Abstract
A gene encoding a new member of the Pur protein family, Purγ, has been detected upstream of, and contrapodal to, the gene encoding the Werner syndrome helicase, Wrn, at human chromosome band 8p11–12. Both the PURG and WRN genes initiate transcription at multiple sites, the major clusters of which are ∼90 bp apart. A segment containing this region strongly promotes transcription of a reporter gene in both directions. Both promoters are TATA-less and CAAT-less and both are positively regulated by Sp1 elements. While promoter elements for the two genes are interleaved, in the contrapodal direction, certain elements critical for each gene are distinct. Sequencing of cDNAs for Purγ mRNA reveals that two alternative coding sequences are generated from a single gene, resulting in different Purγ C-termini. PURG-A mRNA consists of a single intronless transcript of ∼3 kb. PURG-B mRNA results from transcription through the PURG-A polyadenylation site and splicing out of an intron of >30 kb. In this unique example of a switch, splicing of a single intron either occurs or does not occur depending upon differential termination/polyadenylation. PURG-B is the primary PURG transcript detected in testis, but it is undetectable in all members of a normal adult tissue cDNA panel. PURG-A levels are low or undetectable in the normal tissue panel, but they are greatly elevated in all members of a tumor tissue panel. PURG-B is detected in several tumor panel members.
INTRODUCTION
Pur family members are multifunctional proteins reported to function in both DNA and RNA metabolism (1–4). Pur family members studied thus far are sequence-specific single-stranded DNA- and RNA-binding proteins. The ability of Purα, the first family member cloned (GenBank accession no. NM005859) (1), to regulate viral DNA transcription and replication in mammalian cells may provide a model for its cellular roles. Purα binds the protein Tat, encoded by HIV-1, and enhances transcription of the HIV-1 genome in a manner dependent on Purα also binding to the resulting RNA transcript (2). Purα binds to the TAR RNA element near the 5′-end of the HIV-1 transcript and to Tat, which binds TAR near the Purα-binding site. Purα binds to a similar element in JC virus (JCV) DNA in the viral replication/transcription regulatory region and Purα and Tat act synergistically at this element to stimulate JCV late gene transcription (5). In mouse cells Purα stimulates transcription of the BC1 gene and binds to the resulting transcript (6), providing another example of a DNA- and RNA-binding pattern similar to that seen with HIV-1. In human cells Purα binds to 7SL RNA, which is related to mouse BC1 RNA (7). Purα has now been implicated in transcriptional regulation of a variety of additional cellular genes (8–12). Purα has been reported to associate with another Pur family member, Purβ, the resulting complex being involved in regulation of the vascular smooth muscle actin gene (13). The sequence of human Purβ cDNA has now been reported (14) (GenBank accession no. AY039216). Increased levels of Purα protein inhibit cell cycle progression at G1 or G2 checkpoints (15,16). Simultaneous deletions of the PURA and PURB genes occur at a high frequency in acute myelogenous leukemia (14). The PUR gene family is strongly conserved from plants to humans. Whereas the human genome contains at least three family members, the Drosophila genome contains only one (GenBank accession no. AF021259).
Each Pur family member contains three class I amino acid repeats which are highly conserved and are related to repeats in human Myb proteins and the yeast protein Rap1. All Pur proteins also contain two class II repeats separating the class I repeats. The class I repeats in Purα are crucial for its ability to bind its single-stranded PUR recognition element (17). To identify new members of the Pur family of proteins, a short Purα sequence corresponding to the class I repeat was used to screen for homologous DNA sequences registered in the GenBank database using the tBLASTn program (18). A significant Purα homolog was found to be present in human genomic DNA upstream of the Werner syndrome gene, WRN. This sequence comprised a complete open reading frame (ORF) for a characteristic Pur protein, which we have designated in the present report as Purγ. The gene is designated as PURG. In characterizing the WRN promoter, investigators had previously deleted regions of PURG, but they did not note the presence of the PURG ORF (19,20). Intriguingly, the PURG ORF is in the opposite orientation to the WRN ORF.
Werner syndrome is a rare autosomal recessive disorder causing several characteristics of premature aging and certain types of cancers (21–23). Werner syndrome patients are prone to juvenile cataracts, atrophy of the skin, graying and loss of hair, diabetes, arteriosclerosis, osteoporosis and neoplasia characterized by multiple sarcomas (22,23). The WRN gene is located at chromosome band 8p11–12 and encodes a protein of 1432 amino acids with significant homology in its helicase domain to that of the Escherichia coli RecQ DNA helicase (24). To date, five human RecQ-type helicase genes, i.e. RecQL, WRN, BLM, RecQ4 and RecQ5, have been identified. Mutations in the WRN, BLM and RecQ4 genes have been shown to be responsible for the autosomal disorders known as Werner syndrome, Bloom syndrome and Rothmund–Thomson sydrome, respectively (25). Most mutations in WRN affect helicase activity (25). The mechanism by which mutations in the WRN gene cause the phenotypes in Werner syndrome patients remains unknown.
The head-to-head orientation of WRN and PURG raises the question of whether the two genes could share promoter sequences and whether their expression could be co-regulated. Therefore, we have sought here to define the PURG transcription unit and to characterize its promoter, the structure of the gene and its expression. Surprisingly, we have found unique aspects of both the initiation and termination of the PURG gene. A novel termination/polyadenylation switch allows the gene to encode either of two proteins, with different C-termini.
MATERIALS AND METHODS
Cell culture
HeLa cells were maintained in Dulbecco’s modified Eagle’s medium with l-glutamine and 4.5 g/l glucose (Cellgro Mediatech) supplemented with 10% (v/v) fetal bovine serum (Gibco BRL) at 37°C in a humidified atmosphere of 5% CO2, 95% air.
Two-step RT–PCR and 5′-RACE of human testis PURG mRNA
Human testis mRNA (Clontech) was reverse transcribed with C.therm. Polymerase (Roche Molecular Biochemicals) and the products were purified with a Qiagen PCR purification kit. PCR was done with an Expand High-Fidelity PCR System (Roche) including 1.5 M betaine and 5% DMSO. Performance of 5′-RACE (rapid amplification of cDNA ends) (26) was carried out according to the kit manufacturer’s instructions (Roche) with some modifications. Human testis mRNA was reverse transcribed with a PURG-specific primer and C.therm. Polymerase and the products treated with RNase H before being purified with a Qiagen PCR purification kit. The single-stranded cDNA was then tailed at its 3′-end, corresponding to the 5′-end of the original mRNA, with dATP and terminal deoxynucleotide transferase. The cDNA was then amplified, first using an oligo(dT) anchor primer and a nested PURG-specific primer and second using the anchor primer and a further nested PURG-specific primer. The second PCR products were phosphorylated at their 5′-ends with polynucleotide kinase, and the ends completed with the Klenow fragment of DNA polymerase I. Products were then digested with SalI and ligated into the SalI and SmaI sites of pBK-CMV (Stratagene). Sequencing was carried out by the DNA Sequencing Core Facility at the Mount Sinai School of Medicine.
PCR primer sequences employed
The following sequences were employed as primers for PCR amplification for 5′-RACE or for amplification of either the PURG-A or PURG-B cDNAs in expression panels. Numbering is relative to the WRN +1 site as shown in Figure 1. R1 (–1110, –1085), 5′-CTACTCTTGCTTAGGCCAGAGCCCCC-3′; R2 (–390, –366), 5′-GAGGGGGTCTCCGCTCTTCCTCAGT-3′; R3 (–363, –339), 5′-ACTCTCTGACTGAAGCCCGGCGCGT-3′; R4 (–325, –304), 5′-AGTGCGAGGGGACTGGACAGGT-3′; F1 (–905, –925), 5′-GGGCCCGGCGTGGGGGTCGCG-3; F2 (–716, –740), 5′-ATGGCAGAGATGATGTGAGTGCCCG-3′; F3 (–562, –583), 5′-CGATGCCCCTGCCCGGCTGCTC-3′; F4 (–411, –431), 5′-CGGGGGAGGCGGGGTCCACCG-3′; F5 (–261, –285), 5′-AGTGAGAACATTCCCGCCGCCCGGT-3′; F6 (–176, –199), 5′-CTGCCGCTGAGCACCTCGGGAAGT-3′; F7 (+3, –20), 5′-ATCAGCTCCGGGAGACCTACCCG-3′; F8 (+257, +233), 5′-GCCTAAGCGCCTCCACGTGTCTCAG-3′; HPC5, 5′-ATGGAAAGAGCCAGGCGAAGGG-3′; HPC5M, 5′-GTGATGCCTTGGTTCAGCTG-3′; HPCA3, 5′-CTAGTCGAGGCATTCTTGTTC-3′; HPCB3, 5′-GAGGCCTTGCATTCATGATGAAC-3′. HPC5 is at the 5′-end of the Purγ coding sequence. HPC5M is internal in the shared region of both Purγ-A and Purγ-B. HPCA3 is at the 3′-end of the Purγ-A coding sequence. HPCB3 is in the 3′-untranslated region (UTR) of Purγ-B mRNA.
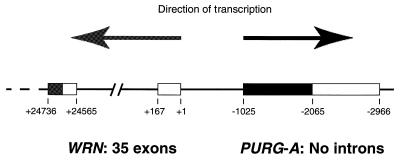
Figure 1.
Genomic organization of the PURG and WRN genes at human chromosome band 8p11–12. Arrows represent the direction of gene transcription. Boxes represent exons of PURG and WRN. Coding regions of the two genes are shown as dark boxes. The most upstream transcription start site of WRN defined by Yamabe et al. (19) is designated position +1. The positions of WRN exons in this number system were calculated from the genomic sequence, GenBank accession no. AF181896. The 5′-UTR of PURG is not shown here.
PGL3 constructs for luciferase assays
Plasmid pGL3-Basic is a luciferase reporter vector with no promoter upstream of the firefly luciferase gene (Promega). DNA fragments of different size in the PURG promoter region were obtained by PCR using the Expand High-Fidelity System (Roche) and appropriate primers. A XhoI and a HindIII restriction site were added to the upstream primer and downstream primer, respectively. PCR products were digested with XhoI and HindIII and cloned into the XhoI and HindIII sites of pGL3-Basic. Insert sequences in all pGL3 constructs were verified at the Mount Sinai School of Medicine DNA Sequencing Core Facility.
Transient transfections and luciferase assays
Monolayers of HeLa cells were allowed to reach 50–80% confluence 16–20 h before transfection. Each well of a 6-well plate was transfected with an equimolar amount of each of the different pGL3 constructs and 0.3 µg pSV-β-galactosidase control vector (Promega) using the FuGene6 transfection reagent (Roche) according to the manufacturer’s instructions. After 48 h, cells in each well were lysed in 150 µl of Reporter Lysis Buffer and stored at –70°C. An aliquot of 20 µl was assayed for luciferase activity and an aliquot of 70 µl was assayed for β-galactosidase activity as recommended (Promega). Light emissions in luciferase assays were determined with a Top Count NXT Microscintillation and Luminescence Counter (Packard Instrument Co.). All measurements were in the linear range of detection. Transfection for each pGL3 construct was performed at least twice. The luciferase activities of pGL3 constructs were normalized for differences in transfection efficiencies using the β-galactosidase activity. Luciferase activities were expressed as fold stimulation above the activity observed for pGL3-Basic, the parental vector for the different pGL3 constructs.
RESULTS
Head-to-head orientation of genes encoding a new Pur family member, Purγ, and the Werner syndrome helicase, Wrn, at chromosome band 8p11–12
By searching for homologous DNA sequences in the GenBank database with a Purα amino acid sequence comprising a class I repeat using the tBLASTn program, we have discovered a Purα homolog within the promoter region of the Werner syndrome gene, WRN. This sequence contains an ORF of 1044 bp which is located between 1025 and 2068 bp upstream of the most 5′ transcription start site of the WRN gene, WRN +1 (Fig. 1). Numbering for this region of the WRN gene is that of Yamabe et al. (19). The new ORF encodes a peptide sequence of 347 amino acids which has high homology to the human Purα and Purβ protein sequences. Especially high homology is observed in regions corresponding to the three class I repeats, two class II repeats and the ‘psycho’ motif of Purα. To find out whether this ORF is transcribed in vivo, we performed a tBLASTn search of its amino acid sequence against the dbEST database. A human cDNA clone from pooled libraries of fetal lung, testis and B cells was identified. The 106 bp sequence tag of that cDNA clone is exactly the same as a part of the ORF. That cDNA clone, designated here pHPURG1, was obtained from the American Type Culture Collection and the entire insert of 1925 bp was sequenced. The insert sequence goes from codon 14 to the stop codon of the ORF, followed by an 897 bp 3′-UTR and a poly(A) tail. It corresponds to the genomic sequence between 1064 and 2966 bp upstream of the WRN transcriptional start site, WRN +1 (Fig. 1). Thus, pHPURG1 contains an incomplete protein coding sequence and an intact 3′-UTR. These data, together with data obtained from the following experiments, indicate that a new Pur family gene, PURG, is located upstream of the WRN gene. Because, as described later in this paper, there exists another isoform of Purγ, we designate the transcription unit shown in Figure 1 as PURG-A. Sequencing of the complete PURG-A cDNA (GenBank accession no. AF195513) is described later in this paper after identification of the 5′-UTR. WRN and PURG are organized in a head-to-head configuration, such that they are transcribed in opposite directions, as shown in Figure 1. Contemplation of this figure raises two immediate questions. Where does transcription of PURG initiate? Do the two genes share essential promoter elements? We set out to answer these questions.
5′-RACE identifies a long 5′-UTR of Purγ mRNA with multiple transcriptional start sites
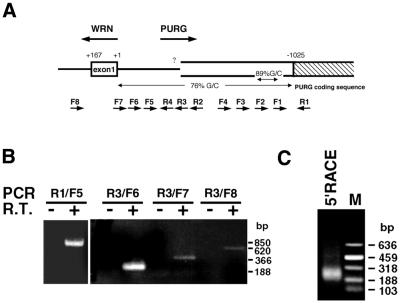
We noticed that the G+C content of the upstream DNA sequence of PURG, i.e. the sequence between WRN +1 and –1025, is high (76%, Fig. 2A). In particular, that of a 202 bp region, encompassing sites –755 and –956, is extremely high (89%, Fig. 2A). No specific products were obtained in an initial 5′-RACE experiment using nested PURG reverse primers downstream of the 202 bp region. If the 5′-end of Purγ mRNA is located upstream of this region, we could perform 5′-RACE using reverse primers that are located upstream of this region. We thus employed RT–PCR to roughly define the position of PURG transcription start site(s). C.therm. Polymerase was used as the reverse transcriptase in the RT step because it functions well at temperatures as high as 70°C, enabling successful synthesis of cDNA from G+C-rich mRNA templates. A human testis mRNA preparation (Clontech) was chosen for both RT–PCR and 5′-RACE because it was determined to contain Purγ mRNA. A reverse primer located downstream of, but near to, the 5′-end of the Purγ coding sequence in pHPURG1 (R1, Fig. 2A) was used in reverse transcription and R1 and each of the forward primers (F1–F5, Fig. 2A) were used in PCR. PCR with R1 and F5 yields a single 850 bp product, exactly the size of genomic DNA located between R1 and F5 (Fig. 2B, left), a result deemed surprising since it indicated a long 5′-UTR. This product was not seen in the negative control in which the RT step had an incubation time of 0 (Fig. 2B, left). These data indicate that the 5′-end of Purγ mRNA is located upstream of the position of the primer F5 and that there is no intron between F5 and R1. RT–PCR with reverse primer R3, located downstream of F5, and one forward primer located upstream of F5 (F6, F7 or F8, Fig. 2A) revealed that some PURG transcripts start from sites located upstream of primer F8, well into intron 1 of the WRN gene (Fig. 2B, right). There was a sharp decrease in band intensity when F7, instead of F6, was used together with R3 in the RT–PCR, suggesting that major PURG transcripts start from sites between F6 and F7 while minor transcripts start from sites further upstream. Thus, while human PURG might have more than one promoter, it is likely that one promoter governs the vast bulk of transcripts. The size of each of these PCR products is equal to that of the genomic DNA between each pair of the primers used in the PCR (data not shown), indicating that there is no intron in the 5′-end of PURG up to the position of F8.
Figure 2.
Identification of transcription start sites of human PURG. (A) Schematic diagram of genomic region around the 5′-ends of PURG and WRN and positions of primers used in RT–PCR and 5′-RACE. The numerical system used here is the same as in Figure 1. The region with 89% G+C encompasses positions –755 to –956. The position of each primer is shown as follows: R1, –1110 to –1085; R2, –390 to –366; R3, –363 to –339; R4, –325 to –304; F1, –905 to –925; F2, –716 to –740; F3, –562 to –583; F4, –411 to –431; F5, –261 to –285; F6, –176 to –199; F7, +3 to –20; F8, +257 to +233. (B) A human testis mRNA preparation (Clontech) was reverse transcribed with reverse primer R1 (left) or R3 (right) and was then PCR amplified with R1 and F5 (left) or R3 and F6, F7 or F8 (right). For negative controls there was no incubation time in the reverse transcription step. (C) The 5′-RACE of PURG was performed as described in Materials and Methods. Nested primers R2, R3 and R4 were used in the reverse transcription, first PCR and second PCR, respectively. The second PCR products were separated in a 1.2% agarose gel.
We then performed 5′-RACE (26) using nested PURG reverse primers R2, R3 and R4, which are located downstream of F5 but upstream of the 202 bp region (Fig. 2A). C.therm. Polymerase was used instead of AMV reverse transcriptase in the 5′-RACE kit (Roche) to ensure complete synthesis of first-strand cDNA from G+C-rich Purγ mRNA. The second PCR products in the 5′-RACE are shown in Figure 2C. The size of major PCR products ranged from ∼188 to ∼318 bp, while some minor PCR products were >318 bp. These different sized products were specifically derived from Purγ mRNAs with different transcription start sites, as confirmed by cloning and sequencing of these products. They were cloned into the pBK-CMV plasmid (Stratagene). Twenty clones containing inserts were selected, out of which eight were sequenced, including the two clones with the longest and shortest inserts. The inserts in all the eight clones were derived from Purγ. They share the same 3′-end, i.e. the end of reverse primer R4, but have different 5′-ends, i.e. –92, –104, –119, –122, –130, –158, –185 and –193 with respect to WRN +1. The 5′-RACE results indicated that Purγ was transcribed primarily from a cluster of initiation sites roughly between –92 and –193 with respect to WRN +1, and much less frequently from further upstream positions. This is consistent with the RT–PCR results shown in Figure 2B, in which the data suggest that Purγ starts transcription mainly from sites located between F7 and F6, i.e. between +3 and –176 with respect to WRN +1, and less frequently from further upstream sites. Analysis of the mRNA sequence upstream of the 1044 bp ORF indicates that this ORF represents the complete Purγ coding sequence. The median Purγ mRNA 5′-UTR identified here is ∼900 bp in length. Such a long 5′-UTR is uncommon among eukaryotic mRNAs (27).
Characterization of the PURG promoter reveals limited co-regulation with WRN
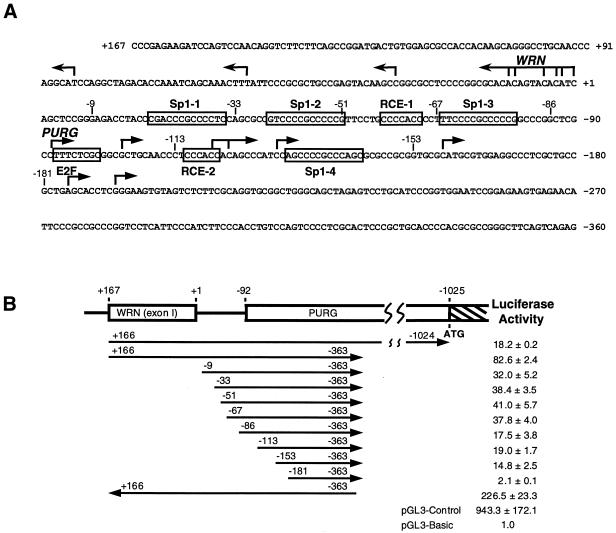
As illustrated in Figure 3A, the human PURG and WRN genes are organized such that their major clusters of transcription start sites are separated by ∼90 bp. Their minor transcription start sites are located upstream of their major transcription start sites, well into the contraposed gene (20). The WRN gene reportedly has two promoters (19,20). Luciferase reporter assays performed in several cell lines revealed that the activity of the WRN downstream promoter was 2–10-fold stronger than its upstream promoter (20). The region encompassing positions –67 and +160 was shown to be the minimal region required for full activity of the WRN downstream promoter (19). These investigators have demonstrated that two Sp1 elements (Sp1-1 and Sp1-2) and one RCE motif (RCE-1) (Fig. 3A) in the 67 bp region upstream of WRN +1 are indispensable for WRN downstream promoter activity. This short region also lies proximal to the downstream transcriptional start sites of PURG. In order to find out whether the human PURG transcription unit shares cis-acting regulatory elements with that of WRN, we performed 5′ progressive deletion analyses of the PURG downstream promoter in luciferase reporter assays in HeLa cells. These cells were chosen because they had previously been used in characterization of the WRN promoter (19). A series of luciferase reporter constructs were generated by inserting PURG upstream sequences with progressively deleted 5′-ends into the pGL3-Basic vector (Promega). These constructs were transiently transfected into HeLa cells and the relative promoter activities of the various inserts were assessed by comparing the luciferase activities of these constructs with that of the parental vector pGL3-Basic. As revealed in Figure 3B, the DNA fragment from +166 to –363 with respect to WRN +1 had a strong promoter activity, ∼36% of the promoter activity of the same DNA fragment in the reverse direction. Deletion of 175 bp from the 5′-end (from +166 to –9) reduced promoter activity by ∼60%. This same region was also shown to be required for WRN promoter activity in HeLa cells (19). Thus this region is important for the transcriptional regulation of both genes. Scanning of this region using MatInspector, a program that searches for transcription factor-binding sites (28), did not reveal any clearly relevant candidates for regulatory elements. Further deletions of Sp1-1 (from –9 to –33), Sp1-2 (from –33 to –51) and RCE-1 (from –51 to –67), which play key roles in WRN transcription initiation, had no remarkable influence on PURG promoter activity. Truncation from –67 to –86 removed another Sp1 element, Sp1-3 (Fig. 3A), and reduced promoter activity by ∼50% (Fig. 3B). Sp1-3 may thus have a positive modulating effect on PURG transcription initiation. Further truncation to –113 had little effect on promoter activity, although several transcription start sites and a potential E2F-binding site were removed (Fig. 3). The removal of another 40 bp (from –113 to –153) only slightly reduced the promoter activity, leaving significant remaining promoter activity. This deletion removes several other transcription start sites and two potential elements, RCE-2 and Sp1-4 (Fig. 3). Little promoter activity remained when the deletions progressed to position –181 (Fig. 3B). The region between –153 and –181 contains five PUR elements of the form GNGG or GGNG. It is quite conceivable that Purγ could autoregulate its gene, and WRN as well, since Purα has recently been shown to autoregulate its own promoter (29). This region contains no other known cis-regulatory elements, as revealed by MatInspector. It is notable that the region encompassing +52 to –153 also contains multiple PUR elements. The DNA fragment from positions +167 to –1024, the beginning of the Purγ coding sequence, had a promoter activity 20% that of the DNA fragment from +166 to –363, suggesting that there are negative regulatory elements in the long 5′-UTR between –363 to –1024 (Fig. 3B).
Figure 3.
Characterization of the PURG promoter. (A) The PURG and WRN major transcription start sites are indicated by arrowed lines. Several potential cis-regulatory elements described in the text are indicated by open boxes. The numerical system used here is the same as in Figures 1 and 2A. (B) Promoter activities of various DNA fragments were measured as described in Materials and Methods. The arrows with numbers at their ends indicate the locations of various DNA fragments and the directions in which they are cloned into pGL3-Basic (Promega). Plasmid pGL3-Control (Promega) contains SV40 promoter and enhancer sequences that drive strong expression of the firefly luciferase gene, while plasmid pGL3-Basic lacks eukaryotic promoter and enhancer sequences. Numbers presented are relative light emission from the luciferase reaction, detected as described in Materials and Methods. Emission due to plasmid pGL3-Basic promoter activity is assigned the value 1.0.
Two isoforms of Purγ mRNA are expressed from the PURG gene
Results presented thus far indicate that the major transcripts of human PURG-A possess a coding region of 1044 bp, a 5′-UTR as long as 1282 bp and an intact 3′-UTR of 897 bp (GenBank accession no. AF195513). The entire sequence of PURG-A contains no intron, as is also true of PURA and PURB. The 347 amino acid Purγ protein is highly homologous to Purα and Purβ. Searching the dbEST database with the human Purγ coding sequence using the BLAST program yielded a mouse blastocyst cDNA clone, designated here pMPURG1. We obtained the mouse clone and sequenced the entire insert. The insert in pMPURG1 begins with an ORF of 293 codons, followed by an intact 3′-UTR of 806 bp and concludes with a poly(A) tail. The first 258 codon sequence in pMPURG1 is 93% identical to the 258 codon sequence from 31 to 288 in the human Purγ cDNA, while their amino acid sequences are 97% identical. Yet the 35 codons from 259 to 292 and the 3′-UTR in pMPURG1 are completely different to the sequence from codon 289 to the 3′-end in the human Purγ cDNA. The genomic sequence after codon 288 of human Purγ is GTAAGT, matching the consensus sequence for a 5′ splice site of introns in higher eukaryotes. Based on these observations, we hypothesized that an intron located after codon 288 was not spliced out from the precusor RNA of human Purγ, which we named hPURG-A, but that a corresponding intron was spliced out from the precusor RNA of mouse Purγ, which we named mPURG-B, the sequence of which is now available (GenBank accession no. AF479672). We shall present evidence that this hypothesis is correct and that the two isoforms of Purγ mRNA are expressed alternatively from a single PURG gene in both human and mouse.
Human primers HPC5 and HPCA3, corresponding to the 5′- and 3′-ends, respectively, of the 1044 bp hPurγ-A coding sequence, were used to amplify potential mouse Purγ-A (mPurγ-A) coding sequence from mouse genomic DNA. A single product with a size similar to the 1044 bp human DNA was obtained. Sequencing revealed that it was 1053 bp long and 94% identical to the 1044 bp hPurγ-A coding sequence. The protein thus encoded would be 350 amino acids long and 97% identical to the 347 amino acid hPurγ-A protein. A cDNA clone from adult mouse testis has subsequently been identified that contains the 1053 bp PCR product sequence plus an 11 bp 5′-UTR and a 139 bp 3′-UTR, all of which are highly homologous to corresponding regions in hPurγ-A cDNA. Thus this clone contains a partial cDNA sequence of mPurγ-A. The complete coding sequence of mPURG-A is now available (GenBank accession no. AF479673). While the first 258 codons of mPurγ-B in pMPURG1 are identical to codons 34–291 in mPurγ-A cDNA, the codons C-terminal to these are different. Since there is no intron in the 1053 bp mPurγ-A coding sequence, there must be an intron located between codons 258 and 259 of mPurγ-B in pMPURG1. In agreement with this conclusion, the genomic sequence after codon 258 is GTAAGT, the typical 5′ splice site of introns in higher eukaryotes. It is thus evident that two different Purγ mRNAs are transcribed from a single mouse PURG gene.
By searching the dbEST database with the exon 2 sequence of mPurγ-B with the BLAST program, we identified and sequenced a cDNA clone from human fetal lung (GenBank accession no. AI338787). It contains an insert of 789 bp, followed by a poly(A) tail. Since the insert is 82% identical to the 806 bp mPurγ-B 3′-UTR, we surmised that it is the intact 3′-UTR of human Purγ-B (hPurγ-B). RT–PCR amplification from a human testis mRNA preparation (Clontech) with reverse primer HPCB3, derived from the 789 bp insert sequence, in the RT step and HPCB3 and HPC5 in the PCR step indeed generated a partial hPurγ-B cDNA (Fig. 4). As expected, this hPurγ-B cDNA shares only the first 288 codon sequence of hPurγ-A. The following 35 codon sequence encodes 34 amino acids that are 91% identical to the 34 amino acids encoded by exon 2 of mPurγ-B mRNA. The remaining partial 3′-UTR sequence is the same as that of the 789 bp insert initially obtained. The complete sequence of hPURG-B cDNA is now available (GenBank accession no. AV077841). As in mPurγ-B, there is an intron located between codons 288 and 289 of hPurγ-B. There is no other intron after codon 289 because the cDNA sequence is identical to the genomic sequence from nucleotides 16737 to 17629 in the human BAC clone RP11-293D9 (GenBank accession no. AC008066). In summary, the genomic structure and DNA sequence of PURG genes are highly conserved between human and mouse. Figure 5 presents a comparison of the C-termini of the human and mouse Purγ-A and -B isoforms. The position of the apparently spliced-out intron in the mRNAs encoding the B isoforms is indicated by dashed lines. To shed further light on how these isoforms are generated it was necessary to examine chromosome 8 PURG genomic DNA.
Figure 4.

Expression of Purγ-A and Purγ-B mRNAs in human testis. Human Purγ-A or Purγ-B mRNA in a human testis mRNA preparation (Clontech) was reverse transcribed with either primer HPCA3 or HPCB3 and was then PCR amplified with either primers HPCA3 and HPC5 or primers HPCB3 and HPC5. For negative controls there was no incubation time in the reverse transcription step.
Figure 5.
Comparison of the C-termini of human and mouse Purγ-A and Purγ-B. Residues that are different in human and mouse Purγ proteins are underlined. Dashed lines indicate the position of the spliced-out intron in Purγ-B mRNA. Asterisks represent the stop codons for Purγ-A and Purγ-B.
Diffferential termination/polyadenylation accounts for two different PURG mRNAs encoding distinct protein isoforms
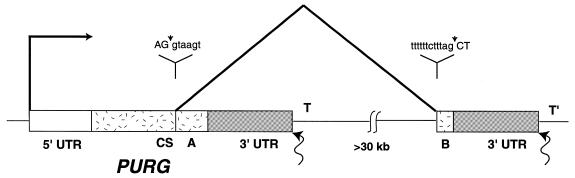
The human chromosome 8 BAC clone RP11-293D9 contains a 16736 bp partial intron sequence which is located upstream of exon 2 of hPURG-B. The sequence at the 3′-end of the intron is TTTTTTCTTTAG, matching the consensus sequence for the 3′ splice site of introns in higher eukaryotes. The genomic sequence upstream of WRN (GenBank accession no. AF181896) contains exon 1 and the following 12427 bp partial intron sequence of hPURG-B. It does not overlap the 16736 bp partial intron sequence in RP11-293D9. Human chromosome 8 BAC clone RP11-473A17 (GenBank accession no. AC084736) overlaps these two clones of the intron, but there are unsequenced regions between them. Together these data indicate that exon 2, encoding the hPurγ-B C-terminus, does in fact lie downstream of hPURG-A on chromosome 8. The data also indicate that the size of the hPURG-B intron is >30 kb. Figure 6 illustrates the structure of the human genomic PURG locus. PURG-A and PURG-B cDNAs were both generated from poly(A)+ mRNA molecules. Thus PURG-A and PURG-B polyadenylation sites are located >30 kb apart on genomic DNA. The only plausible mechanism that can explain the transcription and processing of PURG-A and -B mRNAs is that PURG transcription terminates at two different regions, one near the PURG-A poly(A) site, the other near the PURG-B poly(A) site. One alternative mechanism is extremely unlikely but not presently completely ruled out. It would posit that PURG transcription terminates at only one region, i.e. 3′ to the PURG-B poly(A) site. Cleavage and polyadenylation would then occur either near the 3′-terminus or, alternatively, >30 kb upstream of the terminus. Such cleavage is unprecedented and no mechanism thus far elucidated can account for cleavage and polyadenylation so far upstream of transcriptional termination. This is further considered in the Discussion. There are several instances of possible alternative termination in the literature. Nonetheless, transcription of the PURG gene is unique in that it is the only reported instance in which splicing of a single intron either does or does not occur, depending on differential termination/polyadenylation, resulting in different protein C-termini.
Figure 6.
Genomic structure of human PURG-A and PURG-B. The open box represents the 5′-UTR of PURG-A and PURG-B. The gray boxes represent the 3′-UTRs of PURG-A and PURG-B. The coding sequences (CS) of Purγ-A and Purγ-B are represented by stippled boxes. DNA sequences at the exon–intron boundaries are shown, with exon sequences in upper case letters and intron sequences in lower case letters. Waved arrows indicate the polyadenylation sites for PURG-A and PURG-B mRNAs. T and T′ indicate the regions of transcription termination for PURG-A and PURG-B, respectively. The pitched lines indicate the splicing that generates Purγ-B mRNA.
Differences in Purγ-A and Purγ-B mRNA expression in various organs and tumor cell lines
RT–PCR with reverse primer HPCA3 in the RT step and HPCA3 and HPC5 in the PCR step yielded the hPurγ-A coding sequence from the same testis mRNA preparation shown to yield hPurγ-B (Fig. 4). Thus both isoforms of Purγ mRNA are expressed in human testis. Sequencing of RT–PCR products confirms this. Given similarity in amplification, hPurγ-B mRNA appears more abundant than hPurγ-A in testis (Fig. 4). They are both transcribed primarily from sites located between F6 and F7 and less frequently from further upstream sites (data not shown), suggesting that they are under the same control of transcription initiation.
By identifying Purγ cDNA clones in the dbEST database, we found that hPurγ-A was expressed in glioblastoma (BF245424), hPurγ-B was expressed in fetal lung (AI338787), hPurγ (unknown whether A or B) was expressed in hippocampus (BI550191) and in ovarian fibrotheoma (BF434119), mPurγ-A was expressed in testis (AK015630), mPurγ-B was expressed in blastocyst (AA793440) and in kidney (AI931378), and mPurγ (unknown whether A or B) was expressed in skin (AA079942) and in mammary tumor (BE554692).
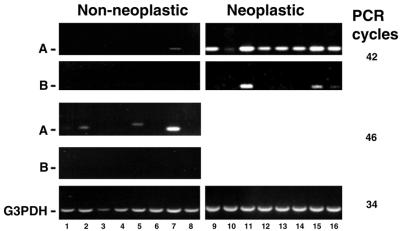
Human Purγ mRNA expression was examined using commercially available cDNA panels from eight normal organs [Human Multiple Tissue cDNA (MTC) Panel I; Clontech] and eight tumor cell lines (Human Tumor MTC Panel; Clontech) by PCR amplification of hPurγ-A and hPurγ-B cDNAs from each of the 16 cDNA preparations (Fig. 7). When 34 PCR cycles were employed, no hPurγ products were detected (not shown). While 46 cycles were necessary to detect hPurγ PCR products in several normal tissues, 42 cycles were sufficient to detect hPurγ mRNA expression in all of the tumor cells studed. Human Purγ-A mRNA was expressed at different, but quite significant, levels in all of the eight tumor cell lines. Human Purγ-B mRNA was detected in three of the tumor cell lines and its expression level was lower than that of the corresponding hPurγ-A mRNA in each of these. In contrast, hPurγ-A mRNA was only detected in three of the eight normal organs and no hPurγ-B mRNA was detected in any of the eight normal organs. Thus far, hPurγ-B mRNA has only been detected in certain fetal cells, testis and tumor cells. Human Purγ-A mRNA, on the other hand, has been detected in a broader range of normal tissues and it may be expressed ubiquitously in tumor cells. The overall Purγ mRNA expression levels in the eight normal organs were much lower than those in eight tumor cell lines. In particular, no hPurγ-A or hPurγ-B mRNA was detected in lung or pancreas, but hPurγ-A mRNA was detected in the two lung carcinoma cell lines and the pancreas adenocarcinoma cell line (Fig. 7). These data suggest a potential relation between the up-regulation of Purγ expression and tumorigenesis.
Figure 7.
Expression of human Purγ-A and Purγ-B mRNAs in normal tissues and tumor cell lines. Human MTC Panel I (lanes 1–8) and Human Tumor MTC Panel (lanes 9–16) were obtained from Clontech Laboratories. Purγ-A and Purγ-B cDNAs were PCR amplified with primer sets HPC5M/HPCA3 and HPC5M/HPCB3, respectively. The sizes of the products are 356 and 355 bp, respectively. G3PDH expression in all samples was examined as an internal control. The 16 samples are as follows: lane 1, heart; lane 2, brain; lane 3, placenta; lane 4, lung; lane 5, liver; lane 6, skeletal muscle; lane 7, kidney; lane 8, pancreas; lane 9, breast carcinoma GI-101; lanes 10 and 11, colon adenocarcinoma CX-1 and GI-112; lanes 12 and13, lung carcinoma LX-1 and GI-117; lane 14, pancreatic adenocarcinoma GI-103; lane 15, prostatic adenocarcinoma PC3; lane 16, ovarian carcinoma GI-102.
DISCUSSION
Overlapping, contrapodal promoters regulate initiation of transcription of PURG and WRN
The contrapodal orientation of PURG and WRN raises questions regarding potential co-regulation of the two genes. For that reason we have defined the transcription unit and characterized the promoter for PURG. That has previously been done for WRN (19,20). It has been reported that WRN initiates transcription from multiple sites (19,20). Here 5′-RACE indicates that PURG also initiates from multiple sites (Fig. 2). In each case the 5′-most initiation sites lie within the transcription unit of the contraposed gene, suggesting that regulatory sequences may exist within the contraposed gene. The vast bulk, however, of transcripts for each gene initiate from closely spaced sites separated by ∼90 bp of intervening regulatory sequence. This separating regulatory region has previously been evaluated for transcription of WRN and evaluated in the present publication for PURG by us. In both cases the promoters have been evaluated in HeLa cells, in which both proteins have been observed to be expressed by immunohistochemistry (data not shown). We report that the intervening potential regulatory region functions in HeLa cells to highly promote transcription in both directions. In both cases the promoters are TATA-less and CAAT-less and contain many potentially bi-directional SP1 transactivational binding sites. Important distinctions, however, indicate that these two genes are not strictly co-regulated. Several of the SP1 sites deemed critical for WRN transcription (19) do not, when deleted, affect PURG transcription. While transcript levels for both genes are relatively high in testis and brain, PURG transcript levels in all normal tissues are low overall. Thus, while both genes may be housekeeping genes, there are important elements that regulate their expression levels in different tissues. The possibility of autoregulation of the PURG gene must be noted. All SP1 elements are also PUR elements and there are many more PUR elements in the common promoter region as well as in the PURG 5′-leader sequence. Thus, Purγ or its related family members may bind to this region to influence transcription. Autoregulation of the PURA gene in this fashion by Purα has recently been described (29). In that study increasing levels of Purα reduced transcription from the PURA gene. Likewise, binding of Pur proteins to the PURG–WRN promoter could affect WRN transcription. It is conceivable in that case that expression of the Purγ protein could directly regulate transcription of the WRN gene. It would thus be interesting to determine whether deregulation of PURG ever occurs in patients with Werner syndrome.
It is now known that several human gene pairs are organized in contrapodal fashion with central, intervening regulatory regions (30–43). In most cases regulatory elements critical for transcription in each direction differ (35,42), whereas in certain cases bi-directional regulatory elements critical for both genes have been reported (39,40). Even in those cases, however, additional regulatory elements differed between the two genes. What evolutionary benefit is there for the presence of head-to-head overlapping promoters if transcription of the two contraposed genes is not co-regulated? One possibility is that two such genes might not be normally co-regulated, but that in response to certain cellular signals it would be beneficial to simultaneously up- or down-regulate both of them. All of the promoters referenced above are highly G+C-rich and may be targets for DNA methylation. The PURG–WRN promoter region is also extremely G+C-rich. It will be interesting to determine whether, and how, DNA methylation affects expression of these genes.
Results from scanning a panel of human tissues reveal that WRN mRNA exists in all 24 tissues examined (data not shown), suggesting that it is a housekeeping gene, in keeping with previous speculation. Housekeeping genes generally have a CpG island of some 200–1000 bp spanning the promoter region, frequently extending into the first exon (44). These promoters are not only rich in G/C, but are also characterized by a CpG/GpC ratio that is >0.6 (45). The PURG–WRN promoter region has these features. Similarly, the two genes each have no TATA or CAAT boxes. Human PURG may also be a housekeeping gene, but, if so, its levels must be quite low in certain cells or under certain conditions, as indicated by the expression levels in Figure 7.
There are several potential ATG translation start sites in the hPURG 5′-UTR prior to the one at which translation actually initiates. This is unusual among gene 5′-UTRs. Note, however, that WRN also has two ATG codons upstream of its translation start site. In each case, for either gene, the upstream ATGs do not have favorable surrounding consensus sequences for translation initiation (27) and they are followed shortly thereafter by stop codons.
Purγ protein isoforms relate structural features of Pur family members to neoplasia
Figure 7 reveals that the PURG gene is highly expressed in many tumor cell lines. In cells in which the PURG-B form is expressed the PURG-A form is also present at high levels. The results suggest that enhanced expression of PURG may be regulated at the level of initiation of transcription. It remains to be determined whether or not increased PURG transcription is coupled to increased cell proliferation. The present results likely extend the Pur family to comprise at least four distinct proteins. Purα and Purβ are both single-stranded DNA-binding proteins that bind specifically to repeats of GGN (1,13). The repeat region of Purα is essential for both DNA (17) and RNA (46) binding. Given the high degree of homology of Purγ repeats to those of the other Pur proteins, it is likely that the Purγ DNA- and RNA-binding properties are similar, although this remains to be demonstrated. Outside the repeat motifs there are important distinctions between Pur proteins. Both the Purγ-A and -B forms contain the sequence QGSRRRQKHS, a potential nuclear localization signal absent from Purα and Purβ. Thus, while Purα access to, and exclusion from, the nucleus is highly regulated by multiple amino acid motifs (15), Purγ may be more stably directed there. All human Pur proteins contain a variation of the Purα psycho motif, located C-terminal to the repeats and implicated in Rb binding (47). Part of this motif in Purα includes a potential casein kinase II Ser phosphorylation site in the form FCKYSEEM. This differs in the hPur family members as follows:
FCKYSEEM Purα
FCRYADEM Purβ
FIKYEEEM Purγ-A
In Purα a negative charge may be conferred on the S by phosphorylation. In Purβ this is not possible at the A. In Purγ-A the E residue specifies a permanent negative charge. Given that this is reportedly in a protein–protein interaction region (17), this change could be functionally significant, although further work must substantiate this. It is most notable that Purγ-B completely lacks the psycho motif. Thus, while Rb binding may be altered among Purα, Purβ and Purγ-A, it may not be possible at all for Purγ-B. In this regard, Purγ-B may be a dominant-negative form of Pur protein, since it contains the conserved DNA- and RNA-binding repeats. Its preferential expression in certain tumor cells could conceivably be related to the propensity of such cells to evade Rb-controlled cell cycle checkpoints. The PURG B-form is expressed in colon adenocarcinoma and prostatic adenocarcinoma (Fig. 7). It would be interesting to know whether such expression occurs early or late in the progression to neoplasia.
Novel aspects of transcription termination/polyadenylation control expression of hPurγ isoforms
A novel mechanism involving differential termination/polyadenylation controls the expression of mRNAs for the two Purγ isoforms in both human and mouse. Little is known about the control of transcriptional termination by RNA polymerase II in higher eukaryotes, but it is widely held that termination occurs 3′, and quite proximal, to a polyadenylation signal. The precise mechanism by which synthesis of the two PURG mRNAs is regulated are not known, but one observation is clear: PURG-A and PURG-B transcripts are polyadenylated at different sites which are greatly distant in the genome. Therefore, it can be concluded that some aspect of termination/polyadenylation differentially controls the expression of PURG-A and PURG-B mRNAs. In yeast certain proteins controlling termination and those controlling polyadenylation interact (48). In higher eukaryotes it has been reported that specific G-rich sequences near to, and downstream of, a polyadenylation signal induce transcriptional pausing, leading to the cleavage that precedes polyadenylation (49). This tight coupling of termination and polyadenylation strongly mitigates against a termination site >30 kb 3′ to a polyadenylation signal. Therefore, it is likely that a factor, or factors, controlling coupled termination/polyadenylation is responsible for differential expression of the two Purγ isoforms.
There is a canonical polyadenylation signal AATAAA located 14 bp upstream of the hPURG-B poly(A) site. In mouse this signal is 15 bp upstream of the mPURG-B poly(A) site. There is no such sequence near the hPURG-A poly(A) site, yet PURG-A comprises the bulk of the expressed PURG in most cells. There are several sequences similar to AATAAA in the vicinity of the 3′-end of PURG-A and it is conceivable that a cryptic polyadenylation signal is employed. There is also an ATTAAA element ∼300 nt from the 3′-end of PURG-A and it is also possible that it is functional.
The detection of PURG-B expression primarily in tumor cells raises the possibility that a polyadenylation signal near the 3′-end of PURG-A could be mutated, thus allowing transcription to proceed through the PURG-B exon. We cannot currently state that this never happens, but, thus far, sequencing of the 3′-UTR from mRNAs from several tumor tissues and lines has not revealed any mutations or deletions. Thus, if PURG transcription is aberrant in cancer, as suggested in Figure 7, it is likely to be due to alterations in the termination/polyadenylation mechanism.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Yayoi Kinoshita for excellent technical assistance. This work was supported by grants from the NCI and NINDS to E.M.J.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +1 212 241 7510; Fax: +1 212 534 7491; Email: edward.johnson@mssm.edu AF195513.2, AV077841, AF479672, AF479673
REFERENCES
- 1.Bergemann A.D., Ma,Z.W. and Johnson,E.M. (1992) Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol. Cell. Biol., 12, 5673–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chepenik L.G., Tretiakova,A.P., Krachmarov,C.P., Johnson,E.M. and Khalili,K. (1998) The single-stranded DNA binding protein, Pur-alpha, binds HIV-1 TAR RNA and activates HIV-1 transcription. Gene, 210, 37–44. [DOI] [PubMed] [Google Scholar]
- 3.Gallia G.L., Johnson,E.M. and Khalili,K. (2000) Puralpha: a multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res., 28, 3197–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itoh H., Wortman,M.J., Kanovsky,M., Uson,R.R., Gordon,R.E., Alfano,N. and Johnson,E.M. (1998) Alterations in Pur(alpha) levels and intracellular localization in the CV-1 cell cycle. Cell Growth Differ., 9, 651–665. [PubMed] [Google Scholar]
- 5.Krachmarov C.P., Chepenik,L.G., Barr-Vagell,S., Khalili,K. and Johnson,E.M. (1996) Activation of the JC virus Tat-responsive transcriptional control element by association of the Tat protein of human immunodeficiency virus 1 with cellular protein Pur alpha. Proc. Natl Acad. Sci. USA, 93, 14112–14117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi S., Agui,K., Kamo,S., Li,Y. and Anzai,K. (2000) Neural BC1 RNA associates with pur alpha, a single-stranded DNA and RNA binding protein, which is involved in the transcription of the BC1 RNA gene. Biochem. Biophys. Res. Commun., 277, 341–347. [DOI] [PubMed] [Google Scholar]
- 7.Tretiakova A., Gallia,G.L., Shcherbik,N., Jameson,B., Johnson,E.M., Amini,S. and Khalili,K. (1998) Association of Puralpha with RNAs homologous to 7 SL determines its binding ability to the myelin basic protein promoter DNA sequence. J. Biol. Chem., 273, 22241–22247. [DOI] [PubMed] [Google Scholar]
- 8.Du Q., Tomkinson,A.E. and Gardner,P.D. (1997) Transcriptional regulation of neuronal nicotinic acetylcholine receptor genes. A possible role for the DNA-binding protein Puralpha. J. Biol. Chem., 272, 14990–14995. [DOI] [PubMed] [Google Scholar]
- 9.Herault Y., Chatelain,G., Brun,G. and Michel,D. (1993) The PUR element stimulates transcription and is a target for single strand-specific binding factors conserved among vertebrate classes. Cell. Mol. Biol. Res., 39, 717–725. [PubMed] [Google Scholar]
- 10.Kelm R.J. Jr, Sun,S., Strauch,A.R. and Getz,M.J. (1996) Repression of transcriptional enhancer factor-1 and activator protein-1-dependent enhancer activity by vascular actin single-stranded DNA binding factor 2. J. Biol. Chem., 271, 24278–24285. [DOI] [PubMed] [Google Scholar]
- 11.Ohashi S., Kobayashi,S., Omori,A., Ohara,S., Omae,A., Muramatsu,T., Li,Y. and Anzai,K. (2000) The single-stranded DNA- and RNA-binding proteins pur alpha and pur beta link BC1 RNA to microtubules through binding to the dendrite-targeting RNA motifs. J. Neurochem., 75, 1781–1790. [DOI] [PubMed] [Google Scholar]
- 12.Zambrano N., De Renzis,S., Minopoli,G., Faraonio,R., Donini,V., Scaloni,A., Cimino,F. and Russo,T. (1997) DNA-binding protein Pur alpha and transcription factor YY1 function as transcription activators of the neuron-specific FE65 gene promoter. Biochem. J., 328, 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelm R.J. Jr, Elder,P.K., Strauch,A.R. and Getz,M.J. (1997) Sequence of cDNAs encoding components of vascular actin single-stranded DNA-binding factor 2 establish identity to Puralpha and Purbeta. J. Biol. Chem., 272, 26727–26733. [DOI] [PubMed] [Google Scholar]
- 14.Lezon-Geyda K., Najfeld,V. and Johnson,E.M. (2001) Deletions of PURA, at 5q31, and PURB, at 7p13, in myelodysplastic syndrome and progression to acute myelogenous leukemia. Leukemia, 15, 954–962. [DOI] [PubMed] [Google Scholar]
- 15.Barr S.M. and Johnson,E.M. (2001) Ras-induced colony formation and anchorage-independent growth inhibited by elevated expression of Puralpha in NIH3T3 cells. J. Cell. Biochem., 81, 621–638. [DOI] [PubMed] [Google Scholar]
- 16.Stacey D.W., Hitomi,M., Kanovsky,M., Gan,L. and Johnson,E.M. (1999) Cell cycle arrest and morphological alterations following microinjection of NIH3T3 cells with Pur alpha. Oncogene, 18, 4254–4261. [DOI] [PubMed] [Google Scholar]
- 17.Johnson E.M., Chen,P.L., Krachmarov,C.P., Barr,S.M., Kanovsky,M., Ma,Z.W. and Lee,W.H. (1995) Association of human Pur alpha with the retinoblastoma protein, Rb, regulates binding to the single-stranded DNA Pur alpha recognition element. J. Biol. Chem., 270, 24352–24360. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 19.Yamabe Y., Shimamoto,A., Goto,M., Yokota,J., Sugawara,M. and Furuichi,Y. (1998) Sp1-mediated transcription of the Werner helicase gene is modulated by Rb and p53. Mol. Cell. Biol., 18, 6191–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L., Hunt,K.E., Martin,G.M. and Oshima,J. (1998) Structure and function of the human Werner syndrome gene promoter: evidence for transcriptional modulation. Nucleic Acids Res., 26, 3480–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epstein C.J., Martin,G.M., Schultz,A.L. and Motulsky,A.G. (1966) Werner’s syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore), 45, 177–221. [DOI] [PubMed] [Google Scholar]
- 22.Goto M., Tanimoto,K., Horiuchi,Y. and Sasazuki,T. (1981) Family analysis of Werner’s syndrom: a survey of 42 Japanese families with a review of the literature. Clin. Genet., 19, 8–15. [DOI] [PubMed] [Google Scholar]
- 23.Goto M., Miller,R.W., Ishikawa,Y. and Sugano,H. (1996) Excess of rare cancers in Werner syndrome (adult progeria). Cancer Epidemiol. Biomarkers Prev., 5, 239–246. [PubMed] [Google Scholar]
- 24.Yu C.E., Oshima,J., Fu,Y.H., Wijsman,E.M., Hisama,F., Alisch,R., Matthews,S., Nakura,J., Miki,T., Ouais,S., Martin,G.M., Mulligan,J. and Schellenberg,G.D. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]
- 25.Oshima J. (2000) The Werner syndrome protein: an update. Bioessays, 22, 894–901. [DOI] [PubMed] [Google Scholar]
- 26.Frohman M.A., Dush,M.K. and Martin,G.R. (1988) Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl Acad. Sci. USA, 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak M. (1984) Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res., 12, 857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muralidharan V., Sweet,T., Nadraga,Y., Amini,S. and Khalili,K. (2001) Regulation of Puralpha gene transcription: evidence for autoregulation of Puralpha promoter. J. Cell. Physiol., 186, 406–413. [DOI] [PubMed] [Google Scholar]
- 30.Wright K.L., White,L.C., Kelly,A., Beck,S., Trowsdale,J. and Ting,J.P. (1995) Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J. Exp. Med., 181, 1459–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinya E. and Shimada,T. (1994) Identification of two initiator elements in the bidirectional promoter of the human dihydrofolate reductase and mismatch repair protein 1 genes. Nucleic Acids Res., 22, 2143–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmiedeknecht G., Buchler,C. and Schmitz,G. (1997) A bidirectional promoter connects the p14.5 gene to the gene for RNase P and RNase MRP protein subunit hPOP1. Biochem. Biophys. Res. Commun., 241, 59–67. [DOI] [PubMed] [Google Scholar]
- 33.Saito T., Matsuda,Y., Ishii,H., Watanabe,F., Mori,M., Hayashi,A., Araki,R., Fujimori,A., Fukumura,R., Morimyo,M., Tatsumi,K., Hori,T. and Abe,M. (1998) Mouse cdc21 only 0.5 kb upstream from dna-pkcs in a head-to-head organization: an implication of co-evolution of ATM family members and cell cycle regulating genes. Mamm. Genome, 9, 769–772. [DOI] [PubMed] [Google Scholar]
- 34.Lennard A.C. and Fried,M. (1991) The bidirectional promoter of the divergently transcribed mouse Surf-1 and Surf-2 genes. Mol. Cell. Biol., 11, 1281–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S.G. and Song,K. (2000) Identification and characterization of a bidirectional promoter from the intergenic region between the human DDX13 and RD genes. Mol. Cells, 10, 47–53. [DOI] [PubMed] [Google Scholar]
- 36.Ikeda S., Mochizuki,A., Sarker,A.H. and Seki,S. (2000) Identification of functional elements in the bidirectional promoter of the mouse Nthl1 and Tsc2 genes. Biochem. Biophys. Res. Commun., 273, 1063–1068. [DOI] [PubMed] [Google Scholar]
- 37.Igaki H., Nakagawa,K., Aoki,Y., Ohtomo,K., Kukimoto,I. and Kanda,T. (2001) Characterization of the bi-directional transcriptional control region between the human UFD1L and CDC45L genes. Biochem. Biophys. Res. Commun., 283, 569–576. [DOI] [PubMed] [Google Scholar]
- 38.Guarguaglini G., Battistoni,A., Pittoggi,C., Di Matteo,G., Di Fiore,B. and Lavia,P. (1997) Expression of the murine RanBP1 and Htf9-c genes is regulated from a shared bidirectional promoter during cell cycle progression. Biochem. J., 325, 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujii H., Shinya,E. and Shimada,T. (1992) A GC box in the bidirectional promoter is essential for expression of the human dihydrofolate reductase and mismatch repair protein 1 genes. FEBS Lett., 314, 33–36. [DOI] [PubMed] [Google Scholar]
- 40.Fischer G., Schmidt,C., Opitz,J., Cully,Z., Kuhn,K. and Poschl,E. (1993) Identification of a novel sequence element in the common promoter region of human collagen type IV genes, involved in the regulation of divergent transcription. Biochem. J., 292, 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dimitrov S., Brennerova,M. and Forejt,J. (2001) Expression profiles and intergenic structure of head-to-head oriented Brca1 and Nbr1 genes. Gene, 262, 89–98. [DOI] [PubMed] [Google Scholar]
- 42.Ame J.C., Schreiber,V., Fraulob,V., Dolle,P., de Murcia,G. and Niedergang,C.P. (2001) A bidirectional promoter connects the poly(ADP-ribose) polymerase 2 (PARP-2) gene to the gene for RNase P RNA. Structure and expression of the mouse PARP-2 gene. J. Biol. Chem., 276, 11092–11099. [DOI] [PubMed] [Google Scholar]
- 43.Kaneko K.J. and DePamphilis,M.L. (2000) Soggy, a spermatocyte-specific gene, lies 3.8 kb upstream of and antipodal to TEAD-2, a transcription factor expressed at the beginning of mouse development. Nucleic Acids Res., 28, 3982–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bird A.P. (1986) CpG-rich islands and the function of DNA methylation. Nature, 321, 209–213. [DOI] [PubMed] [Google Scholar]
- 45.Razin A. and Kafri,T. (1994) DNA methylation from embryo to adult. Prog. Nucleic Acid Res. Mol. Biol., 48, 53–81. [DOI] [PubMed] [Google Scholar]
- 46.Wortman M.J., Krachmarov,C.P., Kim,J.H., Gordon,R.G., Chepenik,L.G., Brady,J.N., Gallia,G.L., Khalili,K. and Johnson,E.M. (2000) Interaction of HIV-1 Tat with Puralpha in nuclei of human glial cells: characterization of RNA-mediated protein–protein binding. J. Cell. Biochem., 77, 65–74. [DOI] [PubMed] [Google Scholar]
- 47.Ma Z.W., Bergemann,A.D. and Johnson,E.M. (1994) Conservation in human and mouse Pur alpha of a motif common to several proteins involved in initiation of DNA replication. Gene, 149, 311–314. [DOI] [PubMed] [Google Scholar]
- 48.Calvo O. and Manley,J.L. (2001) Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation and termination. Mol. Cell, 7, 1013–1023. [DOI] [PubMed] [Google Scholar]
- 49.Yonaha M. and Proudfoot,N.J. (2000) Transcriptional termination and coupled polyadenylation in vitro. EMBO J., 19, 3770–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]