Abstract
DNA methylation is an essential epigenetic modification that plays a crucial role in regulating gene expression and maintaining genomic stability. With the advancement in sequencing technology, methylation studies have provided valuable insights into the diagnosis of rare diseases through the various identification of episignatures, epivariation, epioutliers, and allele-specific methylation. However, current methylation studies are not without limitations. This mini-review explores the current understanding of DNA methylation in rare diseases, highlighting the key mechanisms and diagnostic potential, and emphasizing the need for advanced methodologies and integrative approaches to enhance the understanding of disease progression and design more personable treatment for patients, given the nature of rare diseases.
Keywords: DNA methylation, rare diseases, review, epioutliers, allele specific methylation, epivariation
1. Introduction
Rare diseases (RDs) are medical conditions affecting fewer than 200,000 individuals in the United States and less than 1 in 2000 in the European Union [1]. While individual RDs affect a limited number of the population, on aggregate, RDs encompass more than 10,000 unique disorders [2] and impact approximately 300 million people [3,4]. The majority of RDs are Mendelian genetic disorders [5,6] and comprehensive genetic testing, using exome sequencing and more recently, genome sequencing, is often used to identify causal variation. Sequencing technology advancements have also led to increased use of multi-omic approaches for variant identification, interpretation, and prioritization, which may include phenomics, transcriptomics, proteomics, metabolomics, and epigenomics [7]. While most rare diseases are associated with genetic factors, there is growing recognition of epigenetic involvement, particularly DNA methylation (DNAm), as a significant contributor to a subset of RDs. This is exemplified by Mendelian disorders of the epigenetic machinery (MDEM) [8], also referred to as chromatinopathies [9]. These insights highlight the complementary relationship between genetic and epigenetic factors, offering new avenues for understanding pathogenesis and potential therapeutic interventions [8].
DNA methylation is a crucial epigenetic modification involved in the regulation of gene expression. The most common and best-described methylation of DNA is the addition of a methyl group to carbon 5 of cytosine bases, forming 5-methylcytosine or 5mC [10] (Figure 1A). It is known that 5mC is often, but not exclusively, maintained in the context of cytosine-phosphate-guanine (CpG) dinucleotides [10]. DNA methylation patterns are commonly established and maintained by groups of DNA methyltransferases, such as the DNMT family and associated proteins [11]. The ten-eleven translocation (TET) family proteins mediate the reversal of 5mC methylation through iterative oxidation of 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), 5-carboxylcytosine (5caC) and a complete reversal is achieved by thymine-DNA glycosylase (TDG)-mediated abasic site (AP) and base excision repair (BER) [11]. It is believed that genetic variants that alter the functions of these regulatory proteins can lead to aberrant DNA methylation patterns, including hypermethylation (excessive methylation) or hypomethylation (reduced methylation), subsequently affecting gene expression patterns and chromatin structure [12]. In summary, DNA methylation is one of the primary epigenetic mechanisms that coordinates gene activity at the transcriptional level and regulates critical developmental and physiological pathways. As such, dysregulation of methylation is an important contributor to the manifestation of RDs [13,14].
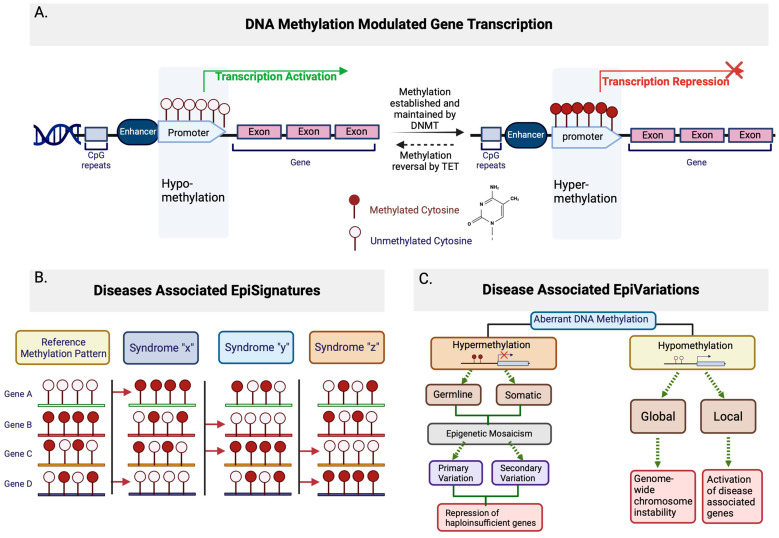
Figure 1.
Schematic of DNA methylation modulated gene transcription and associated episignatures and epivariations. (A) General representation of the association between DNA methylation state and gene transcription. Hypomethyaltion in promoter generally leads to transcription activation, where the establishment and maintenance of DNA methylation through DNMT family proteins leads to hypermethylation in the promoter, which represses transcription. The methylation state can be reversed in a process initiated by the TET family proteins. (B) General representation of disease-associated episignatures. Left most represents the reference methylation patterns, with methylated or unmethylated CpGs within a gene. Hypothetical syndromes are shown with red arrows to indicate DNA methylation patterns at specific genes that differ from the reference methylation patterns. (C) General representation of disease-associated epivariations. Aberrant DNA methylation can be either hyper- or hypo-methylation. Hypermethylation can be either germline or somatic, primary or secondary, and ultimately leads to the repression of haploinsufficient genes. Hypomethylation occurs globally (genome-wide) or locally, global hypomethylation leads to chromosomal instability, while local hypomethylation leads to the activation of disease-associated genes.
2. DNA Methylation and Rare Diseases
DNA methylation profiles have been extensively investigated using DNA methylation microarrays to identify epigenetic dysregulation in many RDs [15]; reviewed in [16]. The resulting distinct and stable DNA methylation signatures that are induced by pathogenic variants in disease-causing genes, e.g., [5,12,17,18,19], coined “episignatures”, have been associated with more than 70 Mendelian conditions [7,20] (Figure 1B). The number of novel diagnostic episignatures is on the rise, with many identified for RDs, including α-thalassemia mental retardation syndrome (MIM: 300032) [21], Kabuki syndrome (MIM: 602113 and 300128) [22], CHARGE syndrome (MIM: 214800) [22], Sotos syndrome (MIM: 606681) [23], Floating Harbor syndrome (MIM: 611421) [24], Coffin–Siris syndrome, and other BAFopathies (MIM: 135900, 614607, 614608, 614609, and 615866) [25]. Episignatures have shown the capabilities to distinguish between different genetic conditions [26], support variant classification by defining phenotypic specificity, and are effective diagnostic modalities for rare Mendelian conditions following inconclusive testing [27,28]. As Sadikovic et al. (2021) reported, the validation rate for VUS reclassification in a selected cohort of patients with previous ambiguous/inconclusive genetic findings can reach 35% using methylation signature (EpiSign) analysis [28].
Episignatures are typically discovered using epigenome-wide association studies (EWAS) investigating DNA methylation patterns from microarrays, followed by the application of a multiclass machine learning classifier [29]. Moreover, the use of artificial intelligence tools for epigenomic studies in RDs has increased over the last decade [30]. For example, Aref-Eshgi et al. (2019) introduced a computational model using whole genome methylation data to aid in diagnosing 14 neurodevelopmental disorders characterized by known episignatures [24]. The model effectively identified methylation profiles suggestive of specific Mendelian conditions for 31% (21/67) of individuals with uncertain diagnosis, including cases where conventional molecular testing failed to identify any candidate variants. In addition, Turinsky et al. (2020) created EpigenCentral, a free portal to interactively classify and analyze epigenome data for known disease-associated episignatures [29]. Further optimization of episignature classifiers is important to improve model sensitivity and molecular diagnosis of Mendelian disorders of the epigenetic machinery. Walsh et al. (2024) introduced a machine learning method that uses an age- and sex-stratified methylation model for outlier detection, which significantly reduces false negatives in array-based methylation signature analysis. By accounting for age- and sex-related methylation changes, this approach improved the classification of samples with potential methylation-associated congenital disorders [31]. Oexle et al. (2023) trained episignature classifiers to robustly detect low-level mosaics while also revoking erroneous exome calls of mosaicism to highlight improved diagnostic yield for RDs [32].
In addition to episignatures, epivariations and epimutations, can also be detected by methylation microarrays. The term “epimutation” was originally intended to describe epigenetic changes that occur without alterations in the underlying DNA sequence [33]. Over time, it has been applied more broadly to various epigenetic changes [34]. The term “epivariations” was later introduced to describe rare epigenetic aberrations [30,35]. While the two terms are now often used interchangeably, “epivariation” generally refers to regions exhibiting aberrant methylation patterns, characterized by significant enrichment in epimutations, which are abnormal mutational changes that do not change the DNA sequence [35,36] (Figure 1C).
Epivariations, or in some literature, epimutations, can be subdivided into primary or secondary types based on their origin [37]. Primary epivariations are thought to arise from stochastic (random) errors in the establishment or maintenance of the epigenome by the DNA methyltransferase proteins family [38]. These errors are sporadic and not necessarily linked to the changes in the DNA sequences, such as certain types of imprinting anomalies seen in Prader–Willi (MIM: 176270) and Angelman Syndromes (MIM: 105830) [33]. On the other hand, secondary epivariations derive from underlying changes in local DNA sequence, including copy number variations (CNVs), where segments of DNA are duplicated or deleted, or single nucleotide variations (SNVs), which are single base changes in the DNA sequence [37] at differentially methylated loci. Additionally, mutations that disrupt regulatory elements [38] and expansions of CpG-rich tandem repeats (STR) [39]—repetitive sequences rich in CpG sites—are believed to contribute to changes in the local DNA sequence. For example, Fragile X syndrome (MIM: 309550) is caused by secondary epivariations in the FMR1 gene [40]. Both epivariation types are found in patients with RDs.
It is important to note that next-generation sequencing (NGS) technologies, including targeted (e.g., reduced representation bisulfite sequencing or RRBS), offer high specificity for targeted CpG-rich regions and sensitivity for detecting small changes in methylation levels at individual CpG sites. Whole genome (e.g., whole genome bisulfite sequencing or WGBS, enzymatic methyl sequencing or EMseq) provides comprehensive coverage for CpG sites across the entire genome, which allows higher sensitivity for detecting methylation changes even in non-CpG regions. Additionally, long-read sequencing (LRS) provides higher resolution of complex genomic regions and improves assembly quality, generating methylation profiling data that can help clarify variant pathogenicity and aid in the diagnosis of RDs [14]. Moreover, the usage of optical genome mapping (OGM) has enhanced the detection of large structural variants (SVs), CNVs, and repetitive sequence motifs at the single-cell level, enabling a more detailed characterization of cellular heterogeneity [41].
Several published studies have illustrated epigenomic approaches for improving the diagnosis of rare diseases and shortening diagnostic journeys for patients. Gatto et al. (2017) identified a rare pathogenic variant in DNMT3B by studying the methylation profiles in immunodeficiency-centromeric instability-facial anomalies syndrome 1 (ICF1; MIM: 242860) through RRBS [42]. Sun et al. (2014) studied genome-wide DNA methylation profiles of hereditary sensory and autonomic neuropathy type 1E (HSAN1E) patients with DNMT1 mutations using WGBS and discovered all chromosomes hypomethylated with enrichment of NAD+/NADH pathway-associated genes in differentially methylated regions (DMRs) [43]. More recent examples include Smith et al. (2021) which found that loss-of-function variants of DNMT3A lead to decreased global DNA methylation in Tatton–Brown–Rahman syndrome (TBRS; MIM: 615879) [44]. Zhu et al. (2022) utilized WGBS to identify autism spectrum disorder (ASD)-associated methylation changes with an enrichment of DMRs in ASD-associated genes [45]. Miller et al. (2020) leveraged targeted long-read sequencing to identify the cause of altered GNAS exon A/B methylation in autosomal dominate pseudohypoparathyroidism type 1b (PHP1B; MIM: 603233) [46]. In summary, multiple studies have demonstrated the power of DNA methylation profiling as a valuable functional tool to aid in the diagnosis of RD cases, complementing standard genomic sequencing to unambiguously identify and interpret variation [47,48,49].
3. Limitations
The reported diagnostic yield among patients with neurodevelopmental disorders varies by method, with chromosomal microarray achieving a yield of 15–20% [50] and 30–40% for exome sequencing [51]. When compared to these benchmarks, DNA methylation profiling has demonstrated a diagnostic yield of approximately 30% in a selected cohort exhibiting features suggestive of rare neurodevelopmental conditions [28,52]. While promising, the described episignature and epivariation detection methods are not without limitations. With the number of identified episignatures and epivariations steadily growing, the specificity of the signature for different cell types and tissues remains to be defined. Epigenetic profiles vary between tissues and show cell-type heterogenicity which hamper EWAS studies that are primarily conducted using data derived from whole blood specimens [53,54]. For tissues that are not always easily accessible, such as the brain, using blood as a surrogate may not fully capture the tissue-specific methylation pattern due to variability in correlation, as observed in previous studies [55,56]. In addition, the lack of consensus methods to correct for population heterogeneity (e.g., genetic background, environmental exposures, and demographic factors) in disease cohort samples limits the reproducibility of analysis across methods and impacts subsequent result interpretation [57]. For example, genetic background can influence baseline methylation patterns, making it challenging to distinguish disease-associated changes from population-specific variations. Similarly, environmental exposures, such as smoking or diet, can alter methylation profiles independently of disease status. Demographic factors, like age and sex, are associated with dynamic and tissue-specific methylation changes. These challenges have been reported in studies [57,58] describing the influence of experimental design, training data size, normalization method, and effect size as limitations of episignature generation. It was observed that the lack of consensus methodology led to the generation of different episignatures for similar pathologies [17,23,59]. Moreover, although improvements have been made to enhance the detection of genetic mosaicism [32,60], existing DNA methylation array technologies remain susceptible to molecular misdetection and fail to detect low-level mosaicism, with the reported resolution limit for detecting mosaicism being 10–15% [60]. There is also a reliance on pre-existing characterized episignatures to identify Mendelian disorders, together reducing diagnostic sensitivity [61]. Additionally, the expansion on syndrome-specific episignatures should be considered, as exemplified by the paralogous genes CREBBP and EP300. Loss-of-function variants in either gene cause Rubinstein–Taybi syndrome (RSTS), whereas pathogenic gain-of-function missense and in-frame indel variants in exons 30 and 31 lead to Menke–Hennekam syndrome (MKHK). While the current MKHK subtype categorizations are gene-specific (subtype 1 for CREBBP; subtype 2 for EP300), the observed distinct domain-specific episignatures in MKHK subtypes suggest the need for a more nuanced, domain-specific categorization [62,63]. The clinical interpretation of rare epivariants can also be challenging, especially within intragenic regions or genes not yet associated with the patient’s phenotype [64].
4. Methylation for Single Patient (N = 1) Rare Disease Studies
In addition to the limitations discussed above, conventional group vs. group or multigroup comparisons with appropriate cohort sizes to meet statistical significance in standard randomized control trials are often not feasible for studying RDs [65,66]. Rare diseases often have very few reported patients, resulting in small and heterogeneous cohorts where canonical group comparison method assumptions are not met, particularly in DNA methylation studies that utilize bisulfite sequencing. These group comparison methods, such as t-tests or ANOVA, assume independence of observations, normality of data, homogeneity of variances, and sufficient sample size to detect statistically meaningful differences in methylation patterns [67]. Despite ongoing sequencing efforts, the lack of publicly available population epigenome datasets as a benchmark to study changes in DNA methylation creates analytical challenges [64]. In addition, without adequate control groups, the ability to capture interpatient heterogeneity, including disease presentation, progression, genetic makeup, and environmental exposures, is often complicated. Within the context of rare diseases, similarities in aberrant CpG methylation patterns have been observed in certain neurodegenerative diseases when targeting a defined gene set [68]. However, disease-associated aberrant DNA methylation at a specific locus in an individual is presumably significantly different from individuals with unrelated phenotypes or control group cohorts [69]. Outlier strategies for identifying potentially causal findings in transcriptomic studies have improved rare disease diagnosis and discovery [70,71]. More recently, investigating methylation levels to understand outlier effects has facilitated the diagnosis of unsolved rare disease cases [31,72,73].
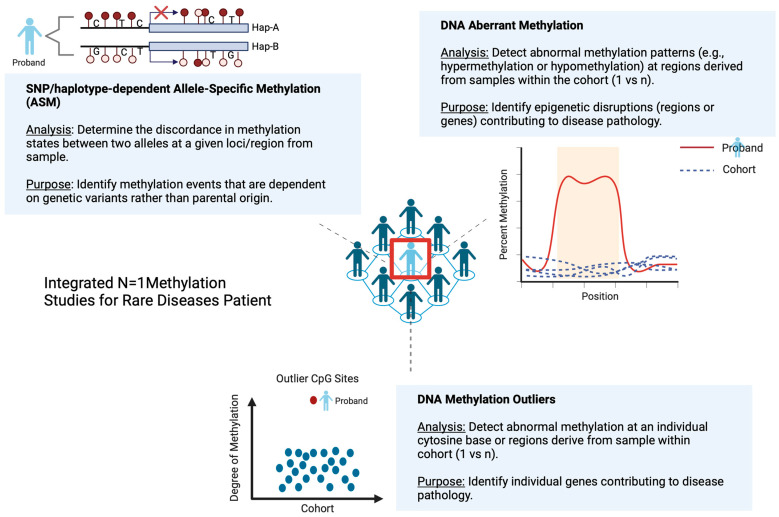
Cheung et al. (2023) proposed that screening for outlier methylation events genome-wide could aid in the identification of coding and noncoding, functional rare SNVs and structural variants (SVs) in unsolved rare cases to improve the diagnosis rate [74]. The authors further demonstrated that rare methylation outliers are heritable and proximally linked to causal rare noncoding or complex SV events using long-read sequencing. Further, Oliver et al. (2021) presented BOREALIS, a tool to identify outlier methylation events using sequence-based methylation data as a novel avenue of exploration in undiagnosed cases of rare disease [66]. These studies highlight the potential of methylation outlier detection methods as a complementary approach to increase diagnostic rates in rare disease patients (Figure 2).
Figure 2.
Schematic representation of integrated N = 1 methylation studies for rare disease patients. Possible integrated approaches include SNP/haplotype-dependent Allele-Specific Methylation (ASM), aberrant DNA methylation, and DNA methylation outlier studies. The purpose of the integration is to capture the epigenetic landscape of the proband within a rare disease cohort, which could provide useful insight into rare disease pathology, leading to improved diagnosis and management.
Allele-specific methylation (ASM) is another phenomenon that may contribute to rare disease diagnosis. ASM events are reportedly increased in cancers, including lymphoma and myeloma, due to global allele-specific CpG hypomethylation [75]. ASM is considered a hallmark of both genomic imprinting, where the methylation of an allele is determined by its parent-of-origin, and non-imprinted status, or haplotype-dependent ASM (hap-ASM).
Hap-ASM refers to the difference in methylation state between two alleles in a heterozygous individual. This phenomenon occurs when cis-acting polymorphisms, such as single-nucleotide polymorphisms (SNPs), influence the methylation status of nearby CpGs. These SNPs can alter transcription factor (TF) binding, which affects the recruitment of methyltransferases and subsequently modulates the methylation landscape of one allele compared to the other. In hap-ASM regions, the presence of heterozygous SNPs is commonly observed, they are responsible for genetic differences between the two homologous chromosomes [76,77,78].
Traditionally, hap-ASM was assessed directly by WGBS or methylation quantitative trait loci (mQTL) analysis, which correlates the net methylation of single CpGs with genotypes at nearby SNPs [79]. For these methods, simultaneous observation of the DNA methylation state within each genetic variant of a haplotype (i.e., allele) and the allele of origin are needed [80]. The lack of ASM detection tools has historically limited research into its potential as a diagnostic method; however, the recent development of several ASM detection tools—MethPipe [81,82], MONOD2 [83], MethHaplo [84], DAMEfinder [85], and CpelAsm [86]—have expanded the possibilities for genome-wide ASM analysis. These advancements have made it possible to identify ASM events, potentially opening the door for the using ASM in the diagnosis of complex cases in RD patients.
Identification of methylation outliers or ASM events alone may not be sufficient to assess the causal relationship between a variant and an RD phenotype. Understanding the allelic specificity of these methylation events is critical in attributing observation to a suspected RD diagnosis. Cheung et al. (2023) reported that 80% of rare hypermethylation events from an RD cohort appeared to be allele-specific [74]. Non-diagnostic exome sequencing is typically followed by genome sequencing and transcriptome profiling, where expression outlier identification is often coupled with allele-specific expression (ASE) and structural variant analysis to determine allelic specificity and identify pathogenic SVs [64]. As methylation studies are increasingly used for RD diagnosis, similar strategies using correlated evidence should be pursued (Figure 2).
The limited number of tools available to study methylation in the scope of RDs and the dependency on accurately phased data to distinguish maternal and paternal alleles impose significant challenges. Most current phasing methods do not use parental data [87]; instead, the calculation of alleles from each chromosome, or subchromosomal phase block, are grouped into two haplotypes. Subsequent ASM events are then inferred from the identified haplotypes without a clear understanding of the allele or parent of origin. This creates a burden in terms of analysis and result interpretation when sequence data from at least one parent are not available. The emerging use of long-read sequencing has begun to overcome this issue, as tools such as MethPhaser [88] and PatMat [87], improve the quality of phasing through parental data inclusion. Epigenomics has the potential to significantly expand and improve clinical testing modalities in rare disease patients; however, the development of guidelines and standardized workflows is critical in a rapidly growing field.
5. Future Perspectives
There is increasing global awareness and attention to the impact of rare diseases on human health, with studies demonstrating the importance of early and unambiguous diagnosis. Despite significant advancements in sequence-based testing over the last decade, most patients suspected of having a rare genetic disease remain undiagnosed. As answers are sought for undiagnosed patients, studies are evaluating the intricate interplay between epigenetic mechanisms and disease pathogenesis. Methylation studies may provide valuable data to complement traditional genomic testing and significantly advance diagnostic and therapeutic strategies. Such studies will benefit from increased adoption of long-read technologies, where methylation signals can be obtained directly from the genomic sequencing of a sample. However, before DNA methylation profiling can be broadly implemented, several gaps require further attention and exploration. The presence of comprehensive, open reference databases containing epigenomes of individuals across different tissues and cell types is needed. Loyfer et al. (2023) highlighted the importance of addressing this deficiency, emphasizing the need for robust reference datasets that accurately capture the epigenomic landscape of healthy individuals [89]. Methylation reference datasets serve as invaluable resources for researchers seeking to compare epigenetic patterns observed in individuals affected by rare diseases with those in healthy populations. The availability of large-scale, well-annotated reference datasets will enable broader accessibility, establish a comprehensive benchmark, and foster interdisciplinary collaboration to train complex algorithms to improve clinical utility and diagnostics.
It is also critical to collaboratively invest in developing standardized analytical pipelines for epigenomic data to ensure the reproducibility and reliability of findings derived from individual rare diseases. Harmonized data generation, processing, and interpretation across research laboratories will facilitate cross-study comparisons and meta-analyses. DNA methylation analysis has the utility to become another critical care tool for RD patients. Early diagnosis using epigenome profiling may support the implementation of interventions through behavioral therapy, specialized learning programs, and individualized medicine [16].
In conclusion, technological advancements have not only significantly advanced our understanding of the role of epigenetics in rare diseases, but they have also provided valuable insights into the diagnosis of rare diseases through the various identification of episignatures, epivariation, epioutliers, and ASMs. Identifying the links between these epigenetic factors and the underlying disease phenotype requires robust basic research efforts. Moreover, overcoming the limitations outlined depends on sustained funding for current and future rare disease research programs, as the associated costs can accumulate for both the researchers and patients.
Looking forward, that with the future establishment of comprehensive reference databases, promotion of standardization efforts, and encouragement of data sharing and collaboration will unlock the full potential of epigenomic studies to support the diagnosis and management of rare diseases. These developments will improve diagnostic precision and support the creation of more effective, personalized (targeted) therapies for individuals affected by rare diseases.
Author Contributions
J.W.T. wrote and composed the manuscript, E.J.B. and J.D.F. edited and revised the manuscript. E.W.K. supervised and contributed significantly to the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Funding Statement
This study was supported by the Mayo Clinic Center for Individualized Medicine.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Danese E., Lippi G. Rare diseases: The paradox of an emerging challenge. Ann. Transl. Med. 2018;6:329. doi: 10.21037/atm.2018.09.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguengang Wakap S., Lambert D.M., Olry A., Rodwell C., Gueydan C., Lanneau V., Murphy D., Le Cam Y., Rath A. Estimating cumulative point prevalence of rare diseases: Analysis of the Orphanet database. Eur. J. Hum. Genet. 2020;28:165–173. doi: 10.1038/s41431-019-0508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira C.R. The burden of rare diseases. Am. J. Med. Genet. Part A. 2019;179:885–892. doi: 10.1002/ajmg.a.61124. [DOI] [PubMed] [Google Scholar]
- 4.Health T.L.G. The landscape for rare diseases in 2024. Lancet Glob. Health. 2024;12:e341. doi: 10.1016/S2214-109X(24)00056-1. [DOI] [PubMed] [Google Scholar]
- 5.Ekins S. Industrializing rare disease therapy discovery and development. Nat. Biotechnol. 2017;35:117–118. doi: 10.1038/nbt.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy M.A., McConkey H., Kerkhof J., Barat-Houari M., Bargiacchi S., Biamino E., Bralo M.P., Cappuccio G., Ciolfi A., Clarke A. Novel diagnostic DNA methylation episignatures expand and refine the epigenetic landscapes of Mendelian disorders. Hum. Genet. Genom. Adv. 2022;3:100075. doi: 10.1016/j.xhgg.2021.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smirnov D., Konstantinovskiy N., Prokisch H. Integrative omics approaches to advance rare disease diagnostics. J. Inherit. Metab. Dis. 2023;46:824–838. doi: 10.1002/jimd.12663. [DOI] [PubMed] [Google Scholar]
- 8.Fahrner J.A., Bjornsson H.T. Mendelian disorders of the epigenetic machinery: Tipping the balance of chromatin states. Annu. Rev. Genom. Hum. Genet. 2014;15:269–293. doi: 10.1146/annurev-genom-090613-094245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copping N.A., McTighe S.M., Fink K.D., Silverman J.L. Emerging Gene and Small Molecule Therapies for the Neurodevelopmental Disorder Angelman Syndrome. Neurotherapeutics. 2021;18:1535–1547. doi: 10.1007/s13311-021-01082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 11.Shen L., Song C.-X., He C., Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu. Rev. Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velasco G., Francastel C. Genetics meets DNA methylation in rare diseases. Clin. Genet. 2019;95:210–220. doi: 10.1111/cge.13480. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Delgado B., Barrero M.J. Epigenomic approaches for the diagnosis of rare diseases. Epigenomes. 2022;6:21. doi: 10.3390/epigenomes6030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kernohan K.D., Boycott K.M. The expanding diagnostic toolbox for rare genetic diseases. Nat. Rev. Genet. 2024;25:401–415. doi: 10.1038/s41576-023-00683-w. [DOI] [PubMed] [Google Scholar]
- 15.Fahrner J.A., Bjornsson H.T. Mendelian disorders of the epigenetic machinery: Postnatal malleability and therapeutic prospects. Hum. Mol. Genet. 2019;28:R254–R264. doi: 10.1093/hmg/ddz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu M.P., Merrill S.M., Sharma M., Gibson W.T., Turvey S.E., Kobor M.S. Rare diseases of epigenetic origin: Challenges and opportunities. Front. Genet. 2023;14:1113086. doi: 10.3389/fgene.2023.1113086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aref-Eshghi E., Kerkhof J., Pedro V.P., Barat-Houari M., Ruiz-Pallares N., Andrau J.-C., Lacombe D., Van-Gils J., Fergelot P., Dubourg C. Evaluation of DNA methylation episignatures for diagnosis and phenotype correlations in 42 Mendelian neurodevelopmental disorders. Am. J. Hum. Genet. 2020;106:356–370. doi: 10.1016/j.ajhg.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haghshenas S., Levy M.A., Kerkhof J., Aref-Eshghi E., McConkey H., Balci T., Siu V.M., Skinner C.D., Stevenson R.E., Sadikovic B. Detection of a DNA methylation signature for the intellectual developmental disorder, X-linked, syndromic, armfield type. Int. J. Mol. Sci. 2021;22:1111. doi: 10.3390/ijms22031111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciolfi A., Aref-Eshghi E., Pizzi S., Pedace L., Miele E., Kerkhof J., Flex E., Martinelli S., Radio F.C., Ruivenkamp C.A. Frameshift mutations at the C-terminus of HIST1H1E result in a specific DNA hypomethylation signature. Clin. Epigenet. 2020;12:7. doi: 10.1186/s13148-019-0804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris J.R., Gao C.W., Britton J.F., Applegate C.D., Bjornsson H.T., Fahrner J.A. Five years of experience in the Epigenetics and Chromatin Clinic: What have we learned and where do we go from here? Hum. Genet. 2024;143:607–624. doi: 10.1007/s00439-023-02537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirza-Schreiber N., Zech M., Wilson R., Brunet T., Wagner M., Jech R., Boesch S., Škorvánek M., Necpál J., Weise D. Blood DNA methylation provides an accurate biomarker of KMT2B-related dystonia and predicts onset. Brain. 2022;145:644–654. doi: 10.1093/brain/awab360. [DOI] [PubMed] [Google Scholar]
- 22.Schenkel L.C., Kernohan K.D., McBride A., Reina D., Hodge A., Ainsworth P.J., Rodenhiser D.I., Pare G., Bérubé N.G., Skinner C. Identification of epigenetic signature associated with alpha thalassemia/mental retardation X-linked syndrome. Epigenet. Chromatin. 2017;10:10. doi: 10.1186/s13072-017-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butcher D.T., Cytrynbaum C., Turinsky A.L., Siu M.T., Inbar-Feigenberg M., Mendoza-Londono R., Chitayat D., Walker S., Machado J., Caluseriu O. CHARGE and Kabuki syndromes: Gene-specific DNA methylation signatures identify epigenetic mechanisms linking these clinically overlapping conditions. Am. J. Hum. Genet. 2017;100:773–788. doi: 10.1016/j.ajhg.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aref-Eshghi E., Bend E.G., Colaiacovo S., Caudle M., Chakrabarti R., Napier M., Brick L., Brady L., Carere D.A., Levy M.A. Diagnostic utility of genome-wide DNA methylation testing in genetically unsolved individuals with suspected hereditary conditions. Am. J. Hum. Genet. 2019;104:685–700. doi: 10.1016/j.ajhg.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hood R.L., Schenkel L.C., Nikkel S.M., Ainsworth P.J., Pare G., Boycott K.M., Bulman D.E., Sadikovic B. The defining DNA methylation signature of Floating-Harbor Syndrome. Sci. Rep. 2016;6:38803. doi: 10.1038/srep38803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aref-Eshghi E., Rodenhiser D.I., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Paré G., Hood R.L., Bulman D.E., Kernohan K.D. Genomic DNA methylation signatures enable concurrent diagnosis and clinical genetic variant classification in neurodevelopmental syndromes. Am. J. Hum. Genet. 2018;102:156–174. doi: 10.1016/j.ajhg.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.der Laan L.V., Rooney K., Trooster T.M., Mannens M.M., Sadikovic B., Henneman P. DNA methylation episignatures: Insight into copy number variation. Epigenomics. 2022;14:1373–1388. doi: 10.2217/epi-2022-0287. [DOI] [PubMed] [Google Scholar]
- 28.Sadikovic B., Levy M.A., Kerkhof J., Aref-Eshghi E., Schenkel L., Stuart A., McConkey H., Henneman P., Venema A., Schwartz C.E. Clinical epigenomics: Genome-wide DNA methylation analysis for the diagnosis of Mendelian disorders. Genet. Med. 2021;23:1065–1074. doi: 10.1038/s41436-020-01096-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turinsky A.L., Choufani S., Lu K., Liu D., Mashouri P., Min D., Weksberg R., Brudno M. EpigenCentral: Portal for DNA methylation data analysis and classification in rare diseases. Hum. Mutat. 2020;41:1722–1733. doi: 10.1002/humu.24076. [DOI] [PubMed] [Google Scholar]
- 30.Brasil S., Neves C.J., Rijoff T., Falcão M., Valadão G., Videira P.A., dos Reis Ferreira V. Artificial intelligence in epigenetic studies: Shedding light on rare diseases. Front. Mol. Biosci. 2021;8:648012. doi: 10.3389/fmolb.2021.648012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh J.R., Sun G., Balan J., Hardcastle J., Vollenweider J., Jerde C., Rumilla K., Koellner C., Koleilat A., Hasadri L., et al. A supervised learning method for classifying methylation disorders. BMC Bioinform. 2024;25:66. doi: 10.1186/s12859-024-05673-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oexle K., Zech M., Stühn L.G., Siegert S., Brunet T., Schmidt W.M., Wagner M., Schmidt A., Engels H., Tilch E. Episignature analysis of moderate effects and mosaics. Eur. J. Hum. Genet. 2023;31:1032–1039. doi: 10.1038/s41431-023-01406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holliday R. Mutations and epimutations in mammalian cells. Mech. Mutagen. 1991;250:351–363. doi: 10.1016/0027-5107(91)90192-Q. [DOI] [PubMed] [Google Scholar]
- 34.Buiting K., Groß S., Lich C., Gillessen-Kaesbach G., El-Maarri O., Horsthemke B. Epimutations in Prader-Willi and Angelman syndromes: A molecular study of 136 patients with an imprinting defect. Am. J. Hum. Genet. 2003;72:571–577. doi: 10.1086/367926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garg P., Jadhav B., Rodriguez O.L., Patel N., Martin-Trujillo A., Jain M., Metsu S., Olsen H., Paten B., Ritz B. A survey of rare epigenetic variation in 23,116 human genomes identifies disease-relevant epivariations and CGG expansions. Am. J. Hum. Genet. 2020;107:654–669. doi: 10.1016/j.ajhg.2020.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gentilini D., Cavagnola R., Possenti I., Calzari L., Ranucci F., Nola M., Olivola M., Brondino N., Politi P. Epigenetics of autism spectrum disorders: A multi-level analysis combining epi-signature, age acceleration, epigenetic drift and rare Epivariations using public datasets. Curr. Neuropharmacol. 2023;21:2362–2373. doi: 10.2174/1570159X21666230725142338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horsthemke B. DNA Methylation: Development, Genetic Disease and Cancer. Springer; Berlin/Heidelberg, Germany: 2006. Epimutations in human disease; pp. 45–59. [DOI] [Google Scholar]
- 38.Barbosa M., Joshi R.S., Garg P., Martin-Trujillo A., Patel N., Jadhav B., Watson C.T., Gibson W., Chetnik K., Tessereau C. Identification of rare de novo epigenetic variations in congenital disorders. Nat. Commun. 2018;9:2064. doi: 10.1038/s41467-018-04540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaCroix A.J., Stabley D., Sahraoui R., Adam M.P., Mehaffey M., Kernan K., Myers C.T., Fagerstrom C., Anadiotis G., Akkari Y.M. GGC repeat expansion and exon 1 methylation of XYLT1 is a common pathogenic variant in Baratela-Scott syndrome. Am. J. Hum. Genet. 2019;104:35–44. doi: 10.1016/j.ajhg.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willemsen R., Levenga J., Oostra B.A. CGG repeat in the FMR1 gene: Size matters. Clin. Genet. 2011;80:21425. doi: 10.1111/j.1399-0004.2011.01723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warburton P.E., Sebra R.P. Long-Read DNA Sequencing: Recent Advances and Remaining Challenges. Annu. Rev. Genom. Hum. Genet. 2023;24:109–132. doi: 10.1146/annurev-genom-101722-103045. [DOI] [PubMed] [Google Scholar]
- 42.Gatto S., Gagliardi M., Franzese M., Leppert S., Papa M., Cammisa M., Grillo G., Velasco G., Francastel C., Toubiana S. ICF-specific DNMT3B dysfunction interferes with intragenic regulation of mRNA transcription and alternative splicing. Nucleic Acids Res. 2017;45:5739–5756. doi: 10.1093/nar/gkx163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Z., Wu Y., Ordog T., Baheti S., Nie J., Duan X., Hojo K., Kocher J.-P., Dyck P.J., Klein C.J. Aberrant signature methylome by DNMT1 hot spot mutation in hereditary sensory and autonomic neuropathy 1E. Epigenetics. 2014;9:1184–1193. doi: 10.4161/epi.29676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith A.M., LaValle T.A., Shinawi M., Ramakrishnan S.M., Abel H.J., Hill C.A., Kirkland N.M., Rettig M.P., Helton N.M., Heath S.E., et al. Functional and epigenetic phenotypes of humans and mice with DNMT3A Overgrowth Syndrome. Nat. Commun. 2021;12:4549. doi: 10.1038/s41467-021-24800-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Y., Gomez J.A., Laufer B.I., Mordaunt C.E., Mouat J.S., Soto D.C., Dennis M.Y., Benke K.S., Bakulski K.M., Dou J. Placental methylome reveals a 22q13.33 brain regulatory gene locus associated with autism. Genome Biol. 2022;23:46. doi: 10.1186/s13059-022-02613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller D.E., Hanna P., Galey M., Reyes M., Linglart A., Eichler E.E., Jüppner H. Targeted Long-Read Sequencing Identifies a Retrotransposon Insertion as a Cause of Altered GNAS Exon A/B Methylation in a Family with Autosomal Dominant Pseudohypoparathyroidism Type 1b (PHP1B) J. Bone Miner. Res. 2020;37:1711–1719. doi: 10.1002/jbmr.4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montano C., Britton J.F., Harris J.R., Kerkhof J., Barnes B.T., Lee J.A., Sadikovic B., Sobreira N., Fahrner J.A. Genome-wide DNA methylation profiling confirms a case of low-level mosaic Kabuki syndrome 1. Am. J. Med. Genet. Part A. 2022;188:2217–2225. doi: 10.1002/ajmg.a.62754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferilli M., Ciolfi A., Pedace L., Niceta M., Radio F.C., Pizzi S., Miele E., Cappelletti C., Mancini C., Galluccio T. Genome-wide DNA methylation profiling solves uncertainty in classifying NSD1 variants. Genes. 2022;13:2163. doi: 10.3390/genes13112163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paparella A., Squeo G.M., Di Venere E., Cardea E., Mazza T., Castellana S., Kerkhof J., McConkey H., Sadikovic B., Sinibaldi L. Genome-wide DNA methylation profiling and exome sequencing resolved a long-time misdiagnosed case. J. Hum. Genet. 2022;67:547–551. doi: 10.1038/s10038-022-01043-y. [DOI] [PubMed] [Google Scholar]
- 50.Miller D.T., Adam M.P., Aradhya S., Biesecker L.G., Brothman A.R., Carter N.P., Church D.M., Crolla J.A., Eichler E.E., Epstein C.J., et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srivastava S., Love-Nichols J.A., Dies K.A., Ledbetter D.H., Martin C.L., Chung W.K., Firth H.V., Frazier T., Hansen R.L., Prock L. Meta-analysis and multidisciplinary consensus statement: Exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet. Med. 2019;21:2413–2421. doi: 10.1038/s41436-019-0554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kerkhof J., Rastin C., Levy M.A., Relator R., McConkey H., Demain L., Dominguez-Garrido E., Kaat L.D., Houge S.D., DuPont B.R., et al. Diagnostic utility and reporting recommendations for clinical DNA methylation episignature testing in genetically undiagnosed rare diseases. Genet. Med. 2024;26:101075. doi: 10.1016/j.gim.2024.101075. [DOI] [PubMed] [Google Scholar]
- 53.Jaffe A.E., Irizarry R.A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15:R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng S.C., Breeze C.E., Beck S., Teschendorff A.E. Identification of differentially methylated cell types in epigenome-wide association studies. Nat. Methods. 2018;15:1059–1066. doi: 10.1038/s41592-018-0213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hannon E., Lunnon K., Schalkwyk L., Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: Implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10:1024–1032. doi: 10.1080/15592294.2015.1100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Braun P.R., Han S., Hing B., Nagahama Y., Gaul L.N., Heinzman J.T., Grossbach A.J., Close L., Dlouhy B.J., Howard M.A., III, et al. Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Transl. Psychiatry. 2019;9:47. doi: 10.1038/s41398-019-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chater-Diehl E., Goodman S.J., Cytrynbaum C., Turinsky A.L., Choufani S., Weksberg R. Anatomy of DNA methylation signatures: Emerging insights and applications. Am. J. Hum. Genet. 2021;108:1359–1366. doi: 10.1016/j.ajhg.2021.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giuili E., Grolaux R., Macedo C.Z., Desmyter L., Pichon B., Neuens S., Vilain C., Olsen C., Van Dooren S., Smits G. Comprehensive evaluation of the implementation of episignatures for diagnosis of neurodevelopmental disorders (NDDs) Hum. Genet. 2023;142:1721–1735. doi: 10.1007/s00439-023-02609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choufani S., Cytrynbaum C., Chung B., Turinsky A., Grafodatskaya D., Chen Y., Cohen A., Dupuis L., Butcher D., Siu M. NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niceta M., Ciolfi A., Ferilli M., Pedace L., Cappelletti C., Nardini C., Hildonen M., Chiriatti L., Miele E., Dentici M.L. DNA methylation profiling in Kabuki syndrome: Reclassification of germline KMT2D VUS and sensitivity in validating postzygotic mosaicism. Eur. J. Hum. Genet. 2024;32:819–826. doi: 10.1038/s41431-024-01597-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wojcik M.H., Reuter C.M., Marwaha S., Mahmoud M., Duyzend M.H., Barseghyan H., Yuan B., Boone P.M., Groopman E.E., Délot E.C. Beyond the exome: What’s next in diagnostic testing for Mendelian conditions. Am. J. Hum. Genet. 2023;110:1229–1248. doi: 10.1016/j.ajhg.2023.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levy M.A., Relator R., McConkey H., Pranckeviciene E., Kerkhof J., Barat-Houari M., Bargiacchi S., Biamino E., Palomares Bralo M., Cappuccio G., et al. Functional correlation of genome-wide DNA methylation profiles in genetic neurodevelopmental disorders. Hum. Mutat. 2022;43:1609–1628. doi: 10.1002/humu.24446. [DOI] [PubMed] [Google Scholar]
- 63.Haghshenas S., Bout H.J., Schijns J.M., Levy M.A., Kerkhof J., Bhai P., McConkey H., Jenkins Z.A., Williams E.M., Halliday B.J., et al. Menke-Hennekam syndrome; delineation of domain-specific subtypes with distinct clinical and DNA methylation profiles. Hum. Genet. Genom. Adv. 2024;5:100287. doi: 10.1016/j.xhgg.2024.100287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marwaha S., Knowles J.W., Ashley E.A. A guide for the diagnosis of rare and undiagnosed disease: Beyond the exome. Genome Med. 2022;14:23. doi: 10.1186/s13073-022-01026-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grolaux R., Hardy A., Olsen C., Van Dooren S., Smits G., Defrance M. Identification of differentially methylated regions in rare diseases from a single-patient perspective. Clin. Epigenet. 2022;14:174. doi: 10.1186/s13148-022-01403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oliver G.R., Jenkinson W.G., Olson R.J., Schultz-Rogers L.E., Klee E.W. BOREALIS: An R/Bioconductor package to detect outlier methylation from bisulfite sequencing data. F1000Research. 2022;11:1538. doi: 10.12688/f1000research.128354.1. [DOI] [Google Scholar]
- 67.Wreczycka K., Gosdschan A., Yusuf D., Grüning B., Assenov Y., Akalin A. Strategies for analyzing bisulfite sequencing data. J. Biotechnol. 2017;261:105–115. doi: 10.1016/j.jbiotec.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 68.Sanchez-Mut J.V., Heyn H., Vidal E., Moran S., Sayols S., Delgado-Morales R., Schultz M.D., Ansoleaga B., Garcia-Esparcia P., Pons-Espinal M., et al. Human DNA methylomes of neurodegenerative diseases show common epigenomic patterns. Transl. Psychiatry. 2016;6:e718. doi: 10.1038/tp.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seeboth A., McCartney D.L., Wang Y., Hillary R.F., Stevenson A.J., Walker R.M., Campbell A., Evans K.L., McIntosh A.M., Hägg S. DNA methylation outlier burden, health, and ageing in Generation Scotland and the Lothian Birth Cohorts of 1921 and 1936. Clin. Epigenet. 2020;12:49. doi: 10.1186/s13148-020-00838-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frésard L., Smail C., Ferraro N.M., Teran N.A., Li X., Smith K.S., Bonner D., Kernohan K.D., Marwaha S., Zappala Z. Identification of rare-disease genes using blood transcriptome sequencing and large control cohorts. Nat. Med. 2019;25:911–919. doi: 10.1038/s41591-019-0457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferraro N.M., Strober B.J., Einson J., Abell N.S., Aguet F., Barbeira A.N., Brandt M., Bucan M., Castel S.E., Davis J.R. Transcriptomic signatures across human tissues identify functional rare genetic variation. Science. 2020;369:eaaz5900. doi: 10.1126/science.aaz5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jenkinson G., Li Y.I., Basu S., Cousin M.A., Oliver G.R., Klee E.W. LeafCutterMD: An algorithm for outlier splicing detection in rare diseases. Bioinformatics. 2020;36:4609–4615. doi: 10.1093/bioinformatics/btaa259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mertes C., Scheller I.F., Yépez V.A., Çelik M.H., Liang Y., Kremer L.S., Gusic M., Prokisch H., Gagneur J. Detection of aberrant splicing events in RNA-seq data using FRASER. Nat. Commun. 2021;12:529. doi: 10.1038/s41467-020-20573-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheung W.A., Johnson A.F., Rowell W.J., Farrow E., Hall R., Cohen A.S., Means J.C., Zion T.N., Portik D.M., Saunders C.T. Direct haplotype-resolved 5-base HiFi sequencing for genome-wide profiling of hypermethylation outliers in a rare disease cohort. Nat. Commun. 2023;14:3090. doi: 10.1038/s41467-023-38782-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Do C., Dumont E.L., Salas M., Castano A., Mujahed H., Maldonado L., Singh A., DaSilva-Arnold S.C., Bhagat G., Lehman S. Allele-specific DNA methylation is increased in cancers and its dense mapping in normal plus neoplastic cells increases the yield of disease-associated regulatory SNPs. Genome Biol. 2020;21:153. doi: 10.1186/s13059-020-02059-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kerkel K., Spadola A., Yuan E., Kosek J., Jiang L., Hod E., Li K., Murty V.V., Schupf N., Vilain E. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat. Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 77.Shoemaker R., Deng J., Wang W., Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–889. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tycko B. Allele-specific DNA methylation: Beyond imprinting. Hum. Mol. Genet. 2010;19:R210–R220. doi: 10.1093/hmg/ddq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ferguson-Smith A.C. Genomic imprinting: The emergence of an epigenetic paradigm. Nat. Rev. Genet. 2011;12:565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- 80.Do C., Lang C.F., Lin J., Darbary H., Krupska I., Gaba A., Petukhova L., Vonsattel J.-P., Gallagher M.P., Goland R.S. Mechanisms and disease associations of haplotype-dependent allele-specific DNA methylation. Am. J. Hum. Genet. 2016;98:934–955. doi: 10.1016/j.ajhg.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abante J., Fang Y., Feinberg A., Goutsias J. Detection of haplotype-dependent allele-specific DNA methylation in WGBS data. Nat. Commun. 2020;11:5238. doi: 10.1038/s41467-020-19077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song Q., Decato B., Hong E.E., Zhou M., Fang F., Qu J., Garvin T., Kessler M., Zhou J., Smith A.D. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS ONE. 2013;8:e81148. doi: 10.1371/journal.pone.0081148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song Q., Decato B., Kessler M., Fang F., Qu J., Garvin T., Zhou M., Smith A. The Smithlab DNA Methylation Data Analysis Pipeline (MethPipe) The Smith Lab; Hiroshima, Japan: 2021. [(accessed on 17 August 2024)]. Available online: http://smithlabresearch.org/software/methpipe/ [Google Scholar]
- 84.Guo S., Diep D., Plongthongkum N., Fung H.-L., Zhang K., Zhang K. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat. Genet. 2017;49:635–642. doi: 10.1038/ng.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou Q., Wang Z., Li J., Sung W.-K., Li G. MethHaplo: Combining allele-specific DNA methylation and SNPs for haplotype region identification. BMC Bioinform. 2020;21:451. doi: 10.1186/s12859-020-03798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Orjuela S., Machlab D., Menigatti M., Marra G., Robinson M.D. DAMEfinder: A method to detect differential allele-specific methylation. Epigenet. Chromatin. 2020;13:25. doi: 10.1186/s13072-020-00346-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Akbari V., Hanlon V.C., O’Neill K., Lefebvre L., Schrader K.A., Lansdorp P.M., Jones S.J. Parent-of-origin detection and chromosome-scale haplotyping using long-read DNA methylation sequencing and Strand-seq. Cell Genom. 2023;3:100233. doi: 10.1016/j.xgen.2022.100233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fu Y., Aganezov S., Mahmoud M., Beaulaurier J., Juul S., Treangen T.J., Sedlazeck F.J. MethPhaser: Methylation-based haplotype phasing of human genomes. Nat. Commun. 2024;15:5327. doi: 10.1038/s41467-024-49588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Loyfer N., Magenheim J., Peretz A., Cann G., Bredno J., Klochendler A., Fox-Fisher I., Shabi-Porat S., Hecht M., Pelet T. A DNA methylation atlas of normal human cell types. Nature. 2023;613:355–364. doi: 10.1038/s41586-022-05580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]