis. It is shown that hOGG1 is preferentially associated with chromatin and the nuclear matrix during interphase and becomes associated with the condensed chromatin during mitosis. Chromatin-bound hOGG1 was found to be phosphorylated on a serine residue in vivo as revealed by staining with an anti-phosphoserine-specific antibody. Chromatin-associated hOGG1 was co-precipitated with an antibody against protein kinase C (PKC), suggesting that PKC is responsible for the phosphorylation event. Both purified and nuclear matrix-associated hOGG1 were shown to be substrates for PKC-mediated phosphorylation in vitro. This appears to be the first demonstration of a post-translational modification of hOGG1 in vivo.
INTRODUCTION
Organisms are continuously exposed to reactive oxygen species generated as by-products of the respiratory chain, during inflammation and by exposure of cells to ionising radiation or oxidative stress conditions. The reaction of these molecules with DNA yields a wide variety of damage including strand breaks, abasic (AP) sites and oxidised bases. From a human health perspective, oxidative DNA damage has been implicated in mutagenesis, carcinogenesis and ageing (1,2). Therefore, cells have developed specific repair mechanisms to counteract the biological effects of these lesions and the base excision repair pathway is the major pathway for correcting oxidative DNA damage (3). Among the base lesions induced by oxidative stress, 7,8-dihydro-8-oxoguanine (8-oxoG) is probably the most critical residue due to its high abundance and ability to mispair with adenine, which can lead to a high frequency of spontaneous G:C → T:A transversions (4,5).
In Escherichia coli, three different enzymes cooperate to suppress spontaneous mutagenesis caused by 8-oxoG: (i) the DNA glycosylase/AP lyase Fpg (MutM) removes 8-oxoG from 8-oxoG:C base pairs (6,7); (ii) the DNA glycosylase MutY specifically removes A mispaired with 8-oxoG, introduced when 8-oxoG remains in the template during DNA replication (8); and (iii) MutT is an 8-oxo-dGTPase that prevents DNA polymerase-mediated incorporation of 8-oxo-dGTP opposite A in the template strand (9). Both MutY and MutT homologues have been identified in humans but are not apparent in Saccharomyces cerevisiae (10,11). Yeast and human cells contain OGG1, a sequence-unrelated functional homologue of the E.coli Fpg DNA glycosylase that exhibits a high specificity for 8-oxoG paired with C (12,13). Both S.cerevisiae OGG1 and E.coli Fpg deficiency give rise to a spontaneous mutator phenotype (14,15). Homozygous ogg–/– mice show a 10-fold elevated G:C → T:A spontaneous transversion frequency in DNA from liver cells (16,17). G:C → T:A transversions are characteristic of mutational spectra exhibited by human tumours with a defect in the tumour suppressor p53 (18). Interestingly, mutations and polymorphisms in the human OGG1 gene have been found to be associated with tumours from head, neck, lung and kidney cancers (19–21).
hOGG1 has been purified to apparent homogeneity and the catalytic properties have been extensively investigated (13,22,23). In addition, the X-ray structures of both the free hOGG1 enzyme as well as a catalytically inactive mutant bound to an 8-oxoG:C-containing DNA have recently been solved (24,25). Structural analyses have shown that upon binding to DNA, the enzyme undergoes extensive local conformational changes associated with its strong preference for C in the opposite strand. Altogether, these studies have provided a detailed molecular understanding of the enzyme’s biochemical properties. However, relatively little is known about the enzyme’s in vivo functions, intracellular distribution and mode of expression.
In the present study, we demonstrate, using biochemical methods and immunofluorescence analysis, that during cell cycle interphase, hOGG1 becomes associated both with the chromatin and the nuclear matrix. The nuclear matrix is a dynamic RNA–protein network that focally assembles many nuclear processes such as RNA transcription and splicing, DNA replication and repair (26). During mitosis, hOGG1 follows the condensed chromatin. Furthermore, we show that hOGG1 is phosphorylated in vivo on a serine residue and is subject to protein kinase C (PKC)-mediated phosphorylation in vitro. The chromatin-associated hOGG1 co-immunoprecipitates with PKC; this provides the first evidence for a post-translational modification of this repair enzyme.
MATERIALS AND METHODS
Cell culture and localisation studies
HeLa S3 cells were maintained in monolayer cultures in DMEM 4.5 g/l glucose medium (BioWhittaker) supplemented with 10% fetal calf serum (Integro), 0.3 mg/ml glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin (BioWhittaker), at 37°C in 5% CO2 atmosphere. HeLa S3 cells constitutively expressing the EGFP (HeLa EGFP) or the hOGG1–EGFP fusion protein expressing the nuclear form of hOGG1 consisting of exon 7 but not exon 8 (HeLa hOGG1–EGFP) were prepared as described (13; L.Luna, V.Rolseth, M.Otterlei, M.Bjørås, F.Dantzer, I.Toft and E.Seeberg, manuscript in preparation). These cells were cultivated as above supplemented with 0.7 mg/ml geneticin (Gibco BRL).
For localisation studies, cells were grown on glass coverslips, washed twice with PBS, fixed with ice-cold methanol/acetone (1:1) for 10 min and washed again with PBS. The cellular DNA was stained with 0.25 µg/ml DAPI for 10 min at room temperature. Coverslips were mounted with DAKO fluorescent mounting medium (DAKO) and visualised with a Zeiss Axioplan 2 fluorescence microscope (Carl Zeiss, Jena, Germany).
Nuclear matrix preparation
Nuclear matrix proteins were fractionated from the indicated cells according to the method of He et al. (27). Cells were washed twice in PBS and treated with 0.5 ml (per 100 mm dish) of CSK buffer (10 mM PIPES pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EDTA, 1 µg/ml leupeptin and pepstatin, 1 mM PMSF and 0.1% Triton X-100) for 10 min on ice. The monolayer was collected and centrifuged at 5000 g for 2 min. The soluble cytoplasmic fraction was removed and the pellet resuspended in 200 µl of CSK buffer containing 100 U RNase-free DNase I. After 15 min at 37°C, ammonium sulfate was added to a final concentration of 0.25 M. The samples were rotated for 5 min at room temperature and centrifuged as above. The soluble chromatin fraction was removed and the pellet extracted in CSK buffer with 2 M NaCl for 5 min at 4°C. After another centrifugation, the 2 M NaCl wash was removed and the nuclear matrix pellet resuspended in 50 µl dilution buffer (20 mM HEPES KOH pH 7.4, 100 mM KCl, 1 mM EDTA, 2 mM DTT, 0.1 µg/ml BSA and 20% glycerol). All fractions were then dialysed twice against the dilution buffer. For western blot analyses, equal cell equivalents from each fraction were subjected to 8% SDS–PAGE and probed with appropriate antibodies: rabbit polyclonal anti-green fluorescent protein (GFP) antibody (ab 290–50, abcam), goat polyclonal anti-lamin A/C antibody (N-18, Santa Cruz Biotechnology), goat polyclonal anti-histone H1 antibody (C-17, Santa Cruz Biotechnology), monoclonal anti-human Ogg1 antibody (clone 7E2, IBL) and rabbit polyclonal anti-phosphoserine antibody (poly-Z-PS1, Zymed Laboratories).
For in situ extraction of the nuclear matrix, cells grown on coverslips were extracted using the same method as described above, fixed with ice-cold methanol/acetone (1:1) and incubated for 16 h with an anti-lamin A/C antibody (Santa Cruz Biotechnology) diluted 1/200 in PBS, 0.1% Tween, 1% BSA. After three washes with PBS, 0.1% Tween, cells were incubated for 3 h with an Alexa 594-conjugated anti-goat antibody (Molecular Probes) diluted 1/1000 in PBS, 0.1% Tween, 1% BSA. After DNA staining with 0.25 µg/ml DAPI for 10 min at room temperature, coverslips were mounted with DAKO and visualised as described above.
Assays for 8-oxoG DNA glycosylase activity
hOGG1 DNA glycosylase activity was measured essentially as described (13). Briefly, a 24 bp oligonucleotide containing a single 8-oxoG placed at position 14 was 32P-labelled at the 5′ terminus by T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (3000 Ci/mmol, Amersham), and subsequently annealed to the complementary strand containing a cytosine residue opposite 8-oxoG. In a standard reaction (10 µl final volume), 100 fmol of the 8-oxoG:C-labelled duplex was incubated in reaction buffer (10 mM MOPS, 0.2 mM DTT, 20 µg/ml BSA and 1% glycerol) together with equal cell equivalents from each fraction and/or 10 ng of purified hOGG1 for 30 min at 37°C (16). Where indicated, 25 ng of HAP1 (AP endonuclease) was added to the reaction mixture and samples were incubated for an additional 30 min at 37°C. Reaction products were analysed on 20% denaturating polyacrylamide gels using a PhosphorImager (Molecular Dynamics).
Immunoprecipitation experiments
Equal cell equivalents from each fraction were diluted to a final volume of 200 µl with dilution buffer containing 0.5 mM PMSF, 1 µg/ml leupeptin and pepstatin, pre-cleared for 1 h at 4°C with 20 µl protein A–Sepharose beads (Pharmacia Biotech), and incubated for 2 h at 4°C with either the polyclonal anti-GFP antibody or the monoclonal anti-PKC antibody (MC5, Santa Cruz Biotechnology). Immunocomplexes were precipitated by the addition of 20 µl protein A–Sepharose beads for 2 h at 4°C, washed three times in 50 mM Tris–HCl pH 8, 120 mM NaCl, 0.5% NP-40 and separated by 10% SDS–PAGE. Proteins were transfered onto nitrocellulose membranes and probed with either anti-GFP or anti-PKC antibody.
In vitro phosphorylation reactions
The PKC reaction was assayed using the PKC assay kit (Stratagene) according to the manufacturer’s instructions. Reactions contained either 5 µg of purified hOGG1 or equal cell equivalents from each biochemical fraction, and were initiated by the addition of a 17.5 ng mixture containing α, β and γ PKC isoforms. After 10 min at 30°C, the samples were divided and either applied on an 8% SDS–PAGE after addition of 2× SDS buffer, or used for 8-oxoG:C DNA glycosylase activity assays as described above except that the glycosylase reaction buffer was supplemented with 5 mM EDTA.
Protein phosphatase assays
Dephosphorylation assays were performed in a total volume of 15 µl containing 50 mM Tris–HCl, 0.1 mM Na2EDTA, 5 mM DTT, 0.01% Brij 35, 2 mM MnCl2, equal cell equivalents from subcellular fractions or 10 ng of purified hOGG1 and 200 U of lambda protein phosphatase (λ-PPase, Biolabs). The reaction was incubated for 1 h at 30°C and one-third of each reaction sample was then tested for hOGG1 enzymatic activity as described above except that the glycosylase reaction buffer contained 5 mM EDTA.
RESULTS
hOGG1 co-localises with condensed mitotic chromosomes
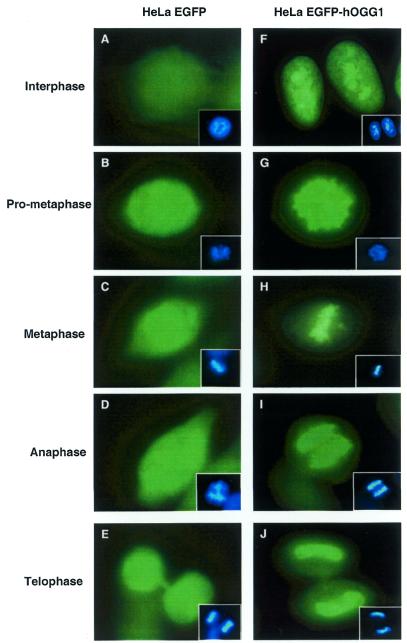
We have described previously a cDNA construct of hOGG1 fused to EGFP that expresses the nuclear form of hOGG1 (13). This and the corresponding ‘empty’ EGFP vector were used to transfect HeLa S3 cells and cell line derivatives were isolated showing stable expression of the fusion protein (hOGG1–EGFP) or EGFP. Examination of the stable hOGG1–EGFP transfectant showed exclusive sorting of hOGG1–EGFP to the nucleus with a stronger signal observed in the nucleoli of interphase cells (Fig. 1F). During mitosis, from pro-metaphase to telophase, the hOGG1–EGFP signal was localised to the condensed chromatin (Fig. 1G–J). The sorting was dependent on the presence of the hOGG1 sequence as cells expressing EGFP protein alone showed fluorescence evenly distributed throughout the cells (Fig. 1A–E). Attempts to detect endogenous hOGG1 by immunofluorescence using anti-hOGG1 antibodies were unsuccessful. Similar localisation dynamics to condensed chromosomes have been reported with chromatin-associated proteins such as ATRX, BRG and hbrm (human SWI/SNF-like proteins), HA95 (homologue to the human PKA anchoring protein AKAP95) and DNA topoisomerase IIα, which are all involved in various aspects of transcriptional regulation and DNA replication, DNA repair and mitotic recombination (28–31).
Figure 1.
Subcellular distribution of hOGG1. HeLa cells expressing EGFP (A–E) or the hOGG1–EGFP fusion protein (F–J) were grown on coverslips, fixed and analysed using an Axioplan 2 fluorescence microscope (Carl Zeiss). DNA (inserts) was labelled with DAPI staining.
hOGG1 is bound to the nuclear matrix in interphase cells
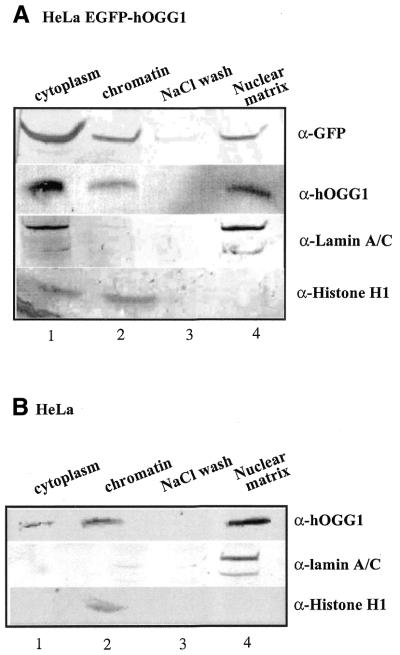
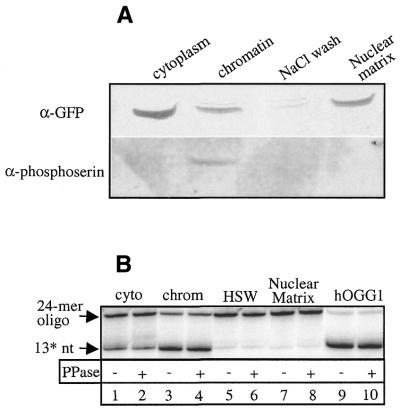
As many proteins associated with the condensed chromatin during mitosis also bind to the core nuclear matrix, we asked if hOGG1 also would be associated with the matrix component. Asynchronously dividing HeLa hOGG1–EGFP cells were sequentially extracted as described in the Materials and Methods to obtain cytoplasm, soluble chromatin and the nuclear matrix (Fig. 2A). The protein extracts were analysed by western blotting. The fractionation procedure was controlled by immunoblotting with antibodies directed against lamin A/C, a nuclear matrix-associated protein, and histone H1, a chromatin-associated protein. Immunoblotting with anti-GFP and anti-hOGG1 antibodies, respectively, revealed hOGG1–EGFP to be present in both the soluble chromatin and nuclear matrix fractions (Fig. 2A, lanes 2 and 4). No hOGG1–EGFP was detected in the high salt wash employed prior to obtaining the matrix fraction (Fig. 2A, lane 3). The localisation of hOGG1–EGFP in the cytoplasmic fraction (Fig. 2A, lane 1) probably reflects protein leakage from the nucleus during the cell lysis procedure as the cellular fluorescence studies showed hOGG1–EGFP to be exclusively nuclear (Fig. 1) (13). In contrast, the EGFP protein was only detectable in the cytoplasmic fraction following the first Triton X-100 treatment (data not shown). To exclude any unspecific localisation due to the overexpression of the hOGG1–EGFP fusion protein, similar experiments were also carried out scoring the endogenous hOGG1 protein in non-transfected HeLa S3 cells (Fig. 2B). Similar to the fusion protein, endogenous hOGG1 was also detected both in the chromatin and in the nuclear matrix fractions as judged by western blotting using an anti-hOGG1 antibody (Fig. 2B, lanes 2 and 4). hOGG1 was absent in the 2 M NaCl wash (Fig. 2B, lane 3) but leakage of the protein to the cytoplasmic fraction was observed also in this case (Fig. 2B, lane 1). Thus, the results obtained with the fused and the free hOGG1 proteins were essentially identical, implying that the intracellular distribution of hOGG1 is not significantly affected by the C-terminal EGFP fusion extension. Similar results were also obtained when nuclear matrices were prepared using an alternative fractionation protocol according to Reyes et al. (29) (data not shown).
Figure 2.
hOGG1 is associated with the chromatin and the nuclear matrix. HeLa cells expressing the hOGG1–EGFP fusion protein (A) or non-transfected HeLa cells (B) were extracted to obtain the cytoplasmic fraction (lane 1), the whole chromatin fraction (lane 2), a high salt wash (lane 3) and the core nuclear matrix (lane 4). Proteins from equal cell equivalents from each fraction were analysed by western blotting with the indicated antibodies.
hOGG1 is detected after in situ extraction of the core nuclear matrix
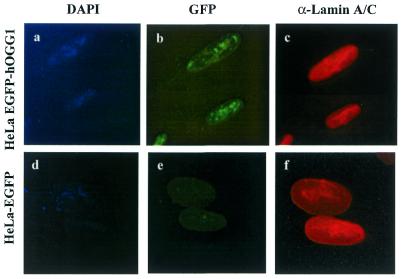
To further investigate the association of hOGG1 to the nuclear matrix, HeLa hOGG1–EGFP cells were grown on coverslips and subjected to sequential extractions in situ using Triton X-100, DNase I, ammonium sulfate and high salt in order to obtain the nuclear matrix (Fig. 3). The efficiency of chromatin digestion was indicated by the disappearance of the DAPI stain (Fig. 3a and d), while the integrity of the nuclear matrix was verified by immunostaining with lamin A/C antibody (Fig. 3c and f). DNase treatment of the cells partly removed the intranuclear localisation of hOGG1–EGFP, thus confirming its association with the DNase-sensitive chromatin (Fig. 2A). However, a nuclear speckled pattern of hOGG1–EGFP remained even after high salt treatment indicating that a significant proportion of the fusion protein was associated with the nuclear matrix (Fig. 3b). No staining was detected for the HeLa EGFP cells subjected to identical treatment (Fig. 3e). EGFP staining was removed immediately after detergent treatment (data not shown). These results confirm that hOGG1 in vivo is associated with both the chromatin and the nuclear matrix in interphase cells.
Figure 3.
In situ extraction of the core nuclear matrix. HeLa cells expressing hOGG1–EGFP (a–c) or EGFP (d–f) were grown on coverslips, extracted sequentially as described in Figure 2, fixed and processed for immunofluorescence imaging. (a and d) The validity of the procedure is shown by the absence of DAPI-stained nuclear DNA. (b and e) GFP (green) and (c and f) Alexa 594-labelled lamin A/C.
The chromatin-associated hOGG1 shows 8-oxoG DNA glycosylase/AP lyase activity
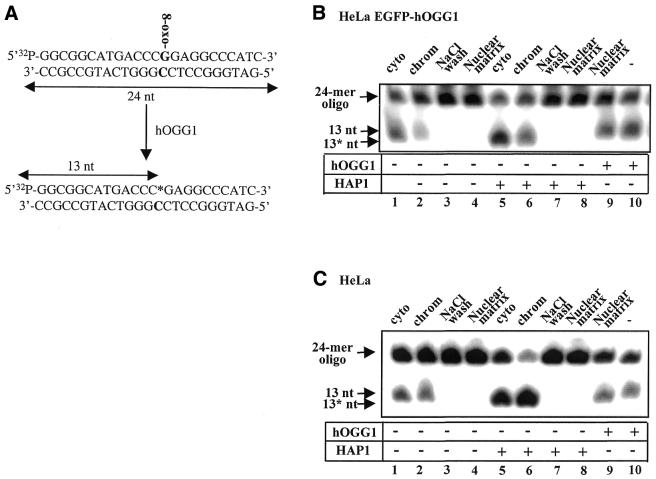
hOGG1 enzyme activity was assayed in the cytoplasmic, chromatin and nuclear matrix fractions of HeLa hOGG1–EGFP cells (Fig. 4B). hOGG1 removes 8-oxoG from DNA by its glycosylase activity and cleaves the DNA strand at the 3′ side of the resulting abasic site by its AP lyase activity (β elimination). This results in strand cleavage of a double-stranded oligonucleotide containing an 8-oxoG:C base pair (Fig. 4A). hOGG1 activity was observed in the HeLa hOGG1–EGFP-derived cytoplasmic and chromatin fractions (Fig. 4B, lanes 1 and 2) but no cleavage of the 8-oxoG:C substrate was detected in either the high salt wash (Fig. 4B, lane 3) or the nuclear matrix fractions (Fig. 4B, lane 4), despite the presence of high amounts of hOGG1 in the latter (Fig. 2A, compare lanes 2 and 4). As the AP lyase activity associated with hOGG1 is quite weak, HAP1 endonuclease was added to the reaction mixture to force strand cleavage at AP sites in the DNA (Fig. 4B, lanes 5–8). HAP1 cleaves DNA specifically at the 5′ side of abasic sites, generating fragments containing a 3′-OH that migrate faster than the products of the AP lyase cleaving at the 3′ side (Fig. 4B, compare lanes 5 and 6 with lanes 1 and 2). However, despite the increased cleavage efficiency observed in the cytoplasmic and chromatin fractions (Fig. 4B, lanes 5 and 6) no hOGG1 activity was detected in the high salt wash and the nuclear matrix fractions upon the addition of HAP1 (Fig. 4B, lanes 7 and 8). Similar results were obtained for the non-transfected HeLa-derived extracts (compare Fig. 4B and C). Addition of purified hOGG1 to the nuclear matrix fraction restores DNA glycosylase/AP lyase activity (Fig. 4B and C, compare lanes 9 and 10) thus excluding the possibility that the absence of hOGG1 activity is due to the presence of inhibitors, or the absence of specific partners or essential cofactors that may be necessary for efficient glycosylase activity. However, the lack of 8-oxoG:C DNA glycosylase activity in the nuclear matrix fraction was found to be caused by insolubility of hOGG1 in the matrix fraction effected by the extraction conditions. The hOGG1 could be solubilised by NP-40 extraction, but the enzyme was not recovered in an active form (data not shown).
Figure 4.
Comparative 8-oxoG cleavage activity in chromatin and nuclear matrix fractions. (A) Schematic representation of hOGG1 enzymatic activity. A 24 bp double-stranded oligonucleotide containing an 8-oxoG:C base pair at position 14 (in bold) was prepared as described (13) and 32P-labelled at the 5′ end of the 8-oxoG-containing strand. hOGG1 activity excises the 8-oxoG and cleaves the 32P-labelled 8-oxoG-containing strand 3′ of the lesion (β-elimination). The addition of HAP1 endonuclease induces subsequent cleavage of the terminal sugar phosphate. Both lyase and endonuclease activities produce a 13 nt 32P-end-labelled product when the reaction is run on denaturating polyacrylamide gels. (B) HeLa cells expressing hOGG1–EGFP or (C) non-transfected HeLa cells were fractionated as described in the Material and Methods and shown in Figure 2. Equivalent amounts of each fraction were incubated with the 8-oxoG:C-containing double-stranded oligonucleotide. Arrows indicate the positions of the 5′ radiolabelled substrate (24 bp) and reaction products as follows: 13 nt, 13mer product produced by hOGG1 strand nicking activity (lanes 1–4); 13* nt, 13 mer with a 3′-OH produced by the addition of HAP1 endonuclease where indicated (lanes 5–8). Ten nanograms of purified hOGG1 was added in control reaction mixtures with (lane 9) or without (lane 10) nuclear matrix extract. Cleavage products were analysed by PhosphorImaging after 20% denaturating PAGE.
The chromatin-associated hOGG1–EGFP fusion protein is phosphorylated on a serine residue in vivo
Cell cycle-dependent changes in subcellular localisation have often been correlated with changes in protein phosphorylation patterns as demonstrated for a number of proteins including ATRX, BRG and hbrm, and the Rb protein (28,32,33). The hOGG1 protein has several predicted phosphorylation sites for cAMP and cGMP-dependent kinases, casein kinase II and PKC and this prompted us to examine whether the localisation of hOGG1 could be related to the protein phosphorylation status. In vivo phosphorylation was examined by western blot analysis of each HeLa hOGG1–EGFP fraction using an anti-phosphoserine antibody (Fig. 5A). Although the anti-GFP antibody did not reveal any obvious differences in the electrophoretic mobility of the hOGG1–EGFP fusion protein in the various fractions, the anti-phosphoserine antibody detected a phosphorylated form of hOGG1–EGFP, however, only in the chromatin fraction. Using an anti-phosphotyrosine antibody, no phosphorylation could be detected in any of the fractions (data not shown). Treatment of the purified hOGG1 protein or the different biochemical fractions with λ-PPase did not have any significant effect on the hOGG1 enzymatic activity (Fig. 5B).
Figure 5.
The chromatin-associated hOGG1–EGFP is phosphorylated on a serine residue in vivo. (A) HeLa cells expressing the hOGG1–EGFP fusion protein were extracted to obtain the cytoplasmic fraction, the whole chromatin fraction, a high salt wash and the core nuclear matrix. Proteins from equal cell equivalents of each fraction were analysed by western blotting with the indicated antibodies. (B) Equal cell equivalents from each fraction (lanes 1–8) or 10 ng of hOGG1 (lanes 9 and 10) were mock treated (lanes 1, 3, 5, 7 and 9) or treated with λ-PPase (lanes 2, 4, 6, 8 and 10) before measuring hOGG1 activity in the presence of HAP1 endonuclease as described in Figure 4. 13* nt, 13 mer with a 3′-OH produced by the addition of HAP1 after hOGG1-strand nicking activity.
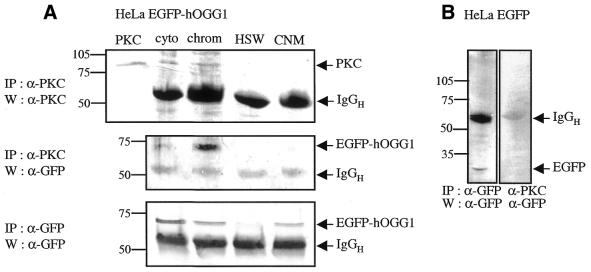
The chromatin-associated hOGG1–EGFP interacts with PKC
Previous studies have suggested a role for PKC phosphorylation in controlling protein interactions between the nuclear envelope and the chromatin (34,35). As hOGG1 contains potential PKC consensus sequences (S44FR, S211AR and T295NK), we investigated possible interactions between PKC and hOGG1 by attempts at co-immunoprecipitation using a monoclonal antibody against PKC (Fig. 6A). PKC was immunoprecipitated from the cytoplasmic and chromatin fractions using the monoclonal anti-PKC antibody, while hOGG1–EGFP was immunoprecipitated by the polyclonal anti-GFP antibody from the cytoplasmic, chromatin and nuclear matrix fractions. However, hOGG1–EGFP was specifically co-immunoprecipitated with PKC from the chromatin fraction. Weak co-immunoprecipitation could also occasionally be observed from the cytoplasmic fraction. In contrast, no co-immunoprecipitation was observed with EGFP (Fig. 6B, cytoplasmic fraction that contains all the EGFP). These results demonstrate that PKC and hOGG1–EGFP can be co-immunoprecipitated from a low salt, DNase-treated chromatin extract of HeLa hOGG1–EGFP cells.
Figure 6.
The chromatin-associated hOGG1–EGFP is co-immunoprecipitated with PKC in vivo. Equal cell equivalents from HeLa hOGG1–EGFP (A) or HeLa EGFP (B) biochemical fractions were subjected to immunoprecipitation with either anti-GFP or anti-PKC antibody as indicated. Immunoprecipitates were then analysed by immunoblotting with the appropriate antibodies.
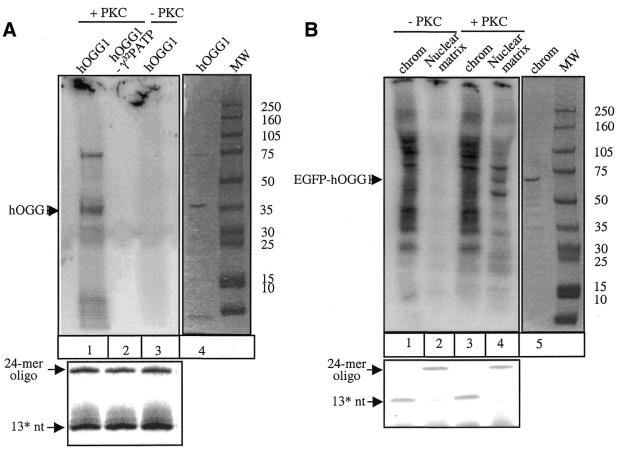
Purified hOGG1 and nuclear matrix-associated hOGG1–EGFP are phosphorylated by PKC in vitro
We also investigated if hOGG1 could be phosphorylated by PKC in vitro and, if so, whether such modification would affect its catalytic activity. Purified recombinant hOGG1 did not demonstrate any autophosphorylation activity (Fig. 7, lane 3) but was a substrate for phosphorylation by a mixture of α, β and γ PKC isoforms in vitro (Fig. 7, lane 1). The phosphorylation did not appear to have any significant effect on hOGG1 enzymatic activity as assayed by the cleavage of the 8-oxoG:C-containing double-stranded oligonucleotide (Fig. 7A, lower panel, compare lanes 2 and 3 with lane 1).
Figure 7.
PKC phosphorylates purified hOGG1 and the nuclear matrix-associated hOGG1–EGFP in vitro. (A) Purified recombinant hOGG1 protein was incubated in the presence (lanes 1 and 2) or in the absence (lane 3) of PKC together with [γ-32P]ATP (added in lanes 1 and 3) (left panel). The position of the phosphorylated protein was identified by comparing with purified hOGG1 run on a Coomassie-stained gel (lane 4, right panel). An equivalent of 10 ng of protein was assayed for hOGG1 enzymatic activity as described in Figure 4 (lower panel). (B) Cell extract equivalents from chromatin (lanes 1 and 3) and nuclear matrix fractions (lanes 2 and 4) from HeLa hOGG1–EGFP cells were incubated in the presence (lanes 3 and 4) or in the absence (lanes 1 and 2) of PKC together with [γ-32P]ATP (left panel). The position of phosphorylated hOGG1–EGFP was identified by Coomassie staining of the treated fraction (right panel). Following the phosphorylation reaction, samples were assayed for hOGG1 enzymatic activity as described in Figure 4 (lower panel).
We next investigated whether the hOGG1 protein obtained by the cellular fractionation was a substrate for PKC. HeLa cells expressing hOGG1–EGFP were fractionated as described (i.e. see Fig. 2) and each fraction was incubated with PKC isoforms mixture in vitro (Fig. 7B). hOGG1–EGFP was identified by the staining of the co-migrating band with Coomassie Blue (Fig. 7B, lane 5). No phosphorylation of hOGG1–EGFP was observed in the chromatin fraction although the labelling pattern clearly suggested the presence of kinase activities (Fig. 7B, compare lanes 1 and 3). In contrast, no endogenous kinase activity was detected in the nuclear matrix fraction (Fig 7B, lane 2). However, the resident hOGG1–EGFP fusion protein was indeed a substrate for in vitro phosphorylation by PKC (Fig. 7B, lane 4). We also tested if phosphorylation of the fractionated hOGG1–EGFP protein would recover any enzymatic activity. Treatment of the chromatin-associated hOGG1–EGFP with PKC did not have any significant effect on the enzymatic activity of the fusion protein (Fig 7B, lower panel, compare lanes 1 and 3). Furthermore, as expected, enzymatic activity of the insoluble nuclear matrix-associated hOGG1–EGFP (Fig. 7B, lane 2) could not be restored by PKC phosphorylation (Fig. 7B, lane 4).
DISCUSSION
In order to gain further insights into the in vivo function of the hOGG1 protein, we have examined its subcellular distribution in form of a fusion to EGFP (hOGG1–EGFP). We report that hOGG1 is localised to condensed chromatin at the onset of mitosis from pro-metaphase to telophase. Co-localisation of hOGG1 with mitotic chromosomes resembles that of several other nuclear proteins including the SWI/SNF-like protein ATRX, the heterochomatin-binding protein HP1 and DNA topoisomerase IIα. These proteins are all involved in transcriptional regulation through an effect on chromatin remodelling, and are also believed to be involved in DNA replication and repair (28,36,37). Our findings suggest that hOGG1 may be important during mitosis as part of a mitotic mutation avoidance system similar to that described previously for the mismatch repair proteins in S.cerevisiae (38) and the nucleotide excision repair proteins in Schizosaccharyomyces pombe (39).
The nuclear matrix is viewed as a dynamic RNA–protein network, which not only hosts transcription and replication complexes but also proteins involved in DNA repair. Among the multitude of enzymes involved in repair processes, PARP, mutant p53 and DNA topoisomerase IIα have been shown to preferentially localise to the matrix (40–43). Furthermore, several proteins including Ku70, Fanconi Anemia proteins and BLM in complex with hRAD51 have been shown to bind to the nuclear matrix in a DNA damage and cell cycle-regulated manner (44–46). In this report, we show that hOGG1 is associated with the DNase-sensitive chromatin as well as the nuclear matrix during interphase. The chromatin-associated hOGG1 displays 8-oxoG:C DNA glycosylase/AP lyase activity and therefore most probably works in vivo to repair the oxidation-associated lesions in genomic DNA. The functional significance of the association of hOGG1 with the nuclear matrix is more difficult to interpret. We were unable to measure any 8-oxoG:C DNA glycosylase activity associated with the nuclear matrix but this can be ascribed to precipitation and inactivation of the hOGG1 protein during the extraction procedure. It is still likely that hOGG1 remains active in the nuclear matrix in vivo and participates in base excision repair of oxidative damage that could arise for instance at scaffold/matrix (S/MAR) regions. Repair activity at these sequences was suggested previously for PARP (40). More recent observations demonstrating that S/MARs are hypersensitive to DNA breakage also support the possibility of excision repair going on in these sequences (47). It has also been proposed that the nuclear matrix is a site where damaged ends meet to be repaired (48).
The observed association of hOGG1 with the nuclear matrix also raises the question about other roles of the protein, for instance in mRNA synthesis and processing and/or replication. Several recent genetic and biochemical studies provide evidence for a dual role of DNA glycosylases in both transcriptional control and DNA repair. The human uracil DNA glycosylase UDG was shown to negatively regulate E2F-mediated transcriptional activity (49). More recently, Missero et al. (50) described an unexpected role for the human T:G mismatch specific thymine DNA glycosylase TDG as a transcriptional repressor. In addition, the three-dimensional structure of the E.coli DNA glycosylase AlkA revealed the presence of an N-terminal β-sheet/α-helix domain very similar to the structure of the conserved tandem repeat of the TFIID TATA-binding protein (51). In comparative analysis, we observed that AlkA and hOGG1 show very similar three-dimensional structures despite limited sequence similarity. This observation, taken together with the present study showing the localisation of hOGG1 to putative transcription sites (i.e. condensed chromatin and nuclear matrix), are consistent with an as yet undescribed role of hOGG1 in transcriptional regulation.
The possibility that a post-translational modification could be correlated with the dynamic localisation of hOGG1 to either the chromatin or the nuclear matrix during interphase was investigated using anti-phosphoserine or anti-phosphotyrosine antibodies. The results show that chromatin-associated hOGG1 is phosphorylated on a serine residue whereas the nuclear matrix-associated hOGG1 appears unphosphorylated. Phosphorylation experiments showed that the nuclear matrix-associated hOGG1 could be a substrate for PKC in vitro whereas the chromatin-associated hOGG1 was not. In addition, immunoprecipitation experiments demonstrated that the expressed hOGG1 binds to PKC in the chromatin extract. These findings suggest that the phosphorylation of the chromatin-associated hOGG1 could be mediated by PKC. PKC has been reported to phosphorylate and modulate the enzymatic functions of a number of DNA repair proteins including DNA polymerase β, RPA, DNA topoisomerase IIα and HAP1 (52–55). However, we could not detect any significant alteration of enzymatic function correlated with the phosphorylation event, suggesting that phosphorylation could be involved in protein re-localisation rather than in modulation of the enzymatic function. On the other hand, it is difficult to assess the extent of the phosphorylation reaction and more detailed kinetic studies are needed to conclude firmly on the effects on the enzymatic reactions. Nevertheless, the present study provides clear evidence for the existence of a phosphorylated subfraction of hOGG1 in the cell, which preferentially associates with interphasic chromatin.
In conclusion, we have shown that hOGG1 is dynamically regulated with respect to its nuclear localisation, such that the DNase-sensitive chromatin as well as the nuclear matrix appears to be the preferential sites of association during interphase, while the protein re-localises to condensed chromatin during mitosis. These results suggest that the 8-oxoG DNA glycosylase hOGG1 may be involved in more aspects of DNA metabolism and repair than previously assumed and possibly also have other functions than just removing 8-oxoG from DNA. In this context, it might be worth mentioning that hOGG1 has a strong affinity for C/C mismatches in DNA (I.Morland, M.Bjørås and E. Seeberg, unpublished data). We have also shown that hOGG1 becomes phosphorylated in vivo and that PKC is likely to be responsible for this phosphorylation reaction. Phosphorylation may direct compartmentalisation of hOGG1 although further experiments are required to be more conclusive about the exact role of the phosphorylation event. Nevertheless, this appears to be the first demonstration of a post-translational modification of hOGG1, similarly as has been described for many other proteins being involved in DNA repair.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Tor Lea for providing the anti-phosphotyrosine antibody (clone 4G10) and to Philippe Collas and Kathy Baynton for a critical reading of the manuscript. This work was supported by the International Agency of Cancer Research (IARC, Lyon, France), the Norwegian Cancer Society, the Research Council of Norway and an EU contract (QLRT-1999-02002).
REFERENCES
- 1.Marnett L.J. (2000) Oxyradicals and DNA damage. Carcinogenesis, 21, 361–370. [DOI] [PubMed] [Google Scholar]
- 2.Raha S. and Robinson,B.H. (2000) Mitochondria, oxygen free radicals diseases and ageing. Trends Biochem. Sci., 25, 502–508. [DOI] [PubMed] [Google Scholar]
- 3.Seeberg E., Eide,L. and Bjørås,M. (1995) The base excision repair pathway. Trends Biochem. Sci., 20, 391–397. [DOI] [PubMed] [Google Scholar]
- 4.Cheng K.C., Cahill,D.S., Kasai,H., Nishimura,S. and Loeb,L.A. (1992) 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G-T and A-C substitutions. J. Biol. Chem. 267, 166–172. [PubMed] [Google Scholar]
- 5.Moriya M. (1993) Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G.C → T.A transversions in simian kidney cells. Proc. Natl Acad. Sci. USA, 90, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boiteux S., O’Connor,T.R. and Laval,J. (1987) Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J., 6, 3177–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tchou J., Kasai,H., Shibutani,S., Chung,M.H., Laval,J., Grollman,A.P. and Nishimura,S. (1991) 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl Acad. Sci. USA, 88, 4690–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michaels M.L., Pham,L., Nghiem,Y., Cruz,C. and Miller,J.H. (1990) MutY, an adenine glycosylase active on G-A mispairs, has homology to endonuclease III. Nucleic Acids Res., 18, 3841–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maki H. and Segikuchi,M. (1992) Mut T protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature, 355, 273–275. [DOI] [PubMed] [Google Scholar]
- 10.Sakumi K., Furuichi,M., Tsuzuki,T., Kakuma,T., Kawabata,S.I., Maka,H. and Sekiguchi,M. (1993) Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem., 268, 23524–23530. [PubMed] [Google Scholar]
- 11.Slupska M.M., Baikalov,C., Luther,W.M., Chiang,J.H., Wei,Y.F. and Miller,J.H. (1996) Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J. Bacteriol., 178, 3885–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Kemp P.A., Thomas,D., Barbey,R., de Oliveira,R. and Boiteux,S. (1996) Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which encodes a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl Acad. Sci. USA, 93, 5197–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bjørås M., Luna,L., Johnsen,B., Hoff,E., Haug,T., Rognes,T. and Seeberg,E. (1997) Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J., 16, 6314–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michaels M.L. and Miller,J.H. (1992) The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol., 174, 6321–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas D., Scott,A., Barbey,R., Padula,M. and Boiteux,S. (1996) Inactivation of OGG1 increases the incidence of G.C → T.A transversions in Saccharomyces cerevisiae: evidence for endogenous oxidative damage to DNA in eukaryotic cells. Mol. Gen. Genet., 254, 171–178. [DOI] [PubMed] [Google Scholar]
- 16.Klungland A., Rosewell,I., Hollenbach,S., Larsen,E., Daly,G., Epe,B., Seeberg,E., Lindhal,T. and Barnes,D.E. (1999) Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl Acad. Sci. USA, 96, 13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minowa O., Arai,T., Hirano,M., Monden,Y., Nakai,S., Fukuda,M., Itoh,M., Takano,H., Hippou,Y., Aburatani,H., Masumura,K., Nohmi,T., Nishimura,S. and Noda,T. (2000) Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl Acad. Sci. USA, 97, 4156–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollstein M., Shomer,B., Greenblatt,M., Soussi,T., Hovig,E., Montesano,R. and Harris,C.C. (1996) Somatic point mutations in the p53 gene of human tumors and cell lines: updated compilation. Nucleic Acids Res., 24, 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chevillard S., Radicella,J.P., Levalois,C., Lebeau,J., Poupon,M.F., Oudard,S., Dutrillaux,B. and Boiteux,S. (1998) Mutations in OGG1, a gene involved in the repair of oxidative DNA damage, are found in human lung and kidney tumors. Oncogene, 16, 3083–3086. [DOI] [PubMed] [Google Scholar]
- 20.Blons H., Radicella,J.P., Laccourreye,O., Brasnu,D., Beaune,P., Boiteux,S. and Laurent-Puig,P. (1999) Frequent allelic loss at chromosome 3p distinct from genetic alterations of the 8-oxoguanine DNA glycosylase 1 gene in head and neck cancer. Mol. Carcinogen., 26, 254–260. [PubMed] [Google Scholar]
- 21.Dherin C., Radicella,J.P., Dizdaroglu,M. and Boiteux,S. (1999) Excision of oxidatively damaged DNA bases by the human alpha-hOgg1 protein and the polymorphic alpha-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res., 27, 4001–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vidal A.E., Hickson,I.D., Boiteux,S. and Radicella,J.P. (2001) Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res., 29, 1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zharkov D.O., Rosenquist,T.A., Gerchman,S.E. and Grollman,A.P. (2000) Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J. Biol. Chem., 275, 28607–28617. [DOI] [PubMed] [Google Scholar]
- 24.Bruner S., Norman,D.P.G. and Verdine,G. (2000) Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature, 403, 859–866. [DOI] [PubMed] [Google Scholar]
- 25.Bjørås M., Seeberg,E., Luna,L., Pearl,L.H. and Barrett,T.E. (2002) Reciprocal ‘flipping’ underlies substrate recognition and catalytic activation by the human 8-oxo-guanine DNA glycosylase. J. Mol. Biol., 317, 171–177. [DOI] [PubMed] [Google Scholar]
- 26.Lamond A.I. and Earnshaw,W.C. (1998) Structure and function in the nucleus. Science, 280, 547–553. [DOI] [PubMed] [Google Scholar]
- 27.He D., Jeffrey,A., Nickerson,J. and Penman,S. (1990) Core filament of the nuclear matrix. J. Cell Biol., 110, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berube N.G., Smeenk,C.A. and Picketts,D.J. (2000) Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum. Mol. Genet., 9, 539–547. [DOI] [PubMed] [Google Scholar]
- 29.Reyes J.C., Muchardt,C. and Yaniv,M. (1997) Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol., 146, 29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martins S.B., Eide,T., Steen,R.L., Jahnsen,T., Skålhegg,B.S. and Collas,P. (2000) HA95 is a protein of the chromatin and the nuclear matrix regulating nuclear envelope dynamics. J. Cell Science, 113, 3703–3713. [DOI] [PubMed] [Google Scholar]
- 31.Mo Y.-Y. and Beck,W. (1999) Association of human DNA topoisomerase IIα with mitotic chromosomes in mammalian cells is independent of its catalytic activity. Exp. Cell Res., 252, 50–62. [DOI] [PubMed] [Google Scholar]
- 32.Muchardt C., Reyes,J.C., Boucharot,B., Leguoy,E. and Yaniv,M. (1996) The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condense chromosomes during mitosis. EMBO J., 15, 3394–3402. [PMC free article] [PubMed] [Google Scholar]
- 33.Mittnacht S. and Weinberg,R.A. (1991) G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell, 65, 381–393. [DOI] [PubMed] [Google Scholar]
- 34.Dreger M., Otto,H., Neubauer,G., Mann,M. and Hucho,F. (1999) Identification of phosphorylation sites in native lamina-associated polypeptide 2 beta. Biochemistry, 38, 9426–9434. [DOI] [PubMed] [Google Scholar]
- 35.Collas P., Thompson,L., Fields,A.P., Poccia,D.L. and Courvalin,J.C. (1997) Protein kinase C-mediated interphase lamin B phosphorylation and solubilization. J. Biol. Chem., 272, 21274–21280. [DOI] [PubMed] [Google Scholar]
- 36.Furuta K., Chan,E.K.L., Kiyosawa,K., Reimer,G., Luderschmidt,C. and Tan,E.M. (1997) Heterochromatin protein HP1Hsβ (p25β) and its localisation with centromeres in mitosis. Chromosoma, 106, 11–19. [DOI] [PubMed] [Google Scholar]
- 37.Gibbons R.J., McDowell,T.L., Raman,S., O’Rourke,D.M., Garrick,D., Ayyub,H. and Higgs,D.R. (2000) Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nature Genet., 24, 368–371. [DOI] [PubMed] [Google Scholar]
- 38.Harfe B.D. and Jinks-Robertson,S. (2000) Mismatch repair proteins and mitotic genome stability. Mutat. Res., 451, 151–167. [DOI] [PubMed] [Google Scholar]
- 39.Hohl M., Christensen,O., Kunz,C., Naegeli,H. and Fleck,O. (2001) Binding and repair of mismatched DNA mediated by Rhp14, fission yeast homologue of human XPA. J. Biol. Chem., 176, 30766–30772. [DOI] [PubMed] [Google Scholar]
- 40.Zaalishvili T.M., Gabriadze,I.Y., Margiani,D.O., Philauri,V.R. and Surguladze,N.M. (2000) Participation of poly(ADP-ribose)-polymerase of nuclear matrix in DNA repair. Biochemistry, 65, 659–661. [PubMed] [Google Scholar]
- 41.Deppert W., Gohler,T., Koga,H. and Kim,E. (2000) Mutant p53: ‘Gain of function’ trough perturbation of nuclear structure and function ? J. Cell. Biochem., 35, 115–122. [PubMed] [Google Scholar]
- 42.Ivanova E.C., Donev,R.M. and Djondjurov,L.P. (1999) Localisation of DNA topoisomerase II alpha in mouse erythroleukemia cells. Mol. Cell, 9, 309–313. [PubMed] [Google Scholar]
- 43.Lambert J.M. and Fernandes,D.J. (2000) Topoisomerase II cleavable complex formation within DNA loop domains. Biochem. Pharmacol., 60, 101–109. [DOI] [PubMed] [Google Scholar]
- 44.Yu E., Song,K., Moon,H., Maul,G.G. and Lee,I. (1998) Characteristic immunolocalisation of Ku protein as nuclear matrix. Hybridoma, 17, 413–419. [DOI] [PubMed] [Google Scholar]
- 45.Giao F., Moss,A. and Kupfer,G.M. (2001) Fanconi Anemia proteins localize to chromatin and the nuclear matrix in a DNA damage- and cell cycle-regulated manner. J. Biol. Chem., 276, 23391–23396. [DOI] [PubMed] [Google Scholar]
- 46.Bischof O., Kim,S.-H., Irving,J., Beresten,S., Ellis,N.A. and Campisi,J. (2001) Regulation and localisation of the bloom syndrome protein in response to DNA damage. J. Cell Biol., 153, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Legault J., Tremblay,A. and Mirault,M.E. (1997) Preferential localisation of DNA damage induced by depurination and bleomycin in a plasmid containing a scaffold-associated region. Biochem. Cell Biol., 75, 369–375. [PubMed] [Google Scholar]
- 48.Bode J., Benham,C., Ernst,E., Knopp,A., Marschalek,R., Strick,R. and Strissel,P. (2000) Fatal connections: when DNA ends meet on the nuclear matrix. J. Cell. Biochem., 35, 3–22. [DOI] [PubMed] [Google Scholar]
- 49.Walsh M.J., Shue,G., Spidoni,K. and Kapoor,A. (1995) E2F-1 and a cyclin-like DNA repair enzyme, uracil-DNA glycosylase, provide evidence for an autoregulatory mechanism for transcription. J. Biol. Chem., 270, 5289–5298. [DOI] [PubMed] [Google Scholar]
- 50.Missero C., Pirro,M.T., Simeone,S., Pischetola,M. and Di Lauro,R. (2001) The DNA glycosylase T:G mismatch-specific thymine DNA glycosylase represses thyroid transcription factor-1-activated transcription. J. Biol. Chem., 276, 33569–33575. [DOI] [PubMed] [Google Scholar]
- 51.Labahn J., Scharer,O.D., Long,A., Ezaz-Nikpay,K., Verdine,G.L. and Ellenberger,T.E. (1996) Structural basis for the excision repair of alkylation-damaged DNA. Cell, 86, 321–329. [DOI] [PubMed] [Google Scholar]
- 52.Tokui T., Inagaki,M., Nishizawa,K., Yatani,R., Kusagawa,M., Ajiro,K., Nishimoto,Y., Date,T. and Matsukage,A. (1991) Inactivation of DNA polymerase beta by in vitro phosphorylation with protein kinase C. J. Biol. Chem., 266, 10820–10824. [PubMed] [Google Scholar]
- 53.Brush G.S. and Kelly,T.J. (2000) Phosphorylation of the replication protein A large subunit in the Saccharomyces cerevisiae checkpoint response. Nucleic Acids Res., 28, 3725–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wells N.J., Fry,A.M., Guano,F., Norbuty,C. and Hickson,I.D. (1995) Cell cycle phase-specific phosphorylation of human topoisomerase IIα. J. Biol. Chem., 270, 28357–28363. [DOI] [PubMed] [Google Scholar]
- 55.Hsieh M.M., Hegde,V., Kelley,M.R. and Deutsch,W.A. (2001) Activation of APE/Ref-1 redox activity is mediated by reactive oxygen species and PKC phosphorylation. Nucleic Acids Res., 29, 3116–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]